To address the clinical need for nodal involvement risk assessment in patients with triple‐negative breast cancer (TNBC), this study performed 3?UTR profiling on publicly available microarrays from a large number of patients with pathologically‐confirmed lymph node status. This is the first study to demonstrate nodal involvement risk assessment capability of an integrative 3?UTR7‐based model in TNBC.
Keywords: Prediction modeling, 3′ untranslated region, Alternative polyadenylation, Triple‐negative breast cancer, Lymph node
Abstract
Background.
Sentinel lymph node biopsy is the standard surgical staging approach for operable triple‐negative breast cancer (TNBC) with clinically negative axillae. In this study, we sought to develop a model to predict TNBC patients with negative nodal involvement, who would benefit from the exemption of the axillary staging surgery.
Materials and Methods.
We evaluated 3′ untranslated region (3′UTR) profiles using microarray data of TNBC from two Gene Expression Omnibus datasets. Samples from GSE31519 were divided into training set (n = 164) and validation set (n = 163), and GSE76275 was used to construct testing set (n = 164). We built a six‐member 3′UTR panel (ADD2, COL1A1, APOL2, IL21R, PKP2, and EIF4G3) using an elastic net model to estimate the risk of lymph node metastasis (LNM). Receiver operating characteristic and logistic analyses were used to assess the association between the panel and LNM status.
Results.
The six‐member 3′UTR‐panel showed a high distinguishing power with an area under the curve of 0.712, 0.729, and 0.708 in the training, validation, and testing sets, respectively. After adjustment by tumor size, the 3′UTR panel retained significant predictive power in the training, validation, and testing sets (odds ratio = 4.93, 4.58, and 3.59, respectively; p < .05 for all). A combinatorial analysis of the 3′UTR panel and tumor size yielded an accuracy of 97.2%, 100%, and 100% in training, validation, and testing set, respectively.
Conclusion.
This study established an integrative 3′UTR‐based model as a promising predictor for nodal negativity in operable TNBC. Although a prospective study is needed to validate the model, our results may permit a no axillary surgery option for selected patients.
Implications for Practice.
Currently, sentinel lymph node biopsy is the standard approach for surgical staging in breast cancer patients with negative axillae. Prediction estimation for lymph node metastasis of breast cancer relies on clinicopathological characteristics, which is unreliable, especially in triple‐negative breast cancer (TNBC)—a highly heterogeneous disease. The authors developed and validated an effective prediction model for the lymph node status of patients with TNBC, which integrates 3′UTR markers and tumor size. This is the first 3′UTR‐based model that will help identify TNBC patients with low risk of nodal involvement who are most likely to benefit from exemption axillary surgery.
摘要
背景。前哨淋巴结活检是针对可手术的三阴性乳腺癌 (TNBC)(临床显示腋窝淋巴结阴性)的标准手术分期方法。在本研究中,我们致力于研发一种模型,以预测阴性淋巴转移的 TNBC 患者,使他们从免除腋窝分期手术中获益。
材料和方法。我们使用来自 2 个基因表达综合数据库的 TNBC 微阵列数据对 3' 非翻译区 (3'UTR) 概况进行了评估。我们将 GSE31519 中的样本划分为训练集 (n = 164) 和验证集 (n =163) 并将 GSE76275 中的样本用于构建测试集 (n =164)。我们利用弹性网络模型组建了一个 6 成员 3’UTR 小组(ADD2、COL1A1、APOL2、IL21R、PKP2和 EIF4G3),以评估淋巴结转移 (LNM) 的风险。受试者手术特征和逻辑分析用于评估小组和 LNM 状态之间的关系。
结果。6 成员 3'UTR 小组显示出高辨别能力,在训练集、验证集和测试集中的曲线下面积分别为 0.712、0.729 和 0.708。在根据肿瘤大小进行校正之后,3’UTR 小组在训练集、验证集和测试集中仍保留了显著的预测能力( 比值比分别为 4.93、4.58 和 3.59;所有p < 0.05)。针对 3'UTR 小组和肿瘤大小的组合分析在训练集、验证集和测试集中分别达到了 97.2%、100% 和 100% 的准确度。
结论。本研究建立了一个基于 3'UTR 的整合模型,该模型是可手术的 TNBC 淋巴结阴性的有希望的预测因素。尽管需要实施前瞻性研究来验证此模型,但是,我们的研究结果可能允许选定的患者不选择腋窝手术。
对临床实践的提示
目前,前哨淋巴结活检是针对腋窝淋巴结阴性的乳腺癌患者的标准手术分期方法。乳腺癌淋巴结转移的预测估计依赖于临床病理特征,该特征具有不可靠性,特别是在三阴性乳腺癌(TNBC,高度异质性疾病)中更是如此。作者开发并验证了一种针对 TNBC 患者淋巴结状态的有效预测模型,该模型将 3'UTR 标记物和肿瘤大小整合在一起。这是第一个基于 3'UTR 的模型,有助于识别淋巴结转移低风险的 TNBC 患者,此类患者最有可能从免除腋窝手术中获益。
Introduction
Triple‐negative breast cancer (TNBC), lacking expression of estrogen receptor (ER), progesterone receptor, and human epidermal growth receptor 2, represents 15%–20% of breast cancer incidences [1]. TNBC is a heterogeneous disease with diverse clinical courses, and pathological, molecular, and genetic features [2]. Molecular insights into TNBCs revealed intrinsic subtypes with different gene expression signatures [3], [4]. There is a major need to better manage the patients with TNBC using the genetics or epi‐genetics information.
Currently, sentinel lymph node biopsy (SLNB) is the standard surgical approach for invasive breast cancer patients with clinically negative axillae. However, SLNB is time‐consuming and costly due to the intraoperative pathological assessment. Furthermore, as an invasive surgical procedure, SLNB can cause pain, swelling, bruising, and lymphedema at the surgical site and increase the risk of infection and skin or allergic reactions. A previous study conducted in a large cohort from multicenters reported that 77.2% patients with early TNBC had negative lymph node involvement [5]. Thus, most TNBC patients receive overtreatment via axillary surgery, which increases the surgical comorbidity without survival benefit. Several clinical trials have been conducted that have shown that axillary surgery can be avoided in older patients with cT1cN0 breast cancer [6], [7]. SOUND (Sentinel node vs. Observation after axillary Ultra‐souND) trial is an ongoing perspective randomized study in which patients with “low‐risk” breast cancer (≤2 cm and negative preoperative axillary ultrasound) are randomized to SLNB ± axillary dissection or no axillary surgical staging [8]. Although the results have not been released, Royal Marsden experience of 10.4 years median follow‐up with a policy of no axillary surgery revealed axillary surgery could be spared in selected “low‐risk” group [9], [10]. That provides additional support for the hypothesis that some patients may have axillary surgery omitted as they will be node negative, and hence avoid morbidity. However, both these studies pertain mainly to ER‐positive disease. In contrast with non‐TNBC, which has a consistent increase in lymph node metastasis (LNM) incidence according tumor size, the nodal involvement risk for TNBC is decoupled from tumor size [11]. Hence, there is a need to develop a predictive model and identify molecular markers that can select patients at low risk for LNM in TNBC who may not benefit from axillary surgery.
Recently, studies of alternative polyadenylation (APA) have provided insight into 3′UTR length dynamics in human diseases including cancer, and in various physiological processes, such as cell proliferation, differentiation, and development [12], [13], [14]. An emerging role of APA dynamics in human cancer has been identified [15], [16], [17]. Selected APA events can be used as prognostic markers in multiple cancers, which adds superior prognostic power beyond common clinical and mRNA variables [13]. The value of gene‐based assays have lately been acknowledged and incorporated into the eighth edition of the primary tumor, lymph node, and metastasis (TNM) classification of the American Joint Commission of Cancer (AJCC) for breast cancer [18]. Thus, we hypothesized that the 3′UTR dynamics might be responsible for lymph node status in TNBC and the 3′UTR landscape profiled by microarrays could be used as powerful prediction biomarkers.
To address the clinical need for nodal involvement risk assessment in TNBC patients, we performed 3′UTR profiling on publicly available microarrays from a large number of TNBC patients with pathologically confirmed lymph node status. Herein, we propose a six‐member 3′UTR‐panel that robustly discriminates patients at different risks for LNM, and combining the panel and tumor size shows a high accuracy for negative node prediction. To the best of our knowledge, this is the first study that demonstrates nodal involvement risk assessment capability of an integrative 3′UTR‐based model in TNBC.
Materials and Methods
Data Collection and Processing
Microarray data collection, normalization, and 3′UTR profiling were described in our earlier paper [19]. Briefly, a previous study [20] identified 327 TNBC samples with follow‐up data and essential clinical information (age, tumor size, and lymph node status) and deposited the data in Gene Expression Omnibus (GEO) under Accession Number GSE31519. An independent dataset of 198 TNBC microarrays (GSE76275) [4] was recently released, and the essential clinical information is available in 143 samples. After initial quality check, the two datasets were downloaded. R package “ERI‐expr” [21] was used to profile the 3′UTR landscape. As previously described [21], the expression ratio index (ERI) was defined as the signal intensity ratio of the 5′ and 3′ probe sets of the APA sites, which correlates to the ratio of short and long 3′UTR isoforms. Combat [22] was used to adjust the batch effects when pooling the batches of microarray data. The adjusted combined ERI data were used in subsequent analysis.
Development and Validation of LNM Prediction Model for TNBC
Three hundred twenty‐seven primary TNBCs with follow‐up data and essential clinical information from GSE31519 were randomly categorized into either a training set (n = 164) or a validation set (n = 163) according to chip batch stratification. We introduced an independent dataset (GSE76275) to test the robustness of the model. However, the GSE76275 cohort had a high node‐positive rate of 50.3%, which may introduce the heterogeneity to the datasets and lower the model performance. Thus, a matched pairs design was used to overcome the above disadvantages. For each subject of the training set (n = 164), we randomly sampled the paired subject in GSE76275 with the same age (≤50 years vs. >50 years), tumor size (≤2 cm vs. >2 cm), and lymph node involvement (negative vs. positive) categories with replacement and constructed a testing set (n = 164).
Using univariate logistic regression, we filtered out noisy features of the 3′UTR ERI data to reduce feature dimensionality. A cutoff of 0.2 was set for the p values, and 3′UTRs with a Wald p value <0.2 were kept in subsequent prediction modeling. Then, the elastic net model [23] was used to discover the 3′UTRs related to LNM and to train the 3′UTR panel with the selected predictors in the training cohort. We used R package “glmnet” [24] to perform the elastic net analysis. To obtain a parsimonious model with a modest predictive accuracy, leave‐one‐out cross‐validation was used to determine the penalty parameter λ, and selected λ using one standard error (1SE) criteria [25]. The risk score function was a linear combination of ERI of the selected 3′UTRs via the elastic net modeling. For the sake of intuitiveness, the z‐score (standard risk score) was also reported for each patient.
To determine the threshold that divided the sample into the high‐ or low‐risk categories, we analyzed each value and chose an optimal threshold with the maximal likelihood according to the logistic regression. To assess the robustness of the 3′UTR panel, we computed risk scores of the patients in the validation and testing sets and allocated them to the high‐ or low‐risk groups according to the risk function and the corresponding threshold determined in the training set. Receiver operating characteristic (ROC) analysis and logistic regression were used to evaluate the predictive accuracy of the 3′UTR panel in training, validation, and testing cohorts.
MKI67 Expression Profiling.
The microarray CEL files were processed with Bioconductor package “affy” [26] using RMA background correction and quantile normalization. As Ki‐67 immunochemistry level is not available, MKI67 expression level was analyzed in all samples, after adjustment of the batch effect using Combat. All samples with MKI67 expression higher than the median MKI67 in the training set were classified as high MKI67, whereas others were classified as low MKI67.
TNBC Subtyping
A Lehmann TNBC subtyping system was proposed after analyzing 587 TNBC gene expression signature from 21 publicly available datasets [3]. The authors developed a web‐based tool “TNBCtype” [27] for classifying TNBC samples (http://cbc.mc.vanderbilt.edu/tnbc/). Using this tool, we obtained the Lehmann subtypes of all samples in this study. The Burstein TNBC classification system was established by analyzing 198 TNBC tumors using non‐negative matrix factorization method [4]. The Burstein subtype information of all samples in the testing set was acquired from the GEO Accession Viewer (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?ac c=GSE76275).
Statistical Analysis
We used Pearson's chi‐square test to compare the categorical variables and Student's t test for continuous variables. We preferentially used relapse‐free survival as the endpoint for event‐free survival. Reverse Kaplan‐Meier method was used to compute the median follow‐up time.
Unadjusted and adjusted odds ratios (OR) with 95% confidence intervals (95% CI) were computed using logistic regression analysis. ROC curves were used to illustrate and evaluate the 3′UTR panel and clinicopathological factors to predict the risk of LNM in patients with TNBC. R package “pROC” was used to calculate the area under the ROC curve (AUC).
This study was conducted from November 2016 to April 2018. All statistical analyses were completed using R software version 3.2.3 (R Development Core Team, Vienna, Austria) and SPSS Statistics (SPSS Inc., Chicago, IL). All reported p values were two‐sided, and p < .05 was considered statistically significant.
Results
Patient Characteristics
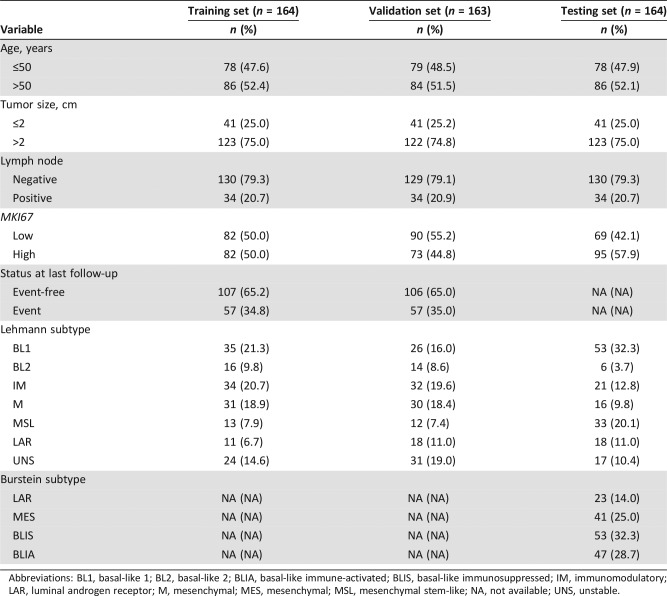
We collected 470 TNBC samples from two GEO datasets (GSE31519 and GSE76275) with essential clinical information. To obtain a robust model, 327 TNBC microarrays from GEO dataset GSE31519 were allocated into training (n = 164) and validation (n = 163) sets using stratified random sampling by microarray batches. However, the disparity of baseline positive lymph node rate (20.8% in GSE31519 and 50.3% in GSE76275) may introduce heterogeneity and lower the validation performance. To overcome this disadvantage, using a matched pairs design, we randomly sampled the paired subject in GSE76275 with the same clinical variable categories with replacement and constructed a testing set (n = 164). The average age of patients was 51.9 years (standard deviation [SD] 11.9, median 51, range 29–80), 53.2 years (SD 12.9, median 52, range 30–84), and 54.2 years (SD 12.6, median 51, range 26–86) in training, validation, and testing cohorts, respectively. The median follow‐up time was 88 months in the training and validation sets. The survival data of the testing set were not available. Table 1 and supplemental online Table 1 depict the clinical characteristics of the subjects.
Table 1. Characteristics of the training, validation, and testing sets.
Abbreviations: BL1, basal‐like 1; BL2, basal‐like 2; BLIA, basal‐like immune‐activated; BLIS, basal‐like immunosuppressed; IM, immunomodulatory; LAR, luminal androgen receptor; M, mesenchymal; MES, mesenchymal; MSL, mesenchymal stem‐like; NA, not available; UNS, unstable.
Establishment and Performance of the 3′UTR Panel to Predict Lymph Node Status in TNBC Patients
The R package “ERI‐expr” was used to analyze the APA profile in microarrays from either HG‐U133A or HG‐U133 plus 2.0 platforms. As a result, 3,210 APA sites in 1,933 unique genes and 6,045 APA sites in 3,542 genes were identified in HG‐U133A and HG‐U133 plus 2.0, respectively. If a gene had multiple APA sites, we reported the site closest to the 5′ end because 90% significant APA dynamic events occurs at the first APA site [28].
Using the ERI values, we developed a prediction model of the lymph node status in TNBC patients with an elastic net approach to the training set after the initial feature filtering via univariate logistic regression analysis. We calculated the risk scores for all patients as a weighted sum of the selected six 3′UTRs, and the optimal penalty parameter was chosen by leave‐one‐out cross validation via 1SE criteria, which chooses the simplest model whose accuracy is comparable with the best model. The final model was a linear combination of the features selected via elastic net. Specifically:
| (1) |
The coefficient is the estimated weight of the information contributed by each predictor, and the risk score gives the logarithmic odds of LNM. For the sake of intuitiveness, we computed the standardized risk scores (z‐scores) using the parameters of linear transformation determined by the training set. As shown in supplemental online Figure 1, the distribution of the standardized risk score was unimodal with similar peaks among the three sets. To divide patients into high‐ and low‐risk categories, we went through all risk scores, and risk score −0.965 (standard risk score 0.870) with maximal likelihood according to the logistic regression was selected as threshold. Patients with standardized risk scores >0.870 were classified as being at high risk of LNM (high‐risk group), whereas those with standardized risk scores ≤0.870 were categorized as being at low risk of LNM (low‐risk group).
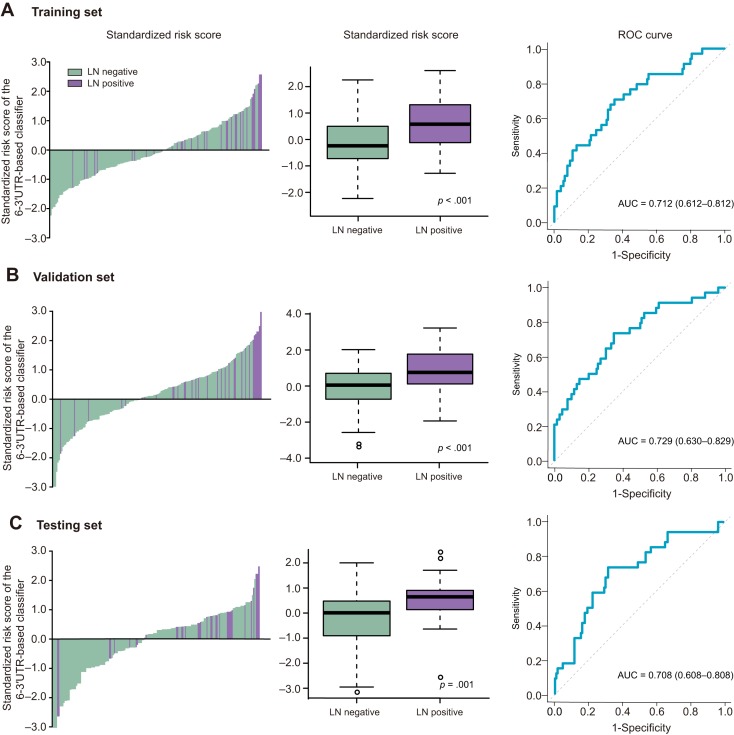
In the training cohort, patients at high risk had a higher risk of LNM (p < .001; OR 5.25, 95% CI 2.25–12.2). The 3′UTR panel showed a high accuracy in predicting negative lymph node status, with a negative predictive value (NPV) of 85.6% in the training set. The distribution of standardized risk scores was assessed in the training set for lymph node‐positive and ‐negative cancers, respectively (Fig. 1A). ROC curve analyses were used to evaluate the predictive power of the six‐member 3′UTR panel to discriminate the lymph node status of TNBC patients, resulting in an AUC of 0.712 (95% CI 0.612–0.812; Fig. 1A).
Figure 1.
Risk score by the 6‐3′UTR‐based classifier and ROC curves. (A) Training set. (B) Validation set. (C) Testing set.
Abbreviations: 3′UTR, 3′ untranslated region; LN, lymph node; ROC, receiver operator characteristic.
In the validation and testing sets, we applied the same threshold and split the patients into two LNM risk groups to test the robustness of the panel. Logistic analysis showed a significant difference between high‐ and low‐risk groups in validation (p = .001; OR 3.86, 95% CI 1.62–9.16) and testing sets (p = .028; OR 2.63, 95% CI 1.11–6.23). The NPV was 85.4% and 82.7% in the validation and testing set, respectively. ROC curve revealed an AUC of 0.729 (95% CI 0.630–0.829) in the validation set and an AUC of 0.708 (95% CI 0.608–0.808) in the testing set (Fig. 1B, 1C).
Next, we investigated whether there was any correlation between the six‐member 3′UTR‐panel (ADD2, COL1A1, APOL2, IL21R, PKP2, and EIF4G3) and the available clinical factors (age, tumor size, lymph node, MKI67, and status at follow‐up). Pearson's chi‐square test confirmed a significant correlation between the risk category and the lymph node status in all sets (p < .05 for all; supplemental online Table 2). There was no significant correlation between the six‐member 3′UTR‐panel and age, tumor size, or status at follow‐up (p > .05 for all), and no association was observed between MKI67 and the risk category in the training and validation sets (p > .05; supplemental online Table 2). As represented in supplemental online Figure 2, ADD2, APOL2, IL21R, and PKP2 displayed 3′UTR shortening in the nodal‐positive samples, whereas COL1A1 and EIF4G3 showed a preference for 3′UTR lengthening in cases with nodal involvement. Using the median ERI of the training set as a threshold, the panel member genes were classified into 3′UTR shortening or lengthening groups. The APA status of APOL2, COL1A1, ADD2, IL21R, PKP2 was correlated with the lymph node status in at least one set (supplemental online Tables 3–7). The association between the lengthening of EIF4G3 and nodal positivity is of borderline significance (supplemental online Table 8). No significant correlation between the ERI of panel members and MKI67 expression was observed (supplemental online Fig. 3).
The 3′UTR Panel Adds Significant Predictive Information to Clinical Variables
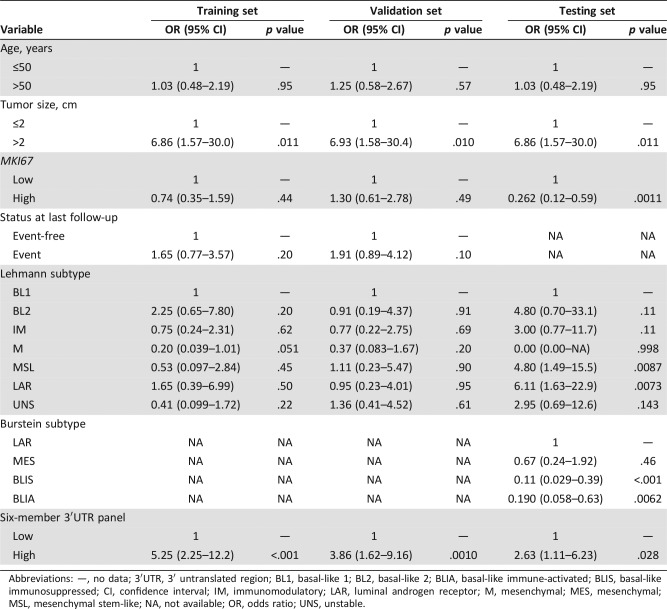
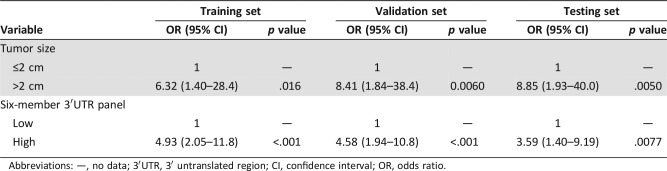
We assessed the additional predictive power of the six‐member 3′UTR‐based panel compared with clinical factors (age at diagnosis, tumor size, MKI67, and status at last follow‐up) and two TNBC subtypes (Lehmann and Burstein subtypes) using a univariate and multivariate logistic regression. Univariate analysis revealed that the six‐member 3′UTR panel and tumor size were significant predictive factors in the training, validation, and testing sets throughout (all p < .05; Table 2). Multivariate logistic regression analysis revealed that the six‐member 3′UTR‐based panel retained significant predictive accuracy (Training set: OR 4.93, 95% CI 2.05–11.8, p < .001; Validation set: OR 4.58, 95% CI 1.94–10.8, p < .001; Testing set: OR 3.59, 95% CI 1.40–9.19, p = .0077) after adjustment by tumor size (Table 3). The 3′UTR panel and tumor size were shown to be independent predictive factors for LNM.
Table 2. Univariate analysis of clinicopathological variables and six‐member 3′UTR panel.
Abbreviations: —, no data; 3′UTR, 3′ untranslated region; BL1, basal‐like 1; BL2, basal‐like 2; BLIA, basal‐like immune‐activated; BLIS, basal‐like immunosuppressed; CI, confidence interval; IM, immunomodulatory; LAR, luminal androgen receptor; M, mesenchymal; MES, mesenchymal; MSL, mesenchymal stem‐like; NA, not available; OR, odds ratio; UNS, unstable.
Table 3. Multivariate analysis of tumor size and six‐member 3′UTR panel.
Abbreviations: —, no data; 3′UTR, 3′ untranslated region; CI, confidence interval; OR, odds ratio.
Furthermore, we investigated the relationship between tumor size and APA status of six panel members and found the 3′UTR shortening status of all panel genes except APOL2 had no significant correlation with tumor size (supplemental online Tables 3–8), which supports that the 3′UTR panel provides additional predictive information of lymph node status.
Integrative 3′UTR‐Based Model Predicts Negative Lymph Node Status in TNBC Patients
Next, we assessed if combining the 3′UTR panel and tumor size (comprehensive model) could improve the predictive accuracy of negative nodal status. A combination of the 3′UTR panel and tumor size classified the patients into four subgroups: low‐risk/≤2cm, low‐risk/>2cm, high‐risk/≤2cm, and high‐risk/>2cm. Table 4 represents the lymph node status among different risk subgroups in all three sets. Low‐risk/≤2cm group achieved a higher NPV of 97.2%, 100%, and 100% in training, validation, and testing sets, respectively, which indicated that the comprehensive model could successfully identify TNBC patients with low LNM risk. Fisher's exact test confirmed that the comprehensive model reclassified the patients into different LNM risk categories (all p < .001).
Table 4. Comparisons of lymph node status among different risk groups.
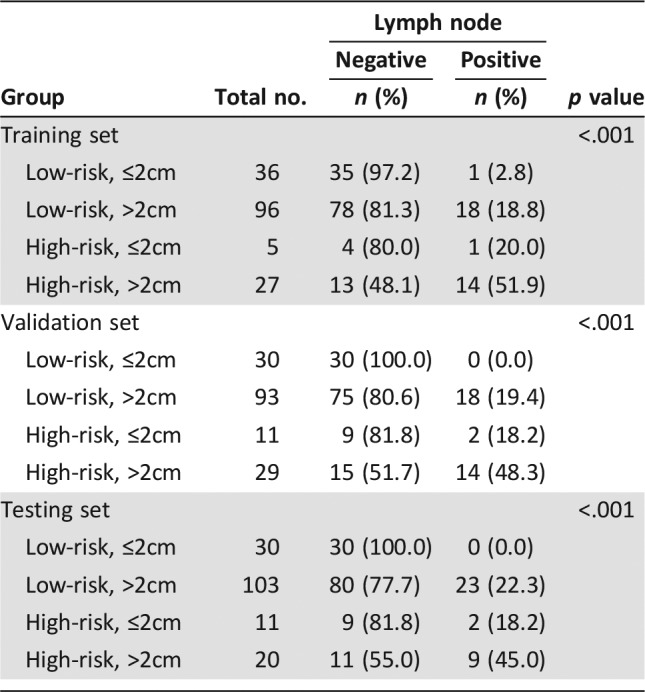
Moreover, we investigated the relationship between TNBC subtyping approaches and LNM. As shown in supplemental online Table 9, the lymph node status was different among the Lehmann subtypes in the training (p = .048) and testing set (p = .006), but no significant correlation was observed in the validation set (p > .05). Besides, COL1A1 and APOL2 were correlated to Lehmann subtypes as well (p < .05 in all sets; supplemental online Tables 4, 5). The Burstein subtypes were significantly correlated with lymph node status (p < .001; supplemental online Table 10), and ADD2, COL1A1, APOL2, IL2R, and PKP2 were correlated to the Burstein subtypes (all p < .05; supplemental online Tables 3–7). These findings suggest classification patterns related to the various ways of subtyping of TNBC may be potential approaches for LNM risk evaluation. However, because we did not have the Burstein subtype information in the training and validation sets, the results need to be further validated in other independent cohorts.
Discussion
In this study, we retrospectively analyzed publicly available TNBC microarrays and profiled the 3′UTR dynamics using annotated APA data. We constructed and validated a novel model composed of six‐member (ADD2, COL1A1, APOL2, IL21R, PKP2, and EIF4G3) 3′UTRs to improve the axillary lymph node status prediction for patients with operable TNBC. Moreover, combining the 3′UTR panel and tumor size could reliably predict patients with negative nodal involvement who can avoid axillary surgery. The integrative 3′UTR‐based model retained high prediction accuracy in the training, validation, and testing sets. We chose TNBC patients as our study population because this subtype of breast cancer includes highly heterogeneous diseases with diverse clinical courses and relapse risk [2], [29]. To the best of our knowledge, this is the first study to propose a 3′UTR panel in TNBC for nodal involvement prediction.
In recent years, great progress has been made in axillary management of breast cancer patients. According to the latest guidelines from the American Society of Clinical Oncology, women with one or two metastatic sentinel lymph nodes who are planning to undergo breast‐conserving surgery with whole‐breast radiotherapy should not undergo axillary lymph node dissection (in most cases) in light of the results of the ACOSOG Z0011 trial [30]. The SOUND trial is questioning whether axillary surgery is needed in “low‐risk” breast cancer patients with positive ER staining [8]. A retrospective cohort from Royal Marsden revealed the long‐term outcomes of patients with “low‐risk” breast cancers who omitted axillary surgery (postmenopausal, <20 mm grade 1 or <15 mm grade 2, lymphovascular invasion negative and ER positive). The cohort achieved a favorable axillary recurrence (AR) rate and distant disease‐free survival (DDFS). The SOUND‐eligible subset had a 5‐ and 10‐year AR rate of 1.6% and 2.7%, respectively, and DDFS was 100% and 95.8% at 5 and 10 years, respectively [10]. It suggests carefully selected breast cancer patients with favorable biology might be safely spared any axillary surgery. In this study, we aimed to identify TNBC patients with low risk of axillary lymph node involvement who would benefit from avoiding SLNB. A systematic review of 69 trials of SLNB (8,059 patients) showed that sentinel lymph nodes could be identified in 95% of patients with an average false‐negative rate of 7.3% (range 0%–29%) [31]. The integrative 3′UTR‐based model proposed in this study achieved false negative rates of 2.8%, 0%, and 0% in the training, validation, and testing set, respectively. Thus, the NPV of the model is comparable to SLNB. As more patient information is not available from the GEO database (ultrasound, mammogram, fine needle biopsy or core needle biopsy results, etc.), we had limited clinical variables (age, tumor size, MKI67) for prediction modeling. With carefully preoperative evaluation of the axillae nodal status (cN staging), we could rule out the patients with clinically positive nodes and further elevated the accuracy of our model. In this study, we combined transcriptome data with traditional clinical variable (tumor size) to evaluate the LNM risk of breast cancer patients. Recently, the role of biological factors and gene expression assays have been increasingly recognized in breast cancer management. In the eighth edition of the AJCC TNM staging system for breast cancer, tumor biomarkers and low Oncotype DX recurrence scores can alter prognosis and stage, which addresses the importance of tumor biology [18]. Our results showed that shortening of four 3′UTRs (ADD2, APOL2, IL21R, and PKP2) predicted LNM in patients with TNBC. This is in agreement with the emerging role of 3′UTR shortening, which allows the escape of oncogenes from microRNA repression, thus enhancing tumor progression [12], [15]. As expected, most of the members with shortened 3′UTR in the panel were tumor‐associated genes. ADD2 has conserved 3′UTR isoforms, and the DNA methylation status of ADD2 is a screening marker of colorectal cancer [32], [33]. IL21R is highly expressed in TNBC and enhances the invasion and migration of IL21R+ breast cancer cells in a dose‐dependent manner [34]. PKP2 harbors driver mutations that affect transcriptional factors regulating metastasis gene signatures in metastatic breast cancer [35]. The remaining two genes (COL1A1 and EIG4G3) harbored lengthened 3′UTRs in nodal‐positive cases. COL1A1 was identified as a key gene for noninflammatory breast cancer [36]. Our recent work [37] established the competing endogenous RNA (ceRNA) network of 3′UTR dynamics in cancer and found the microRNA response elements (MRE) harbored in the 3′UTR can interact with each other and affect gene expression via ceRNA mechanisms. We presume the additional MRE in lengthened 3′UTR results in downregulated gene expression, leading to abnormalities in multiple signaling pathways and tumor metastasis.
Traditionally, clinical palpability of the mass in axilla, tumor size, and the tumor grade have been recognized to be correlated to LNM. Numerous markers, including polysomy of chromosome 7, nm23, and HRad17, are correlated to the lymph node status [38]. However, no single marker or combination of markers is sufficient to obviate surgical axillary staging. In this study, we found basal‐like immunosuppressed and basal‐like immune‐activated of Burstein subtype predicted a lower risk of LNM. Because Burstein subtype information was only available in the testing cohort, the results need to be further validated in other independent cohorts. Recently, accessory examinations have been used to increase the predicted probability of nodal involvement in clinical practice. For instance, the ultrasonic features of lymph nodes can help differentiate diagnosis of nodal status in breast cancer and support clinical decision‐making [39]. Despite the high positive predictive value for 18F‐fluorodeoxyglucose (FDG)‐positron emission tomography (PET) of nodal involvement in breast cancer, FDG‐PET evaluation is not a sufficient indicator for nodal involvement; thus, axillary surgery cannot be avoided in node‐PET‐negative patients [40]. Models based on microarray data for prediction of LNM were previously reported in bladder cancer [41] and hepatocellular carcinoma [42]. However, few studies have reported risk prediction tools for LNM using genomic scale data in breast cancer.
Limitations should be acknowledged for this study. First, in this retrospective study, we tested the associations between the integrative 3′UTR‐based model with the LNM status rather than true prediction. To evaluate the performance of the model, the data from an independent TNBC cohort, with much higher proportion of nodal involvement (50.3%) than that in real world (22.8%) [5], was used in this study. The disparity in baseline LNM rate may introduce heterogeneity and lower the performance of the model. In the GSE76275 cohort, the AUC of the 3′UTR panel was 0.643 (95% CI 0.553–0.733) and the NPV was 84.2% (supplemental online Fig. 4), which was inferior to that in the training and validation sets. In this study, a matched pairs design was used, and the subjects from the GSE76275 cohort were randomly sampled with replacement to construct a new cohort (testing set), of which the baseline characteristics were comparable to those of the training set. Thus, a superior NPV of 100%, comparable to SLNB, was achieved in the testing set, which merits a prospective study to validate the model. The preoperative node status (cN stage) and treatment information (surgery, chemotherapy, and radiotherapy) of the recruited patients were not known. Future studies should only include patients with clinically negative lymph nodes, and the stratification analysis is suggested to be performed according to the surgical procedures (breast‐conserving surgery or mastectomy). Second, because the microarrays involved in this study were profiled through Affymetrix HG‐133A or HG‐U133 plus 2.0, which represent 3′UTRs from ∼9.7% of human protein‐coding genes (1,933 genes), the 3′UTRs identified here may not represent the complete 3′UTR dynamics of the whole transcriptome. We chose these platforms mainly because of their wide use in current studies, and the number of accessible chip data with clinicopathological information was sufficient for predictive analysis and modeling. Besides, the computational results of “ERI‐expr” algorithms we used in this work were validated by reverse transcription polymerase chain reaction in a recent paper [43]. Despite “ERI‐expr,” there are several APA detection algorithms for different platforms, including APADetect, Dapars, and 3′‐seq. It remains to validate the model across platforms in the future. Besides, experimental studies of the selected 3′UTRs are needed to provide the functional and mechanism insights of 3′UTR dynamics in breast cancer.
Conclusion
This study presents a powerful 3′UTR‐based model by integrating and profiling publicly available microarray data. This novel model may permit a no axillary surgery option for selected patients and guide the personalized decision of axillary surgery in TNBC patients. Once further validated in a larger independent cohort, the prediction tool could benefit patients with low risk of LNM and facilitate individualized therapy of TNBC.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81602316, 81672601, 31671380, 81572583), the Shanghai Committee of Science and Technology Funds (15410724000), the Ministry of Science and Technology of China (MOST2016YFC0900300, National Key R&D Program of China), and the Research Fund for the Doctoral Program of Higher Education of China (20130071110057). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Xin Hu, Email: xinhu@fudan.edu.cn.
Peng Wang, Email: wangpeng@picb.ac.cn.
Zhi‐Ming Shao, Email: zhimingshao@yahoo.com.
Author Contributions
Conception/design: Lei Wang, Xin Hu, Peng Wang, Zhi‐Ming Shao
Provision of study material or patients: Lei Wang, Xin Hu, Peng Wang, Zhi‐Ming Shao
Collection and/or assembly of data: Lei Wang, Peng Wang
Data analysis and interpretation: Lei Wang, Peng Wang
Manuscript writing: Lei Wang, Xin Hu, Peng Wang, Zhi‐Ming Shao
Final approval of manuscript: Lei Wang, Xin Hu, Peng Wang, Zhi‐Ming Shao
Disclosures
The authors indicated no financial relationships.
References
- 1.Metzger Filho O, Tutt A, de Azambuja E et al. Dissecting the heterogeneity of triple‐negative breast cancer. JClin Oncol 2012;30:1879–1887. [DOI] [PubMed] [Google Scholar]
- 2.Bianchini G, Balko JM, Mayer IA et al. Triple‐negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;13:674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann BD, Bauer JA, Chen X et al. Identification of human triple‐negative breast cancer subtypes and preclinical models for selection of targeted therapies. JClin Invest 2011;121:2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burstein MD, Tsimelzon A, Poage GM et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple‐negative breast cancer. Clin Cancer Res 2015;21:1688–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houvenaeghel G, Sabatier R, Reyal F et al. Axillary lymph node micrometastases decrease triple‐negative early breast cancer survival. Br J Cancer 2016;115:1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martelli G, Boracchi P, Ardoino I et al. Axillary dissection versus no axillary dissection in older patients with t1n0 breast cancer: 15‐year results of a randomized controlled trial. Ann Surg 2012;256:920–924. [DOI] [PubMed] [Google Scholar]
- 7.International Breast Cancer Study Group , Rudenstam CM, Zahrieh D et al. Randomized trial comparing axillary clearance versus no axillary clearance in older patients with breast cancer: First results of international breast cancer study group trial 10‐93. JClin Oncol 2006;24:337–344. [DOI] [PubMed] [Google Scholar]
- 8.Gentilini O, Veronesi U. Abandoning sentinel lymph node biopsy in early breast cancer? A new trial in progress at the European Institute of Oncology of Milan (SOUND: Sentinel node vs Observation after axillary UltraSouND). Breast 2012;21:678‐681. [DOI] [PubMed] [Google Scholar]
- 9.della Rovere GQ, Bonomi R, Ashley S et al. Axillary staging in women with small invasive breast tumours. Eur J Surg Oncol 2006;32:733–737. [DOI] [PubMed] [Google Scholar]
- 10.O'Connell RL, Rusby JE, Stamp GF et al. Long term results of treatment of breast cancer without axillary surgery ‐ Predicting a SOUND approach? Eur J Surg Oncol 2016;42:942–948. [DOI] [PubMed] [Google Scholar]
- 11.Moon HG, Han W, Kim JY et al. Effect of multiple invasive foci on breast cancer outcomes according to the molecular subtypes: A report from the Korean Breast Cancer Society. Ann Oncol 2013;24:2298–2304. [DOI] [PubMed] [Google Scholar]
- 12.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat Rev Genet 2013;14:496–506. [DOI] [PubMed] [Google Scholar]
- 13.Xia Z, Donehower LA, Cooper TA et al. Dynamic analyses of alternative polyadenylation from RNA‐seq reveal a 3′‐UTR landscape across seven tumour types. Nat Commun 2014;5:5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkon R, Drost J, van Haaften G et al. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol 2012;13:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009;138:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris AR, Bos A, Diosdado B et al. Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res 2012;18:5256–5266. [DOI] [PubMed] [Google Scholar]
- 17.Fu Y, Sun Y, Li Y et al. Differential genome‐wide profiling of tandem 3′UTRs among human breast cancer and normal cells by high‐throughput sequencing. Genome Res 2011;21:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giuliano AE, Connolly JL, Edge SB et al. Breast cancer‐Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67:290–303. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Hu X, Wang P et al. The 3′UTR signature defines a highly metastatic subgroup of triple‐negative breast cancer. Oncotarget 2016;7:59834–59844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rody A, Karn T, Liedtke C et al. A clinically relevant gene signature in triple negative and basal‐like breast cancer. Breast Cancer Res 2011;13:R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lembo A, Di Cunto F, Provero P. Shortening of 3′UTRs correlates with poor prognosis in breast and lung cancer. PLoS One 2012;7:e31129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics 2007;8:118–127. [DOI] [PubMed] [Google Scholar]
- 23.Zou H, Hastie T. Regularization and variable selection via the elastic net. JR Stat Soc Series B Stat Methodol 2005:20. [Google Scholar]
- 24.Simon N, Friedman J, Hastie T. Regularization paths for Cox's proportional hazards model via coordinate descent. JStat Softw 2011:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krstajic D, Buturovic LJ, Leahy DE et al. Cross‐validation pitfalls when selecting and assessing regression and classification models. JCheminform 2014;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gautier L, Cope L, Bolstad BM et al. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307–315. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Li J, Gray WH et al. TNBCtype: A subtyping tool for triple‐negative breast cancer. Cancer Inform 2012;11:147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lianoglou S, Garg V, Yang J et al. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue‐specific expression. Genes Dev 2013;27:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foulkes WD, Smith IE, Reis‐Filho JS. Triple‐negative breast cancer. N Engl J Med 2010;363:1938–1948. [DOI] [PubMed] [Google Scholar]
- 30.Lyman GH, Somerfield MR, Bosserman LD et al. Sentinel lymph node biopsy for patients with early‐stage breast cancer: American Society of Clinical Oncology clinical practice guideline update. JClin Oncol 2017;35:561–564. [DOI] [PubMed] [Google Scholar]
- 31.Kim T, Giuliano AE, Lyman GH. Lymphatic mapping and sentinel lymph node biopsy in early‐stage breast carcinoma: A metaanalysis. Cancer 2006;106:4–16. [DOI] [PubMed] [Google Scholar]
- 32.Wei J, Li G, Dang S et al. Discovery and validation of hypermethylated markers for colorectal cancer. Dis Markers 2016;2016:2192853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan M, Iaconcig A, Muro AF. Identification of 3′ gene ends using transcriptional and genomic conservation across vertebrates. BMC Genomics 2012;13:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang LN, Cui YX, Ruge F et al. Interleukin 21 and its receptor play a role in proliferation, migration and invasion of breast cancer cells. Cancer Genomics Proteomics 2015;12:211–221. [PubMed] [Google Scholar]
- 35.Lee JH, Zhao XM, Yoon I et al. Integrative analysis of mutational and transcriptional profiles reveals driver mutations of metastatic breast cancers. Cell Discov 2016;2:16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chai F, Liang Y, Zhang F et al. Systematically identify key genes in inflammatory and non‐inflammatory breast cancer. Gene 2016;575:600–614. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Wang D, Xue M et al. 3′UTR shortening identifies high‐risk cancers with targeted dysregulation of the ceRNA network. Sci Rep 2014;4:5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patani NR, Dwek MV, Douek M. Predictors of axillary lymph node metastasis in breast cancer: A systematic review. Eur J Surg Oncol 2007;33:409–419. [DOI] [PubMed] [Google Scholar]
- 39.Qiu SQ, Zeng HC, Zhang F et al. A nomogram to predict the probability of axillary lymph node metastasis in early breast cancer patients with positive axillary ultrasound. Sci Rep 2016;6:21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujii T, Yajima R, Tatsuki H et al. Prediction of extracapsular invasion at metastatic sentinel nodes and non‐sentinel lymph nodal metastases by FDG‐PET in cases with breast cancer. Anticancer Res 2016;36:1785–1789. [PubMed] [Google Scholar]
- 41.Seiler R, Lam LL, Erho N et al. Prediction of lymph node metastasis in patients with bladder cancer using whole transcriptome gene expression signatures. JUrol 2016;196:1036–1041. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Xiang ZL, Zeng ZC et al. A microrna‐based prediction model for lymph node metastasis in hepatocellular carcinoma. Oncotarget 2016;7:3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miles WO, Lembo A, Volorio A et al. Alternative polyadenylation in triple‐negative breast tumors allows NRAS and c‐JUN to bypass pumilio posttranscriptional regulation. Cancer Res 2016;76:7231–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]