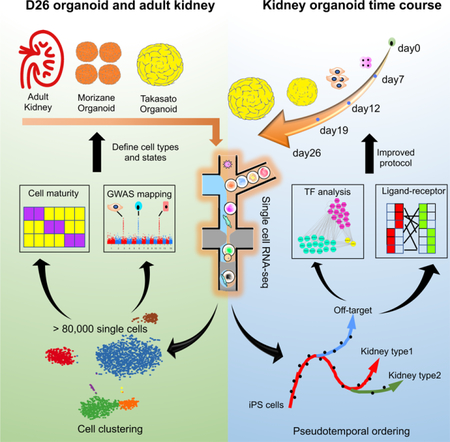
Summary
Kidney organoids derived from human pluripotent stem cells have great utility for investigating organogenesis and disease mechanisms, and potentially as a replacement tissue source, but how closely organoids derived from current protocols replicate adult human kidney is undefined. We compared two directed differentiation protocols by single-cell transcriptomics of 83,130 cells from 65 organoids with single cell transcriptomes of fetal and adult kidney cells. Both protocols generate a diverse range of kidney cells, with differing ratios, but organoid-derived cell types are immature and 10–20% of cells are non-renal. Reconstructing lineage relationships by pseudotemporal ordering identified ligands, receptors, and transcription factor networks associated with fate decisions. Brain-derived neurotrophic factor (BDNF) and its cognate receptor NTRK2 was expressed in the neuronal lineage during organoid differentiation. Inhibiting this pathway improved organoid formation by reducing neurons by 90% without affecting kidney differentiation, highlighting the power of single cell technologies to characterize and improve organoid differentiation.
Graphical Abstract
Introduction
Chronic kidney disease affects 26 – 30 million adults in the United States and 11% of individuals with stage 3 CKD will eventually progress to end stage renal disease (ESRD) - requiring dialysis or kidney transplantation (Coresh et al., 2007). In 2015, 18,805 kidney transplants were performed in the United States, but 83,978 patients were left waiting for a transplant due to a shortage of organs (System, 2017). New treatments to slow progression of kidney disease are desperately needed but progress has been slow in part because the kidney is a complex organ but also because the relevance of rodent kidney models to human kidney disease is debated (de Caestecker et al., 2015).
In this context, the emergence of methods to direct the differentiation of pluripotent human stem cells (PSC) to kidney organoids has been received with great excitement (Lam et al., 2013; Morizane and Bonventre, 2017; Taguchi and Nishinakamura, 2017; Takasato et al., 2015; Xia et al., 2013). Over the last four years several groups have published stepwise protocols, all based upon kidney development during embryogenesis, resulting in generation of kidney tissue in vitro (Morizane and Bonventre, 2017; Taguchi and Nishinakamura, 2017; Takasato et al., 2016; Xia et al., 2014). These protocols modulate activity of several signaling pathways, principally Wnt and Fgf, to generate renal progenitor populations that ultimately self-organize. Mature organoids contain up to hundreds of nephron structures including glomeruli, properly segmented tubules and interstitial cell types.
The ability to grow kidney organoids from patient-derived tissue offers unprecedented opportunities for the investigation of human kidney development, homeostasis and disease. For example, kidney organoids have been used to successfully model and screen for modifiers of autosomal dominant polycystic kidney disease (Czerniecki et al., 2018; Freedman et al., 2015), acute kidney injury (Morizane et al., 2015) and vascularization of the glomerular tuft (Sharmin et al., 2016). A long-term goal is to generate transplantable kidneys grown in the laboratory though many challenges remain. Bulk RNA-sequencing has suggested that kidney organoids are most similar to first trimester kidney (Takasato et al., 2015) and recent marker analysis indicates that organoid nephrons are in the late capillary loop stage (Przepiorski et al., 2018), so improving organoid maturation is one such challenge. Yet no comprehensive analysis of exactly which cells kidney are generated by these protocols, their degree of maturation with respect to adult and the extent to which off target cells contaminate organoids has been undertaken to date. This information is a prerequisite for optimizing differentiation protocols in order to ultimately leverage kidney organoids for investigation of the most common adult kidney diseases such as CKD, diabetic nephropathy and acute kidney injury.
Here we have used scRNA-seq and single nucleus RNA-seq (snRNA-seq) to generate comprehensive molecular maps describing kidney organoid cell diversity in two separate, commonly employed differentiation protocols and two separate pluripotent cell lines, as well as in adult human kidney. Our analysis reveals new insights including: 1, both protocols generate at least 12 separate kidney cell types; 2, off-target non-renal cell types are present in all kidney organoids at similar ratios in human induced pluripotent stem cells (iPSC) vs. human embryonic stem cells (hESC); 3, lineage relationships revealed through pseudotemporal ordering during kidney organoid differentiation; 4, kidney organoid cell types are immature when benchmarked against fetal and adult human single cell datasets and 5, brain derived neurotrophic factor (BDNF) inhibition reduces off target neuronal populations by 90% without altering kidney differentiation. These datasets provide a framework for evaluating and improving organoid differentiation protocols using single cell transcriptomics.
Results
Single Cell RNA-seq Defines Cell Diversity in Kidney Organoids
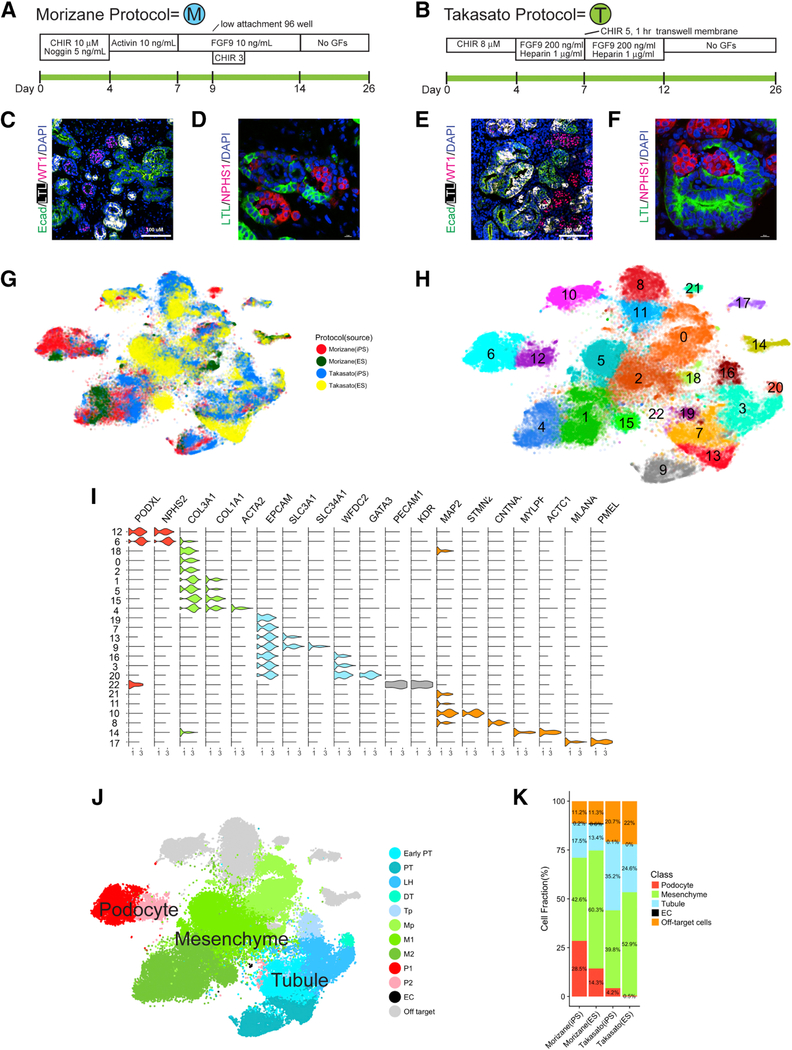
We used the hESC line H9 and the iPSC line BJFF.6, the latter created from newborn male foreskin fibroblasts and reprogrammed with Sendai virus. We confirmed that the BJFF.6 line could efficiently generate kidney organoids using both the protocol described by Takasato et al. (Takasato et al., 2016; Takasato et al., 2015), and the protocol described by Morizane et al. (Morizane and Bonventre, 2017; Morizane et al., 2015) (Figures 1A and 1B, hereafter referred to as the Takasato or Morizane protocol, respectively). Each protocol generated nephron-like structures that closely resembled published reports (Figures 1C–1F).
Figure 1: Comprehensive single-cell RNA sequencing demonstrates development of a spectrum of cell types in kidney organoids.
(A,B) Diagram of human iPS directed differentiation protocols. (C-F) Immunofluorescence analysis of day 26 organoid for proximal tubule (LTL), distal tubule (ECAD), and podocytes (WT1 and NPHS1) from Morizane protocol (C,D) and Takasato protocol, scale bar, 50 μm. (E,F). (G) tSNE projection of all day 26 organoid cells according to protocol (Morizane or Takasato) and cell line (iPS or ES). (H) Unsupervised clustering of all organoid cells reveals 23 separate clusters. (I) Violin plot showing cluster-specific expression of marker genes. (J) Major kidney cell populations depicted after semi-supervised analysis. (K) Proportions of kidney and off-target cell types according to protocol and cell source.
Using DropSeq, we isolated and sequenced mRNA from a total of 71,390 cells harvested from day 26 organoids. Organoids were generated using both hESC and iPSC. The Takasato protocol generated larger organoids, so we sequenced one or two each from separate batches. For the smaller Morizane protocol organoids, we combined 12 organoids each from separate batches. We detected about ~1,930 unique transcripts from ~1,115 genes for each cell (Table S1). After correcting for batch effects by matching mutual nearest neighbors (Haghverdi et al., 2018), we reduced dimensionality by running principal component (PC) analysis on the most highly variable genes, and then performed graph-based clustering on the significant PCs and finally visualized distinct cell sub-groups using t-distributed stochastic neighbor embedding (tSNE). To examine protocol-dependent effects, as well as differences between hESC and iPSC-derived organoids, we projected cells according to protocol and cell source. This revealed co-clustering of cells predominantly based on the protocol used, with less difference attributable to hESC or iPSC source (Figure 1G).
Unsupervised clustering of the entire pooled dataset identified 23 transcriptionally distinct populations present in organoids generated from either the Morizane or the Takasato protocol and derived from either hESC or iPSC (Figure 1H). We annotated broad cluster classes by comparing unique transcript expression with existing RNA-seq datasets and the literature. In Figure 1I, violin plots show expression of marker genes across these clusters. There were four broad classes of cell types in the pooled analysis – podocytes, mesenchyme, tubular epithelia and off-target cells (Figure 1J). The fraction of these cell classes differed both according to protocol, and according to cell source. For example, podocytes made up 28.5% of Morizane organoids derived from iPSC, but only 14.3% derived from hESC. Off-target cell types, by contrast, were similar at about 11% of both iPSC or hESC-derived Morizane organoids, whereas they represented about 21% of both iPSC or hESC Takasato organoids (Figure 1K).
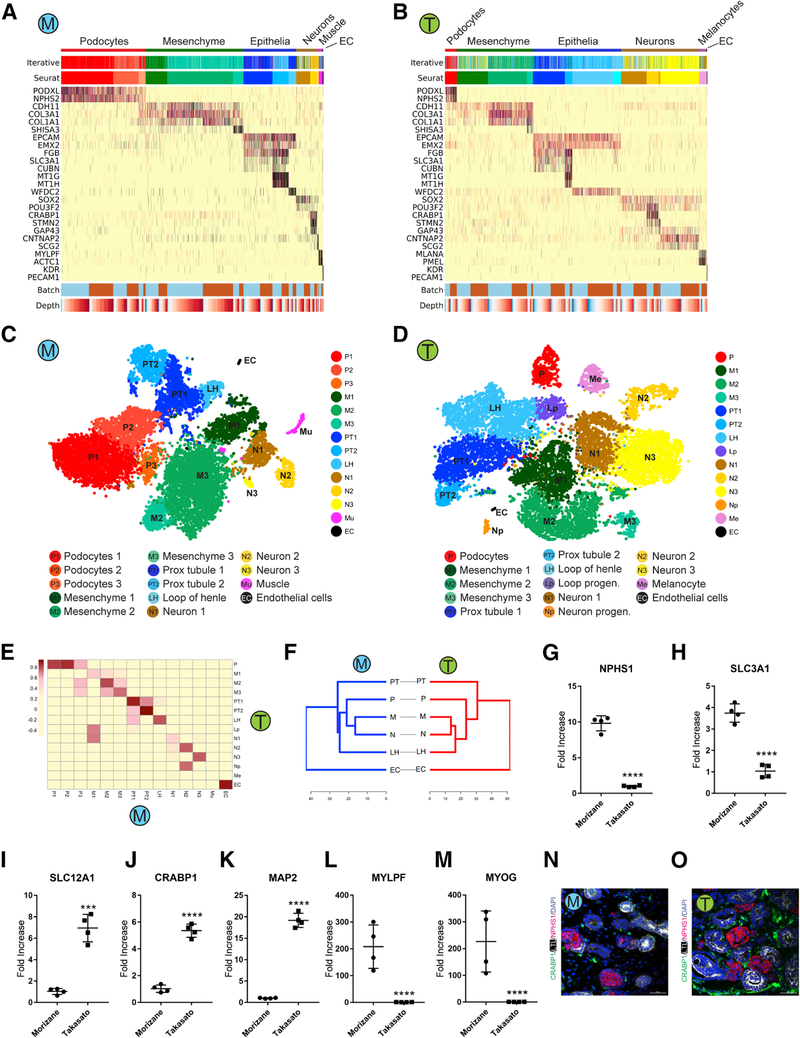
Variations in cell composition between hESC and iPSC-derived organoids complicated efforts to reveal subtle distinctions between cells from the two protocols. We therefore analyzed organoids derived from iPSC and hESC separately in order to evaluate differences between the two protocols. In the iPSC-derived organoids, we analyzed 29,922 single cell transcriptomes from two batches of the Morizane (15,951 cells) or Takasato (14,731 cells) organoids. To examine potential batch effects and to quantify variability among organoids, we projected cells from different batches of iPSC-derived organoids onto the same tSNE diagram, which showed that cells were intermixed regardless of batch (Figures S1A and S1B). Furthermore, cluster-based correlation analysis on both protocols revealed that the correlation for the cells in the same cell cluster from different batches was always greater than the correlation for cells in the same batch from different cell clusters (Figures S1C and S1D). Proportions of cell clusters from different batches was also similar (Figures S1E and S1F). An alternative clustering approach, iterative hierarchical clustering (Baron et al., 2016), identified the same major organoid cell populations (Figures 2A and 2B).
Figure 2: Comparison of kidney cell types and differentiation state in iPS-derived kidney organoids generated with both protocols.
(A,B) Heatmap of all cells clustered by recursive hierarchical clustering and Louvain-Jaccard clustering (Seurat), showing selected marker genes for every population of Morizane protocol (A) and Takasato protocol (B). The bottom bars indicate the batch of origin (“Batch”) and number of UMI detected/cell (“Depth”). (C,D) tSNE plot of cells based on the expression of highly variable genes for the day 26 organoids from Morizane protocol (C) and Takasato protocol (D). The detected clusters are indicated by different colors. (E) Heatmap indicating Pearson’s correlations on the averaged profiles among common cell types for Morizane and Takasato organoids. (F) Dendrogram showing relationships among the cell types in Morizane (left) and Takasato organoid (right). The dendrogram was computed using hierarchical clustering with average linkage on the normalized expression value of the highly variable genes. (G-M) Quantitative PCR comparing cell marker expression for podocyte (NPHS1), PT (SLC3A1), LOH (SLC12A1), neuron (CRABP1 and MAP2), and muscle (MYLPF and MYOG) between organoid protocols. ***p<0.001 and ****p<0.0001. (N,O) Immunofluorescence analysis of neural marker CRABP1 expression (green) in Morizane (N) and Takasato (O) protocols. Cells were co-stained with PT (LTL, white) and podocyte (NPHS1, red) markers. Scale bar, 50 μm.
Clustering on iPSC-derived organoids alone revealed a similar variability in cell frequency between Morizane and Takasato protocols as observed in the global clustering analysis (Figures 2A and 2B). For example, Morizane organoids contained more podocytes, which were marked by the expression of PODXL and NPHS2 (Schwarz et al., 2001), whereas the Takasato protocol produced more tubular epithelial cells based on the expression of EPCAM, SLC3A1 and WFDC2 (Figures 2A–2D) (Litvinov et al., 1994). Despite this variability, both protocols generated very similar cell types since each pair of cell types were highly correlated (Figure 2E). Additionally, dendrograms of analogous cell types from both cell types revealed very similar cell relationships (Figure 2F). We observed substantial numbers of non-renal cell types in both protocols. Morizane organoids contained three neuronal clusters and one muscle cluster. Organoids generated using the Takasato protocol contained four neuronal clusters and one cluster that we could not annotate but that expressed some melanocyte markers such as MLANA and PMEL (Kawakami et al., 1994). Similar findings were observed from separate clustering analysis on the hESC-derived organoids, except that muscle cells were common off-target cells present in both protocols (Figures S1G-S1M).
We confirmed differences in relative abundance of both renal and non-renal cell types by comparing marker gene expression for podocytes (NPHS1) and loop of henle (SLC12A1) as well as muscle (MYLPF, MYOG) and neuronal (CRABP1, MAP2) by qPCR (Figures 2G–2M). To localize neuronal cells, we performed immunostaining for CRABP1, a gene expressed in neuronal clusters from both protocols. CRABP1 protein expression localized to spindly cells present in the interstitium (Figures 2N and 2O). We could identify co-expression of MAP2, a microtubule-associated protein expressed exclusively in neurons (Dehmelt and Halpain, 2005), in many CRABP1+ interstitial cells (Figure S2A), further supporting a neuronal lineage. However, we could also detect coexpression of MEIS1 – a marker of kidney stroma (Chang-Panesso et al., 2018) – in some CRABP1+ cells as well (Figure S2B). Gene imputation analysis showed CRABP1 expression to be present in a small subset of mesenchymal cells from both protocols (Figures S2C and S2D). Expression of CRABP1 and MAP2 was low at earlier timepoints but rose substantially by day 26 (Figures S2F-S2I). Re-analysis of the bulk RNA-seq data in Takasato et al. (Takasato et al., 2015)  and very few of the top 50 stromal progenitor genes were coexpressed in the CRABP1 cluster (Figures S2K and S2L). These analyses confirm the predominant neural identity of CRABP1 expressing cells in the kidney organoid. ). These analyses confirm the predominant neural identity of CRABP1 expressing cells in the kidney organoid. ). These analyses confirm the predominant neural identity of CRABP1 expressing cells in the kidney organoid.
Cell Cycle Analysis
During kidney development, progenitor cell populations are characterized by rapid cell cycle progression that slows progressively with differentiation (Short et al., 2014). We analyzed cell cycle status as a proxy for degree of differentiation (Kowalczyk et al., 2015). We scored all cells from both protocols based on cell cycle gene expression and assigned a cell cycle phase (G2M, S or G1). The total fraction of cells in G2M, S or G1 was similar between the protocols and cell source at 39.1% and 43.9% in the Morizane and Takasato organoids derived from iPSC (Figures S3A and S3B), and 37.7% and 45.1% in hESC-derived organoids, respectively (Figures S3C and S3D). However, in the Morizane organoids, cells in G2M were limited to two cell clusters – a mesenchymal and neuronal cluster. By contrast, in Takasato organoids, G2M phase cells were present as a subset of 6 separate clusters (two mesenchymal, two neuron-like, one unidentified and an epithelial cluster). Cell cycle gene expression was not driving cluster identity. Both the Morizane M1 cluster and the Takasato Lp cluster expressed high levels of the cell cycle gene MKI67, but showed divergent expression of COL3A1 (M1) and POU3F3 (Lp) (Figure S3F).
We interpreted the broader proliferative distribution of the Takasato organoid to potentially reflect that the organoid had been harvested before it was fully differentiated. Indeed, cell cycle analysis on the cells collected from different time points using Takasato protocol revealed that the proportion of cycling cells decreased along the kidney organoid differentiation process (Figure S3E), suggesting that the degree of cell differentiation or cell type maturity might be negatively correlated with the proliferative capacity.
Kidney Organoid Cell Subsets
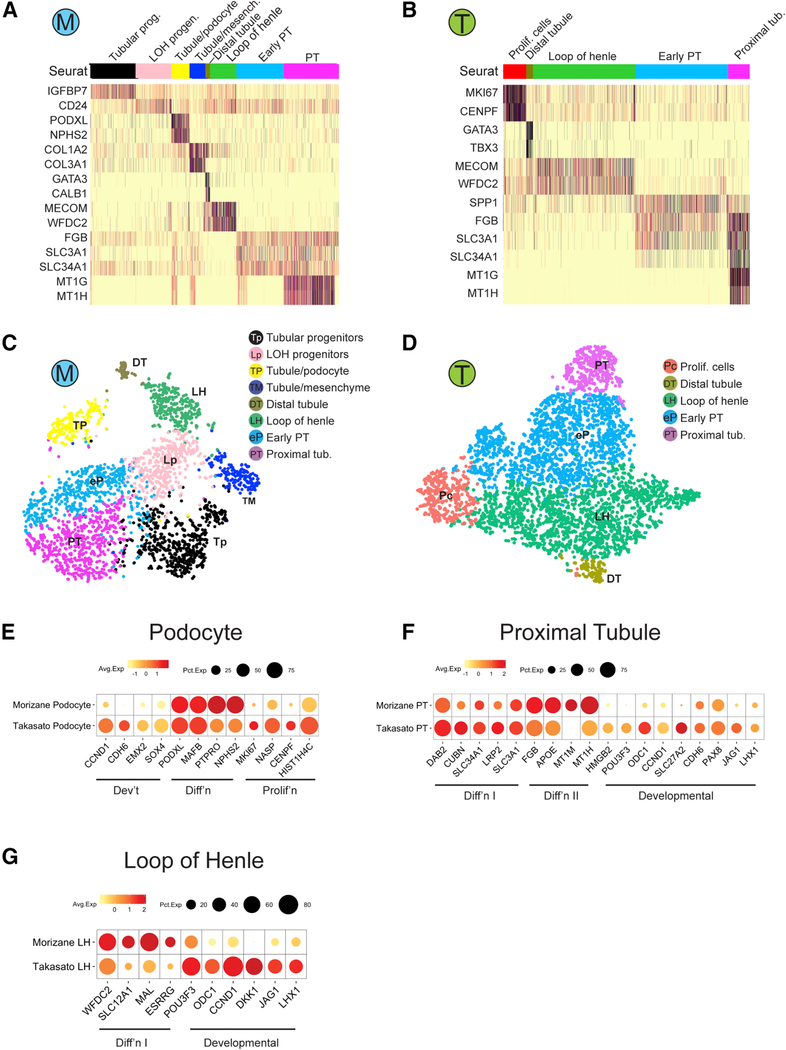
Re-clustering of tubular cells identified additional cell clusters in both protocols. We detected 8 and 5 tubular subtypes in Morizane and Takasato organoids, respectively (Figures 3A-3D). This includes a subpopulation that expressed the ureteric bud marker GATA3 (Labastie et al., 1995) in both protocols. Prior reports have suggested that kidney organoids contain derivatives of both major progenitor populations, the metanephric mesenchyme and the ureteric bud (Takasato et al., 2015). However, several lines of evidence suggest that the GATA3 cluster is actually metanephric mesenchyme-derived distal tubule. First, the GATA3 cell cluster did not express mature collecting duct markers (e.g. AQP2, AQP4), though this could also be explained by immaturity. Second, in addition to principal cells, GATA3 is also expressed in distal convoluted tubule and connecting segment in both human and mouse kidney (Figures S4A and S4B). Third, the Morizane organoid GATA3 cluster also expressed calbindin, a marker of distal tubule (Bindels et al., 1991). We verified that in P1 mouse kidney scRNA-seq, calbindin mRNA is expressed in distal tubule and the ureteric bud tip whereas in our adult kidney snRNA-seq data, calbindin mRNA was exclusively expressed in the distal convoluted tubule and the connecting segment, and not in principal cells (Figures S4A and S4B). Finally, comparison of the GATA3 cluster to adult kidney cell types shows that it is equally or more similar to distal tubule and connecting segment as to principal cells by Pearson’s correlation (Figure 4C). These findings raise significant doubts that ureteric-bud and its derivatives are generated at all using either the Morizane or the Takasato protocol.
Figure 3: Human kidney organoids contain subclasses of tubular epithelial cells.
(A,B) Heatmap showing selected marker genes for every tubular subpopulation of Morizane protocol (A) and Takasato protocol (B) generated from iPS cells. (C,D) tSNE plot of tubular subclusters in kidney organoid from Morizane protocol (C) and Takasato protocol (D). The detected clusters are indicated by different colors. (E-G) Dotplot comparing the expression of cell type signature and developmental/proliferating genes on podocytes (E), proximal tubule (F), and LOH (G) between the two protocols.
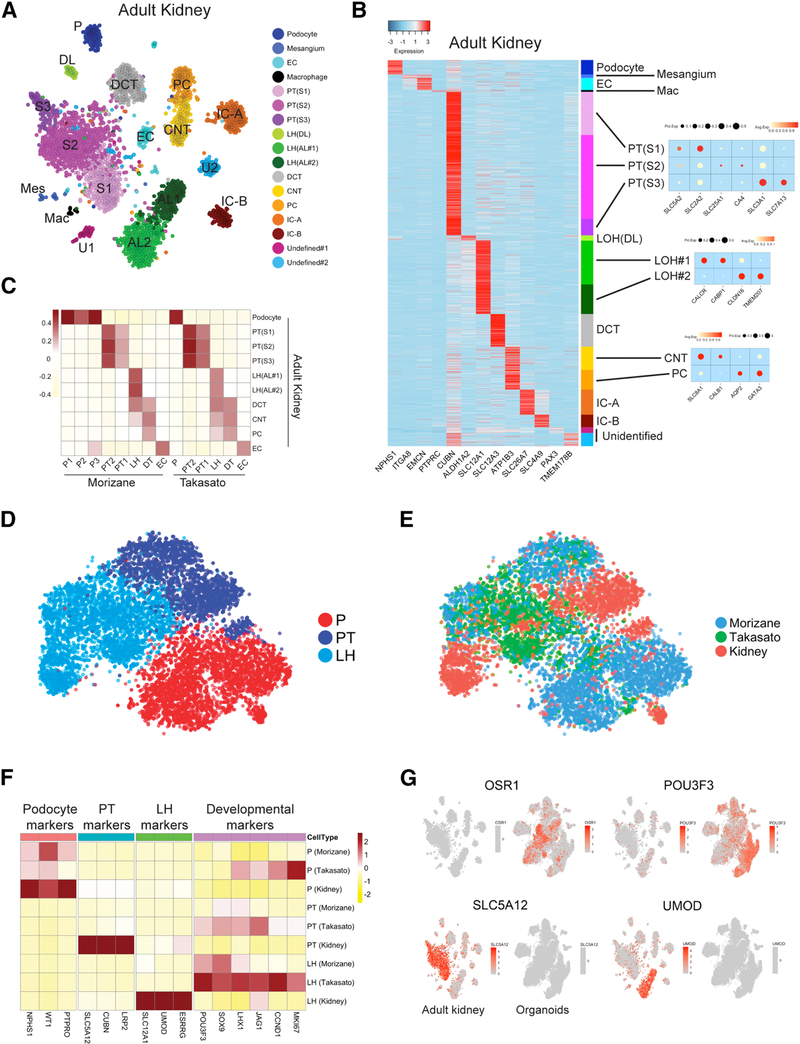
Figure 4: Organoid cell types are immature compared to benchmarked adult kidney cell types.
(A) Unsupervised clustering of snRNA-seq of adult human kidney identified 17 distinct cell types in human adult kidney. That includes 11 tubular cell types, podocytes, mesangium, endothelial cells and macrophages. (B) Heatmap showing uniquely expressed genes for each cluster. (C) Pearson correlation analysis comparing the organoid cell types and their endogenous counterparts in human kidney. Color bar indicates the correlation score. (D) Reclustering of podocytes, proximal tubule (S1-S2) and loop of henle cells derived from both organoids and adult kidney, analyzed using canonical component analysis. (E) Cellular origins (Morizane, Takasato or adult kidney) visualized in the tSNE reveal poor overlap between organoid-derived and adult-derived cells within in each cluster. (F) Comparison of the average expression of marker genes and developmental genes between organoid cell types and adult kidney cell types. Expression value was scaled by z-score. (G) Expression of developmental factors OSR1 and POU3F3 is strong in organoids but almost undetectable in adult kidney. Expression of S1 marker SLC5A12 and loop of henle marker UMOD is strong in adult kidney and undetectable in organoids.
We next compared a panel of developmental and differentiation genes in podocyte, proximal tubule and loop of henle cell clusters across protocols. This revealed higher expression of kidney developmental markers CDH6, EMX2 and SOX4 in the Takasato podocytes (Brunskill et al., 2008). Morizane podocytes had somewhat higher expression of podocyte differentiation markers and lower expression of proliferation markers (Figure 3E). For proximal tubule, both Morizane and Takasato organoids had similar expression of differentiation markers and but Takasato proximal tubule had higher expression of developmental markers. Morizane proximal tubule had higher expression of genes that were difficult to interpret, including metal binding genes MT1M and MT1H (Figure 3F). Loop of henle was more differentiated in the Morizane organoids (Figure 3G).
Quantifying Organoid Kidney Cell Maturity
A critical question is the degree to which kidney organoid cell types resemble their native counterparts in molecular terms. We addressed this question in three ways. First, we compared organoid cell type gene signatures with a recent mouse P1 single-cell RNA-seq dataset (Adam et al., 2017). organoid datasets (Figures S4C and S4D). For both organoid protocols, the M1 mesenchymal clusters showed medium correlation to cap mesenchyme in addition to stroma. Notably, none of the off target clusters (neural, muscle, melanocyte-like) correlated to cell types found in P1 kidney.
We also compared organoid cell clusters to a recently generated single cell dataset generated from human week 16 kidney (Lindstrom et al., 2018b). This analysis revealed excellent correlation of organoid kidney cell types to fetal kidney cell types either by Pearson’s correlation or using a multiclass random forest classifier (Habib et al., 2017) (Figures S4E-S4I). Notably, the off target cell clusters did not map to any human fetal kidney cell types with the sole exception of the Takasato N1 cluster, which showed some correlation to a fetal kidney cluster annotated as cycling. N1 also expresses a strong cell cycle gene signature, likely explaining the correlation (Figure S4H).
Finally, we compared kidney organoid cell types with their adult human counterparts. Attempts at single cell RNA-seq failed, however we were successful in generating adult human kidney single nucleus RNA-seq data from a 62-year-old white male with a serum creatinine of 1.03 mg/dL using the 10X Chromium platform. We sequenced 4,524 nuclei to a similar depth (Table S1) as the organoid datasets, and identified 12 distinct epithelial cell clusters, including podocytes, proximal tubule (S1–3), loop of henle (descending and ascending), distal tubule, connecting segment, principal cells and intercalated cells (type A and type B) (Figures 4A and 4B). The absence of stromal or leukocyte populations most likely reflects either dissociation bias and/or a cell frequency below our limit of detection (Wu et al., 2018).
Single cell RNA-seq measures transcripts from both cytoplasm and nucleus whereas single nucleus RNA-seq measures only nuclear transcripts. Nuclei contain only a fraction of total cell RNA and while nuclear and cytoplasmic mRNAs correlate highly (Barthelson et al., 2007), some protein-coding mRNAs are retained in the nucleus (Bahar Halpern et al., 2015). Despite these differences, single cell and single nucleus RNA-seq datasets predict cell types comparably and with high concordance (Habib et al., 2017; Lake et al., 2017). Therefore, we next correlated all kidney organoid epithelial cell types with their corresponding endogenous counterpart from human adult kidney. We observed an expected correlation between corresponding cell types of organoid and human kidney (Figure 4C). Organoid loop of henle correlated with adult loop of henle, but also distal tubule and collecting duct – likely reflecting their developmental immaturity.
Although organoid and adult kidney epithelial cell types correlated well, our prior analysis suggested that organoid derived cells expressed developmental markers. To visualize overall similarities and differences in cellular transcriptomes from specific organoid and adult cell types, we performed unsupervised clustering of podocytes, proximal tubule and loop of henle clusters from both organoids and adult after batch effects corrected by canonical correlation analysis (Butler et al., 2018), and projected the data by tSNE (Figure 4D). As expected, the analysis revealed three separate clusters. When we then projected cellular origin onto the tSNE, however, there was relatively poor overlap between the organoid-derived vs adult kidney cells within each cluster (Figure 4E). Further emphasizing the transcriptional differences between organoid-derived cells and adult kidney, expression of differentiation markers was much higher in adult cell types, whereas developmental marker expression was much higher in organoid cell types (Figure 4F). For some of these developmental markers (OSR1, POU3F3), expression was high in many organoid cell types in the pooled dataset but undetectable in adult kidney. In the same way, some differentiation markers (SLC5A12, UMOD) were strongly expressed in adult clusters, but undetectable in organoids (Figure 4G). These results indicate that organoid cell types are substantially immature compared to their adult counterparts.
We identified 123 receptors and 97 cognate ligands and mapped their expression to specific adult human kidney cells. Most of these mapped to a single predominant cell type (Figures S5A and S5B). This allowed for development of a simple connectome model for how mature kidney cells might intercommunicate during homeostasis (Figure S5C). Current protocols incubate organoids without any growth factors after 12–14 days, but the expression of so many ligands in mature kidney suggests a possible need to include soluble factors during the maintenance phase of organoid maturation.
Because transcription factors regulate cell state, we next tested the hypothesis that organoid cell immaturity might reflect partial expression of the gene regulatory network present in mature kidney cells. We identified 54 transcription factors present in adult human proximal tubule, over half of which have not been reported previously (Table S2). For example, High mobility group nucleosome-binding domain-containing protein 3 (HMGN3) is a thyroid hormone binding receptor that regulates gene expression, is strongly expressed in proximal tubule, and thyroid hormone is known to regulate renal fluid and electrolyte handling, suggesting HMGN3 may mediate thyroid hormone actions in the proximal tubule (Michael et al., 1972). In human adult podocytes, we identified 38 transcription factors, 25 of which we believe have not been reported previously. We validated expression of six of these transcription factors at the protein level (Figures S5D-S5I). Many of the proximal tubule transcription factors are expressed solely in proximal tubule, whereas podocyte transcription factors are more widely expressed across kidney cell types (Mendeley http://dx.doi.org/10.17632/m4rfg9wb29.1).
Both proximal tubule cells and podocytes derived from organoids expressed only a fraction of the transcription factors we identified in the adult cell types. For example, Takasato protocol proximal tubule expressed 11/54 adult proximal tubule transcription factors, and Morizane proximal tubule only 9/54 (Table S2). Similarly, both Takasato and Morizane protocol podocytes expressed 7/38 adult podocyte transcription factors (Table S3). This result suggests that organoid cells, despite expressing some markers of differentiated cells, are fundamentally different from their terminally differentiated adult counterparts. Collectively, these results identify lineage specific expression of genes that likely regulate cell specification, differentiation and proliferation during kidney organoid maturation.
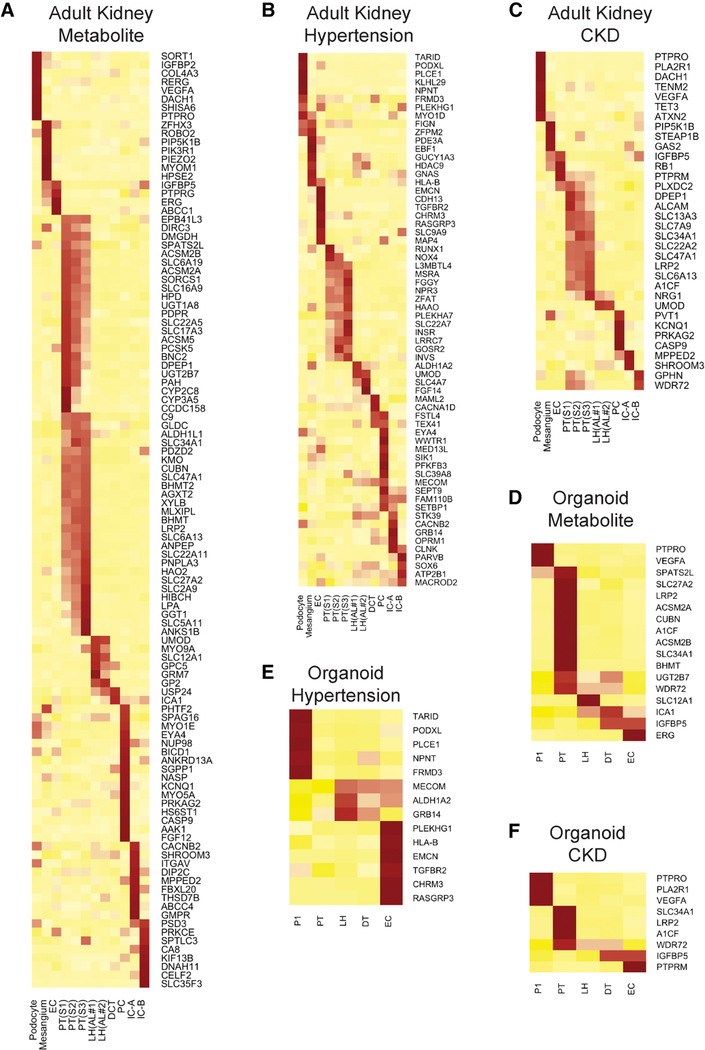
Disease-related Genes Predicted by GWAS are Expressed in Single Cell Types in Adult and Organoid Kidney
Human kidney organoids are already being used to model monogenic human kidney diseases. However there are many more complex trait disease genes that have been identified by Genome-Wide Association Studies (GWAS). Recently Park et al. reported that many human monogenic and complex trait genes are expressed predominantly in a single mouse kidney cell type (Park et al., 2018). In order to gauge how useful kidney organoids might be for modeling disease-relevant genes, we next compared our ability to detect gene expression of GWAS hits in adult kidney vs. organoids. We used established GWAS gene lists including 117 genes for chronic kidney diseases, 275 genes for hypertension and 777 genes for plasma metabolite levels.
We could map expression of 207 of these genes to cell types in our adult kidney single snRNA-seq dataset (Figures 5A–5C). Of these 207 mapped genes, we could only detect expression of 40 of them (19%) in the correct corresponding organoid cell types (Figures 5D–5F). In most cases, we confirmed that these GWAS genes are expressed in only a single kidney cell type (Figure 5). Unexpectedly, podocytes and mesangial cells expressed a substantial number of hypertension genes. These glomerular cell types are not widely believed to play important roles in regulating blood pressure. Consistent with their central role in secretion and reabsorption, proximal tubule had the highest number of genes associated with plasma metabolite levels.
Figure 5: Cell specific expression of disease-relevant genes in adult kidney compared to organoids.
Cell specific expression of genes reported in CKD related GWAS (A), hypertension related GWAS (B), and plasma metabolite levels related GWAS (C) in adult kidney. Each gene reported in a kidney disease related GWAS was assigned to the adult kidney cell type in which it was found to be differentially expressed (likelihood ratio test). Heatmap was used to visualize the z-score normalized average gene expression of the candidate genes for each cell cluster. (D-F) Disease relevant genes identified in (A-C) for which cell-specific expression could also be detected in organoid cell types. Results from both protocols and both cell sources were pooled for the analysis.
As a complementary approach, we used RolyPoly, a regression-based polygenic model that allows prioritization of trait-relevant cell types by combining GWAS and single cell expression datasets (Calderon et al., 2017). We focused on CKD and eGFR GWAS hits, and asked whether trait-relevant cell types identified using the adult kidney dataset were similar to those identified using the organoid datasets. For CKD, RolyPoly identified principal cells, type A intercalated cells, podocytes and proximal tubule as trait-relevant cell types. By contrast, only distal tubule (which might also represent principal cells) was identified using Morizane data and only podocytes using Takasato organoids. Similarly, for eGFR, trait-relevant cell types using adult kidney data included proximal tubule, podocytes, ascending loop of henle and principal cells. The Morizane dataset identified proximal tubule and the Takasato dataset failed to identify any trait-relevant cell types (Figures S5J-S5O).
These results confirm and extend those of Park et al., which was performed in mouse and not human (Park et al., 2018), but also suggest that kidney organoids are limited in their ability to predict trait-relevant cell types in comparison with adult kidney, because many GWAS hits are not expressed in organoid cell types.
Lineage Reconstruction during Kidney Differentiation
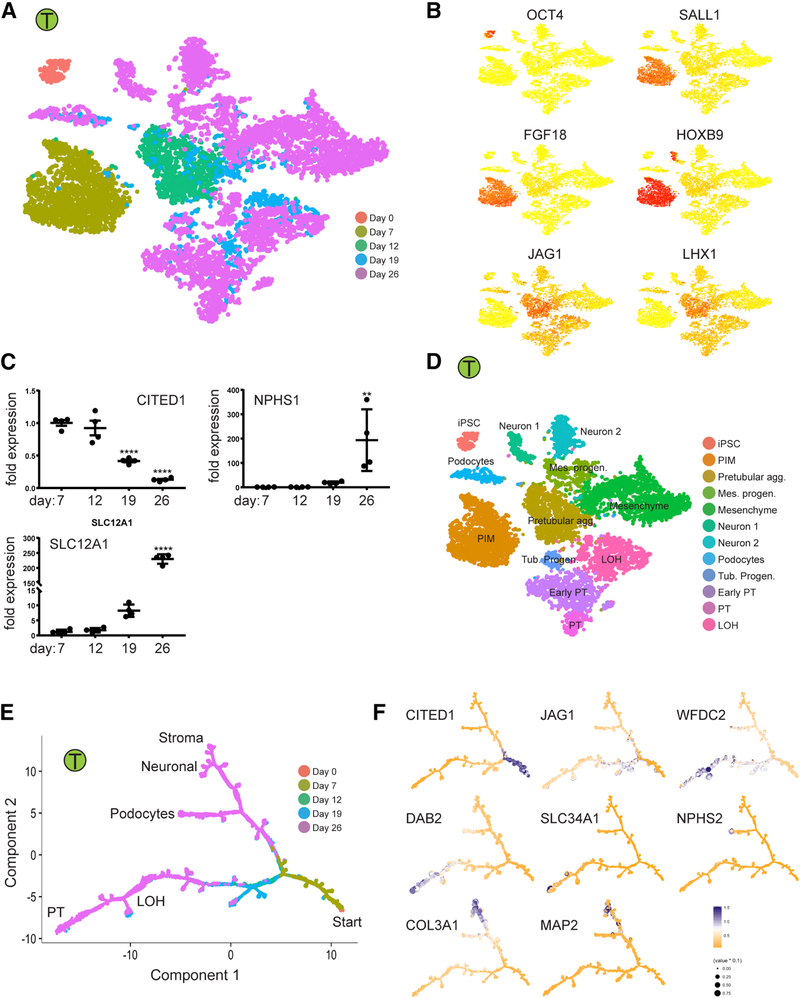
To explore lineage relationships and the mechanisms of cell fate decisions during kidney organoid differentiation, we performed scRNA-seq at separate timepoints during differentiation using the Takasato protocol (days 0, 7, 12, 19 and 26). A total of 9,190 cells from all five timepoints were projected by tSNE, and days 0, 7 and 12 each formed single distinct clusters (Figure 6). Pluripotency gene expression (e.g., POU5F1/Oct4) was completely downregulated by day 7 with upregulation of metanephric mesenchyme markers (SALL1, FGF18 and HOXB9, Figure 6B) (Brunskill et al., 2008). The day 12 cluster most closely resembled the pretubular aggregate, with genes such as JAG1 and LHX1 strongly enriched at this timepoint. Multiple clusters corresponding to differentiating cell types were present at days 19 and 26, and most later clusters contained cells from both time points, reflecting asynchronous differentiation.
Figure 6: Time-course analysis of cells during organoid differentiation reveals lineage relationships.
(A,B) Projecting cells across time-points to the tSNE. Cells were colored by the time point where they were collected (A) or gene expression of stage specific markers (B). (C) Validation of the stage specific marker by qPCR. **p<0.01 and ****p<0.001 versus day 7. (D) Annotation of cell clusters based on gene expression of cell type specific markers. (E,F) Ordering of scRNA-seq expression data according to the pseudotemporal position along the lineage revealed a continuum of gene expression changes from iPSCs to differentiated cell types.
We compared our results with the bulk RNA-seq data from (Takasato et al., 2015) by deconvolving cell frequency across time using a bulk sequence single-cell deconvolution analysis pipeline (Baron et al., 2016). This confirmed downregulation of the pretubular aggregate and posterior intermediate mesoderm and increasing fractions of differentiated cell populations with time (Figures S6A-S6H). While differentiation marker expression generally increased with time in our dataset, and certain progenitor markers such as CITED1 decreased over time (Figure 6C), many genes marking developmental cell types persisted at day 26. Genes reflecting renal vesicle (DKK1) and S-shaped body (JAG1, CCND1, CDH6 and LHX1) continued to be expressed, for example, suggesting an ongoing nephrogenic program (Figures 3E–3G) (Brunskill et al., 2008). Future enhancements to kidney organoid differentiation protocols will need to push maturation of these developmental states towards fully mature kidney cell types.
To detect gene expression changes during organoid differentiation, we reconstructed kidney lineage relationships by performing pseudotemporal ordering using Monocle2 (Qiu et al., 2017). The resulting cell trajectories revealed one major branch point, separating loop of henle and proximal tubule cell fates from podocyte, stromal and neural cell fates (Figure 6E). A second branch point distinguished podocyte from stromal and neural fates. Cell fates were defined by projecting marker gene expression onto the pseudotime trajectories (Figure 6F).
Lineage-Specific Expression of Transcription Factors, Receptors and Ligands During Organoid Differentiation
Although many steps in murine nephrogenesis are well characterized, the transcriptional pathways underlying human kidney development are less well characterized. To identify candidate transcription factors and signaling pathways whose modulation might improve kidney cell maturation and eliminate off-target cell types we performed branched expression analysis modeling (BEAM). We identified a large number (56) of dynamically expressed transcription factors over the course of differentiation (Figure S6I). The analysis assigned expression of these genes to either of the main branches, but it could not resolve single cell cluster expression. We therefore mapped expression of these transcription factors to all 12 major clusters (Figure S6J). Most genes were expressed in only one or a few cell types. We identified five genes (POU2F2, POU3F2, NHLH2, HES6, LHX9) whose expression was limited to a neuronal cluster, and confirmed that their expression corresponded to the neuronal branch by psedotemporal ordering (Figure S6K). A subset of these TFs have previously been implicated in neuronal development. For example, loss of LHX1 prevents formation of the neocortex (Bulchand et al., 2001). Similarly, POU3F2 is required for survival of hypothalamic neural progenitors (Nakai et al., 1995). Thus, the induction of these TFs may be critical for one or more of the neuronal lineages present in kidney organoids. For finer mapping of gene expression changes during fate decisions, we performed BEAM on the podocyte and mesenchyme/neuron and on themesenchyme and neuron branch (Mendeley http://dx.doi.org/10.17632/m4rfg9wb29.1). This analysis included all differentially expressed genes as well as transcription factors alone.
To test whether longer organoid incubation times might improve cell differentiation status, we grew organoids from Takasato protocol out to 34 days and performed scRNA-seq on a total of 6,115 cells (Table S1). We compared expression of differentiation markers across clusters, and discovered that differentiation was generally worse not better at this later timepoint, with loss of endothelial cells, reduced expression of differentiation markers across most clusters, and the emergence of an off-target muscle cell cluster (Figure S7A). We next pooled the day 34 scRNA-seq results with our day 26 results, removed batch effects by matching nearest neighbors (Haghverdi et al., 2018) and reclustered. While there was overlap with kidney cell clusters, the muscle cluster was unrelated to any day 26 cluster (Figures S7B and S7C). Furthermore, we could detect a separate new cluster, also specific only to day 34, that expressed a high percentage of mitochondrial genes indicating that these are unhealthy cells (Figure S7D). Overall, there was a reduction in the fraction of mature kidney cell types, for example mature stroma and proximal tubule, and a substantial increase in off-target cell types at day 34 compared to day 26 (Figure S7E).
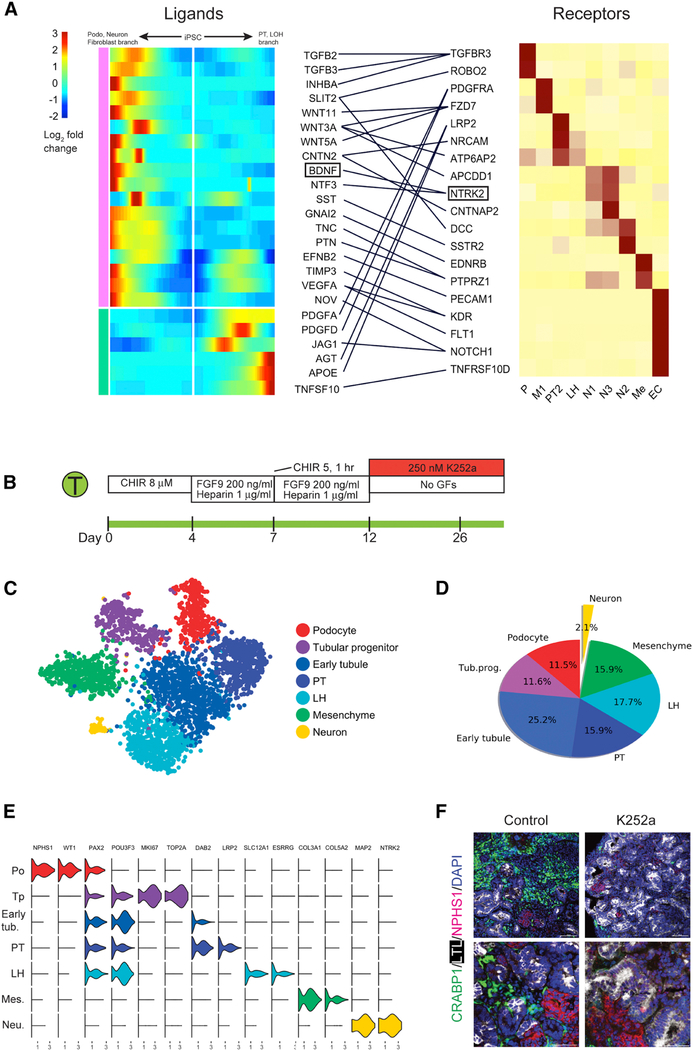
Since organoid differentiation is accomplished by exposure of iPSC to sequential combinations of extrinsic factors, we also searched for ligand and receptor pairs whose expression changed in a lineage specific fashion during organoid differentiation. We identified 19 receptors with 24 cognate ligands in this way and mapped their expression to the major organoid cell types (Figure 7A). NTRK2, which encodes Neurotrophic Tyrosine Kinase Receptor, type 2, was expressed exclusively in neural clusters N1 and N3. Its ligand, brain-derived neurotrophic factor (BDNF) was also strongly induced in the podocyte/neuron/stroma branch.
Figure 7: Reduction in off-target cell differentiation by analysis of cell-specific expression of receptors and ligands during organoid differentiation.
(A) Heat map showing kinetics of branch-dependent ligand expression identified by BEAM (Monocle2) and corresponding cell-specific receptor expression in day 26 organoids from the Takasato protocol. The analysis identified that BDNF expression was induced in the podocyte/mesenchyme/neuron branch and its receptor NTRK2 was exclusively expressed in neurons. (B) Inhibition of the BDNF pathway using K252a (250 nM from days 12 to 26). (C) tSNE of K252a treated organoids showing very small neuronal population. (D) Off target cells made up only 2.1% of the total cells in K252a-treated organoids. (E) Violin plot showing marker gene expression across clusters in K252a-treated organoids. (F) verification of strong reduction in neuronal cells by immunofluorescence staining of an independent organoid batch.
Inhibition of BDNF-NTRK2 Signaling Reduces Off-Target Cells
Since BDNF promotes neuron survival, growth and differentiation (Huang and Reichardt, 2001), we reasoned that inhibition of signaling by its receptor might reduce off target neuron populations in kidney organoids. To test this hypothesis, we first selected a dose of the NTRK2 inhibitor K252a (Tapley et al., 1992) that did not alter gross tubular morphology (Figure S7F). We administered 250nM K252a beginning at day 12 of the Takasato protocol (Figure 7B). Preliminary qPCR data suggested a reduction in off target marker expression (Figures S7G and S7H). We therefore performed single cell RNA-seq on K252a-treated organoids which revealed a 90% reduction in neuronal cells, from 20 – 22% to 2.1%. There was also a decrease in mesenchymal cells from 39.8% to 15.9%, consistent with pseudotemporal ordering that placed neurons and kidney mesenchyme in the same branch. We observed an increase in tubular cells from 35.2% to 70.4% accompanied by an increase in podocytes from 4.2% to 11.5% (Figures 7C–7D). All kidney lineages expressed expected marker genes (Figure 7E). The reduction in neuronal cells was confirmed by immunofluorescence analysis of an independent batch (Figure 7F).
Discussion
Human kidney and kidney organoids are composed of a wide array of cell types, all required for proper development and organ function. Recent studies have begun to characterize these cell types using scRNA-seq revealing important new insights (Adam et al., 2017; Chen et al., 2017; Czerniecki et al., 2018; Der et al., 2017; Lindstrom et al., 2018a; Magella et al., 2018; Park et al., 2018). Fulfillment of the promise of human kidney organoids requires comprehensive characterization of their cell composition, comparison of differing protocols and a better understanding of the degree to which they produce mature, differentiated kidney cell types. Using scRNA-seq, we have established that current protocols generate a remarkable diversity of kidney cell types. We also provide the first direct comparison of separate differentiation protocols, revealing broadly similar outcomes, but important differences in cell ratio and differentiation state. The information provided in this comprehensive dataset will guide future attempts to improve differentiation protocols.
These results will help guide protocol choice for investigators interested in modeling kidney function or disease. Based on increased expression of podocytes with the Morizane protocol, it is better suited for analysis of glomerular biology. Similarly, the Takasato protocol generated more tubular epithelium and is better suited for studying the tubulointerstitium. Unexpectedly, our analysis suggests the apparent absence of ureteric bud-derived cell types. The ureteric bud undergoes branching morphogenesis to form the collecting system, and is required for the formation of an interconnected collecting duct (Costantini and Kopan, 2010). In a recent study, Taguchi and Nishinakamura induced mouse metanephric mesenchyme and ureteric bud progenitors separately, and their recombination led to organoids with much more complex and interconnected collecting duct architecture than previously reported (Taguchi and Nishinakamura, 2017). Similar protocols for human pluripotent stem cell-derived kidney organoids have not yet been established. Our results suggest that establishing conditions that will support growth of the ureteric bud lineage in human kidney organoids is an immediate priority.
One measure of the usefulness of kidney organoids for modeling disease is the degree to which organoid cells express disease-relevant genes. We found that organoid cells expressed about 20% of trait-relevant genes defined by GWAS, when compared with their adult counterparts. Similarly, we could only detect about 20% of the transcription factors present in adult proximal tubule and podocytes, when compared with organoid equivalents. Consistent with these observations, a global comparison of organoid derived cell types with human fetal and adult counterparts revealed that although organoid-derived cells express some markers of terminal differentiation, they are all immature. Unexpectedly, longer organoid incubation did not improve differentiation, but rather caused reduced expression of terminal markers and generated new off-target cells, suggesting kidney cell type dedifferentiation with time, consistent with a recent report (Przepiorski et al., 2018). These results indicate a need to identify conditions that will better support continued organoid maturation.
Off-target cell populations, primarily neural, were present in kidney organoids generated from both protocols. By combining pseudotemporal ordering with lineage-specific expression of transcription factors, ligands and receptors, our analysis provides a roadmap to understand lineage relationships and signaling during differentiation as well as a framework around which to test improvements to the differentiation protocol. The expression of BDNF and its receptor NTRK2 in neural clusters suggested a strategy to reduce these cell types by inhibiting BDNF-NTRK2 signaling. That this strategy reduced off-target cell types by 90% suggests that similar analyses could be applied broadly in the organoid field to reduce unwanted cell types.
We envision that analysis of signaling pathways and transcription factors expressed before and after branch points will suggest other potential strategies to regulate organoid cell fates. Future studies are necessary to determine how modulation of the gene expression patterns revealed here can be used to improve organoid maturation and ultimately better model physiological function.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benjamin D. Humphreys (humphreysbd@wustl.edu).
Experimental Model and Subject Details
iPSC Culture
All experiments utilized the BJFF6 human iPSC line reprogrammed by Sendai virus from human foreskin fibroblasts (Washington University Genome Engineering and iPSC Core). This line is confirmed to be karyotypically normal. BJFF6 cells were maintained in 6-well plates coated with matrigel (Corning) in Essential 8 medium (Thermo Fisher Scientific). iPSC cells were dissociated using ReLeSR (STEMCELL Technologies), confirmed to be mycoplasma free and maintained below passage 50.
Kidney donor
Institutional review board approval for research use of human tissue was obtained from Washington University. Renal cortex from a nephrectomy kidney was obtained and donor anonymity preserved. The donor was a 62 year-old white male with a serum creatinine of 1.03 mg/dL and BUN of 12 mg/dL.
Method Details
Kidney Organoid Differentiation
Kidney organoids were generated following either the protocol described by Takasato et al.(Takasato et al., 2016) or that of Morizane et al.,(Morizane and Bonventre, 2017) with minimal modifications. Briefly, for the Takasato approach, BJFF cells were treated with CHIR (8 uM, Tocris Bioscience) in basal medium - APEL 2 (STEMCELL Technologies) supplemented with 5% Protein Free Hybridoma Medium II (PFHMII, Gibco) - for 4 days, followed by FGF9 (200 ng/mL, R&D Systems) and heparin (1 ug/mL, Sigma-Aldrich) for another 3 days. At day 7, cells were collected and dissociated into single cells using 0.25 % Trypsin-EDTA (Thermo Fisher Scientific). Cells were spun down at 400 g for 3 min to form a pellet and transferred onto a trans-well membrane. Pellets were incubated with CHIR (5 uM) for 1 hour and then cultured with FGF9 (200 ng/mL) and heparin (1 ug/mL) for 5 days. For the next 13 days organoids were cultured basal medium changed every other day. For the Morizane approach, BJFF cells were treated with CHIR (10 uM) and Noggin (5 ng/mL, PeproTech) in basal medium - Advanced RPMI 1640 medium (Gibco) supplemented with 1X L-GlutaMAX (Thermo Fisher Scientific) - for 4 days, followed by 3 days Activin (10 ng/mL, R&D Systems) and 2 days FGF9 (10 ng/mL) treatment. At day 9, the cells were dissociated with Accutase (StemCell technologies) and resuspended in the basic differentiation medium with CHIR (3 µM) and FGF9 (10 ng/mL), and placed in ultra-low attachment 96-well plates. Two days later the medium was changed to basal medium containing FGF9 (10 ng/mL) and cultured for 3 more days. After that, the organoids were cultured in basal medium with no additional factors until harvest at day 26. For ES (H9) cell line, we adjusted the concentration of CHIR to 5uM in Takasato protocol and to 8uM in Morizane protocol at the initial step.
DropSeq single-cell RNA sequencing
Organoids were dissociated using TrypLE Select (Thermo Fisher Scientific) at 37°C with shaking. After 5 min, cells were further dispersed by gentle pipetting and filtered through a 40μm cell strainer (pluriSelect). Single cell suspension was visually inspected under a microscope, counted by hemocytometer (INCYTO C-chip) and resuspended in PBS + 0.01% BSA. Single cells were coencapsulated in droplets with barcoded beads exactly as described (Macosko et al., 2015). In brief, cells were diluted to a concentration of 100 cells/μL, and co-encapsulated with barcoded beads (ChemGenes #Macosko201110), which were diluted to a concentration of 120 beads/µL. Droplets of about 1 nl in size were generated using microfluidic polydimethylsiloxane (PDMS) co-flow devices (FlowJEM Drop-seq chips). Droplets were collected in a 50-mL RNase-free Falcon tube for a total run time of about 15 min. Droplet emulsion was aliquoted into 1 mL each of cells and beads and were broken promptly by perfluorooctanol, following which barcoded beads with captured transcriptomes were washed and spun down at 4 °C. Hybridized RNA was reverse transcribed and ex onuclease-treated using commercial kits (See Key Resources Table). The beads from one run were then equally distributed into individual PCR tubes with populations of 8,000 beads/tube (~400 cells). PCR tubes were separately amplified for 4+9 PCR cycles, and the PCR products were purified by the addition of 0.6× Agencourt AMPure XP beads (Beckman Coulter #A63881). The quality of the amplified cDNA was evaluated by Bioanalyzer (Agilent 2100) on a High Sensitivity DNA chip. Only cDNA with average insertion size >1200 bp were used for downstream library preparation and sequencing. cDNA from an estimated 5,000 cells were prepared and tagmented by Nextera XT (Illumina) using 600 pg of cDNA input. cDNA library was amplified (12 cycles) using custom primers as described (Macosko et al., 2015). Amplified libraries were purified with a 0.6× volume of AMPure XP beads and quality was measured by Bioanalyzer. Libraries with average length of ~500–700 bp were submitted to Genome Technology Access Center (GTAC) of Washington University in St. Louis and sequenced on HiSeq 2500 and NovaSeq 6000 (Illumina). We routinely tested our DropSeq setup by running species mixing experiments prior to running on actual sample to assure that the cell doublet rate was below 5%. Information about experimental replicates and count statistics is specified in Supplementary Table S1.
Nuclei isolation and single-nucleus RNA-seq of human kidney
Nuclei were isolated with Nuclei EZ Lysis buffer (Sigma #NUC-101) supplemented with protease inhibitor (Roche #5892791001) and RNase inhibitor (Promega #N2615, Life Technologies #AM2696). Samples were cut into <2 mm pieces and homogenized using a Dounce homogenizer (Kimble Chase #885302-0002) in 2ml of ice-cold Nuclei EZ Lysis buffer and incubated on ice for 5 min with an additional 2ml of lysis buffer. The homogenate was filtered through a 40-μm cell strainer (pluriSelect #43-50040-51) and then centrifuged at 500 × for 5 min at 4 °C. The pellet was resuspended and washed with 4 ml of the buffer and incubated on ice for 5 min. After another centrifugation, the pellet was resuspended with Nuclei Suspension Buffer (1× PBS, 0.07% BSA, 0.1% RNase inhibitor), filtered through a 20-µm cell strainer (pluriSelect 43-50020-50) and counted. RNA from single nucleus was encapsulated, barcoded and reversed transcribed on a 10× Chromium Single Cell Platform (10× Genomics). The library was sequenced in HiSeq2500.
Immunofluorescence
Organoids were fixed in 4% paraformaldehyde (Electron Microscopy Services), cryoprotected in 30% sucrose solution overnight and embedded in optimum cutting temperature (OCT) compound (Tissue Tek). Organoids were cryosectioned at 7µm thickness and mounted on Superfrost slides (Thermo Fisher Scientific). Sections were washed with PBS (3 times, 5 minutes each), then blocked with 10% normal goat serum (Vector Labs), permeabilized with 0.2% Triton-X100 in PBS and then stained with primary antibody specific for mouse anti-WT1 (1:200, Santa Cruz Biothechnology, #SC-7385), rat anti-ECAD (1:200, Abcam, #ab11512), biotinylated LTL (1:200, Vector Labs, #B-1325), sheep anti-NPHS1 (1:200, R&D Systems, #AF4269) and rabbit anti-CRABP1 (1:200, Cell Signaling, #13163), chicken anti-MAP2 (1:200, Abcam, #ab5392) and mouse anti MEIS1 (1:200, Active Motif, #39795). Secondary antibodies included FITC-, Cy3, or Cy5-conjugated (Jackson ImmunoResearch). Then, sections were stained with DAPI (4′,6′- diamidino-2-phenylindole) and mounted in Prolong Gold (Life Technologies). Images were obtained by confocal microscopy (Nikon C2+ Eclipse; Nikon, Melville, NY).
BDNF inhibitor (K252a) treatment
We treated the iPSC derived kidney organoids differentiated from Takasato protocol with different doses of a BDNF inhibitor, K252a (Sigma-Aldrich #K1639). A dose of 250 nM was selected because the organoid size and tubular morphology were not altered at this concentration. Starting from day 12, we supplemented K252a to the basal medium, and differentiated the organoid to day 26 in the presence of K252a. The medium containing K252a was replaced every 2 days. At day 26, two different batches of organoids were harvested for DropSeq, immunofluorescence and qPCR.
Real Time PCR Experiments
RNA from whole organoids was extracted using the RNeasy Mini Kit (Qiagen) and 600 ng of total RNA was reverse transcribed with iScript (BioRad). Quantitative polymerase chain reactions were carried out with iQ-SYBR Green supermix (BioRad) and the BioRad CFX96 Real Time System with the C1000 Touch Thermal Cycler. Cycling conditions were 95°C for 3 minutes then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute, followed by one cycle of 95°C for 10 seconds. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene. Data was analyzed using the 2−∆∆ct method. The following primers were used: GAPDH: Fw 5‘-GACAGTCAGCCGCATCTTCT −3′; Rv 5‘-GCGCCCAATACGACCAAATC −3´; Cited1: Fw 5´-CCTCACCTGCGAAGGAGGA-3´;Rv 5‘-GGAGAGCCTATTGGAGATCCC −3´; NPHS1: Fw 5´-CTGCCTGAAAACCTGACGGT −3´; Rv 5‘-GACCTGGCACTCATACTCCG −3´; SLC3A1: Fw 5´-CAGGAGCCCGACTTCAAGG −3´; Rv 5‘-GAGGGCAATGATGGCTATGGT −3´ SLC12A1: Fw 5´-AGTGCCCAGTAATACCAATCGC −3´; Rv 5‘-GCCTAAAGCTGATTCTGAGTCTT −3´; CRABP1: Fw 5´-GCAGCAGCGAGAATTTCGAC −3´; Rv 5‘-CGTGGTGGATGTCTTGATGTAGA - 3´; MAP2: Fw 5´-CTCAGCACCGCTAACAGAGG −3´; Rv 5‘-CATTGGCGCTTCGGACAAG −3´; MYLPF: Fw 5´-GAAGGACAGTAGAGGGCGGAA −3´; Rv 5‘-TCTGGTCGATCACAGTGAAGG - 3´; MYOG: Fw 5´-GGGGAAAACTACCTGCCTGTC −3´; Rv 5‘-AGGCGCTCGATGTACTGGAT −3´; MLANA: Fw 5´-GCTCACTTCATCTATGGTTACCC −3´; Rv 5´-GACTCCCAGGATCACTGTCAG - 3´; PMEL: Fw 5´-AGGTGCCTTTCTCCGTGAG Rv 5´-AGCTTCAGCCAGATAGCCACT −3´
Quantification and Statistical Analysis
Preprocessing of DropSeq data
Paired-end sequencing reads were processed as previously described using the Drop-Seq Tools v1.12 software available in McCarroll’s lab (http://mccarrolllab.com/dropseq/). Briefly, each cDNA read (read2) was tagged with the cell barcode (the first 12 bases in read 1) and unique molecular identifier (UMI, the next 8 bases in read 1), trimmed of sequencing adaptors and poly-A sequences, and aligned to the human (GRCh38) or a concatenation of the mouse and human (for the species-mixing experiment) reference genome assembly using STAR v 2.5.3a (Dobin et al., 2013). Cell barcodes were corrected for possible bead synthesis errors using the DetectBeadSynthesisErrors program, and then collapsed to core barcodes if they were within an edit distance of 1 as previously described (Macosko et al., 2015). Digital gene expression (DEG) matrix was compiled by counting the number of unique UMIs for a given gene (as row) in each cell (as column).
Global clustering analysis
To assess the difference in cell composition across differentiation protocols and cell lines, we performed global clustering analysis on ten datasets from Dropseq sequencing of the day 26 organoids. First, we combined the UMI count matrices from different protocols and cell lines into one gene-cell matrix using the merge function in R. We then removed the low-quality cells with less than 500 or more than 4000 detected genes, or if their mitochondrial gene content was > 20%. Genes were filtered out that were detected in less than 10 cells. This filtering step resulted in 24,574 genes X 71,390 cells sampled from four batches Takasato iPS organoids (26,890 cells), three batches Morizane iPS organoids (18,072 cells), two batches Takasato ES organoids (19,380 cells) and one batch Morizane ES organoids (7,048 cells). The gene expression was then natural log transformed and normalized for scaling the sequencing depth to a total of 1e4 molecules per cell. Batch effect was corrected by matching mutual nearest neighbors using a recently published pipeline (incorporated in scran R package) (Haghverdi et al., 2018). Dimensionality reduction and clustering were performed on the batch effect corrected expression value using Seurat R tool kit.
A total of 23 clusters were classified from this analysis, consisting of cells from four broad “classes” defined as mesenchyme, tubule, podocyte and off-target cells based on their marker genes expression. We then performed a post-hoc merging step, where transcriptionally indistinguishable clusters are merged back together. First, the distance between each pair of clusters within the broad cell class was computed based on the averaged expression value of the highly variable genes. Second, hierarchical clustering analysis was performed on the distance matrix using the hclust function from R. We then merged transcriptionally similar clusters that were placed adjacent on the hierarchical tree. With this approach, we identified five transcriptionally distinct subtypes in the tubule class, and 3 distinct subtypes in the mesenchyme class. Finally, we assessed the cell compositions by calculating the number of cells in each cluster or broad class, subdivided by the differentiation protocol and cell line.
Clustering analysis on the organoid cells from different protocols or cell lines
Seurat was used for quality control, dimensionality reduction and cell clustering for the Dropseq datasets generated by each protocol or cell line. In brief, raw DEG matrices from different batches for each protocol on each cell line were combined and loaded into the Seurat. For normalization, the DGE matrix was scaled by total UMI counts, multiplied by 10,000 and transformed to log space. Only genes found to be expressing in >10 cells were retained. Additional filtering was set on the number of detected genes and mitochondrial gene content to remove the low-quality cells or cell doublets. We note that, depending on the sequencing depth and the variations in mitochondrial gene content from dataset to dataset, the cutoffs may need to be set on a case-by-case basis. Before clustering, variants arising from batch effects, library size and percentage of mitochondrial genes were regressed out by specifying the vars.to.regress argument in Seurat function ScaleData. The highly variable genes were identified using the function FindVariableGenes. The expression level of highly variable genes in the cells was scaled and centered along each gene, and was conducted to principal component analysis. We then assessed the number of PCs to be included in downstream analysis by (1) plotting the cumulative standard deviations accounted for each PC using the function PCElbowPlot in Seurat to identify the ‘knee’ point at a PC number after whic successive PCs explain diminishing degrees of variance, and (2) by exploring primary sources of heterogeneity in the datasets using the PCHeatmap function in Seurat. Based on these two methods, we selected first top significant PCs for two-dimensional t-distributed stochastic neighbor embedding (tSNE), implemented by the Seurat software with the default parameters. We used FindCluster in Seurat to identify cell clusters for each protocol. Alternatively, A hierarchical clustering method devised by Baron et al (Baron et al., 2016) was recruited to validate the clusters identified by Seurat. To identify the marker genes, differential expression analysis was performed by the function FindAllMarkers in Seurat with likelihood-ratio test. Differentially expressed genes that were expressed at least in 25% cells within the cluster and with a fold change more than 0.25 (log scale) were considered to be marker genes. Gene expression of selected markers across clusters were visualized using a Python plotting library Matplotlib.
Sub-clustering tubular cells in Morizane protocol (cluster PT1, PT2, and LH) and Takasato protocol (cluster PT1, PT2, LH, and Lp) were first combined, then sub-clustered using the same approach described above. We identified 12 significant PCs in 3,056 tubular cells from Takasato protocol and 10 PCs in 4,933 tubular cells from Morizane protocol, which were further assigned into 9 and 5 sub-clusters, respectively. To further compare the cell types generated from the two protocols, we extracted the expression matrix for PT2 (a more mature PT cluster), LH and podocyte from each protocol, and used the combined matrices as input to Seurat. After regressing out potential protocol effect and library size difference, we performed differential gene analysis on the PT cells between protocols with likelihood-ratio test. Developmental genes were selected based on the anchor/marker genes listed in GUDMAP (McMahon et al., 2008) and visualized by DotPlot function in Seurat.
For the time course Dropseq data, the datasets were preprocessed before being placed in the Seurat package. The matrices from all time points were merged into one single matrix with the merge function in R. To retain the gene expression variants across time course, all genes from each time point were kept in the merged matrix by assigning zeros to the genes for those cells who have missing expression value. 20 significant PCs calculated from 1,345 high variable genes were selected for tSNE and clustering analysis. We identified 14 clusters including key cell types corresponding to the important developmental states. We selected genes that represent each development stages and visualized them in tSNE map. Initially, we used the normalized gene expression value but found that the high dropout events in single cell data obscure the dynamic gene expression changes across time course. We therefore applied a gene imputation approach named MAGIC (van Dijk et al., 2018) to computationally fill in the missing value for all genes in our time course dataset. In brief, the merged time-course count matrix was loaded into the MAGIC pipeline written in Python (https://github.com/pkathail/magic). Gene expression value in all cells were normalized, dimensionally reduced and transformed by the internal algorithms in MAGIC with the parameters: n_pca_components=20, random_pca=True, t=6, k=30, ka=10, epsilon=1, rescale_percent=99. Developmental genes and marker gene expression across timepoints after MAGIC imputation were visualized in the tSNE map constructed by Seurat and on the pseudotemporal trajectory tree constructed by Monocle (see methods below).
sNuc-10X data processing and clustering analysis
We used a newly developed pipeline, zUMIs (Parekh et al., 2018), to process the single nucleus sequencing data from human adult kidney. In brief, we first filtered out the low-quality barcodes or UMIs based on sequence with the internal read filtering algorithm built in zUMIs. We then used zUMIs to map the filtered reads to human reference genome (GRCh38) using STAR 2.5.3a (two-pass mapping mode). Next, zUMIs quantified the reads that were uniquely mapped to exonic, intronic or intergenic region of the genome and inferred the true barcodes that mark cells by fitting a k-dimensional multivariate normal distribution with mclust package. Finally, a UMI count table utilizing both exonic and intronic reads were generated for downstream analysis. The whole data processing was executed by running the script on a HPC cluster with 96×2.3GHz computing cores (http://brc.wustl.edu/?page_id=12) with the following example script: bash zUMIs-master.sh -f R1.fastq.gz -r R2.fastq.gz -c 1–16 -m 17–26 -l 98 -n Human_kidney - g GRCh38_ref_genome -a GRCh38_ref_genes.gtf -p 30. Clustering analysis was performed on Seurat with a similar approach used for analyzing the organoid Dropseq datasets.
Integrated analysis of multiple datasets
To compare the cell types derived from different organoid protocols/cell lines and those from adult human kidney, we performed comparative analysis on multiple datasets by utilizing a recently developed computational strategy for integrated analysis (implemented in Seurat v2.0) (Butler et al., 2018). We first selected the union of the top 3,000 genes with the highest dispersion from all datasets for a canonical correlation analysis (CCA) to identify common sources of variation across the datasets. Then CCA was performed based on the normalized expression value of the highly-dispersed genes. Next, we selected the top dimensions of the CCA by examining a saturation in the relationship between the number of principle components and the percentage of the variance explained using the MetageneBicorPlot function in Seurat. We obtained a new dimensional reduction matrix by aligning the CCA subspaces with the top dimensions computed above. With the new dimensional reduction matrix, we performed clustering analysis on the cells or nuclei from different datasets by setting an optimal clustering parameters. We visualized the cells by their original identity or by their cluster identity classified by this integrated analysis. Differential gene analysis was performed on the cells or nuclei from different datasets but grouped in the same cluster after the alignment analysis. Differential genes were visualized using the FeatureHeatmap or DotPlot function in Seurat. We applied this computational strategy to compare the matched cell types (i.e. podocyte, PT and LH) from organoids and adult human kidney.
Cell cycle analysis
We assigned a cell cycle score on each cell according to its gene expression of G2/M and S phase markers (Tirosh et al., 2016). Based on this scoring system, we classified each cell in either G2M, S or G1 phase using the CellCycleScoring function in Seurat. The cells at different cell cycle classifications were visualized in the tSNE map, and the expression of cell cycle genes were plotted out using FeaturePlot function in Seurat.
Correlation analysis of kidney organoid and mouse/human kidney
To assess the similarity between kidney organoid cell types and embryonic kidney, we re-analyzed two previously published Dropseq datasets from mouse E14.5 kidney (GSE104396) (Magella et al., 2018), P1 kidney (GSE94333) (Adam et al., 2017) and human fetal kidney (Lindstrom et al., 2018b). We used the Seurat clustering parameters described by the authors and reproduced the same cell types from the datasets. We calculated the Pearson correlation based on the expression patterns of highly variable genes between cell populations within the mouse embryonic kidney dataset against the organoid cell types and the UB cell type identified from subclustering analysis. we performed the same analysis to compare organoid cell types to the human adult kidney cell types identified from the snRNA-seq dataset (data from this manuscript). Correlation matrix were visualized by R package pheatmap. Colour keys (and dot sizes) represent the range of the coefficients of determination (r2) in each analysis.
Hierarchical clustering analysis on the cell types from kidney organoids
To reveal the relationship among the matched cell types derived from Morizane and Takasato protocols, we extracted the expression profiles for P1, M3, PT2, LH and N2 subtypes from the Morizane organoid datasets and expression profiles for P, M3, PT2, LH, N2, and EC from the Takasato organoid datasets. Then we normalized the gene expression and computed the distance between each pair of cell types based on their average gene expression. Hierarchical clustering was performed on the distance matrix using hclust function in R and was visualized by dendrogram. We used the dendextend R package to compare the dendrogram from the Morizane organoid dataset and the Takasato organoid dataset.
Random Forest model to map cell types from human fetal kidney
To determine the congruence between cell types obtained from our organoid datasets and those in a recent fetal kidney dataset (Lindstrom et al., 2018b), we trained a multiclass random forest classifier (Habib et al., 2017; Shekhar et al., 2016) on the fetal kidney cell clusters and used it to map the organoid cell type. First, we composed a ‘training set’ by sampling 60%of the cells from 5 fetal kidney clusters representing mesenchyme, LH, PT, podocyte, endothelium and a cycling cell population (defined as mesenchyme progenitor). We next trained a random forest using 1,000 trees on the training set using the R package randomForest. We then used the remaining 40% of the cells from each cluster from the human fetal kidney dataset to validate the performance of the trained classifier. We used this model to assign a class label (one of the 5 human fetal kidney cell types) to each cell from kidney organoids. Finally, we quantified the number of cells that were mapped to each class label and visualized the data using ggplot2 package.
Cell type specific driving force analysis
To identify the key regulators that control the cell states, we performed cell type specific driving force analysis using the SINCERA pipeline (Guo et al., 2015). This approach consists of three main steps. First, the candidate transcription factors (TFs) and their regulatory target genes (TGs) were extracted from the DEG list identified in each cluster. Second, cell type specific transcription regulatory network (TRN) was constructed by establishing the interaction between TF-TF and TF-TG (TG-TF and TF auto-regulations were not considered) as previously described (Lebre, 2009). Finally, the key TFs were selected based on their network node importance. This was accomplished by collecting the value of six node importance metrics including Degree Centrality (DC), Closeness Centrality (CC), Betweenness Centrality (BC), Disruptive Fragmentation Centrality (DFC), Disruptive Connection Centrality (DCC) and Disruptive Distance Centrality (DDC). TFs were ordered by taking the average of the node importance from these six matrices.
Deconvolution of bulk RNA-seq data
To examine the possible use of our single cell data to infer the cell type compositions (deconvolution) from the bulk RNA profiling, we retrieved the previously published RNA-Seq dataset from Takasato et al (GEO accession GSE70101)(Takasato et al., 2015). This dataset contains gene expression profiles of time-course organoids collected at similar time-points (day 7, day 10, day 18 and day 25) as those collected for our time-course single cell study. We then applied a single cell deconvolution algorithm, BSeq-sc (Baron et al., 2016), to estimate the proportion of iPS cells, PIM, pretubular aggregate, PT, LH, podocyte and neurons in each time point from bulk RNA-seq datasets. The identified marker genes for each cell type from our time-course single cell data were used as an input to estimate the cell proportion in each timepoint of the bulk-seq data according to the tutorial from BSeq-sc package (https://shenorrlab.github.io/bseqsc/vignettes/bseq-sc.html).
GWAS analysis
We downloaded GWAS gene lists associated with chronic kidney disease, hypertension and metabolite from the GWAS site (https://www.ebi.ac.uk/gwas/). Each associated gene that was identified as significant in a GWAS (reported gene) was mapped to the cell type marker gene list obtained from Seurat FindAllMarker function. To plot the GWAS genes, we normalized the gene expression value for each cell by z-scores and generated a new gene expression matrix with mean z-scores for each GWAS gene by averaging the z-score value from all individual cells in the same cluster (i.e. a mean z-scores matrix with GWAS genes as rows and cell type ID as columns). Heatmap.2 function in gplots R package was used to create all heatmap graphs in this analysis.
Kidney disease trait association with single cell-based cell types
We used RolyPoly, a polygenic method that identifies trait-involved cell types by analyzing the enrichment of GWAS signal in cell type specific gene expression (Calderon et al., 2017), to associate the kidney disease trait with gene expression profile from our single cell data. Since Rolypoly requires four indispensable components as data input (GWAS summary statistics, expression data, an expression data annotation file, and linkage disequilibrium (LD) information), we prepared each of them as follows. First, we obtained the summary meta-analysis data from the CKDGen consortium (http://ckdgen.imbi.uni-freiburg.de) for the associations between genotype and CKD or eGFR on the basis of serum creatinine (eGFRcrea) based on Li et al’s study (Li et al., 2017; Pattaro et al., 2016). To prepare cell type expression data, we computed the average expression on each cell type from Takasato’s iPS organoid, Morizane’s iPS organoid, and adult human kidney. To link gene expression with the location of GWAS variants, we defined a block as a 10kb window centered around each gene’s transcription start site (TSS) as recommended by rolypoly. The TSS for all known hg19 genes was downloaded from UCSC Genome Browser (https://genome.ucsc.edu/). Finally, the linkage disequilibrium (LD) information was provided by rolypoly based on the calculation using PLINK for 1000g phase 3 genomes filtered for values of R2>0.2 (https://cran.r-project.org/web/packages/rolypoly/vignettes/intro.html). We input all these data into the main rolypoly function call using the default parameters. Significant association between cell type and CKD or eGFR was determined by p value.
Pseudotemporal ordering
We used Monocle2 (Qiu et al., 2017) (default settings) to draw a minimal spanning tree connecting the 9,190 cells collected from time course organoids into multiple lineages. As input into Monocle2, we selected the ordering genes using a semi-supervised approach as described in the Monocle2 tutorial (http://cole-trapnell-lab.github.io/monocle-release/docs_mobile/). Specifically, we first defined the genes that mark the kidney organoid differentiation process and terminal cell lineage. The criteria for the gene selection include: 1) they are putatively reported as markers for the kidney developmental state or terminally kidney cell fate. 2) they are specific marker genes differentially expressed in the clusters identified by Seurat as noted above. We used them to select the ordering genes that co-vary with these markers using the Monocle function markerDiffTable. With this approach, we generated a final gene set containing 1,604 genes. We then reduced the data space to two dimensions with ‘DDRTree’ method and ordered the cells using the orderCells function in Monocle. Individual cells were color-coded based on the time points where they were collected to validate the accuracy of the cell ordering. We further colored the cells with the cell type identity classified by Seurat as noted above to assure that the ordering is meaningful. We plotted the gene expression on the Monocle tree using MAGIC imputed value as described above.
Ligand-receptor interaction analysis
To study the ligand-receptor interaction at single cell level, we used a human ligand-receptor list comprising 2,557 ligand–receptor pairs curated by Database of Ligand−Receptor Partners (DLRP), IUPHAR and Huma n Plasma Membrane Receptome (HPMR) (Hrvatin et al., 2017; Ramilowski et al., 2015). We selected the receptors that were only differentially expressed in each cell type from the day 26 organoid (Takasato protocol), and the ligands that were induced in either branch on the cell trajectory based on the results computed by the branched expression analysis modeling (BEAM) algorithm in Monocle. To determine the ligand-receptor pairs to plot on the heatmap, we required that (i) The ligands are branch-dependent with significant score q-val< 0.01 based on the score table from BEAM analysis; (ii) The receptors are uniquely expressed in each cell type (q-val<0.05 and logFC>0.6); (iii) Each receptor should have at least one corresponding ligand to pair with. We used plot_genes_branched_heatmap function from the Monoce2 package to plot the ligands and heatmap.2 function from gplots package to show the receptors expressed in each cell type. We used the same list of ligand-receptor pairs to identify cell-type specific ligands and receptors in human adult kidney and employed a network approach to visualize the cell type communication based on connections of ligand-receptor pairs (Ramilowski et al., 2015).
Real Time PCR data quantification and statistical analysis
Data were presented as mean ± SEM. ANOVA with post hoc Bonferroni correction was used for multiple group comparison. Student t-test was used to compare 2 different groups. Graph-Pad Prism software, version 6.0c (GraphPad Software Inc., San Diego, CA) and SPSS version 22 were used for statistical analysis. P-value < 0.05 was considered as statistical significant difference.
Experimental Design
The number of replicate organoids is included in Supplementary Table S1. Experimental groups were neither randomized not blinded. No sample-size estimates were made and the inclusion criteria for analyzing adult human kidney was having normal kidney function and no known kidney disease.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-WT1 | Santa Cruz | Cat#SC-7385; RRID: AB_628448 |
| Rat anti-ECAD | Abcam | Cat#ab11512; RRID: AB_298118 |
| Biotinylated LTL | Vector Labs | Cat#B-1325; RRID: AB_2336558 |
| Sheep anti-NPHS1 | R&D Systems | Cat#AF4269; RRID: AB_2154851 |
| Rabbit anti-CRABP1 | Cell Signaling | Cat#13163 |
| Chicken anti-MAP2 | Abcam | Cat#ab5392; RRID: AB_2138153 |
| Mouse anti MEIS1 | Active Motif | Cat#39795 |
| Secondary antibodies included FITC-, Cy3, or Cy5-conjugated |
Jackson ImmunoResearch |
Cat#711-095-152, Cat#712-095-153, Cat#715-165-151, Cat#713-165-147, Cat#016-600-084, Cat#703-165-155 |
| DAPI (4´,6´- diamidino-2- phenylindole) |
Thermo Fisher Scientific |
Cat#D1306 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Barcoded Dropseq beads | ChemGenes | Cat#MACOSKO-2011-10 |
| 10mM Tris-HCl, pH 8.0 | Teknova | Cat#T1173 |
| UltraPure SSC | Invitrogen | Cat#15557036 |
| Ficoll PM-400 | GE Healthcare/Fisher Scientific |
Cat#45-001-745 |
| Sarkosyl | Sigma-Aldrich | Cat#L7414-50mL |
| Exonuclease I | New England Biolabs | Cat#M0293L |
| Perfluoro-1-octanol | Sigma-Aldrich | Cat#370533-25G |
| dNTP mix | Clontech | Cat#639125 |
| Droplet Generation Oil | BioRad | Cat#186-4006 |
| Tris-EDTA buffer, pH 8.0 | Sigma-Aldrich | Cat#93283 |
| 1M DTT | Teknova | Cat#D9750 |
| 2M Tris pH 7.5 | Sigma-Aldrich | Cat#T2944 |
| Tween 20, RNase Free | Promega | Cat#H5152 |
| NxGen RNAse Inhibitor | Lucigen | Cat#30281-2 |
| Nuclei EZ Lysis Buffer | Sigma-Aldrich | Cat#N-3408 |
| RNasin Plus Ribonuclease Inhibitors |
Promega | Cat#N2615 |
| SUPERaseIN RNase Inhibitor | Thermo Fisher Scientific |
Cat#AM2696 |
| cOmplete ULTRA Tablets, Mini, EDTA-free, EASYpack |
Roche | Cat#05 892 791 001 |
| CHIR | Tocris Bioscience | Cat#4423 |
| APEL 2 | STEMCELL Technologies |
Cat#05275 |
| Protein Free Hybridoma Medium II |
Gibco | Cat#12040077 |
| Advanced RPMI 1640 medium | Gibco | Cat#12633012 |
| FGF9 | R&D Systems | Cat#273-F9-025/CF |
| Heparin | Sigma-Aldrich | Cat#H4784-250MG |
| Trypsin-EDTA | Sigma-Aldrich | Cat#25200-114 |
| Noggin | PeproTech | Cat#120-10C |
| GlutaMAX | Thermo Fisher Scientific |
Cat#35050061 |
| Activin | R&D Systems | Cat#338-AC-010 |
| Accutase | STEMCELL Technologies |
Cat#7920 |
| TrypLE Select | Thermo Fisher Scientific |
Cat#12563-029 |
| ReLeSR | STEMCELL Technologies |
Cat#05872 |
| BDNF inhibitor (K252a) | Sigma-Aldrich | Cat#K1639 |
| Prolong Gold | Life Technologies | Cat#P36930 |
| Essential 8™ Medium | Gibco | Cat#A2858501 |
| Matrigel® hESC-Qualified Matrix |
Corning | Cat#354277 |
| Antibiotic-Antimycotic Solution | Corning | Cat#30004CI |
| Critical Commercial Assays | ||
| Maxima H Minus Reverse Transcriptase |
ThermoFisher | Cat#EP0753 |
| KAPA HiFi hotstart readymix | KAPA Biosystems | Cat#KK2602 |
| Single Cell 3′ Library and Gel Bead Kit V2 |
10× Genomics | Cat#120237 |
| Chromium single cell chip kit V2 | 10× Genomics | Cat#120236 |
| Nextera XT DNA Sample Preparation Kit |
Illumina | Cat#FC-131-1096 |
| Agilent High Sensitivity DNA Kit | Agilent | Cat#5067-4626 |
| Agencourt AMPure XP - PCR Purification |
Backman Coulter | Cat#A63881 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE118184 |
| Mendeley Data | This paper | http://dx.doi.org/10.17632/m4rfg9wb29.1 |
| Experimental Models: Cell Lines | ||
| Human iPS cells: BJFF.6 line | GEiC | N/A |
| Human ES cells: H9 line | GEiC | N/A |
| Software and Algorithms | ||
| Drop-seq_tools v1.12 | (Macosko et al., 2015) | http://mccarrolllab.com/dropseq/ |
| STAR v2.5.3a | (Dobin et al., 2013) | https://github.com/alexdobin/STAR |
| R 3.4.1 | R project | https://www.r-project.org |
| Seurat v2.0 | (Butler et al., 2018) | http://satijalab.org/seurat/ |
| Monocle2 | (Qiu et al., 2017) | https://github.com/cole-trapnell-lab/monocle-release |
| zUMIs | (Parekh et al., 2018) | https://github.com/sdparekh/zUMIs |
| Random Forest | CRAN-R | https://cran.r-project.org/web/packages/randomForest/index.html |
| SINCERA | (Guo et al., 2015) | https://github.com/xu-lab/SINCERA |
| BSeq-sc | (Baron et al., 2016) | https://github.com/shenorrLab/bseqsc |
| MNN | (Haghverdi et al., 2018) | https://github.com/MarioniLab/MNN2017/ |
| rolypoly | (Calderon et al., 2017) | https://github.com/dcalderon/rolypoly |
| Cytoscape 3.6.1 | (Shannon et al., 2003) | http://www.cytoscape.org |
| Connectome | (Ramilowski et al., 2015) | https://github.com/Hypercubed/connectome |
| MAGIC | (van Dijk et al., 2018) | https://github.com/KrishnaswamyLab/MAGIC |
| Other | ||
| Nuclei isolation protocol | This study | STAR Methods |
| Morizane organoid differentiation protocol |
(Morizane et al., 2015) | N/A |
| Takasato organoid differentiation protocol |
(Takasato et al., 2015) | N/A |
| Dropseq library preparation protocol |
McCarroll’s lab | http://mccarrolllab.org/dropseq/ |
| 10× library preparation protocol | 10× genomics | https://support.10×genomics.com/single-cell-gene-expression/library-prep/doc/user-guide-chromium-single-cell-3-reagent-kits-user-guide-v2-chemistry |
| Hemocytometer | INCYTO | Cat#DHC-F015 |
| pluriStrainer 40 µm | pluriSelect | Cat#43-50040 |
| pluriStrainer 20 µm | pluriSelect | Cat#43-50020 |
| KONTES Dounce Tissue Grinders |
Kimble Chase | Cat#KT885300-0002 |
| Transwell® polyester membrane cell culture inserts |
Corning | Cat#3460 |
| 96 Well, Clear Round Bottom, Ultra Low |
Corning | Cat# 7007 |
| Syringe pumps | KD Scientific | Legato 100 |
| Magnetic mixing system | VP Scientific | Part #710D2 |
| PDMS Microfluidic Device | FlowJEM | N/A |
| Mouse E14.5 kidney data | (Magella et al., 2018) | GSE104396 |
| Mouse P1 kidney data | (Adam et al., 2017) | GSE94333 |
| Human fetal kidney data | (Lindstrom et al., 2018b) | GSE102596 |
| Organoid bulk RNA-seq data | (Takasato et al., 2015) | GSE70101 |
Acknowledgements
This work was supported by NIH/NIDDK DK103740 and DK107374, by an Established Investigator Award of the American Heart Association and by Grant 173970 from the Chan Zuckerberg Initiative (all to B.D.H). KU was supported by JSPS Postdoctoral Fellowships for Research Abroad.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
Data and Software Availability
The accession number for the RNA sequencing data reported in this paper is NCBI GEO: GSE118184. Additional figures can be assessed at Mendeley Data: http://dx.doi.org/10.17632/m4rfg9wb29.1
References
- Adam M, Potter AS, and Potter SS (2017). Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development 144, 3625–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar Halpern K., Caspi I, Lemze D, Levy M, Landen S, Elinav E, Ulitsky I, and Itzkovitz S (2015). Nuclear Retention of mRNA in Mammalian Tissues. Cell reports 13, 2653–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, Ryu JH, Wagner BK, Shen-Orr SS, Klein AM, et al. (2016). A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst 3, 346–360 e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelson RA, Lambert GM, Vanier C, Lynch RM, and Galbraith DW (2007). Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics 8, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels RJ, Hartog A, Timmermans JA, and van Os CH (1991). Immunocytochemical localization of calbindin-D28k, calbindin-D9k and parvalbumin in rat kidney. Contrib Nephrol 91, 7–13. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Yu J, Grimmond S, et al. (2008). Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell 15, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, and Tole S (2001). LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev 100, 165–175. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon D, Bhaskar A, Knowles DA, Golan D, Raj T, Fu AQ, and Pritchard JK (2017). Inferring Relevant Cell Types for Complex Traits by Using Single-Cell Gene Expression. Am J Hum Genet 101, 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Panesso M, Kadyrov FF, Machado FG, Kumar A, and Humphreys BD (2018). Meis1 is specifically upregulated in kidney myofibroblasts during aging and injury but is not required for kidney homeostasis or fibrotic response. Am J Physiol Renal Physiol [DOI] [PMC free article] [PubMed]
- Chen L, Lee JW, Chou CL, Nair AV, Battistone MA, Paunescu TG, Merkulova M, Breton S, Verlander JW, Wall SM, et al. (2017). Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci U S A 114, E9989–E9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, and Levey AS (2007). Prevalence of chronic kidney disease in the United States. Jama 298, 2038–2047. [DOI] [PubMed] [Google Scholar]
- Costantini F, and Kopan R (2010). Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell 18, 698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniecki SM, Cruz NM, Harder JL, Menon R, Annis J, Otto EA, Gulieva RE, Islas LV, Kim YK, Tran LM, et al. (2018). High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 22, 929–940 e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Caestecker M, Humphreys BD, Liu KD, Fissell WH, Cerda J, Nolin TD, Askenazi D, Mour G, Harrell FE Jr., Pullen N, et al. (2015). Bridging Translation by Improving Preclinical Study Design in AKI. J Am Soc Nephrol 26, 2905–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmelt L, and Halpain S (2005). The MAP2/Tau family of microtubule-associated proteins. Genome Biol 6, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, Kustagi M, Czuppa M, Izmirly P, Belmont HM, et al. (2017). Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Werff RV, et al. (2015). Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nature communications 6, 8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Wang H, Potter SS, Whitsett JA, and Xu Y (2015). SINCERA: A Pipeline for Single-Cell RNA-Seq Profiling Analysis. PLoS Comput Biol 11, e1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14, 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges-Monroy R, Klein AM, et al. (2017). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci [DOI] [PMC free article] [PubMed]
- Huang EJ, and Reichardt LF (2001). Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci 24, 677–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, Yannelli JR, Adema GJ, Miki T, and Rosenberg SA (1994). Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci U S A 91, 6458–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, and Regev A (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res 25, 1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labastie MC, Catala M, Gregoire JM, and Peault B (1995). The GATA-3 gene is expressed during human kidney embryogenesis. Kidney Int 47, 1597–1603. [DOI] [PubMed] [Google Scholar]
- Lake BB, Codeluppi S, Yung YC, Gao D, Chun J, Kharchenko PV, Linnarsson S, and Zhang K (2017). A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Sci Rep 7, 6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AQ, Freedman BS, Morizane R, Lerou PH, Valerius MT, and Bonventre JV (2013). Rapid and Efficient Differentiation of Human Pluripotent Stem Cells into Intermediate Mesoderm That Forms Tubules Expressing Kidney Proximal Tubular Markers. J Am Soc Nephrol [DOI] [PMC free article] [PubMed]
- Lebre S (2009). Inferring dynamic genetic networks with low order independencies. Stat Appl Genet Mol Biol 8, Article 9. [DOI] [PubMed] [Google Scholar]
- Li M, Li Y, Weeks O, Mijatovic V, Teumer A, Huffman JE, Tromp G, Fuchsberger C, Gorski M, Lyytikainen LP, et al. (2017). SOS2 and ACP1 Loci Identified through Large-Scale Exome Chip Analysis Regulate Kidney Development and Function. J Am Soc Nephrol 28, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom NO, De Sena Brandine G, Tran T, Ransick A, Suh G, Guo J, Kim AD, Parvez RK, Ruffins SW, Rutledge EA, et al. (2018a). Progressive Recruitment of Mesenchymal Progenitors Reveals a Time-Dependent Process of Cell Fate Acquisition in Mouse and Human Nephrogenesis. Dev Cell 45, 651–660 e654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom NO, Guo J, Kim AD, Tran T, Guo Q, De Sena Brandine G, Ransick A, Parvez RK, Thornton ME, Baskin L, et al. (2018b). Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. J Am Soc Nephrol 29, 806–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, and Warnaar SO (1994). Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J Cell Biol 125, 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magella B, Adam M, Potter AS, Venkatasubramanian M, Chetal K, Hay SB, Salomonis N, and Potter SS (2018). Cross-platform single cell analysis of kidney development shows stromal cells express Gdnf. Dev Biol 434, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, et al. (2008). GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol 19, 667–671. [DOI] [PubMed] [Google Scholar]
- Michael UF, Barenberg RL, Chavez R, Vaamonde CA, and Papper S (1972). Renal handling of sodium and water in the hypothyroid rat. Clearance and micropuncture studies. J Clin Invest 51, 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R, and Bonventre JV (2017). Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nat Protoc 12, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, and Bonventre JV (2015). Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol 33, 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, et al. (1995). The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev 9, 3109–3121. [DOI] [PubMed] [Google Scholar]
- Parekh S, Ziegenhain C, Vieth B, Enard W, and Hellmann I (2018). zUMIs A fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, and Susztak K (2018). Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, et al. (2016). Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nature communications 7, 10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przepiorski A, Sander V, Tran T, Hollywood JA, Sorrenson B, Shih JH, Wolvetang EJ, McMahon AP, Holm TM, and Davidson AJ (2018). A Simple Bioreactor-Based Method to Generate Kidney Organoids from Pluripotent Stem Cells. Stem Cell Reports 11, 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, et al. (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K, Simons M, Reiser J, Saleem MA, Faul C, Kriz W, Shaw AS, Holzman LB, and Mundel P (2001). Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J Clin Invest 108, 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmin S, Taguchi A, Kaku Y, Yoshimura Y, Ohmori T, Sakuma T, Mukoyama M, Yamamoto T, Kurihara H, and Nishinakamura R (2016). Human Induced Pluripotent Stem Cell-Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation. J Am Soc Nephrol 27, 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323 e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KM, Combes AN, Lefevre J, Ju AL, Georgas KM, Lamberton T, Cairncross O, Rumballe BA, McMahon AP, Hamilton NA, et al. (2014). Global quantification of tissue dynamics in the developing mouse kidney. Dev Cell 29, 188–202. [DOI] [PubMed] [Google Scholar]
- System USRD (2017). 2017 USRDS annual data report: Epidemiology of kidney disease in the United States (Bethesda, MD: ). [Google Scholar]
- Taguchi A, and Nishinakamura R (2017). Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell 21, 730–746 e736. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Chiu HS, and Little MH (2016). Generation of kidney organoids from human pluripotent stem cells. Nat Protoc 11, 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568. [DOI] [PubMed] [Google Scholar]
- Tapley P, Lamballe F, and Barbacid M (1992). K252a is a selective inhibitor of the tyrosine protein kinase activity of the trk family of oncogenes and neurotrophin receptors. Oncogene 7, 371–381. [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. (2018). Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729 e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, Gaut JP, and Humphreys BD (2018). Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol 29, 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Nivet E, Sancho-Martinez I, Gallegos T, Suzuki K, Okamura D, Wu MZ, Dubova I, Esteban CR, Montserrat N, et al. (2013). Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat Cell Biol 15, 1507–1515. [DOI] [PubMed] [Google Scholar]
- Xia Y, Sancho-Martinez I, Nivet E, Rodriguez Esteban C., Campistol JM, and Izpisua Belmonte J.C. (2014). The generation of kidney organoids by differentiation of human pluripotent cells to ureteric bud progenitor-like cells. Nat Protoc 9, 2693–2704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.