Abstract
Background
The evolution of dioecious plants is occasionally accompanied by the establishment of sex chromosomes: both XY and ZW systems have been found in plants. Structural studies of sex chromosomes are now being followed up by functional studies that are gradually shedding light on the specific genetic and epigenetic processes that shape the development of separate sexes in plants.
Scope
This review describes sex determination diversity in plants and the genetic background of dioecy, summarizes recent progress in the investigation of both classical and emerging model dioecious plants and discusses novel findings. The advantages of interspecies hybrids in studies focused on sex determination and the role of epigenetic processes in sexual development are also overviewed.
Conclusions
We integrate the genic, genomic and epigenetic levels of sex determination and stress the impact of sex chromosome evolution on structural and functional aspects of plant sexual development. We also discuss the impact of dioecy and sex chromosomes on genome structure and expression.
Keywords: Dioecy, sex determination, sex chromosomes, genome evolution, epigenetics, transposable elements
INTRODUCTION
While dioecious species represent a minority in angiosperms, there are nevertheless about 15 600 dioecious species (5–6 % of all species) in 987 genera (7 % of all genera) and 175 families (43 % of all families) (Renner, 2014). An important feature of flowering plants is the numerous transitions between breeding systems that have occurred across the phylogeny (Tree of Sex Consortium, 2014). Sex chromosomes have so far been identified in about 40 species and observably different morphology between X (Z) and Y (W) has been confirmed in just 18 species (Ming et al., 2011; Renner, 2014).
The presence of sex chromosomes enables a part of the genome (the non-recombining part of the Y or W) to optimize its function fully for the benefit of one sex and also influences evolution of the genes in the rest of the genome (reviewed by Charlesworth, 2013, 2015). Degeneration of sex chromosomes occurs as a consequence of suppressed recombination, and dosage compensation can be established (Muyle et al., 2017). Degeneration of sex chromosomes has been reported in the following plant species: Silene latifolia (Marais et al., 2008; Muyle et al., 2012; Bergero et al., 2015; Papadopulos et al., 2015), Carica papaya (Wu and Moore, 2015), Mercurialis annua (Ridout, 2017), Rumex hastatulus (Hough et al., 2014; Beaudry et al., 2017; Crowson et al., 2017; Hough et al., 2017) and Rumex rotschildianus (Beaudry et al., 2017). With the exception of C. papaya, the degeneration was found only in the Y-linked regions. In papaya, not only the non-recombining male-specific region of the Y (MSY) but also the corresponding part of the X chromosome (both located close to the centromere) shows signs of degeneration (Wu and Moore, 2015). The X chromosome in papaya has been influenced by a selective sweep that drastically lowered its diversity (VanBuren et al., 2016). A relatively high percentage of deleted sequences from the Y chromosome (tens of per cent) has been detected in S. latifolia (Bergero et al., 2015; Papadopulos et al., 2015) and in R. rotschildianus (Beaudry et al, 2017). In contrast, in M. annua, only 0.2 % of genes appear to be lost. Pseudogenization of the Y-linked genes is more frequent in M. annua (approx. 36 %), suggesting that even in this system there is a tendency to degeneration of the Y-linked alleles. Comparatively, no significant degeneration of the sequences in the non-recombining region has been found in Salix viminalis (Pucholt et al., 2017). Albeit the dioecy in the Salicaceae is of an ancient origin, this fact suggests that the current sex-determining region (SDR) in Salix is relatively young, and switches in sex determination occurred in Salicaceae. These switches could have been caused by a new master gene that took over the sex-determining function (reviewed by Beukeboom and Perrin, 2014). An alternative explanation is that a random ZW recombination occurred in sex-reversed individuals. The occasional XY recombination enabled by periodic sex reversal is known to protect the Y chromosome in frogs from degeneration (Perrin, 2009; Rodrigues et al., 2018). Rare cases of hermaphroditism have also been reported in the genus Salix (reviewed by Mirski, 2014; Nagamitsu and Futamura, 2014). There are, however, no data concerning the possible influence of sexual phenotype on Z–W recombination in Salix. If compared with other taxa, the sex-determining systems in the genus Fragaria (small mobile sex-determining cassette) appear as an efficient method to avoid the spread of W chromosome degeneration (Tennessen et al., 2017). The current data support the view of Bachtrog et al. (2014) that the degeneration of Y and W chromosomes is not inevitable, at least in some systems.
Phenotypic effects of the changes that occur on the sex chromosomes include lowered fitness and fertility, and reduced numbers of one sex in interspecific crosses. The occurrence of these problems in animals and also in plants (Demuth et al., 2014) seems to follow Haldane’s rule (Haldane, 1922: ‘When in the offspring of two different animal races one sex is absent, rare, or sterile, that sex is the heterozygous [heterogametic] sex’. The study of the causes of male rarity and low fertility in male hybrid plants in interspecific crosses (Haldane’s rule) between two dioecious species of Melandrium, i.e. S. diclinis and S. latifolia, by Demuth et al. (2014) has also revealed a possible difference in mechanisms behind Haldane’s rule in Silene and animal models. The dominance theory [supposing that the problems in interspecific hybrids are caused by recessive mutations in X-linked genes as they are only present in one copy in the hybrid of heterogametic sex (reviewed by Turelli and Orr, 1995)] is sufficient to explain male sterility in Silene hybrids, whereas male rarity is more likely to involve faster male evolution (Demuth et al., 2014) or faster heterogametic sex evolution (Delph and Demuth, 2016) but not dominance. Faster male evolution means that males are under stronger sexual selection and so the corresponding genes are evolving faster (Wu and Davis, 1993). Faster heterogametic sex evolution results from parts of Y- and W-linked sequences evolving without recombination and then co-segregating with heterogametic sex. This can lead to a species-specific loss of some Y- or W-linked genes or to the species-specific differentiation of the X- and Y-linked alleles as a result of the conflict in optima between sexes, e.g. in the sex ratio (reviewed by Delph and Demuth, 2016). Male rarity in interspecific crosses between S. diclinis and S. latifolia could also result from the evolution of the mechanisms controlling sex bias in S. latifolia and S. diclinis. This more severe female bias has also been observed in interspecific crosses between S. latifolia and S. dioica (Taylor, 1994a, b). In these systems, the female genomes appear to carry alleles that cause a female bias, whilst the Y chromosomes carry alleles tending to restore the 1:1 sex ratio. This restoration is apparently not sufficiently efficient in the interspecific crosses. From this point of view, the rapid evolution of the Y-linked sex ratio-restoring alleles in response to the female bias-generating alleles in the rest of genome could be the cause of male rarity in interspecific crosses between S. diclinis and S. latifolia. The consequences of the fast evolution of the Y-linked genes are apparent also in interspecific hybrids between dioecious and hermaphrodite species, e.g. in the cross between S. latifolia and S. viscosa (Zluvova et al., 2005).
The possible role of the X chromosome in speciation (large-X effect) in the genus Silene has also been tested. A study by Hu and Filatov (2016) revealed the consequences of the large-X effect for reproductive barriers between S. latifolia and S. dioica. The interspecific gene flow between these species in X-linked sequences is significantly lower than the gene flow in autosomal sequences, suggesting that the X chromosome could play a role in speciation similar to that in animal models. In these models, the hemizygotic status of X reveals the recessive putatively harmful alleles that accumulated on the X chromosome and that are compensated for in the original species but cause problems in interspecific hybrids.
The presence of genes with sex-specific expression on sex chromosomes is widespread in plants. Similar to animal species, there are also autosomal sequences which are sex specifically expressed (Zluvova et al., 2010; Zemp et al., 2014, 2016). In contrast to animals though, sexual dimorphism in autosomal gene expression in plants is mostly associated with the presence of sex-specific organs and tissues, and therefore can be mostly detected in flowering plants. However, cases of sex-specific expression of autosomal genes in vegetative plant tissues have been reported (Zluvova et al., 2010; Zemp et al., 2014, 2016). The detailed study by Zemp et al. (2016) compared the gene expression patterns of both sexes in S. latifolia with those of a hermaphrodite individual of S. vulgaris. The expression patterns in the S. latifolia female differed greatly from the expression of the given genes in the S. vulgaris hermaphrodite. Both higher and lower expression was observed in an S. latifolia female, and sex-biased gene expression was more frequent in sex-linked genes. The results support the possible feminization of the X chromosome and the masculinization of the Y chromosome.
In this review, we summarize the current knowledge concerning the sex-determining mechanism in several model dioecious species including the epigenetic aspects revealed in some species and, subsequently, we review the questions connected with the origin of dioecy and the evolution of the genomes of dioecious plants with a focus on repetitive sequences.
GENETIC MECHANISMS OF SEX DETERMINATION IN FLOWERING PLANTS
Sex-determining systems with largely differentiated sex chromosomes
Species with largely differentiated sex chromosomes show considerable advantages for studies at the cytogenetic level. Heteromorphic sex chromosomes permit the employment of modern techniques such as flow sorting and microdissection to isolate sex-linked sequences directly. On the other hand, it seems to be more complicated to elucidate the sex-determining systems in these species at the molecular genetic level than in species with less differentiated sex chromosomes as too large a number of candidate genes exist in large non-recombining regions.
Genus Silene section Melandrium (Caryophyllaceae)
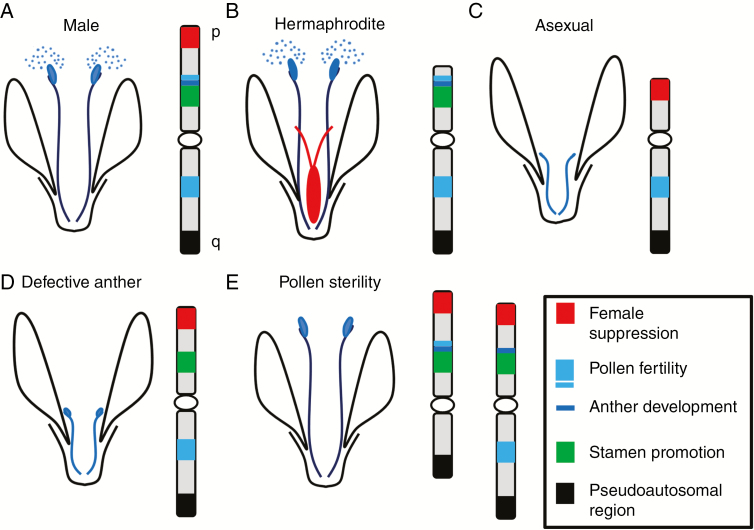
Since the discovery of the first plant XY sex determination system in Silene latifolia (Winge, 1923), plant sex chromosomes have attracted considerable scientific attention. The first attempt to study the role of plant sex chromosomes in developmental processes dates back to the 1940s. As a by-product of experiments with polyploids in S. latifolia, Westergaard created Y deletion mutants with various sexual phenotypes – hermaphroditic and pollen sterile male flowers (Westergaard, 1946). This and later studies on mutants carrying chromosomal aberrations revealed three important, physically separated Y-linked regions: the gynoecium-suppressing functional region (GSF), the stamen-promoting functional region (SPF) on the p arm, and the male fertility functional (MFF) region on the q arm (Westergaard, 1946; Donnison et al., 1996; Farbos et al., 1999) (Fig. 1A–E). Later, large-scale deletions (disruptions) of the Y chromosome were introduced by means of either gamma or X-ray irradiation (Farbos et al., 1999; Lebel-Hardenack et al., 2002). Radiation-induced mutants, in addition to the deletion mutants already observed by Westergaard (1946) [hermaphrodites (Fig. 1B) and infertile males (Fig. 1D, E)], produced asexual plants (Fig. 1C) (containing a Y deletion), confirming Westergaard’s model of the Y chromosome in S. latifolia (Donnison et al., 1996; Barbacar et al., 1997; Lardon et al., 1999). The model of Y chromosome organization was later improved by Zluvova et al. (2007), who showed (based on deletion mapping and the histological studies of anthers in deletion mutants) the existence of further genes involved in anther development and also one male fertility gene close to the stamen promoter region (Fig. 1D, E). Recently, the integration of deletion mutant data from various resources to study SDRs has been improved using the DelMapper software package (based on the resolution of the travelling salesman problem) and fast neutron-induced mutants (Kazama et al., 2016).
Fig. 1.
Schematic functional map of the S. latifolia Y chromosome. Study of individuals with chromosomal aberrations revealed three functional regions on the Y chromosome. (A) Wild-type male. (B) Deletion of a distal section (red) of the p arm led to a hermaphrodite. Thus, this functions as the gynoecium-suppressing functional (GSF) region. (C) A proximal area of the p arm (marked in green) produces an asexual phenotype when deleted. For this reason, it has been designated as the stamen-promoting functional (SPF) region. (D) The region important for further anther development is located near to the SPF region. (E) Deletion of male fertility factors (MFFs) residing mainly on the q arm (light blue) but also in proximity to the SPF region on the p arm leads to male sterility.
However, despite these efforts, the molecular details of the sex determination in S. latifolia remain unknown. In fact, only a few sex-linked genes have been studied in detail in S. latifolia so far. The Class B floral MADS-box gene APETALA3 (SlAP3 – TM6 lineage) was confirmed to be localized on S. latifolia sex chromosomes (Cegan et al., 2010) and, interestingly, expression studies of SlAP3 revealed a different mRNA pattern in relation to sex, with the Y allele expressed predominantly in the stamens and petals, and the X allele expressed in petals, styles and ovaries (Matsunaga et al., 2003). Therefore, SlAP3 represents a unique case of a floral developmental gene where an originally identical pair of alleles residing on autosomes underwent sub-functionalization during sex chromosome evolution. In comparison, in arabidopsis, APETALA3 controls anther development, and a mutant phenotype involves anther replacement by carpels in the third whorl of the flower (Bowman et al., 1991). It is therefore possible that a homologue of this gene confers sex-specific floral development in S. latifolia, even though it is not directly located in one of the functionally described Y-linked regions.
The autosomal genes controlling gynoecium development are probably involved in flower meristem maintenance and in the control of its size. Orthologues of SHOOTMERISTEMLESS (SlSTM1 and SlSTM2) and CUP-SHAPED COTYLEDON (SlCUC) show a male-specific expression pattern in meristematic tissue before any morphological differences between the sexes become apparent (Zluvova et al., 2006). Later, another study showed that SlCLV1 (orthologue of CLAVATA1) might also be engaged in carpel suppression (Koizumi et al., 2010).
Another candidate gene with a possible role in sex-specific development in S. latifolia is the homologue of WUSCHEL (SlWUS1) which has also been reported to contribute to meristem maintenance and anther development in arabidopsis (Deyhle et al., 2007). There are two copies of SlWUS in S. latifolia (SlWUS1 and SlWUS2), one located on the X chromosome with no homologous copy on the Y chromosome (Kazama et al., 2012). Since expression studies have shown no dosage compensation of SlWUS1 in early floral meristem development, SlWUS1 could act as a haploinsufficient gene involved in carpel development. Nevertheless, the proposed functions of SlAP3 and SlWUS1 in S. latifolia are not supported by any direct evidence, since S. latifolia was considered recalcitrant to genetic transformation. Recently, a protocol for gene delivery to S. latifolia was developed (Hudzieczek et al., 2018), allowing functional studies of plant sex-linked genes using targeted mutagenesis and overexpression.
Genus Rumex (Polygonaceae)
Most widely studied species of this genus show either completely dosage-based (X/A system similar to Drosophila; XX/XY1Y2 karyotype) SDRs (e.g. Rumex acetosa: Ono, 1935; Yamamoto, 1938) or modified dosage-based (X/A) sex-determining systems (R. hastatulus). Two races occurring in R. hastatulus differ significantly in their karyotype (North Carolina: XX/XY1Y2 and Texas: XX/XY), and two autosome–heterosome translocations were reported in the North Carolina race (Grabowska-Joachimiak et al., 2015). In spite of these differences, the mechanism of sex determination in both races of R. hastatulus appears similar: an X/A balance mechanism modified in the male direction by the Y chromosome. The Y chromosome, or its fragment, can promote anther development in AAXX plants (Y chromosomes are not involved in gynoecium suppression) and the sex phenotype can be influenced by the environment and/or genetic background (Smith, 1963; Bartkowiak, 1971). Recent efforts in R. acetosa have been focused on the isolation of genes with connections to the female sex programme. A set of genes largely expressed in female floral buds was isolated by cDNA subtraction, and the interesting candidate gene FEM32 underwent molecular and functional analysis (Manzano et al., 2017). This gene encodes a protein with a common architecture in the aerolysin family, acting as β pore-forming toxins. Since genetic analysis of gene function in R. acetosa was not feasible, the FEM32 gene was cloned and studied in transgenic tobacco. The transgenic plants were strongly feminized and their pollen fertility was rather low. The authors concluded that the FEM32 gene may function in R. acetosa by arresting stamen and pollen development probably via programmed cell death (Manzano et al., 2017).
A dominant Y chromosome-based sex-determining system with X and Y chromosomes that appeared indistinguishable when using an optical microscope was found in R. acetosella (Singh and Smith, 1971). Later, electron microscopic observations of whole-mount preparations of synaptonemal complexes (SCs) at pachytene made it possible to detect heteromorphic sex chromosomes in R. acetosella and in the related species R. suffruticosus (Cuñado et al., 2007).
Coccinia grandis (Cucurbitaceae)
Coccinia grandis possesses the largest Y/autosome divergence in flowering plants (Sousa et al., 2013). Its relatively small genome size (0.943 pg/2C in males), large Y chromosome and phylogenetic proximity to the fully sequenced Cucumis sativus (Huang et al., 2009) makes C. grandis a promising model to study sex chromosome evolution. Current RNA sequencing (RNa-seq)-based studies (Devani et al., 2017; Mohanty et al., 2017) support the view that the sex-determining mechanism in Coccinia could be similar to that of C. sativus and Cucumis melo, see below.
Sex-determining systems with small sex-specific regions (homomorphic or non-conspicuously differentiated sex chromosomes)
Progress in the identification of genes responsible for sex determination in dioecious species has increased dramatically due to novel sequencing techniques and the employment of reverse genetic methods. Recently, candidates for sex determination have been discovered in several plant species. For technical reasons, the most significant results have been achieved in species with small SDRs. Here, we are examining several species with non-differentiated sex chromosomes and some with no sex chromosomes.
Genus Diospyros (Ebenaceae)
Persimmon (Diospyros lotus) possesses a very small SDR despite a long evolutionary history of dioecy (Akagi et al., 2014). In the Caucasian persimmon, Diospyros lotus, a dioecious plant with heterogametic males (XY), a male-determining region was identified via genome sequencing and a combination of transcriptomics and phylogenetic analyses. The only sex-determining gene in this region is OGI which encodes a small RNA (sRNA) targeting the autosomal MeGI gene, a homeodomain transcription factor suppressing anther fertility. As no gynoecium-suppressing gene has so far been identified in the Y-linked SDR in D. lotus, the authors consider an alternative model for sex determination in persimmon based on a single master gene (OGI) that could act similarly to the SRY gene in the human (Sinclair et al., 1990) (marked as Sry in other mammals). The SDR Y protein (transcription factor coded by SRY) works as a trigger of male development and so simultaneously indirectly suppresses female development. If the action of the SRY gene is absent, the female developmental programme is realized (reviewed by Kashimada and Koopman, 2010). The male-promoting role of OGI has also been reported in the polygamous D. kaki (Zhang et al., 2016). Taking into account the long evolutionary history of dioecy in the ancestors of Diospyros, we can speculate that dioecy could have evolved via the two-mutation-based model (Charlesworth and Charlesworth, 1978) but later the sex-determining system was transformed into a one master gene-based system as a consequence of the sex dimorphism evolution as suggested by Janousek and Mrackova (2010). Last, but not least, the presence of an unknown Y-linked female suppressor was considered possible (Akagi et al., 2014) and therefore the two-gene model could be applied for D. lotus.
Genus Fragaria (Rosaceae)
This well-studied genus includes gynodioecious, sub-dioecious and dioecious species. Female heterogamety (ZW system) with dominant male sterility alleles was found in sub-dioecious F. virginiana (Spigler et al., 2008, 2010) and dioecious F. chiloensis (Goldberg et al., 2010). Recent studies (Tennessen et al., 2017; Wei et al., 2017) show that the SDR is located in three different positions in different chromosomes from the same homoeologous group in three different octoploid taxa: F. virginiana ssp. platypetala, F. virginiana ssp. virginiana and F. chiloensis.
The results obtained by Tennessen et al. (2017) further show that the SDR (i.e. a conserved SDR cassette) repeatedly changed genomic location across octoploid Fragaria. This is the first unambiguous evidence of SDR translocation in flowering plants. Only two coding genes, annotated as GDP-mannose 3,5-epimerase 2 and 60S acidic ribosomal protein P0, were identified in this mobile SDR cassette. These two genes are expected to have roles in sex determination, but the mechanism of their action in Fragaria has not yet been identified.
Asparagus officinalis (Asparaceae)
Garden asparagus (Asparagus officinalis), a dioecious species with an XX/XY sex determination system, possesses homomorphic sex chromosomes with a hypothetical male activator and female suppressor located in the M locus on the Y chromosome (Telgmann-Rauber et al., 2007). RNA-seq analysis of female (XX), male (XY) and supermale (YY) individuals identified male- and female-biased expression and supported the hypothesis that anther abortion in the female flower started during development of the tapetum and early microspore (Harkess et al., 2015). A further study revealed male and female differentially expressed microRNAs (miRNAs) by sRNA and degradome sequencing (Chen et al., 2016a). Finally, genome sequencing of A. officinalis revealed 13 genes in approx. 1 Mbp of the non-recombining Y region (Harkess et al., 2017). Two genes in this region have been identified as candidates for sex determination. The function of SUPPRESSOR OF FEMALE FUNCTION (SOFF) in the cessation of pistil development has been verified in XY hermaphroditic mutants, while the second candidate gene Asparagus DEFECTIVE IN TAPETUM DEVELOPMENT AND FUNCTION 1 (AspTDF1) is exclusively expressed in males and was previously described as Male Specific Expression 1 (MSE1) (Harkess et al., 2017; Murase et al., 2017). AspTDF1 very probably functions as a male activator, considering that a TDF1 mutant in Arabidopsis thaliana exhibited degeneration of tapetal cells similar to A. officinalis females (Murase et al., 2017). An orthologous sequence to SOFF has also been found in A. conchinchensis (a dioecious species closely related to A. officinalis), but it is not present in the hermaphrodite A. virgatus, suggesting the ancestral role of SOFF in suppression of female function in dioecious species of the genus Asparagus. The role of the AspTDF1 gene is unclear in related A. conchinchensis as it appears pseudoautosomal in this species. However, the fact that SOFF has only a female suppression function suggests that in A. conchinchensis there must be some other cause of male organ suppression in females and thus also another mechanism of restoration of anther development in males. Anyway, these findings strongly indicate that the two-gene model represents the most likely scenario for the evolution of sex determination in Asparagus.
Carica papaya (Caricaceae)
Sex determination in the papaya (Carica papaya) has been subjected to detailed genetic studies in the past. Hofmeyr (1938, 1939) and Storey (1938) proposed three alleles; M, Mh and m, to determine plant sex types (M1, M2 and m according to Hofmeyr, 1938), stressing the dominance of maleness (M) and hermaphroditism (Mh) over the female phenotype in heterozygotes. More recent studies showed that sex determination in the papaya is based on the standard XX/XY system which, however, includes the modified Y chromosome (Yh, corresponding to the Mh allele of Storey, 1938) which is unable to suppress gynoecium development (reviewed by Ming et al., 2007). Plants lacking an X chromosome were absent from the progeny of hermaphrodites, suggesting the presence of a recessive lethal allele on both types of Y chromosome. The exact mechanism of sex determination in the papaya is not yet clear, but it is known that the action of gibberellins is necessary for gynoecium suppression in papaya male plants (Kumar and Jaiswal, 1984). In some plants, the gibberellin inhibitor caused a direct change from male to female. This fact points to a possible difference of the sex-determining mechanism in papaya from the active gynoecium suppressor-based mechanism (also involving a separate Y-linked anther promoter) suggested by Westergaard (1958). On the other hand, increased concentrations of gibberellins do not cause any change in the sexual phenotype in papaya, suggesting that the level of gibberellins is not itself sufficient to control sex determination (Han et al., 2014). Papaya is intensively studied at both the transcriptomic and genomic level (Liu et al., 2004; Ming et al., 2008; Wang et al., 2012; VanBuren et al., 2015). Most of the miRNAs overexpressed in the male flowers were related to auxin signalling pathways, whereas the miRNAs overexpressed in female flowers were the potential regulators of apical meristem identity genes (Aryal et al., 2014). Results of the sequencing and comparisons of the non-recombining regions between hermaphrodites and males suggest that the difference is not present in any coding region and so implies that hermaphroditism may be controlled by changes in gene regulation. Overall, identification of the gene controlling suppression of carpel development in males is quite a challenging task (VanBuren et al., 2015).
Cucumis melo and Cucumis sativus (Cucurbitaceae)
No sex chromosomes have so far been identified in Cucumis melo (mostly monoecious species). Instead, three genes – each located on a different chromosome – are involved in sex determination (Boualem et al., 2008, 2015; Martin et al., 2009). Monoecy in melon (Cucumis melo) is maintained by two dominant alleles of these loci – Andromonoecious (A) and Gynoecious (G) (Kenigsbuch and Cohen, 1990). First, the A locus was reported as a gene encoding 1-aminocyclopropane-1-carboxylic acid synthase (CmACS7), responsible for repressing male organ development (in female flowers) via the ethylene biosynthesis pathway. Therefore, a recessive mutation (genotype aaG–) in this gene results in individuals carrying male and hermaphroditic flowers (Boualem et al., 2008). Secondly, the G allele was identified as a WIP subfamily C2H2 zinc finger transcription factor (CmWIP1) responsible for the arrest of carpel development. In gynoecious individuals (A-gg), the G allele is silenced by heritable epigenetic modification – DNA methylation triggered by the insertion of a transposon in the CmWIP1 promoter region. Furthermore, CmWIP1 indirectly inhibits the expression of the CmACS-7 (Andromonoecious) gene, thus supporting anther development (Martin et al., 2009). Recently, an androecy gene CmACS11 in melon was discovered. CmACS11 is also involved in catalysing ethylene biosynthesis but it negatively regulates the CmWIP1 gene and therefore its disruption leads to the formation of males (Boualem et al., 2015). A recent study by Latrasse et al. (2017) shows that CmACS11 represses the expression of the male-promoting gene CmWIP1 through an epigenetic mechanism (via trimethylation of Lys27 in histone H3). Taken together, the combination of allele variants of CmACS7, CmWIP1 and CmACS11 is responsible for various sexual phenotypes including hermaphrodite, monoecious, gynoecious and androecious individuals. A similar system of sex determination (also based on the ethylene biosynthesis pathway) has been found in Cucumis sativus (Boualem et al., 2009). The melon sex determination system, even without sex chromosomes, illustrates how the evolution of unisexual flowers could take a different evolutionary trail from a ‘two mutations on one chromosome model’.
Genus Populus (Salicaceae)
While the sex chromosomes in all studied species of the genus Populus are homomorphic, archeological data suggest that the genus had (together with the genus Salix) a dioecious ancestor and that the dioecy in Salicaceae is old. In the Salicaceae, almost all species are dioecious, suggesting that sex determination may be as old as 45 million years, based on fossils of Pseudosalix (leaves and flowers; extinct sister genus to Salix and Populus) (Boucher et al., 2003) and fossils of two extinct species of Populus (Manchester et al., 2006).
Geraldes et al. (2015) identified male heterogamety in several cottonwoods (species of sections Tacamahaca and Aigeiros): P. trichocarpa, P. balsamifera, P. deltoides and P. nigra. Male heterogamety has also been found in aspens (P. tremula and P. tremuloides, section Populus) (Kersten et al., 2014; Pakull et al., 2015). At present, it appears that only one Populus species shows female heterogamety (P. alba, section Leuce; Paolucci et al., 2010). Male heterogamety was, however, also reported in this species (reviewed by Kersten et al., 2017). The study by Geraldes et al. (2015) revealed that the SDRs in P. trichocarpa, P. balsamifera, P. deltoides and P. nigra are homologous. In contrast, the SDR in Populus tremuloides (aspen) differs in that it resides on the homologous chromosome (LG 19) but in a pericentromeric position, suggesting that there was at least one switch in sex determination among the species showing male heterogamety (Kersten et al., 2014; Pakull et al., 2015).
In addition, two putative sex-determining genes have been identified in P. trichocarpa and P. balsamifera (Geraldes et al., 2015). The poplar contains sex-linked orthologues of the A. thaliana gene ARABIDOPSIS RESPONSE REGULATOR 17 (ARR17) which mediates the output of cytokinins, and of arabidopsis METHYLTRANSFERASE 1 (MET1) which is involved in DNA methylation. Male-specific DNA methylation patterns in PbRR9 (Populus balsamifera putative response regulator RR, see above) suggest that the methylated allele could be involved in sex determination in Populus (Bräutigam et al., 2017). A more detailed analysis is, however, necessary to confirm this finding and to describe the exact mechanism of the action of PbRR9 in sex determination. One possibility could be that DNA methylation inactivates the Y-linked allele and the gene exhibits haploinsufficiency. Transcription patterns of the X and Y alleles of PbRR9 are, however, so far unknown.
Genus Salix (Salicaceae).
Female heterogamety has previously been described in three species [S. purpurea (Carlson et al., 2017)], S. viminalis (Pucholt et al., 2015) and S. suchowensis (Chen et al., 2016b)]. Carlson et al. (2017) identified TCP-1 (SapurV1A.1538s0020) as one of the few sex-biased genes that exhibited complete null expression in the male shoot tip. TCP transcription factors play pivotal roles in the control of shoot morphogenesis by negatively regulating the expression of boundary-specific genes. TCP24 negatively modulates secondary wall thickening of the anther endothecium in Arabidopsis (Wang et al., 2015). It was suggested that this gene could play a major role in the suppression of the anther endothecium in female S. purpurea catkins (Carlson et al., 2017). It was shown that the SDR in Salix viminalis evolved independently from the SDR present in most of the studied Populus species (Hou et al., 2015; Pucholt et al., 2015). Salix viminalis has also been used to evaluate the relative rate of evolution for sex-biased genes. The results showed slower sequence evolution for male-biased genes expressed in the reproductive tissue compared with unbiased and female-biased genes (Darolti et al., 2018).
Mercurialis annua (Euphorbiaceae).
While initial studies suggested a three-locus model for sex determination in diploid dioecious Mercurialis annua (Louis, 1989; Durand and Durand, 1991), more recent research shows that sex determination in natural populations is based on the standard XX/XY sex-determining system (Russell and Pannell, 2015; Ridout et al., 2017). The current data also reveal that M. annua possesses a relatively large non-recombining region corresponding to about 40 cM on the X chromosome map (Ridout et al., 2017), and this region also began to accumulate specific repetitive sequences (Veltsos et al., 2018). This species is exceptional in variability of breeding systems as the tetraploid races are monoecious and the hexaploid races are androdioecious or monoecious (reviewed by Russell and Pannell, 2015). The hexaploid races are believed to be a result of an interspecific hybridization between the monoecious tetraploid race of M. annua and its diploid dioecious relative M. huetii (Russell and Pannell, 2015).
Section Otites of the genus Silene (Caryophyllaceae).
In contrast to the section Melandrium, the sex-determining systems of the Silene section Otites display much higher dynamics and variability. Very small sex-linked non-recombining regions are present in this section (Mrackova et al., 2008; Slancarova et al., 2013; Balounova et al., 2018), as can be deduced from the small number of sex-linked molecular markers. Interestingly, both female and male heterogamety is present in this section (Mrackova et al., 2008; Slancarova et al., 2013) which is a rare phenomenon in plants. The study of Balounova et al. (2018) suggests that the original sex-determining system in Otites was female heterogamety and that a switch to male heterogamety occurred in S. colpophylla. This switch was also connected to the change of the linkage group from which the sex chromosomes were recruited.
MODIFICATION OF SEX EXPRESSION – ENVIRONMENTAL INFLUENCES AND EPIGENETIC PERSPECTIVE
Gynoecium development in genetic males – epigenetics and environmental influences
Sex expression in dioecious plants with differentiated sex chromosomes is usually genetically determined; however, epigenetic processes may play a role in some species and can cause instability of the sexual phenotype. While gynoecium suppression in S. dioica shows some level of instability even in natural populations, the sexual phenotype in natural European populations of S. latifolia is very stable (van Nigtevecht, 1966). However, modification of sex expression in S. latifolia is possible using the epigenetic drug 5-aza-cytidine to inhibit DNA methyltransferase leading to the hypomethylation of DNA (Janousek et al., 1996). Application of this drug to germinating seeds changes some males to androhermaphrodites, while no apparent phenotypic changes are observed in females. The effect can be enhanced with zebularine, another DNA-hypomethylating drug (V. Bacovsky and R. Hobza, unpubl. res.) (Fig. 2A–D). The DNA methylation pattern is transmitted to subsequent generations in crosses of the androhermaphrodite plant as a pollen donor with wild-type females. Androhermaphroditism is, however, not inherited if a hermaphrodite plant is used as a seed parent (Janousek et al., 1996). It was shown (Janousek et al., 1998) that androhermaphroditism is inherited as an allele tightly linked to the Y chromosome and that the Y chromosome is not inherited through the female gametic line in androhermaphrodite plants. These results suggest that the gynoecium-suppressing gene can be epigenetically regulated via DNA methylation. It is possible to speculate that the rare cases of androhermaphroditism observed in the North American populations of S. latifolia could be connected to epigenetic changes (Miller and Kesseli, 2011). Indeed, the post-invasion evolution of S. latifolia in North America resulted in numerous phenotypic differences between North American and European populations (Blair and Wolfe, 2004), and it is conceivable that at least some of these variations involve epigenetic changes (Blair and Wolfe, 2004). The genetic and/or epigenetic changes in the North American population could also explain the Y chromosome transmissibility through the female gametic line in North American populations.
Fig. 2.
Silene latifolia male and female flower development modification phenomena. Wild-type female (A) and male (B) individual flowers; global view (C) and detail (D) of a flower of a male individual with androhermaphrodite phenotype after zebularine treatment (50 μm). The experimental procedure was otherwise identical to the 5-aza-cytidine experiment by Janousek et al. (1996); flower of female (E) and male (F) individuals infected with M. violaceum.
An analogous example of gene inactivation resulting from the hypomethylation of its surroundings can be found in A. thaliana, where global hypomethylation of the genome (achieved using methylation-deficient mutants and transgenic lines) leads to the hypermethylation and transcriptional inactivation of specific genes [SUPERMAN (Jacobsen et al., 1997, 2000); AGAMOUS (Jacobsen et al., 2000); BONSAI (Saze and Kakutani, 2007)]. Artificial hypomethylation can affect the function of genes in many ways; for example, DNA methylation in introns is important for the correct splicing of genes (To et al., 2015). However, the increase in the penetrance and expressivity of androhermaphroditism in subsequent generations supports the view that the gynoecium suppressor in S. latifolia is influenced in a similar way to the SUPERMAN, AGAMOUS and BONSAI genes. SUPERMAN probably does not play a key role as a gynoecium suppressor in S. latifolia as it is not located on the Y chromosome, but could be involved in stamen suppression during female flower development (Kazama et al., 2009).
Further interesting findings have been obtained in Populus. The instability of the sexual phenotype resulting in andromonoecious plants in P. tomentosa is probably associated with DNA methylation changes influencing miRNA (miRNA172; Song et al., 2015). Geraldes et al. (2015) found that there is a gene present in the small SDRs in Populus balsamifera and P. trichocarpa which codes for DNA methyltransferase. Moreover, recent results indicate that the SDR in P. balsamifera includes a gene with the greatest sex-specific difference in DNA methylation patterns (PbRR9) (Bräutigam et al., 2017). Hypomethylation experiments should be performed to confirm the direct role of correct DNA methylation patterns on the sexual phenotype in Populus.
In males of Carica papaya, gynoecium development can be promoted by cold treatment (Lin et al., 2016). The comparison of the expression profiles between males and cold-induced hermaphrodites revealed a significant increase in the abundance of transcripts connected with the methylation of Lys9 of histone H3 (H3K9) (Lin et al., 2016). Based on the role of methylation of H3K9 in heterochromatin formation (Rivera et al., 2015), the authors suggested that the probable cause of hermaphroditism is the inactivation of the putative gynoecium suppressor present on the Y chromosome (Lin et al., 2016).
Information about the role of resource (water and/or mineral nutrients) availability on gynoecium formation (and/or investment to seed production) has been obtained for several species. Males from strictly dioecious populations of Wurmbea dioica (Colchicaceae) never formed gynoecium. In contrast, the response of males from sub-dioecious populations was dependent on the level of resources, with the gynoecium developed more often in high resource conditions (Vaughton and Ramsey, 2012). In gynodioecious Nemophila menziesii (Boraginaceae), supplemental water increased seed production in hermaphrodites but not in females (Barr, 2004). In sub-dioecious Fragaria virginiana (Rosaceae), Ashman (1999) showed that the fruit set of hermaphrodites, but not of females, increases significantly with plant size, and plant size increases with greater abundance of soil resources. In the sub-dioecious shrub Eurya japonica (Pentaphylacaceae), all possible types of sex changes were observed, but there was a tendency for the transformation from the hermaphrodite state to unisexual (male or female) in unfavourable growth conditions (Wang et al., 2017).
Anther development in genetic females – epigenetics and environmental influences
Asymmetry in the ability to develop the organs of the opposite sex has been observed in many dioecious and sub-dioecious species. Male plants are often more prone to the development of fertile gynoecia, occasionally or under specific environmental influences (Delph and Wolf, 2005). In contrast, female plants are mostly not able to form fertile anthers even if anther development is stimulated by environmental factors. Although stamen arrest in S. latifolia females is stable and insensitive to the exogenous application of phytohormones (Ruddat et al., 1991), there is a natural mechanism modulating sex expression. When female plants are infected by the parasitic smut Microbotryum violaceum, the rudimentary stamens develop into stamens morphologically resembling mature stamens in males (Uchida et al., 2003) (Fig. 2E, F). However, the induced anthers harbour teliospores instead of pollen grains (Antonovics and Alexander, 1992). A recent study showed a strong response of sex-linked genes to pathogen infection (Zemp et al., 2015). This result provides direct biological evidence that genes located on the sex chromosomes are important for male sexual organ development, and their altered expression participates in the defeminization and masculinization of S. latifolia female plants.
As in smut infection, the development of stamens in female plants is also promoted by Ag2S2O3 and AgNO3 treatment (Law et al., 2002), similarly to several other species [Cucumis sativa (Den Nijs and Visser, 1980); Ricinus communis (Mohan Ram and Sett, 1980); Cannabis sativa (Mohan Ram and Sett, 1982); and Coccinia grandis (Devani et al., 2017)]. Although the stamens induced in Silene are not as developed as those of male flowers, anther development proceeds further beyond the anthers induced by M. violaceum infection, and tapetum and non-viable pollen grains are formed. The extension of the flower life span of plants infected by M. violaceum suggests that ethylene inhibition may play a role in the observed morphological changes (Law et al., 2002; Uchida et al., 2003). The role of ethylene signalling in stamen induction in Silene is, however, still unclear as two other ethylene inhibitors did not show this effect (Law et al., 2002).
Interestingly, anther promotion by M. violaceum has also been described in the gynodioecious S. dichotoma, and a related species of M. violaceum – Microbotryum major (previous name Ustilago major) – is able to provoke anther formation in females of S. otites (Correns 1928). This comparability of activity is puzzling as the sex determination systems evolved independently in the section Melandrium and in the section Otites (Mrackova et al., 2009; Slancarova et al., 2013). The parsimonious explanation could be that the actions of these pathogens are able to influence some generally present mechanism controlling gene expression such as ethylene signalling. Similarly, the infection could cause suppression of the nonsense-mediated mRNA decay (NMD) pathway which selectively degrades mRNAs harbouring premature termination codons (PTCs) but also regulates the abundance of a large number of cellular RNAs including coding genes (reviewed by Hug et al., 2016). Suppression of the pathway (i.e. standard reaction of the plant body to the infection) could cause, among other effects, expression of anther-forming genes that would otherwise be silent in the female flower. The fact that other pathogens do not cause stamen formation in Silene females could be explained by the fact that only Microbotryum is able to penetrate deep inside the flower meristem and avoid the complete destruction of this tissue. A connection has been found between ethylene signalling and the NMD pathway (Merchante et al., 2015) and so both pathways could be involved.
EVOLUTION OF DIOECY AND SEX CHROMOSOMES – FROM HERMAPHRODITISM OR MONOECY TO DIOECY AND SEX CHROMOSOMES
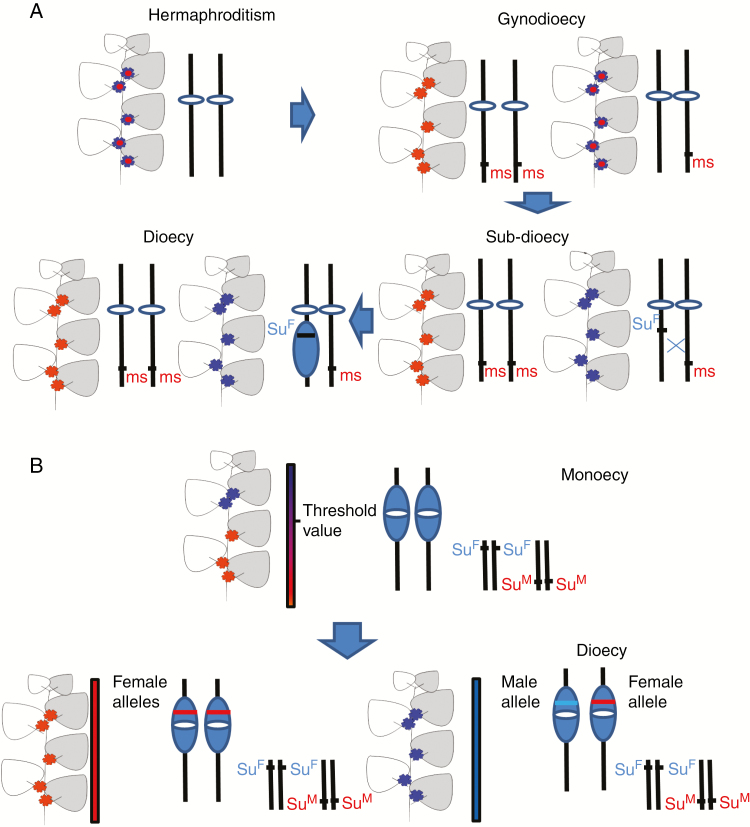
Data based on fossils are still inconclusive as to what original flower type was present in angiosperms – both unisexual and hermaphrodite flowers have been found in very ancient fossils (Friis et al., 2010). Many current models for the evolution of dioecy from hermaphroditism (reviewed by Charlesworth, 2015) include the gynodioecy step, as first suggested by Charlesworth and Charlesworth (1978). This scenario is based on at least two sequential mutations on the same chromosome. The first mutation (most probably recessive) causes male sterility and the second and/or further mutation(s) cause(s) the suppression of gynoecium development in males carrying a proto-Y chromosome (see Fig. 3A). The arrest of recombination between these loci (for the possible mechanisms, see Wright et al., 2016) could subsequently lead to the formation of sex chromosomes. The intermediate stages are characterized by the occurrence of sub-dioecy. Hermaphrodite flowers are present in mostly male individuals, but female plants possess a stable sex phenotype. Charlesworth and Charlesworth (1978) also deduced that the male sterility mutation (if dominant) could subsequently lead to the female heterogamety. This scenario is in accordance with dominant male sterility alleles in F. virginiana (Spigler et al., 2008, 2010) and F. chiloensis (Goldberg et al., 2010). The dominant male sterility allele also probably occurs in Salix purpurea (Carlson et al., 2017). A further supporting fact is that in at least three plant families where several species were tested concerning heterogamety, only female heterogamety has been found: Fragaria (Rosaceae; Spigler et al., 2008, 2010, Goldberg et al., 2010), Cotula (Asteraceae; Lloyd, 1975) and Pistacia (Anacardiaceae; Hormaza et al., 1994; Esfandiyary et al., 2012; Kafkas et al., 2015). The model starting with recessive male sterility has been referred to many times as a possible way to dioecy in plants.
Fig. 3.
Comparison of the evolution of dioecy via one-locus- and two-locus-based models. (A) Model of the origin of dioecy from hermaphroditism via gynodioecy – two-locus model. Recessive male sterility alleles (ms) can be either represented by nuclear recessive mutations or can reflect the absence of the dominant fertility restorer in the case where the male sterility-causing cytoplasm is present in all plants in the population. In the next step, the dominant female-suppressing allele (SuF) is recruited on the proto-Y chromosome. Occasional creation of asexual or hermaphrodite plants via recombination is possible. The cessation of the gynoecium is also influenced by genetic background and environment (sub-dioecy). Finally, the recombination between the female suppressor and the male sterility loci is arrested (as a consequence of divergence due to the accumulation of sexually antagonistic mutations and/or chromosomal inversions) and so the generation of asexual or hermaphrodite plants is avoided. This scheme is based on the original model by Charlesworth and Charlesworth (1978). (B) Alternative model of dioecy evolution from the monoecy – one-locus model. According to this model, the suppressors of gynoecium (SuF) and stamen development (or fertility) (SuM) are already present in the monoecious ancestor and their expression is controlled by intrinsic signals (e.g. hormonal signals connected with apical dominance). Because the strength and direction of the intrinsic signals varies along the plant axis, it results in the upper section carrying male flowers and the bottom part bearing female flowers. In this model, it is thought that there is a threshold value of stimuli separating male and female zones. In this situation, the mutations which are able to modify the effects of internal stimuli can cause an increase or decrease in the size of the male and female zones. Paradioecy (the presence of female flowers in ‘proto-males’ and male flowers in ‘proto-females’) can appear in the intermediate stage (not displayed in this scheme). In the extreme case, one sex can be completely absent in a given plant. Interestingly, if a single master sex-switching locus is recruited near the centromere or other non-recombining region, the sex chromosomes can originate in a single step. This scheme is based on the works by Lloyd (1972a, b, 1975, 1980, 1981), Webb (1999), Renner and Won (2001) and Renner (2016). Note: female flowers are shown in red; male flowers are shown in blue; and hermaphrodite flowers are shown as blue with a red middle part. Sex-linked non-recombining regions are shown as blue ovals.
Animal biologists often rely on the data and theories obtained in plants (mostly on the gynodioecy-based model) in their theories concerning the earliest phases of sex-determining system evolution as there is a lack of suitable model species in the animal kingdom (e.g. Weeks, 2012; Lorenzi and Sella, 2013; Wright et al., 2016). Interestingly, gynodioecy is extremely rare in animals (nine putative cases but only one confirmed) while androdioecy is more common, with 115 species identified (reviewed by Weeks, 2012). As suggested by Charlesworth and Charlesworth (1978), the route via androdioecy to dioecy is not likely in plants because males would need to produce more than twice as much pollen as hermaphrodites in order to invade the population. Indeed, there are just a handful of androdioecious species (five species from different genera and several species in Sagittaria, reviewed by Pannel, 2002). In plants, androdioecy is mostly derived from dioecy, e.g. Datisca glomerata (Wolf et al., 2001; Zhang et al., 2006) and the hexaploid Mercurialis annua (Russell and Pannell, 2015). The possible mechanism promoting the spread of androdioecy in hermaphrodite populations showing sporophytic self-incompatibility has been suggested in Phillyrea angustifolia (Oleaceae) where males are compatible with two reciprocally incompatible types of hermaphrodite (Saumitou-Laprade et al., 2010; Husse et al., 2013).
The original gynodioecy-based model was later slightly modified (Charlesworth, 2015) to reflect the fact that sex-determining loci can evolve in pericentromeric regions with suppressed recombination and that a two-locus-based model can also apply to the evolution of dioecy from monoecy. In this model, the reduction of the proportion of female flowers, i.e. the origin of males, is caused by a dominant mutation (SuF), which also enables the formation of a larger number of male flowers based on resource allocation. Recessive mutations in other genes could cause the replacement of male flowers with female flowers, i.e. the origin of females (Charlesworth, 2015). There is at least one case suggesting that the evolution of dioecy from monoecious ancestors could proceed via a two-locus model (Sagittaria latifolia; Dorken and Barret, 2004).
A less frequently discussed but probably common pathway to dioecy in plants is represented by the so-called monoecy/paradioecy/dioecy pathway (Lloyd, 1980; Webb, 1999; Renner and Won, 2001). In this case, the evolution of dioecy originates from monoecious ancestors. Paradioecy (the presence of male flowers in mostly female plants and vice versa) occurs in the intermediate stage due to the step-wise arrest of the development of the flowers of the opposite sex in both ‘proto-females’ and ‘proto-males’. The fact that there is a pre-existence of pathways for the suppression of both male and female organs in the case of monoecious ancestors has been previously stressed by Renner (2016), based on previous work on the monoecy/paradioecy/dioecy pathway (Lloyd 1972a, b, 1975, 1980, 1981; Webb 1999; Renner and Won, 2001). If this prerequisite is satisfied, mutations in a single gene controlling the expression of the pre-existing sex-determining genes are sufficient to establish dioecy (Renner, 2016).
Two new types of alleles in a developing sex-determining locus can be selected, with each allele shifting the ratio of male and female flowers in the male or female direction, respectively. Alternatively, the reduction of one type of flower (caused by changes in genetic background or by environmental influences) is compensated by the recruitment of a new sex-determining master gene. In this case, even a single mutation in a single gene can lead to dioecy. This scenario can more easily proceed in long-living and/or vegetatively propagating species than in annual species relying on sexual propagation. There is a greater abundance of dioecious species among woody perennials than in plants with herbal growth (Renner and Ricklefs, 1995; Vamosi et al., 2003; Renner, 2014). The chromosomal localization of the sex-determining master gene is important. If this gene (controlling expression of the genes from previously established sex-determining pathways) is recruited in the pericentromeric or other regions demonstrating reduced recombination, the sex chromosome-based sex determination is established in one step (see Fig. 3B). Interestingly, there are two cases of species possessing SDRs in the proximity of centromeres – in Coccinia grandis (Sousa et al., 2016) and in Carica papaya (Yu et al., 2007). An increasing body of evidence supports the theory that non-recombining regions have a tendency to spread. This so-called ‘evolutionary strata’ has been observed in several animal model species and also in three plant models – Silene latifolia (Nicolas et al., 2005; Bergero et al., 2007; Papadopulos et al., 2015), Carica papaya (Wang et al., 2012) and Populus trichocarpa (Pandey and Azad, 2016). It is very difficult to establish if in any particular studied species the sex determination has arisen via a one- or two-locus-based model. A one-locus-based scenario can be erroneously identified as two-locus based or, as switches can occur in sex-determining genes, the two-locus-based sex-determining systems can evolve in systems possessing monogenic sex determination (see Janousek and Mrackova, 2010, for a possible scenario).
Diospyros lotus shows indications of a possible one-locus-based scenario of the evolution of dioecy (Akagi et al., 2014). In addition, a system illustrating a possible way to single ‘master gene-based sex determination has been suggested by Boualem et al. (2015) in C. melo. In this case, the possible scenario would start with a mostly male population with the prevailing genotype homozygous in the CmASC11 null allele. In the next step, a recessive allele would be recruited which has lost the ability to suppress female organ development and, concurrently, prevented male organ development (as it is not able to repress the stamen suppressor, CmACS7 gene).
The only way to affirm the relevance of two- or one-locus-based models in particular taxa is to identify the sex-determining pathways and to perform detailed analysis on the participating genes. Only a limited number of species can be studied to a sufficient extent. The group of chosen species should include both species having hermaphrodite relatives and species having monoecious relatives.
FURTHER EVOLUTION OF SEX CHROMOSOMES AND WHOLE GENOMES OF DIOECIOUS FLOWERING PLANTS – IMPACT OF DIOECY ON GENOME STRUCTURE AND DYNAMICS
The existence of separate sexes and sex chromosomes affects the development of plants. Male and female plants differ in the presence of respective sexual organs as well as in quantitative traits such as the number and shape of flowers and in qualitative traits such as sex-specific expression of some genes even in vegetative tissues (Zluvova et al., 2010). The unique processes shaping sex chromosomes can also, in parallel, influence the rest of the genome, e.g. its size and the distribution of repeats. Dioecious Asparagus species tend to have larger genomes than their hermaphroditic relatives (Kuhl et al., 2005), which is most probably caused by retrotransposon proliferation (Harkess et al., 2016). Similarly, the genome of dioecious S. latifolia is larger than the genome of hermaphroditic S. vulgaris mainly due to the expansion of the Ty3-gypsy long terminal repeat (LTR) retrotransposon Ogre (Cegan et al., 2012). A likely cause of transposable element (TE) proliferation could be the fast diversification of TE families and the subsequent relaxation of epigenetic regulation accompanying the evolution of dioecy. How these processes interact is discussed below.
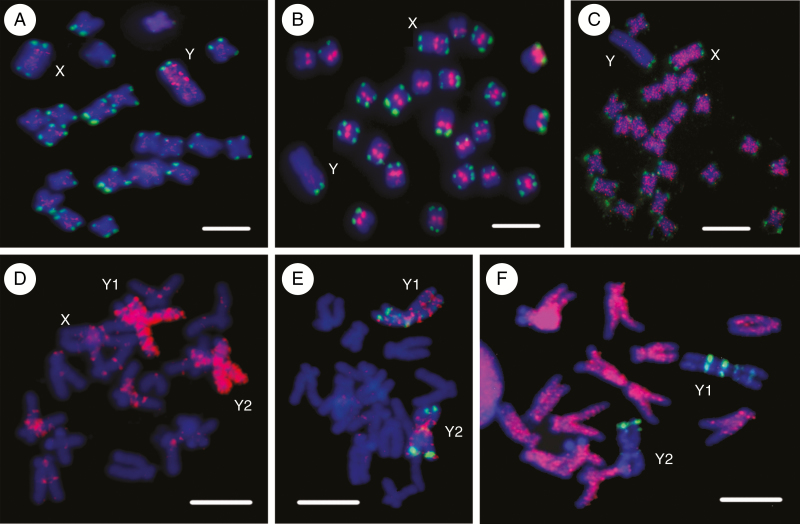
Besides gene degeneration, the main structural characteristic of plant sex chromosomes is the different distribution of repetitive sequences (satellites and TEs) compared with the rest of the genome (Shibata et al., 2000; Navajas-Pérez et al., 2006, 2009; Kubat et al., 2008; Cermak et al., 2008; Kejnovsky et al., 2009, 2013; Mariotti et al., 2009; Steflova et al., 2013; Kralova et al., 2014; Puterova et al., 2017). As an example, differences in the chromosomal localization of repeats on the sex chromosomes of S. latifolia and R. acetosa, both species possessing heteromorphic Y chromosomes, are depicted in Fig. 4A–F. The accumulation of microsatellites on the Y chromosomes in particular (Fig. 4A, D, E) could be a consequence of recombination restriction and may represent an early event shaping the Y chromosome. Microsatellite arrays could then serve as DNA substrates for TE insertions (Kejnovsky et al., 2013), thus accelerating genetic degeneration and size expansion of the Y chromosome (reviewed in Hobza et al., 2015). Moreover, the suppressed recombination between Y chromosomes reduces the rate of concerted evolution and gives rise to the diversification of satellites (Navajas-Pérez et al., 2006) and potentially of TE families. Thus, non-recombining sex chromosomes could serve as a source of new forms of repetitive sequences that could spread into the rest of the genome. The immature epigenetic defence against newly emerged TE families is the most likely cause of the extensive proliferation of TEs and genome expansion in dioecious plants. Simultaneously, an increased outcrossing of dioecious plants can allow TEs to proliferate and spread into the population.
Fig. 4.
Chromosomal localization of repetitive DNA in Silene latifolia (A–C) and Rumex acetosa (D–F) determined by fluorescence in situ hybridization. (A) Microsatellite (CA)15 is accumulated on the Y chromosome (red signal), (B) satellite STAR (red signal) is present in the centromeres of all autosomes and the X chromosome, and satellite X-43.1 (green signal) is gathered in both sub-telomeres of all chromosomes with the exception of the Y chromosome possessing only one sub-telomeric signal, (C) The Ogre LTR retrotransposon (red signal) is ubiquitous on all chromosomes except the Y chromosome. (D) A mixture of all mono-, di- and trinucleotide microsatellites (red signal) shows strong accumulation along both Y chromosomes. (E) The (TA)15 microsatellite (red signal) gives a signal at several discrete loci on both Y chromosomes, and the RAYSI satellite (green signal) is present at the distal regions of both Y chromosomes. (F) The Maximus/SIRE LTR retrotransposon (red signal) covers all autosomes and the X chromosome except telomeres/sub-telomeres, but is absent from both Y chromosomes; the RAYSI satellite is localized at several discrete loci on both Y chromosomes (green signal). Scale bars indicate 10 μm.
A widely accepted hypothesis suggests that repeats are accumulated in the non-recombining region of the Y chromosome (Charlesworth, 1991). Although this is the case for some TEs, many TEs are under-represented on the Y chromosomes of both S. latifolia and R. acetosa (Fig. 4C, F). Ty3/gypsy LTR retrotransposons of Retand, Ogre and Athila families have fewer copies on the Y chromosome than on the X chromosome and on the autosomes of S. latifolia (Kejnovsky et al., 2006; Cermak et al., 2008; Filatov et al., 2009; Kralova et al., 2014), whereas only some Ty1-copia retrotransposons are more abundant on the Y chromosome than on autosomes (Cermak et al., 2008). In addition, the absence of TEs on the Y chromosome is often accompanied by a build-up of TEs on the X chromosomes, while TEs accumulated on the Y chromosome usually have fewer copies on the X chromosomes compared with autosomes (Hobza et al., 2017; Puterova et al., 2018). Based on this observation, three TE groups were classified (Puterova et al., 2018). The first group of TEs tend to be preferentially active in females, the second group of TEs in males and TEs from the third group are equally active in both sexes. Hobza et al. (2017) proposed a testable model of preferential TE proliferation either in males or in females and its impact on the chromosomal distribution of TEs. Irregularity in TE distribution on sex chromosomes is indicative of the existence of hidden mechanisms influencing the transposition of TEs sex specifically, i.e. differently in male and female individuals or germlines.
The most likely candidates are epigenetic TE-repressive mechanisms employing specific classes of sRNA molecules that mediate the silencing of all homologous TE copies in the genome, post-transcriptionally or through the deposition of repressive chromatin modifications. Epigenetic mechanisms are known to inhibit TE activity in vegetative tissues, but they also play a critical role in gametogenesis where they can influence sex-specific transgenerational proliferation of TEs as exemplified by the EVADE (EVD) retrotransposon in A. thaliana. EVD retrotransposons amplify if transmitted through the paternal germline but are suppressed when maternally inherited due to epigenetic silencing in the maternal sporophytic tissues of the flower (Reinders et al., 2013). Since A. thaliana is a hermaphroditic plant, the effect of the silencing on the chromosomal distribution of EVD is not directly apparent. This is in contrast to a number of case studies of the dioecious plant S. latifolia (Cermak et al., 2008; Filatov et al., 2009; Kubat et al., 2014; Puterova et al., 2018).
The precise mode of TE epigenetic silencing, and how the silencing differs for male and female lineages, is still unknown. It is assumed that the epigenetic reprogramming of plant gametes plays a central role. In plants, a germline is not set aside early in embryogenesis, as in animals, and differentiates later from flower tissues. Thus, the epigenetic marks contributing both to plant development and to TE silencing must be deleted in the germline to re-establish the totipotent state in the zygote. It is presumed that epigenetic marks are gradually lost during germline differentiation due to the downregulation of DNA methyltransferases and heterochromatin remodellers as well as to the active effect of Demeter (DME) (Gehring et al., 2009; Hsieh et al., 2009; Calarco et al., 2012). Epigenetic information is then re-established in sperm production and during embryo development. Companion cells of plant gametes, the vegetative cell in pollen and the central cell (CCN) in ovuli, retain low levels of heterochromatic marks causing the active transcription of TEs. Current models propose that RNA transcripts from active TEs are converted to mobile small interfering RNAs (siRNAs) that can migrate into the sperm and embryo, where they reinforce both transcriptional and post-transcriptional silencing of TEs (Slotkin et al., 2009; Ibarra et al., 2012, Martínez et al., 2016). According to the latest studies in arabidopsis and maize, it is primarily TEs which are near genes which are subject to epigenetic reprogramming driven by 24 nucleotide siRNAs, while TEs in TE islands probably remain heterochromatinized even during germline differentiation (Gent et al., 2013, 2014; West et al., 2014; Li et al., 2015; Sigman and Slotkin, 2016; Vergara and Gutierrez, 2017).
An interesting discovery here is that the male germline adopted unique silencing mechanisms which are not present in the female germline. First, miRNA genes have been shown to trigger the production of easiRNAs (epigenetically activated siRNA molecules) that specifically target TE transcripts in the vegetative nucleus of pollen grains (Creasey et al., 2014). Secondly, pollen-specific tRNA-derived siRNA molecules target reactivated TE transcripts. The latter process has been shown to be conserved in plants and may have a role in gamete protection against gametophyte-associated TE reactivation (Martínez et al., 2017). Considering that most of the sex specifically distributed TEs tend to be less abundant on the Y chromosomes of R. acetosa and S. latifolia (Steflova et al., 2013; Puterova et al., 2018), we can hypothesize that the male germline has a stronger epigenetic defence against TEs than the female germline. This could have evolved due to the existence of a vegetative haploid stage – the pollen tube. TEs (especially the most active ones) have to be under tight control in the male germline to decrease the risk of deleterious changes of the genes required for pollen tube growth. In dioecious species with male-specific Y chromosomes, these mechanisms may slow down irreversible genetic and epigenetic degeneration of Y-linked genes and subsequently contribute to a decrease in the rate of gene loss. Loss of Y-linked genes is slower in S. latifolia than in primates, probably due to a combination of several factors such as the minor effect of haploid selection and relatively high effective population size of the former (reviewed in Muyle et al., 2017). Supporting the importance of the male-specific TE silencing mechanisms is the fact that TEs under-represented on the Y chromosome substantially contributed to the recent genome size expansion in S. latifolia (Cegan et al., 2012; Puterova et al., 2018).
All in all, parent-of-origin effects, although likely to be widespread in plants, are hidden in traditional hermaphroditic model species but have a significant manifestation in species carrying sex chromosomes. These mechanisms affect the transposition of specific TE families in a sex-specific manner, which influences not only the male haploid stage (pollen tube) but also some aspects of sex chromosome evolution. Male-specific silencing of TEs in particular can decelerate Y chromosome size expansion as well as the genetic and epigenetic degeneration of Y-linked genes and simultaneously increase the size of the X chromosome (X chromosomes are much larger than any autosome in S. latifolia and R. acetosa). In contrast, female-specific silencing of TEs can contribute to Y chromosome size increase relative to autosomes. Moreover, dioecious plants, when used for studies of sex-specific and sexual organ-specific epigenetic processes in TE–host coevolution, can in some ways be more informative than traditional models due to the existence of separate sexes as well as the presence of unique genomic parts, such as non-recombining Y (W) chromosome(s). Such studies may, for example, include the precise quantification of TE families on particular chromosomes as well as sex-specific transgenerational proliferation of selected TE families with known chromosomal distribution.
PERSPECTIVES
Based on the reviewed data, it can be deduced that there are not only differences among studied species concerning sex-determining systems but also common evolutionary trends, e.g. a varying tendency towards the growth and degeneration of non-recombining regions (this feature is shared even with animal models, as reviewed by Schartl et al., 2016 and Wright et al., 2016). It also appears that the evolution of the controlling pathways, which occurs after various sex-determining systems are established, is to some extent convergent. Even though dioecy has originated independently many times, there are similarities in the role of epigenetic factors, e.g. DNA methylation and histone methylation, in their influence on sexual phenotype. Additionally, the role of the ethylene signalling pathway in sex determination is shared by several unrelated species (Den Nijs and Visser, 1980; Mohan Ram and Sett, 1980, 1982; Boualem et al., 2008, 2009; Devani et al., 2017). As hypothesized in this review, the controlling pathways involved in sex determination in plants evolved de novo but could also have employed controlling mechanisms that were developed for other purposes. As the number of such pathways is limited, the similarities between unrelated taxa are not so surprising. In comparison with mammals, due to the lack of locomotion, plant genomes evolved mechanisms to achieve more genotypic and phenotypic plasticity to adapt passively to stress (reviewed by Kejnovsky et al., 2009) and some of these mechanisms could also influence the phenotypic plasticity of the sexual phenotype in plants. Due to the huge differences in the depth of knowledge in particular plant models, global comparison of many aspects of sex chromosome evolution is limited. There are an increasing number of studies regarding sex-determining mechanisms, the impact of dioecy on genome structure, epigenetic aspects of sex chromosome evolution as well as the interplay between regulation of TEs and non-recombining region formation, but detailed data on the phenotypic or at the population level are still scarce, especially in new models. Continuing study of many varying model species and a complex view is needed to understand fully the complex aspects of sex chromosome evolution.
ACKNOWLEDGEMENTS
This research was supported by the Czech Science Foundation (grants 18-06147S, 13-06264S, 16-08698S, 15-21523Y, 18-00258S and 17-00567S). We would like to thank Dr Alex Oulton, Francesco Muto and Dr Chris Johnson for English corrections.
LITERATURE CITED
- Akagi T, Henry IM, Tao R, Comai L. 2014. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346: 646–650. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Alexander HM. 1992. Epidemiology of anther-smut infection of Silene alba (= S. latifolia) caused by Ustilago violacea: patterns of spore deposition in experimental populations. Proceedings of the Royal Society B: Biological Sciences 250: 157–163. [Google Scholar]
- Aryal R, Jagadeeswaran G, Zheng Y, Yu Q, Sunkar R, Ming R. 2014. Sex specific expression and distribution of small RNAs in papaya. BMC Genomics 15: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashman T-L. 1999. Determinants of sex allocation in a gynodioecious wild strawberry: implications for the evolution of dioecy and sexual dimorphism. Journal of Evolutionary Biology 12: 648–661. [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, et al. 2014. Sex determination: why so many ways of doing it?PLoS Biology 12: e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balounova V, Gogela R, Cegan R, et al. 2018. Evolution of sex determination and heterogamety changes in section Otites of the genus Silene. bioRxivdoi:https://doi.org/10.1101/325068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacar N, Hinnisdaels S, Farbos I, et al. 1997. Isolation of early genes expressed in reproductive organs of the dioecious white campion (Silene latifolia) by subtraction cloning using an asexual mutant. The Plant Journal 12: 805–817. [DOI] [PubMed] [Google Scholar]
- Barr CM. 2004. Soil moisture and sex ratio in a plant with nuclear–cytoplasmic sex inheritance. Proceedings of the Royal Society B: Biological Sciences 271: 1935–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkowiak E. 1971. Mechanism of sex determination in Rumex hastatulus Baldw. Theoretical and Applied Genetics 41: 320–326. [DOI] [PubMed] [Google Scholar]
- Beaudry FEG, Barrett SCH, Wright SI. 2017. Genomic loss and silencing on the Y chromosomes of Rumex. Genome Biology and Evolution 9: 3345–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Forrest A, Kamau E, Charlesworth D. 2007. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175: 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Qiu S, Charlesworth D. 2015. Gene loss from a plant sex chromosome system. Current Biology 25: 1234–1240. [DOI] [PubMed] [Google Scholar]
- Beukeboom N, Perrin N. 2014. The evolution of sex determination. Oxford: Oxford University Press. [Google Scholar]
- Blair AC, Wolfe LM. 2004. The evolution of an invasive plant: an experimental study with Silene latifolia. Ecology 85: 3035–3042 [Google Scholar]
- Boucher LD, Manchester SR, Judd WS. 2003. An extinct genus of Salicaceae based on twigs with attached flowers, fruits, and foliage from the Eocene Green River Formation of Utah and Colorado, USA. American Journal of Botany 90: 1389–99. [DOI] [PubMed] [Google Scholar]
- Boualem A, Fergany M, Fernandez R, et al. 2008. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321: 836–838. [DOI] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Kovalski I, Sari M-A, Perl-Treves R, Bendahmane A. 2009. A conserved ethylene biosynthesis enzyme leads to andromonoecy in two cucumis species. PLoS One 4: e6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Camps C, et al. 2015. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 350: 688–691 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1991. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20. [DOI] [PubMed] [Google Scholar]
- Bräutigam K, Soolanayakanahally R, Champigny M, et al. 2017. Sexual epigenetics: gender-specific methylation of a gene in the sex determining region of Populus balsamifera. Scientific Reports 7: 45388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MTA, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CH, Choi Y, Chan AP, Serapiglia MJ, Town CD, Smart LB. 2017. Dominance and sexual dimorphism pervade the Salix purpurea L. transcriptome. Genome Biology and Evolution 9: 2377–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegan R, Marais GA, Kubekova H, et al. 2010. Structure and evolution of Apetala3, a sex-linked gene in Silene latifolia. BMC Plant Biology 10: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegan R, Vyskot B, Kejnovsky E, et al. 2012. Genomic diversity in two related plant species with and without sex chromosomes – Silene latifolia and S. vulgaris. PLoS One 7: e31898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Kubat Z, Hobza R, et al. 2008. Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Research 16: 961–976. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 1991. The evolution of sex chromosomes. Science 251: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 1978. A model for the evolution of dioecy and gynodioecy. American Naturalist 112: 975. [Google Scholar]
- Charlesworth D. 2013. Plant sex chromosome evolution. Journal of Experimental Botany 64: 405–420. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2015. Plant contributions to our understanding of sex chromosome evolution. New Phytologist 208: 52–65. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng Y, Qin L, et al. 2016. a Identification of miRNAs and their targets through high-throughput sequencing and degradome analysis in male and female Asparagus officinalis. BMC Plant Biology 16: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang T, Fang L, Li X, Yin T. 2016b Confirmation of single-locus sex determination and female heterogamety in willow based on linkage analysis. PLoS One 11: e0147671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correns C. 1928. Bestimmung, Vererbung and Verteilung des geschlechtes bei den hoheren Pflanzen. Handbuch der Vererbungswissenschaft 2: 1–138. [Google Scholar]
- Creasey KM, Zhai J, Borges F, et al. 2014. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowson D, Barrett SCH, Wright SI. 2017. Purifying and positive selection influence patterns of gene loss and gene expression in the evolution of a plant sex chromosome system. Molecular Biology and Evolution 34: 1140–1154. [DOI] [PubMed] [Google Scholar]
- Cuñado N, Navajas-Pérez R, Herrán R, et al. 2007. The evolution of sex chromosomes in the genus Rumex (Polygonaceae): identification of a new species with heteromorphic sex chromosomes. Chromosome Research 15: 825–833. [DOI] [PubMed] [Google Scholar]
- Darolti I, Wright AE, Pucholt P, Berlin S, Mank JE. 2018. Slow evolution of sex-biased genes in the reproductive tissue of the dioecious plant Salix viminalis. Molecular Ecology 27: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delph LF, Demuth JP. 2016. Haldane’s rule: genetic bases and their empirical support. Journal of Heredity 107: 383–391. [DOI] [PubMed] [Google Scholar]
- Delph LF, Wolf DE. 2005. Evolutionary consequences of gender plasticity in genetically dimorphic breeding systems. New Phytologist 166: 119–128. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Flanagan RJ, Delph LF. 2014. Genetic architecture of isolation between two species of Silene with sex chromosomes and Haldane’s rule. Evolution 68: 332–342. [DOI] [PubMed] [Google Scholar]
- Den Nijs APM, Visser DL. 1980. Induction of male flowering in gynoecious cucumbers (Cucumis sativus L.) by silver ions. Euphytica 29: 273–280. [Google Scholar]
- Devani RS, Sinha S, Banerjee J, Sinha RK, Bendahmane A, Banerjee AK. 2017. De novo transcriptome assembly from flower buds of dioecious, gynomonoecious and chemically masculinized female Coccinia grandis reveals genes associated with sex expression and modification. BMC Plant Biology 17: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyhle F, Sarkar AK, Tucker EJ, Laux T. 2007. WUSCHEL regulates cell differentiation during anther development. Developmental Biology 302: 154–159. [DOI] [PubMed] [Google Scholar]
- Donnison IS, Siroky J, Vyskot B, Saedler H, Grant SR. 1996. Isolation of Y chromosome-specific sequences from Silene latifolia and mapping of male sex-determining genes using representational difference analysis. Genetics 144: 1893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorken ME, Barrett SCH. 2004. Sex determination and the evolution of dioecy from monoecy in Sagittaria latifolia (Alismataceae). Proceedings of the Royal Society B: Biological Sciences 271: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Durand R. 1991. Sex determination and reproductive organ differentiation in Mercurialis. Plant Science 80: 49–66. [Google Scholar]
- Esfandiyari B, Davarynejad GH, Shahriari F, Kiani M, Mathe A. 2012. Data to the sex determination in Pistacia species using molecular markers. Euphytica 185: 227–231. [Google Scholar]
- Farbos I, Veuskens J, Vyskot B, et al. 1999. Sexual dimorphism in white campion: deletion on the Y chromosome results in a floral asexual phenotype. Genetics 151: 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA, Howell EC, Groutides C, Armstrong SJ. 2009. Recent spread of a retrotransposon in the Silene latifolia genome, apart from the Y chromosome. Genetics 181: 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis EM, Pedersen KR, Crane PR. 2010. Diversity in obscurity: fossil flowers and the early history of angiosperms. Philosophical Transactions of the Royal Society B: Biological Sciences 365: 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. 2009. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, et al. 2013. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Research 23: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Madzima TF, Bader R, et al. 2014. Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. The Plant Cell 26: 4903–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Hefer CA, Capron A, et al. 2015. Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Molecular Ecology 24: 3243–3256. [DOI] [PubMed] [Google Scholar]
- Goldberg MT, Spigler RB, Ashman TL. 2010. Comparative genetic mapping points to different sex chromosomes in sibling species of wild strawberry (Fragaria). Genetics 186: 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska-Joachimiak A, Kula A, Książczyk T, Chojnicka J, Sliwinska E, Joachimiak AJ. 2015. Chromosome landmarks and autosome–sex chromosome translocations in Rumex hastatulus, a plant with XX/XY1Y2 sex chromosome system. Chromosome Research 23: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. 1922. Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics 12: 101–109. [Google Scholar]
- Han J, Murray JE, Yu Q, Moore PH, Ming R. 2014. The effects of gibberellic acid on sex expression and secondary sexual characteristics in papaya. HortScience 49: 378–383. [Google Scholar]
- Harkess A, Mercati F, Shan HY, Sunseri F, Falavigna A, Leebens-Mack J. 2015. Sex-biased gene expression in dioecious garden asparagus (Asparagus officinalis). New Phytologist 207: 883–892. [DOI] [PubMed] [Google Scholar]
- Harkess A, Mercati F, Abbate L, et al. 2016. Retrotransposon proliferation coincident with the evolution of dioecy in Asparagus. G3 (Bethesda) 6: 2679–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Zhou J, Xu C, et al. 2017. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nature Communications 8: 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobza R, Kubat Z, Cegan R, Jesionek W, Vyskot B, Kejnovsky E. 2015. Impact of repetitive DNA on sex chromosome evolution in plants. Chromosome Research 23: 561–570. [DOI] [PubMed] [Google Scholar]
- Hobza R, Cegan R, Jesionek W, Kejnovsky E, Vyskot B, Kubat Z. 2017. Impact of repetitive elements on the Y chromosome formation in plants. Genes 8: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeyr JDJ. 1938. Genetical studies of Carica papaya L. South African Journal of Science 35: 300–304. [Google Scholar]
- Hofmeyr JDJ. 1939. Sex-linked inheritance in Carica papaya L. South African Journal of Science 36: 283–285. [Google Scholar]
- Hormaza JI, Dollo L, Polito VS. 1994. Identification of a RAPD marker linked to sex determination in Pistacia vera using bulked segregant analysis. Theoretical and Applied Genetics 89: 9–13. [DOI] [PubMed] [Google Scholar]
- Hou J, Ye N, Zhang D, et al. 2015. Different autosomes evolved into sex chromosomes in the sister genera of Salix and Populus. Scientific Reports 5: 9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough J, Hollister JD, Wang W, Barrett SCH, Wright SI. 2014. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proceedings of the National Academy of Sciences, USA 111: 7713–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough J, Wang W, Barrett SCH, Wright SI. 2017. Hill–Robertson interference reduces genetic diversity on a young plant Y-chromosome. Genetics 207: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, et al. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X-S, Filatov DA. 2016. The large-X effect in plants: increased species divergence and reduced gene flow on the Silene X-chromosome. Molecular Ecology 25: 2609–2619. [DOI] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, et al. 2009. The genome of the cucumber, Cucumis sativus L. Nature Genetics 41: 1275–1281. [DOI] [PubMed] [Google Scholar]
- Hudzieczek V, Cegan R, Cermak T, et al. 2018. Agrobacterium rhizogenes-mediated transformation of a dioecious plant model Silene latifolia. New Biotechnology. doi:10.1016/j.nbt.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Hug N, Longman D, Cáceres JF. 2016. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Research 44: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husse L, Billiard S, Lepart J, Vernet P, Saumitou-Laprade P. 2013. A one-locus model of androdioecy with two homomorphic self-incompatibility groups: expected vs. observed male frequencies. Journal of Evolutionary Biology 26: 1269–1280. [DOI] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, et al. 2012. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Meyerowitz EM. 1997. Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277: 1100–1103. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Sakai H, Finnegan EJ, Cao X, Meyerowitz EM. 2000. Ectopic hypermethylation of flower-specific genes in Arabidopsis. Current Biology 10: 179–186. [DOI] [PubMed] [Google Scholar]
- Janousek B, Siroky J, Vyskot B. 1996. Epigenetic control of sexual phenotype in a dioecious plant, Melandrium album. Molecular Genetics and Genomics 250: 483–490. [DOI] [PubMed] [Google Scholar]
- Janousek B, Grant SR, Vyskot B. 1998. Non-transmissibility of the Y chromosome through the female line in androhermaphrodite plants of Melandrium album. Heredity 80: 576–583. [Google Scholar]
- Janousek B, Mrackova M. 2010. Sex chromosomes and sex determination pathway dynamics in plant and animal models. Biological Journal of the Linnean Society 100: 737–752. [Google Scholar]
- Kafkas S, Khodaeiaminjan M, Guney M, Kafkas E. 2015. Identification of sex-linked SNP markers using RAD sequencing suggests ZW/ZZ sex determination in Pistacia vera L. BMC Genomics 16: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashimada K, Koopman P. 2010. Sry: the master switch in mammalian sex determination. Development 137: 3921–3930. [DOI] [PubMed] [Google Scholar]
- Kazama Y, Fujiwara MT, Koizumi A, et al. 2009. A SUPERMAN-like gene is exclusively expressed in female flowers of the dioecious plant Silene latifolia. Plant and Cell Physiology 50: 1127–1141. [DOI] [PubMed] [Google Scholar]
- Kazama Y, Nishihara K, Bergero R, et al. 2012. SlWUS1; an X-linked gene having no homologous Y-linked copy in Silene latifolia. G3 (Bethesda) 2: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama Y, Ishii K, Aonuma W, et al. 2016. A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Scientific Reports 6: 18917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E, Kubat Z, Macas J, Hobza R, Mracek J, Vyskot B. 2006. Retand: a novel family of gypsy-like retrotransposons harboring an amplified tandem repeat. Molecular Genetics and Genomics 276: 254–263. [DOI] [PubMed] [Google Scholar]
- Kejnovsky E, Hobza R, Cermak T, Kubat Z, Vyskot B. 2009. The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity 102: 533–541. [DOI] [PubMed] [Google Scholar]
- Kejnovsky E, Michalovova M, Steflova P, et al. 2013. Expansion of microsatellites on evolutionary young Y chromosome. PLoS One 8: e45519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenigsbuch D, Cohen Y. 1990. The inheritance of gynoecy in muskmelon. Genome 33: 317–320. [Google Scholar]
- Kersten B, Pakull B, Groppe K, Lueneburg J, Fladung M. 2014. The sex-linked region in Populus tremuloides Turesson 141 corresponds to a pericentromeric region of about two million base pairs on P. trichocarpa chromosome 19. Plant Biology 16: 411–418. [DOI] [PubMed] [Google Scholar]
- Kersten B, Pakull B, Fladung M. 2017. Genomics of sex determination in dioecious trees and woody plants. Trees 31: 1113–1125. [Google Scholar]
- Koizumi A, Yamanaka K, Nishihara K, Kazama Y, Abe T, Kawano S. 2010. Two separate pathways including SlCLV1, SlSTM and SlCUC that control carpel development in a bisexual mutant of Silene latifolia. Plant and Cell Physiology 51: 282–93. [DOI] [PubMed] [Google Scholar]
- Kralova T, Cegan R, Kubat Z, et al. 2014. Identification of a novel retrotransposon with sex chromosome specific distribution in Silene latifolia. Cytogenetic and Genome Research 143: 87–95. [DOI] [PubMed] [Google Scholar]
- Kubat Z, Hobza R, Vyskot B, Kejnovsky E. 2008. Microsatellite accumulation on the plant Y chromosome in Silene latifolia. Genome 51: 350–356. [DOI] [PubMed] [Google Scholar]
- Kubat Z, Zluvova J, Vogel I, et al. 2014. Possible mechanisms responsible for absence of a retrotransposon family on a plant Y chromosome. New Phytologist 202: 662–678. [DOI] [PubMed] [Google Scholar]
- Kuhl JC, Havey MJ, Martin WJ, et al. 2005. Comparative genomic analyses in Asparagus. Genome 1060: 1052–1060. [DOI] [PubMed] [Google Scholar]
- Kumar A, Jaiswal VS. 1984. Sex reversal and fruit formation on male plants of Carica papaya L. by ethrel and chlorflurenol. Proceedings of the Indian Academy Sciences (Plant Sciences) 93: 635–641 [Google Scholar]
- Lardon A, Georgiev S, Aghmir A, Le Merrer G, Negrutiu I. 1999. Sexual dimorphism in white campion: complex control of carpel number is revealed by Y chromosome deletions. Genetics 151: 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Rodriguez-Granados NY, Veluchamy A, et al. 2017. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics and Chromatin 10: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law TF, Lebel-Hardenack S, Grant SR. 2002. Silver enhances stamen development in female white campion (Silene latifolia [Caryophyllaceae]). American Journal of Botany 89: 1014–1020. [DOI] [PubMed] [Google Scholar]
- Lebel-Hardenack S, Hauser E, Law TF, Schmid J, Grant SR. 2002. Mapping of sex determination loci on the white campion (Silene latifolia) Y chromosome using amplified fragment length polymorphism. Genetics 160: 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gent JI, Zynda G, et al. 2015. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proceedings of the National Academy of Sciences, USA 112: 14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Liao Z, Zhang L, Yu Q. 2016. Transcriptome analysis of the male-to-hermaphrodite sex reversal induced by low temperature in papaya. Tree Genetics and Genomes 12: 1–14. [Google Scholar]
- Liu Z, Moore PH, Ma H, et al. 2004. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427: 348–352. [DOI] [PubMed] [Google Scholar]
- Lloyd D. 1972a Breeding systems in Cotula L. (Compositae, Anthemideae). I. The array of monoclinous and diclinous systems. New Phytologist 71: 1181–1194. [Google Scholar]
- Lloyd D. 1972b Breeding systems in Cotula L. (Compositae, Anthemideae). II. Monoecious populations. New Phytologist 71: 1195–1202. [Google Scholar]
- Lloyd DG. 1975. Breeding systems in Cotula. III. dioecious populations. New Phytologist 74: 109–123. [Google Scholar]
- Lloyd D. 1980. The distribution of gender in four angiosperm species illustrating two evolutionary pathways to dioecy. Evolution 34: 123–134. [DOI] [PubMed] [Google Scholar]
- Lloyd D. 1981. The distribution of sex in Myrica gale. Plant Systematics and Evolution 138: 29–45. [Google Scholar]
- Lorenzi MC, Sella G. 2013. In between breeding systems: neither dioecy nor androdioecy explains sexual polymorphism in functionally dioecious worms. Integrative and Comparative Biology 53: 689–700. [DOI] [PubMed] [Google Scholar]
- Louis JP. 1989. Genes for the regulation of sex differentiation and male fertility in Mercurialis annua L. Journal of Heredity 89: 104–111. [Google Scholar]
- Manchester SR, Judd WS, Handley B. 2006. Foliage and fruits of early poplars (Salicaceae: Populus) from the Eocene of Utah, Colorado, and Wyoming. International Journal of Plant Sciences 167: 897–908. [Google Scholar]
- Manzano S, Megías Z, Martínez C, et al. 2017. Overexpression of a flower specific aerolysin-like protein from the dioecious plant Rumex acetosa alters flower development and induces male sterility in transgenic tobacco. The Plant Journal 89: 58–72. [DOI] [PubMed] [Google Scholar]
- Marais GAB, Nicolas M, Bergero R, et al. 2008. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Current Biology 18: 545–549. [DOI] [PubMed] [Google Scholar]
- Mariotti B, Manzano S, Kejnovský E, Vyskot B, Jamilena M. 2009. Accumulation of Y-specific satellite DNAs during the evolution of Rumex acetosa sex chromosomes. Molecular Genetics and Genomics 281: 249–59. [DOI] [PubMed] [Google Scholar]
- Martin A, Troadec C, Boualem A, et al. 2009. A transposon-induced epigenetic change leads to sex determination in melon. Nature 461: 1135–1138. [DOI] [PubMed] [Google Scholar]
- Martínez G, Panda K, Köhler C, Slotkin RK. 2016. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nature Plants 2: 16030. [DOI] [PubMed] [Google Scholar]
- Martínez G, Choudury SG, Slotkin RK. 2017. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Research 45: 5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga S, Isono E, Kejnovsky E, et al. 2003. Duplicative transfer of a MADS box gene to a plant Y chromosome. Molecular Biology and Evolution 20: 1062–1069. [DOI] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, et al. 2015. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697. [DOI] [PubMed] [Google Scholar]
- Miller PM, Kesseli RV. 2011. A sex-chromosome mutation in Silene latifolia. Sexual Plant Reproduction 24: 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Yu Q, Moore PH. 2007. Sex determination in papaya. Seminars in Cell and Developmental Biology 18: 401–408. [DOI] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, et al. 2008. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Bendahmane A, Renner SS. 2011. Sex chromosomes in land plants. Annual Review of Plant Biology 62: 485–514. [DOI] [PubMed] [Google Scholar]
- Mirski P. 2014. Exceptions from dioecy and sex lability in genus Salix. Dendrobiology 71: 167–171. [Google Scholar]
- Mohan Ram HY, Sett R. 1980. Induction of male flowers in a pistillate line of Ricinus communis L. by silver and cobalt ions. Planta 149: 413–415. [DOI] [PubMed] [Google Scholar]
- Mohan Ram HY, Sett R. 1982. Induction of fertile male flowers in genetically female Cannabis sativa plants by silver nitrate and silver thiosulphate anionic complex. Theoretical and Applied Genetics 62: 369–375. [DOI] [PubMed] [Google Scholar]
- Mohanty JN, Nayak S, Jha S, Joshi RK. 2017. Transcriptome profiling of the floral buds and discovery of genes related to sex-differentiation in the dioecious cucurbit Coccinia grandis (L.) Voigt. Gene 626: 395–406. [DOI] [PubMed] [Google Scholar]
- Mrackova M, Nicolas M, Hobza R, et al. 2008. Independent origin of sex chromosomes in two species of the genus Silene. Genetics 179: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Shigenobu S, Fujii S, et al. 2017. MYB transcription factor gene involved in sex determination in Asparagus officinalis. Genes to Cells 22: 115–123. [DOI] [PubMed] [Google Scholar]
- Muyle A, Shearn R, Marais GA. 2017. The evolution of sex chromosomes and dosage compensation in plants. Genome Biology and Evolution 9: 627–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Zemp N, Deschamps C, Mousset S, Widmer A, Marais GAB. 2012. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biology 10: e1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamitsu T, Futamura N. 2014. Sex expression and inbreeding depression in progeny derived from an extraordinary hermaphrodite of Salix subfragilis. Botanical Studies 55: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navajas-Pérez R, Schwarzacher T, de la Herrán R, Ruiz Rejón C, Ruiz Rejón M, Garrido-Ramos MA. 2006. The origin and evolution of the variability in a Y-specific satellite-DNA of Rumex acetosa and its relatives. Gene 368: 61–71. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, Schwarzacher T, de la Herrán R, Ruiz Rejón C, Ruiz Rejón M, Garrido-Ramos MA. 2009. Molecular cytogenetic characterization of Rumex papillaris, a dioecious plant with an XX/XY1Y2 sex chromosome system. Genetica 135: 87–93. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Marais G, Hykelova V, et al. 2005. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biology 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T. 1935. Chromosomen und Sexualitat von Rumex Acetosa. Tohoku Imperial University, Science Reports, Series 4 10: 41–210. [Google Scholar]
- Pakull B, Kersten B, Lüneburg J, Fladung M. 2015. A simple PCR-based marker to determine sex in aspen. Plant Biology (Stuttgart, Germany) 17: 256–261. [DOI] [PubMed] [Google Scholar]
- Pandey RS, Azad RK. 2016. Deciphering evolutionary strata on plant sex chromosomes and fungal mating-type chromosomes through compositional segmentation. Plant Molecular Biology 90: 359–373. [DOI] [PubMed] [Google Scholar]
- Pannell JR. 2002. The evolution and maintenance of androdioecy. Annual Review of Ecology and Systematics 33: 397–425. [Google Scholar]
- Papadopulos AST, Chester M, Ridout K, Filatov DA. 2015. Rapid Y degeneration and dosage compensation in plant sex chromosomes. Proceedings of the National Academy of Sciences, USA 112: 13021–13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolucci I, Gaudet M, Jorge V, et al. 2010. Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genetics and Genomes 6: 863–875. [Google Scholar]
- Perrin N. 2009. Sex reversal: a fountain of youth for sex chromosomes?Evolution 63: 3043–3049. [DOI] [PubMed] [Google Scholar]
- Pucholt P, Rönnberg-Wästljung A-C, Berlin S. 2015. Single locus sex determination and female heterogamety in the basket willow (Salix viminalis L.). Heredity 114: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puterova J, Razumova O, Martinek T, et al. 2017. Satellite DNA and transposable elements in seabuckthorn (Hippophae rhamnoides), a dioecious plant with small Y and large X chromosomes. Genome Biology and Evolution 9: 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puterova J, Kubat Z, Kejnovsky E, et al. 2018. The slowdown of Y chromosome expansion in dioecious Silene latifolia due to DNA loss and male-specific silencing of retrotransposons. BMC Genomics 19: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Mirouze M, Nicolet J, Paszkowski J. 2013. Parent-of-origin control of transgenerational retrotransposon proliferation in Arabidopsis. EMBO Reports 14: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS. 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. American Journal of Botany 101: 1588–1596. [DOI] [PubMed] [Google Scholar]
- Renner SS. 2016. Pathways for making unisexual flowers and unisexual plants: moving beyond the ‘two mutations linked on one chromosome’ model. American Journal of Botany 103: 587–589. [DOI] [PubMed] [Google Scholar]
- Renner SS, Ricklefs RE. 1995. Dioecy and its correlates in the flowering plants. American Journal of Botany 82: 596–606. [Google Scholar]
- Renner SS, Won H. 2001. Repeated evolution of dioecy from monoecy in Siparunaceae (Laurales). Systematic Biology 50: 700–712. [DOI] [PubMed] [Google Scholar]
- Ridout KE, Veltsos P, Muyle A, et al. 2017. Hallmarks of early sex-chromosome evolution in the dioecious plant Mercurialis annua revealed by de novo genome assembly, genetic mapping and transcriptome analysis. bioRxiv doi: http://dx.doi.org/10.1101/106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera C, Saavedra F, Alvarez F, et al. 2015. Methylation of histone H3 lysine 9 occurs during translation. Nucleic Acids Research 43: 9097–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N, Studer T, Dufresnes C, Perrin N. 2017. Sex-chromosome recombination in common frogs brings water to the fountain-of-youth. Molecular Biology and Evolution 35: 942–948. [DOI] [PubMed] [Google Scholar]
- Ruddat M, Kokontis J, Birch L, et al. 1991. Interactions of Microbotryum violaceum (Ustilago violacea) with its host plant Silene alba. Plant Science 80: 157–165. [Google Scholar]
- Russell JRW, Pannell JR. 2015. Sex determination in dioecious Mercurialis annua and its close diploid and polyploid relatives. Heredity 114: 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumitou-Laprade P, Vernet P, Vassiliadis C, et al. 2010. A self-incompatibility system explains high male frequencies in an androdioecious plant. Science 327: 1648–1650. [DOI] [PubMed] [Google Scholar]
- Saze H, Kakutani T. 2007. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO Journal 26: 3641–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Schmid M, Nanda I. 2016. Dynamics of vertebrate sex chromosome evolution: from equal size to giants and dwarfs. Chromosoma 125: 553–571. [DOI] [PubMed] [Google Scholar]
- Shibata F, Hizume M, Kuroki Y. 2000. Differentiation and the polymorphic nature of the Y chromosomes revealed by repetitive sequences in the dioecious plant, Rumex acetosa. Chromosome Research 8: 229–236. [DOI] [PubMed] [Google Scholar]
- Sigman MJ, Slotkin RK. 2016. The first rule of plant transposable element silencing: location, location, location. The Plant Cell 28: 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, et al. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346: 240. [DOI] [PubMed] [Google Scholar]
- Singh RB, Smith BW. 1971. The mechanism of sex determination in Rumex acetosella. Theoretical and Applied Genetics 41: 360–364. [DOI] [PubMed] [Google Scholar]
- Slancarova V, Zdanska J, Janousek B, et al. 2013. Evolution of sex determination systems with heterogametic males and females in Silene. Evolution 67: 3669–3677. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, et al. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BW. 1963. The mechanism of sex determination in Rumex hastatulus. Genetics 48: 1265–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Tian M, Ci D, Zhang D. 2015. Methylation of microRNA genes regulates gene expression in bisexual flower development in andromonoecious poplar. Journal of Experimental Botany 66: 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A, Fuchs J, Renner SS. 2013. Molecular cytogenetics (FISH, GISH) of Coccinia grandis: a ca. 3 myr-old species of Cucurbitaceae with the largest Y/autosome divergence in flowering plants. Cytogenetic and Genome Research 139: 107–118. [DOI] [PubMed] [Google Scholar]
- Sousa A, Bellot S, Fuchs J, Houben A, Renner SS. 2016. Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. The Plant Journal 88: 387–396. [DOI] [PubMed] [Google Scholar]
- Spigler RB, Lewers KS, Main DS, Ashman T-L. 2008. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity 101: 507–517. [DOI] [PubMed] [Google Scholar]
- Spigler RB, Lewers KS, Johnson AL, Ashman T-L. 2010. Comparative mapping reveals autosomal origin of sex chromosome in octoploid Fragaria virginiana. Journal of Heredity 101(Suppl 1): S107–S117. [DOI] [PubMed] [Google Scholar]
- Steflova P, Tokan V, Vogel I, et al. 2013. Contrasting patterns of transposable element and satellite distribution on sex chromosomes (XY1Y2) in the dioecious plant Rumex acetosa. Genome Biology and Evolution 5: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey WB. 1938. Segregation of sex types in solo papaya and their application to the selection of seed. Journal of the American Society for Horticultural Science 35: 83–85. [Google Scholar]
- Taylor DR. 1994a Sex ratio in hybrids between Silene alba and Silene dioica: evidence for Y-linked restorers. Heredity 73: 518–526. [DOI] [PubMed] [Google Scholar]
- Taylor DR. 1994b The genetic basis of sex ratio in Silene alba (= S. latifolia). Genetics 136: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telgmann-Rauber A, Jamsari A, Kinney MS, Pires JC, Jung C. 2007. Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. Molecular Genetics and Genomics 278: 221–234. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Wei N, Straub S, Govindarajulu R, Liston A, Ashman TL. 2017. Repeated translocation of a gene cassette drives sex chromosome turnover in strawberries. bioRxiv doi: http://dx.doi.org/10.1101/163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TK, Saze H, Kakutani T. 2015. DNA methylation within transcribed regions. Plant Physiology 168: 1219–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree of Sex Consortium.. 2014. Tree of Sex: a database of sexual systems. Scientific Data 1: 140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. 1995. The dominance theory of Haldane’s rule. Genetics 140: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida W, Matsunaga S, Sugiyama R, Kazama Y, Kawano S. 2003. Morphological development of anthers induced by the dimorphic smut fungus Microbotryum violaceum in female flowers of the dioecious plant Silene latifolia. Planta 218: 240–248. [DOI] [PubMed] [Google Scholar]
- Vamosi JC, Otto SP, Barrett SCH. 2003. Phylogenetic analysis of the ecological correlates of dioecy in angiosperms. Journal of Evolutionary Biology 16: 1006–1018. [DOI] [PubMed] [Google Scholar]
- VanBuren R, Zeng F, Chen C, et al. 2015. Origin and domestication of papaya Y(h) chromosome. Genome Research 25: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R, Wai CM, Zhang J, et al. 2016. Extremely low nucleotide diversity in the X-linked region of papaya caused by a strong selective sweep. Genome Biology 17: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nigtevecht G. 1966. Genetic studies in dioecious Melandrium. II. Sex determination in Melandrium album and Melandrium dioicum. Genetica 37: 307–344. [Google Scholar]
- Vaughton G, Ramsey M. 2012. Gender plasticity and sexual system stability in Wurmbea. Annals of Botany 109: 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltsos P, Cossard G, Beaudoing E, et al. 2018. The size and content of the sex-determining region of the Y chromosome in dioecious Mercurialis annua, a plant with homomorphic sex chromosomes. Genes (Basel) 9: 277. doi: 10.3390/genes9060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara Z, Gutierrez C. 2017. Emerging roles of chromatin in the maintenance of genome organization and function in plants. Genome Biology 18: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Mao Y, Yang J, He Y. 2015. TCP24 modulates secondary cell wall thickening and anther endothecium development. Frontiers in Plant Science 6: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Matsushita M, Tomaru N, Nakagawa M. 2017. Sex change in the subdioecious shrub Eurya japonica (Pentaphylacaceae). Ecology and Evolution 7: 2340–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Na JK, Yu Q, et al. 2012. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proceedings of the National Academy of Sciences, USA 109: 13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Web BCJ. 1999. Empirical studies: evolution and maintenance of dimorphic breeding systems. In: Geber MA, Dawson TE, Delph LF, eds. Gender and sexual dimorphism in flowering plants. Berlin: Springer, 61–95. [Google Scholar]
- Weeks SC. 2012. The role of androdioecy and gynodioecy in mediating evolutionary transitions between dioecy and hermaphroditism in the animalia. Evolution 66: 3670–3686. [DOI] [PubMed] [Google Scholar]
- Wei N, Govindarajulu R, Tennessen JA, Liston A, Ashman T-L. 2017. Genetic mapping and phylogenetic analysis reveal intraspecific variation in sex chromosomes of the Virginian strawberry. Journal of Heredity 108: 731–739. [DOI] [PubMed] [Google Scholar]
- West PT, Li Q, Ji L, et al. 2014. Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS One 9: e105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M. 1946. Aberrant Y-chromosomes and sex expression in Melandrium album. Hereditas 32: 419–443. [DOI] [PubMed] [Google Scholar]
- Westergaard M. 1958. The mechanism of sex determination in dioecious flowering plants. Advances in Genetics 9: 217–281. [DOI] [PubMed] [Google Scholar]
- Winge O. 1923. On sex chromosomes, sex determination and preponderance of females in some dioecious plants. Comptes Rendus des Travaux du Laboratoire Carlsberg 15: 1–26. [Google Scholar]
- Wolf DE, Satkoski JA, White K, Rieseberg LH. 2001. Sex determination in the androdioecious plant Datisca glomerata and its dioecious sister species D. cannabina. Genetics 159: 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Dean R, Zimmer F, Mank JE. 2016. How to make a sex chromosome. Nature Communications 7: 12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Davis AW. 1993. Evolution of postmating reproductive isolation: the composite nature of Haldane’s rule and its genetic bases. American Naturalist 142: 187–212. [DOI] [PubMed] [Google Scholar]
- Wu M, Moore RC. 2015. The evolutionary tempo of sex chromosome degradation in Carica papaya. Journal of Molecular Evolution 80: 265–277. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y. 1938. Karyogenetische Untersuchungen bei der Gattung Rumex. VI. Geschechts- bestimmung bei eu- und aneuploiden Pflanzen von Rumex acetosa L. Memoirs of the College of Agriculture, Kyoto University 43: 1–59. [Google Scholar]
- Yu Q, Shaobin H, Hobza R, et al. 2007. Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Molecular Genetics and Genomics 278: 177–185. [DOI] [PubMed] [Google Scholar]
- Zemp N, Minder A, Widmer A. 2014. Identification of internal reference genes for gene expression normalization between the two sexes in dioecious white Campion. PLoS One 9: e92893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp N, Tavares R, Widmer A. 2015. Fungal infection induces sex-specific transcriptional changes and alters sexual dimorphism in the dioecious plant Silene latifolia. PLoS Genetics 11: e1005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp N, Tavares R, Muyle A, Charlesworth D, Marais GAB, Widmer A. 2016. Evolution of sex-biased gene expression in a dioecious plant. Nature Plants 2: 16168. [DOI] [PubMed] [Google Scholar]
- Zhang L-B, Simmons MP, Kocyan A, Renner SS. 2006. Phylogeny of the Cucurbitales based on DNA sequences of nine loci from three genomes: implications for morphological and sexual system evolution. Molecular Phylogenetics and Evolution 39: 305–322. [DOI] [PubMed] [Google Scholar]
- Zhang P, Yang S, Liu Y, et al. 2016. Validation of a male‐linked gene locus (OGI) for sex identification in persimmon (Diospyros kaki Thunb.) and its application in F1 progeny. Plant Breeding 135: 721–727. [Google Scholar]
- Zluvova J, Lengerova M, Markova M, et al. 2005. The inter-specific hybrid Silene latifolia × S. viscosa reveals early events of sex chromosome evolution. Evolution and Development 7: 327–336. [DOI] [PubMed] [Google Scholar]
- Zluvova J, Nicolas M, Berger A, Negrutiu I, Monéger F. 2006. Premature arrest of the male flower meristem precedes sexual dimorphism in the dioecious plant Silene latifolia. Proceedings of the National Academy of Sciences, USA 103: 18854–18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova J, Georgiev S, Janousek B, Charlesworth D, Vyskot B, Negrutiu I. 2007. Early events in the evolution of the Silene latifolia Y chromosome: male specialization and recombination arrest. Genetics 177: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova J, Zak J, Janousek B, Vyskot B. 2010. Dioecious Silene latifolia plants show sexual dimorphism in the vegetative stage. BMC Plant Biology 10: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]