Abstract
Backgrounds and Aims
The genus Stylosanthes includes nitrogen-fixing and drought-tolerant species of considerable economic importance for perennial pasture, green manure and land recovery. Stylosanthes scabra is adapted to variable soil conditions, being cultivated to improve pastures and soils worldwide. Previous studies have proposed S. scabra as an allotetraploid species (2n = 40) with a putative diploid A genome progenitor S. hamata or S. seabrana (2n = 20) and the B genome progenitor S. viscosa (2n = 20). We aimed to provide conclusive evidence for the origin of S. scabra.
Methods
We performed fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH) experiments and Illumina paired-end sequencing of S. scabra, S. hamata and S. viscosa genomic DNA, to assemble and compare complete ribosomal DNA (rDNA) units and chloroplast genomes. Plastome- and genome-wide single nucleotide variation detection was also performed.
Key Results
GISH and phylogenetic analyses of plastid DNA and rDNA sequences support that S. scabra is an allotetraploid formed from 0.63 to 0.52 million years ago (Mya), from progenitors with a similar genome structure to the maternal donor S. hamata and the paternal donor S. viscosa. FISH revealed a non-additive number of 35S rDNA sites in S. scabra compared with its progenitors, indicating the loss of one locus from A genome origin. In S. scabra, most 5S rDNA units were similar to S. viscosa, while one 5S rDNA site of reduced size most probably came from an A genome species as revealed by GISH and in silico analysis.
Conclusions
Our approach combined whole-plastome and rDNA assembly with additional cytogenetic analysis to shed light successfully on the allotetraploid origin of S. scabra. We propose a Middle Pleistocene origin for S. scabra involving species with maternal A and paternal B genomes. Our data also suggest that variation found in rDNA units in S. scabra and its progenitors reveals differences that can be explained by homogenization, deletion and amplification processes that have occurred since its origin.
Keywords: Stylosanthes scabra, Stylosanthes hamata, Stylosanthes viscosa, allopolyploidy, chloroplast genome, GISH, FISH, rDNA, SNP, INDEL, Middle Pleistocene
INTRODUCTION
The Leguminosae is the third largest family of flowering plants, with enormous importance as both fodder crops and green manures in temperate and tropical regions of the world (Lewis et al., 2005). The genus Stylosanthes Sw. (stylo; Papilionoideae) is amongst the most economically important forage legumes, with species grown worldwide as a pasture crop with grasses, as well as for land reclamation and restoration, soil stabilization and regeneration, particularly in regions with low precipitation. The advantages of growing Stylosanthes species include high nitrogen fixation efficiency, high protein content and drought resistance. Growing these plants can restore soil fertility, improve soil physical properties and provide permanent vegetation cover (Stace and Edye, 1984; Santos et al., 2009a). These attributes have resulted in Stylosanthes being the most economically significant pasture and forage legume in the tropics (Cameron and Chakraborty, 2004). Consequently, agricultural research centres in Brazil, South East Asia and Australia are developing Stylosanthes breeding programmes for their improvement as pasture crops and for green manure, especially in regions of low precipitation.
Stylosanthes is known to be highly diverse and polymorphic, comprising around 50 tropical and sub-tropical species, most of them found in the neotropics. Brazil, with 30 species and 12 endemics, is the centre of diversity of Stylosanthes (Stace and Cameron, 1984; da Costa and Valls, 2010; Santos-Garcia et al., 2012). Preliminary phylogenetic analyses revealed that Stylosanthes belongs to the Dalbergieae clade and is probably sister to the genus Arachis (Cardoso et al., 2013).
Despite its socio-economic importance, genetic characterization of Stylosanthes has been limited, which together with its complex systematics has hampered the development of breeding programmes. Conventional cytogenetic approaches have shown that most species are diploid with 2n = 20 and a few are tetraploid (2n = 40) or hexaploid (2n = 60) (Maass and Sawkins, 2004). Previous molecular studies have shown that polyploids are probably formed by interspecific hybridization (allopolyploidy), mostly involving species in two sections, sect. Stylosanthes and sect. Styposanthes (Maass and Sawkins, 2004). Phylogenetic analyses revealed that these sections are not monophyletic; however, the evolutionary relationships of the group are still poorly understood, and phylogenetic relationships between species remain with limited resolution (Vander Stappen et al., 1999, 2002). Studies based on restriction fragment length polymorphism (RFLP) and sequence-tagged site (STS) analyses identified ten basal genomes, named A to J (Liu et al., 1999; Ma et al., 2004). At least 11 species are thought to have an allopolyploid origin, with most having similar parental A genome donors, resembling the diploids S. hamata (L.) Taub. or S. seabrana B. L. Maass & 't Mannetje (Liu et al., 1999; Liu and Musial, 2001; Ma et al., 2004; Maass and Sawkins, 2004). However, the origins and species relationships of allopolyploids in Stylosanthes remain largely unresolved (Maass and Sawkins, 2004).
Stylosanthes scabra Vogel (2n = 40, AABB) is widely distributed across South America (Williams et al., 1984; Liu, 1997) and is one of the most exploited allotetraploid species in the genus (Cameron and Chakraborty, 2004; Pathak et al., 2004). It has high drought tolerance and is well adapted to infertile, acid, friable or hard-setting, sandy-surfaced soils. It is used predominantly for pasture and soil improvement in Brazil, Australia and South Asian countries, where it shows great potential to spread naturally on some soil types (Nascimento et al., 2001; Chandra, 2013). Stylosanthes scabra is thought to have originated by allopolyploidy involving S. hamata/S. seabrana (A genome) and S. viscosa (L.) Sw. (B genome) (Liu et al., 1999; Liu and Musial, 2001; Vander Stappen et al., 2002; Chandra, 2013), though there is no conclusive cytogenetic evidence for its hybrid origin and genome constitution. The involvement of S. hamata as the maternal parent and donor of the A genome is questioned by a number of studies supporting a role for the diploid S. seabrana in the origin of S. scabra (Liu et al. 1999; Liu and Musial, 2001; Chandra and Kaushal, 2009). Nevertheless Liu and Musial (2001) reported that chloroplast DNA of S. scabra corresponded to that of S. seabrana not that of S. hamata. However, this approach was based on a single chloroplast clone of 499 bp in length, which might provide insufficient resolution to discriminate between S. seabrana and S. hamata. Moreover, the taxonomic status of S. seabrana (Maass and Mannetje, 2002) is dubious, since two recent studies have suggested that S. seabrana is a synonym for S. scabra (Vanni and Fernandez, 2011; Vanni, 2017).
Genomic in situ hybridization (GISH) and locus-specific fluorescence in situ hybridization (FISH) are powerful tools to investigate the origin of allopolyploids (Chester et al., 2010). The use of GISH can facilitate the detection of the parental genome of an allopolyploid species using genomic DNA from putative progenitor species as probes. The distribution patterns of nuclear ribosomal DNA (rDNA) sites, and the unit structure of rDNA, can shed further light on the evolutionary origin and patterns of divergence in allopolyploids compared with related diploids (Volkov et al., 2007; Kovarik et al., 2008; Ferreira and Pedrosa-Harand, 2014).
rDNA units occur in tandem arrays at one or multiple loci and comprise rRNA genes separated by internally transcribed (ITS) and intergenic (IGS) spacers, the latter containing both non-transcribed (NTS) and externally transcribed (ETS) sequences (Srivastava and Schlessinger, 1991). Because of the relatively low selection pressures acting on non-coding spacer sequences, these regions can have a high degree of variation, even between closely related species (Kovarik et al., 2008). However, rDNA may undergo concerted evolution via unequal crossing over and gene conversion, promoting relatively high intragenomic homogeneity of the repeat units (Ganley and Kobayashi, 2007). Despite the intragenomic homogeneity of coding regions being typically high, recent high-throughput sequencing has shown that there can be a high frequency of variation within non-coding spacer sequences, even within the same plant species (Matyasek et al., 2012; Song et al., 2012; Lunerová et al., 2017). These features make rDNA sequence analysis a valuable tool for characterizing allopolyploids.
Organelle inheritance is strictly maternal for most angiosperm species (Reboud and Zeyl, 1994; Greiner et al., 2014). This property makes the sequence of organelle genomes ideal for identifying patterns of maternal genome inheritance in hybrid species (Gastony and Yatskievych, 1992; Jankowiak et al., 2005). Indeed, Liu and Musial (2001) have demonstrated that chloroplast inheritance in Stylosanthes can be used to identify putative maternal genome donors for 16 allopolyploid taxa. The combination of both plastid DNA and rDNA sequences is therefore useful for recognizing not only progenitor species of allopolyploids, but also which is the maternal or paternal genome donor (Soltis et al., 2008; Cires et al., 2014).
This study aims to better characterize the allopolyploid origin of S. scabra and to shed new light on divergence patterns in rDNA sequences between the species and its putative progenitor diploids. We combined both cytogenetic (FISH and GISH) and bioinformatic approaches to provide further evidence that S. scabra is indeed an allotetraploid, with a maternal A genome donor (S. hamata/S. seabrana) and a paternal B genome donor (S. viscosa). Because of the controversial taxonomical status of S. seabrana and the previous studies showing that its genome is the same as that found in the well-established S. hamata, we decided to use the latter as the A genome representative in the present study.
MATERIALS AND METHODS
DNA isolation
Plant tissue (young leaves, fresh 5–20 g each) of Stylosanthes hamata ‘LC 7666’, S. viscosa ‘A-01’ and S. scabra ‘CPAC-5234’ was collected from five plants from each accession growing in the greenhouse of the Laboratory of Genetic Resources for DNA isolation and next-generation sequencing (NGS). Total genomic DNA (gDNA) was extracted with a modified cetyltrimethylammonium bromide (CTAB) protocol (Doyle and Doyle, 1987). The quality of extracted DNA was checked on 1 % (w/v) agarose gels. The gDNA concentration was measured using a NanoDrop 2000 photometer (Thermo Scientific).
Slide preparation
For cytogenetic analysis, seeds from accessions ‘LC 7666’ and ‘CPAMIG 1454’ of Stylosanthes hamata; ‘A-01’, ‘EMB’ and ‘274’ of S. viscosa; and ‘1489’, ‘1500’, ‘2254’, ‘2257’ and ‘CPAC-5234’ of S. scabra were germinated and root tips were collected and pre-treated with 8-hydroxyquinoline for 20 h at 10 °C, fixed in ethanol:acetic acid (3:1; v/v) from 2 to 24 h at room temperature and stored at –20 °C. The fixed roots were washed in distilled water and digested in 2 % cellulase (Onozuka) and 20 % pectinase (Merck) at 37 °C for 90 min. Then apical meristems were squashed in 45 % acetic acid under a coverslip. The coverslip was removed in liquid nitrogen.
Probe labelling and in situ hybridization
In order to localize the rDNA sites to chromosomes, a 500 bp 5S rDNA clone (D2) of Lotus japonicus was labelled with Cy3-dUTP (GE Healthcare) and a 6.5 kb 18S–5.8S–25S clone (R2) from Arabidopsis thaliana was labelled with digoxigenin-11-dUTP (Merck) (Pedrosa et al., 2002). These labelled probes were used for rDNA-FISH. For GISH, gDNAs of S. hamata (A genome) and S. viscosa (B genome) were labelled with Cy3-dUTP and digoxigenin-11-dUTP, respectively, and used in a 1:1 ratio in the hybridization mixture. Alternatively, the genomic probe from S. hamata together with blocking DNA of S. viscosa were added to the hybridization mixture in the concentration ratio of 1:20, respectively, and hybridized to S. scabra chromosomes. Blocking DNA was produced by boiling gDNA of S. viscosa and checking on an agarose gel for appropriate DNA length. All probes were labelled by nick translation (Merck). Digoxigenin-labelled probes were detected with sheep anti-digoxigenin–fluorescein isothiocyanate (FITC) conjugate (Merck) and amplified with rabbit anti-sheep–FITC conjugate (Bio-Rad).
In situ hybridization was performed according to Marques et al. (2015). The hybridization mix contained 50 % formamide (v/v), 10 % dextran sulphate (w/v), 2× SSC and 50 ng of each probe. The final hybridization stringency was estimated to be 76 %. The slides were mounted with 4’,6-diamidino-2-phenylindole (DAPI, 4 μg mL–1)/Vectashield (Vector) 1:1 (v/v) and analysed under an epifluorescence microscope (Leica DMLB) equipped with DAPI, FITC and Cy3 filters. Images were recorded using a Cohu CCD camera and software Leica QFISH before editing with the software Adobe Photoshop CS3 version 10.0. In total at least 20 cells per sample per experiment were analysed.
Genome size estimation using flow cytometry
Nuclear DNA contents were estimated following the one-step flow cytometry procedure described by Dolezel et al. (2007). Briefly, approx. 1 cm2 of leaf material from young leaves (from the same accessions used for NGS) together with the appropriate calibration standard, either Solanum lycopersicum ‘Stupicke polki tyckove rane’ (2C = 1.96 pg) or Raphanus sativus ‘Saxa’ (2C = 1.11 pg) (Dolezel et al., 1992), were chopped using a new razor blade and mixed together in a Petri dish containing 1 mL of ‘WPB buffer’ (Loureiro et al., 2007). A further 1 mL of buffer was added, the resulting suspension was filtered through a 30 μm nylon mesh and the nuclei were stained with 100 μL of propidium iodide (1 mg mL–1). Samples were kept at 37 °C for 20 min and the relative fluorescence of 5000 particles was then recorded using a Partec Cyflow SL3 flow cytometer (Partec GmbH, Münster, Germany) fitted with a 100 mW green solid-state laser (532 nm, Cobolt Samba, Solna, Sweden). Three leaves from different individuals were measured separately for each species and three replicates of each leaf were processed. The output histograms were analysed with the FlowMax software v. 2.4 (Partec GmbH).
DNA library preparation and sequencing
For construction of the sequencing library, a total of 5 μg of gDNA was used as input material for library preparation. Briefly, DNA was mechanically fragmented to generate fragments of, on average, 300–500 bp, and the fragments were ligated with adaptors enabling barcoding. The library was size-selected with a SYBR Gold-stained (ThermoFisher) electrophoresis gel. Fragment size distribution and DNA concentration were evaluated using an Agilent BioAnalyzer High Sensitivity DNA Chip and the Qubit DNA Assay Kit in a Qubit 2.0 Flurometer (Life Technologies). DNA clusters generated using Illumina cBot were sequenced with Illumina PhiX library as internal controls using Illumina HiSeq 2500 paired-end sequencing (2 × 101 bp). Sequencing was performed in the Central Laboratory of High Performance Technologies in Life Sciences (LacTad) of the State University of Campinas, São Paulo, Brazil. The raw Illumina paired-end sequence reads of each sample were deposited in GenBank with the following Sequence read Archive (SRA) accession number: SRP127621.
De novo plastome and rDNA assembly
The total number of raw paired-end reads obtained for S. hamata, S. viscosa and S. scabra are listed in Supplementary Data Table S1. De novo plastome assembly of reads was performed by NOVOPlasty v2.6.3 (Dierckxsens et al., 2017) using default parameters and the Arachis hypogaea plastome reference sequence (GenBank accession no. KJ468094). As NOVOPlasty does not need quality trimming of the reads, all reads for each species were used. NOVOPlasty was able to assemble a single circularized contig for each species, representing the whole plastome including all regions: long single copy (LSC), short single copy (SSC) and both inverted repeats (IRs). The contigs obtained were imported into Geneious v. 9.1.8 and the assembly checked by mapping the raw reads to the contigs using the Geneious mapper with low sensitivity. The plastomes of the three species were annotated using the Geneious annotation tool, guided by the available A. hypogaea plastome annotation. Annotations were manually checked to correct misannotated regions. The plastome maps were generated using OrganellarGenomeDraw (OGDraw v1.2) (Lohse et al., 2013). Complete annotated plastomes were deposited in GenBank with the following accession numbers: MG735673 for S. hamata, MG735674 for S. scabra and MG735675 for S. viscosa.
To obtain the sequence of 5S and 35S rDNA units, we have filtered the reads to PHRED scores 30 over 100 % of the read length, resulting in 28.5, 31 and 18 million reads for S. hamata, S. viscosa and S. scabra, respectively. The remaining reads of each species were assembled using the software RepeatExplorer (Novak et al., 2013). rDNA contigs were identified using BLAST searches. Both 5S and 35S rDNA 3ʹ end and 5ʹ end regions were annotated based on comparison with other rRNA genes in GenBank. Furthermore, graph-based clustering using RepeatExplorer was able to assemble different 5S rDNA variants for each species, which were assembled in separate contigs. These contigs were then used for further analysis (see below).
Graph-based clustering of rDNA reads and interactive visualization
Circular contigs comprising entire rDNA units were used as reference for retrieving all rDNA-containing reads using the Geneious mapper tool. Reads retrieved were used as input for comparative graph-based clustering with RepeatExplorer. Interactive visualization of 5S and 35S rDNA graphs was performed with the R package SeqGrapheR, which provides a simple graphical user interface for interactive visualization of sequence clusters. SeqGrapheR enabled the selection of species-specific reads from rDNA graphs, allowing simultaneous viewing of the graph layout (Novak et al., 2010).
Alignment and phylogenetic sequence comparison of assembled rDNA and plastomes of Stylosanthes
Alignment of whole plastomes, 5S and 35S rDNA units was performed with MAFFT v7.222 (Katoh and Standley, 2013) as a Geneious v. 9.1.8 plugin (Kearse et al., 2012). Phylogenetic relationships were inferred using the Bayesian inference (BI) approach implemented in MrBayes v.3.2.6. (Ronquist et al., 2012). As the outgroup for (1) plastome phylogenetic comparisons, we used the finished plastome of A. hypogaea (KJ468094); (2) 5S rDNA comparisons, we used the Arachis duranensis 5S rDNA unit obtained from the PeanutBase (https://peanutbase.org/); and (3) 35S rDNA comparisons, the SRA file accession no. DRR056349 from A. hypogaea was used, where the assembly of 35S rDNA was performed as described above for Stylosanthes.
Plastome, rDNA and genome-wide single nucleotide variation (SNV) detection: single nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs)
For reference-based intragenomic SNV calling of rDNA sequences, filtered whole genomic reads (see above) were mapped against reference rDNA consensus contigs assembled by RepeatExplorer. For estimating the total number of reads resembling 5S rDNA variants, we subjected the reads of each species to mapping at the same time as the consensus 5S variants found in each species. All mappings were conducted using highly strict parameters as follows: maximum mismatches per read 5 %, maximum gap size 3, word length 24, index word length 14. Thus, reads of each species competed for matching a single most similar variant. Finally, SNV detection was conducted on mapped variants using Geneious v.9.1.8. SNVs were identified with a minimum variant frequency of 0.05 and minimum coverage of 100. Adjacent variations were considered as a single variation. For the 35S rDNA unit, the IGS region was removed from the SNV analysis because of its highly repetitive nature.
For comparative whole-plastome and genome-wide reference-free SNV detection, we used discoSnp++ (Uricaru et al., 2015; Peterlongo et al., 2017) with default parameters. The software discoSnp++ is designed for discovering isolated SNPs and INDELs from raw sets of reads obtained with NGS without the need for a reference genome. The software is composed of two modules: the first module, kissnp2, detects SNPs from read sets; the second module, kissreads2, enhance the kissnp2 results by computing per read set and, for each variant found (1) its mean read coverage and (2) the (phred) quality of reads generating the polymorphism. Finally, a VCF file is also created (for details, see Peterlongo et al., 2017). VCF files generated by discoSnp++ were compared using vcf-compare from VCFtools (Danecek et al., 2011) and Venn diagrams were drawn using the R library VennDiagram (Chen and Boutros, 2011). For comparative analysis, NGS reads from A. hypogaea were obtained from the SRA file DRR056349.
Analysis of microsatellites
The Perl script MISA (MIcroSAtellite) (Thiel et al., 2003) was employed to search for simple sequence repeat (SSR) loci in the plastid genome sequence, with threshold values for the repeat number set at ≥5 for dinucleotide repeats, ≥4 for trinucleotide repeats and ≥3 for tetranucleotide, pentanucleotide and hexanucleotide repeats. Afterwards, the identified SSRs were checked for interspecific polymorphisms with the software Geneious v.9.1.8 (Kearse et al., 2012).
Estimating the age of Stylosanthes scabra
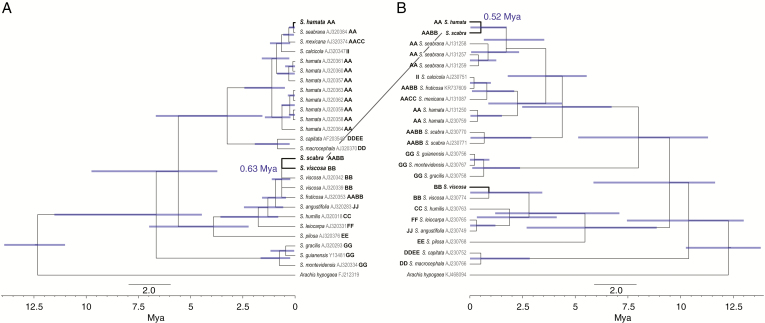
To estimate the age of S. scabra, we performed a molecular phylogeny of the genus Stylosanthes based on the ITS (ITS1–5.8S–ITS2) of nuclear rDNA and the plastid spacer trnL–trnF. We estimated the divergence of the S. scabra ITS in relation to its putative paternal progenitor S. viscosa (Fig. 7A) and the divergence of the S. scabra plastid trnL–trnF sequence in relation to its putative maternal progenitor S. hamata (Fig. 7B). We sampled 126 specimens of 17 taxa of the genus Stylosanthes, including as outgroup A. hypogaea (Fig. 7). All sequences were obtained from GenBank (Vander Stappen et al., 1999, 2002). Additionally, we have also added the DNA sequences from S. hamata, S. scabra and S. viscosa resulting from the plastome and rDNA assemblies from the present study. All the sequences were aligned using MUSCLE (Edgar, 2004) as a plugin in Geneious v.9.1.8 (Kearse et al., 2012) with subsequent manual adjustments.
Fig. 7.
Chronogram of Stylosanthes, with focus on the age of S. scabra, based on BEAST analysis using nuclear ITS (A) and plastid trnL–trnF (B). Blue bars indicate 95 % highest posterior density intervals. GenBank accession numbers are given as grey letters on the side of each sample name. Genome structure is given in bold upper case letters after Liu et al. (1999) or Mass and Sawkins (2004).
We used jModelTest v.2.1.6 to assess the best model of DNA substitution for each individual locus (Darriba et al., 2012) through the Akaike information criterion (Akaike, 1974). The best fitting model was GTR + G for both ITS and trnL–trnF. Phylogenetic relationships were inferred using the BI approach implemented in MrBayes v.3.2.6. (Ronquist et al., 2012). All analyses were performed for each region separately. Four independent runs with four Markov Chain Monte Carlo (MCMC) runs were conducted, sampling every 1000 generations for 3 000 000 generations. Each run was evaluated in Tracer v.1.6 (Rambaut et al., 2014) to determine that the estimated sample size (ESS) for each relevant parameter was >200, and a burn-in of 25 % was applied. The majority rule consensus tree and posterior probability (PP) were visualized and edited in FigTree v.1.4.2 (Rambaut, 2014).
Divergence time estimates were performed in BEAST v.1.8.3 (Drummond and Rambaut, 2007; Drummond et al., 2012) fixing the tree topology of the Bayesian analyses. An uncorrelated relaxed lognormal clock (Drummond et al., 2006) and a Yule Process speciation model (Gernhard, 2008) were applied. Two independent runs of 10 000 000 generations each were performed, sampling every 10 000 generations for each ITS and plastid trnL–trnF. In order to verify the effective sampling of all parameters and assess the convergence of independent chains, we examined their posterior distributions in Tracer v.1.6, and the MCMC sampling was considered sufficient at an ESS >200. After removing 25 % of samples as burn-in, the independent runs were combined and a maximum clade credibility (MCC) tree was constructed using TreeAnnotator v.1.8.2. (Drummond et al., 2012). Calibrations were performed using the secondary calibrations of Särkinen et al. (2012) for the Arachis/Stylosanthes divergence approx. 12.4 million years ago (Mya).
RESULTS
In situ hybridization confirms the allopolyploid origin of S. scabra
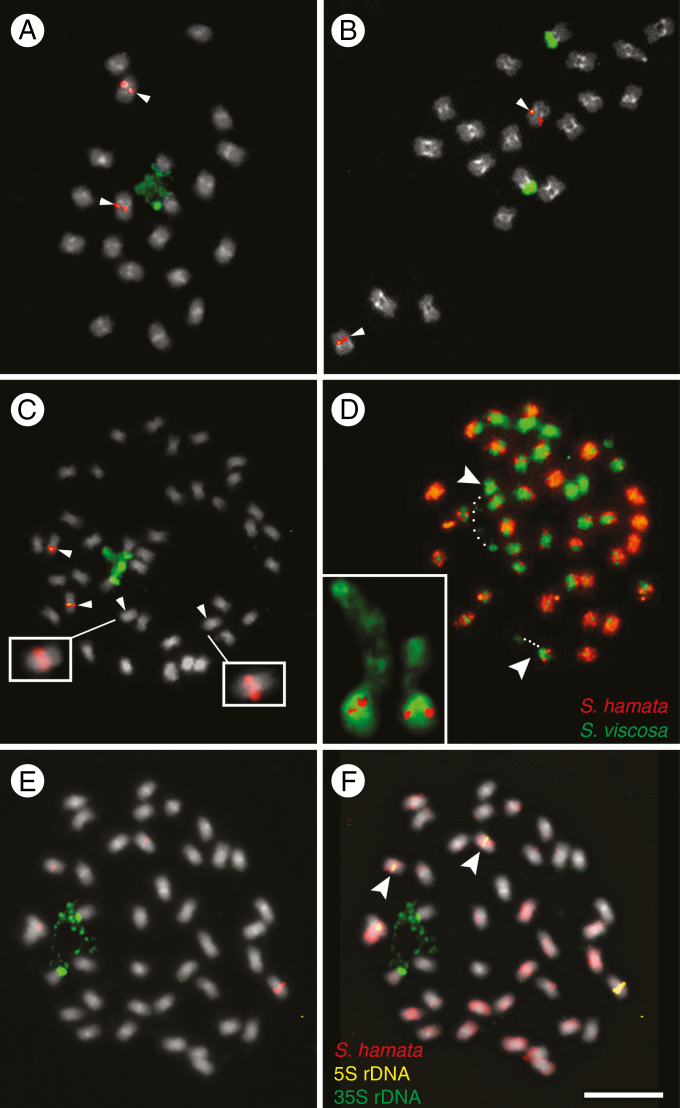
To test the hypothesis that S. scabra is an allotetraploid originating from progenitor species closely related to S. hamata (A genome representative) and S. viscosa (B genome representative), we performed FISH with rDNA probes and GISH with genomic probes from these diploid species. rDNA FISH revealed one site of 5S rDNA in the proximal region of a chromosome pair and one site of 35S rDNA in the terminal region of another chromosome pair in both diploids S. hamata (2n = 20) and S. viscosa (2n = 20) (Fig. 1A, B). FISH results for samples identified as S. seabrana wer similar to those for S. hamata (data not shown). In S. scabra, two pairs of chromosomes showing proximal 5S rDNA sites were observed (one strongly labelled and one weakly labelled site: Fig. 1C). Stylosanthes scabra, despite having a tetraploid chromosome number (2n = 40), showed only one pair of 35S rDNA sites (Fig. 1C).
Fig. 1.
FISH and GISH in the Stylosanthes scabra complex. (A–C, E) FISH with 5S (red) and 35S (green) rDNA probes in S. viscosa (A), S. hamata (B) and S. scabra (C, E). Arrowheads in (C) show chromosomes harbouring the 5S rDNA. Insets in (C) show overexposed S. scabra chromosomes with weak 5S rDNA signals. (D) GISH in S. scabra using the genomic probes of S. hamata (orange) and S. viscosa (green). (F) GISH in S. scabra using the gDNA of S. hamata as probe (red) and the gDNA of S. viscosa as blocking DNA (grey). (F) The same cell as in (E) after sequential rDNA-FISH and GISH. Arrowheads and the inset in (D) show the NOR-harbouring chromosomes labelled with S. viscosa probe in green. Note the position of assumed decondensed 35S rDNA (dotted lines) connecting a satellite to the rest of the chromosome (dotted lines). Note also that in (F) the 35S rDNA-harbouring chromosomes are not labelled with the S. hamata probe, while the chromosome pair with the weak 5S rDNA signal is labelled in red (arrowheads).
Heterochromatin which is DAPI positive in a (peri)centromeric location was observed on chromosomes of all three species, although sometimes the signals were weak, especially in S. scabra (Fig. 1A–C). GISH with S. hamata and S. viscosa genomic probes revealed differential labelling of the chromosomes (Fig. 1D), with approximately half the chromosomes labelled predominantly with one genomic probe and half with the other. However, there were equivocal parental genome identities for several chromosomes, arising because of strong signal from both probes, which could be caused by cross-hybridization of probes or by intergenomic translocations. To increase the reliability of GISH, we hybridized the chromosomes of S. scabra with a labelled S. hamata genomic probe and blocked DNA of S. viscosa in the concentration ratio of 1:20, respectively. Indeed, this increased chromosome signal differentiation and reduced cross-hybridization at (peri)centromeric regions (Fig. 1F). One pair of predominantly green chromosomes of presumed S. viscosa origin, and which carried pericentromeric signal labelling with the S. hamata probes, also carried satellite chromatin, which is probably isolated from the rest of the chromosome by a secondary constriction caused by the decondensation of 35S rDNA (Fig. 1D). Thus, to check the parental origin of rDNA-harbouring chromosomes in S. scabra, we performed sequential rDNA-FISH and GISH. Indeed, one pair of chromosomes harbouring the stronger 5S rDNA and another pair harbouring the 35S rDNA locus found in S. scabra was not labelled with the genomic probe of S. hamata, while the chromosome pair harbouring the weak 5S rDNA locus was labelled (Fig. 1E, F, arrowheads).
Whole-plastome assembly recognizes the maternal genome donor of S. scabra
Illumina HiSeq 2500 paired-end sequencing yielded about 38 million, 42 million and 28 million paired reads for S. hamata, S. viscosa and S. scabra, respectively. This is equivalent to about 4 Gb of sequence for the first two species and 2.8 Gb for the latter species. Based on the estimated genome size for S. hamata (1C = 880 Mbp), S. viscosa (1C = 665 Mbp) and S. scabra (1C = 1398 Mbp), our Illumina sequencing had a coverage of around ×4.4, ×6.4 and ×2, respectively (Supplementary Data Table S1).
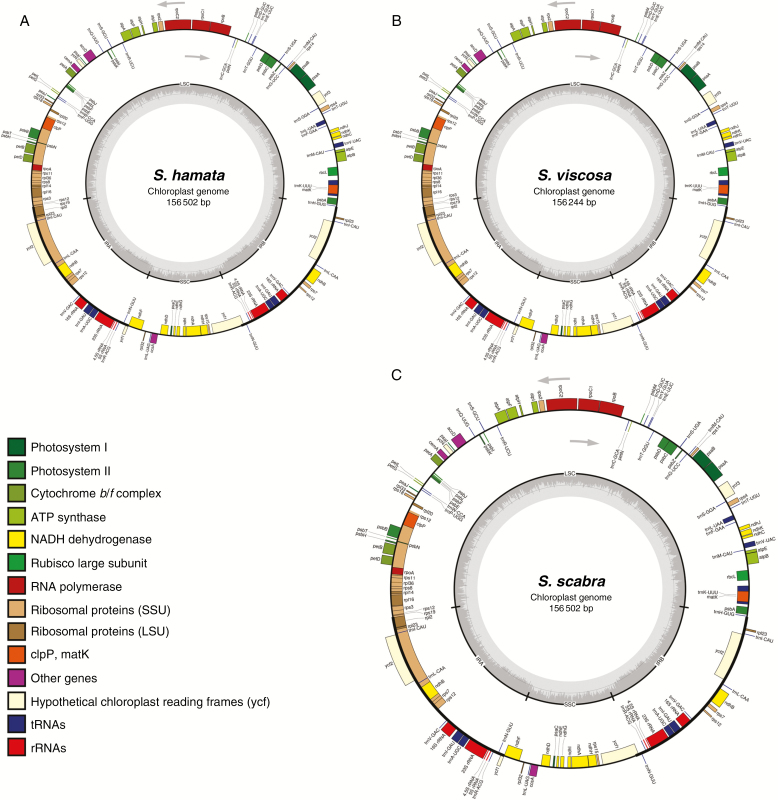
To shed more light on the origin of S. scabra and to check which genome, A or B, is maternally inherited, we performed whole plastome unit assemblies. Whole plastome lengths of S. scabra (156 502 bp) (Fig. 2A) and S. hamata (156 502 bp) (Fig. 2B) were identical, and S. viscosa (156 244 bp) (Fig. 2C) was 258 nucleotides smaller. General features of Stylosanthes plastomes are depicted in Table 1. Pairwise comparisons of the plastome sequence alignments of the three study species revealed the highest similarity (99.8 % pairwise identity) between plastomes of S. scabra and S. hamata, with 314 polymorphic sites, of which 178 were INDELs and 136 SNPs (Table 2). Bayesian phylogenetic analysis based on whole-plastome alignment confirmed the close relationship of S. hamata and S. scabra (Fig. 3A). Plastome gene annotation revealed 125 annotated regions in all three species, which include 36 tRNAs, two copies of four different rRNA genes and 81 protein-coding genes, distributed into 17 groups (Fig. 2; Supplementary Data Table S2). These results indicate that the inherited plastome (and thus the maternal genome as well) in S. scabra was derived from a progenitor with S. hamata-like chloroplast DNA.
Fig. 2.
Schematic representation of the annotated Stylosanthes plastomes: (A) S. hamata, (B) S. viscosa and (C) S. scabra.
Table 1.
Summary of plastid genome characteristics of Stylosanthes
| S. hamata | S. viscosa | S. scabra | |
|---|---|---|---|
| Total size (bp) | 156 502 | 156 244 | 156 502 |
| LSC size in bp | 86 047 | 85 912 | 86 064 |
| SSC size in bp | 18 749 | 18 710 | 18 734 |
| IR length in bp | 25 853 | 25 811 | 25 852 |
| Size of coding regions in bp | 96 948 | 96 914 | 96 962 |
| Size of protein-coding regions in bp | 76 134 | 76 107 | 76 140 |
| Size of rRNA in bp | 9062 | 9062 | 9062 |
| Size in bp of tRNA | 2735 | 2735 | 2735 |
| Size in bp of intergenic regions | 52 223 | 51 999 | 52 209 |
| No. of genes | 118 | 118 | 118 |
| No. of protein-coding genes | 81 | 81 | 81 |
| No. of tRNA genes | 36 | 36 | 36 |
| No. of rRNA genes | 8 | 8 | 8 |
| No. of genes duplicated by IR | 15 | 15 | 15 |
| No. of genes with introns | 11 | 11 | 11 |
| Overall % GC content | 36.6 | 38.8 | 36.6 |
| % GC content in protein-coding regions | 37.6 | 37.6 | 37.6 |
| % GC content in IGSs | 30.8 | 30,8 | 30.8 |
| % GC content in rRNA | 55.5 | 55.5 | 55.5 |
| % GC content in tRNA | 53.2 | 53.2 | 53.2 |
| Plastome coverage | 1100.1 | 1208.5 | 653.9 |
Table 2.
Whole-plastome intergenomic SNP calling and pairwise identity (%) among the three species of Stylosanthes
| Species | S. hamata | S. viscosa | ||||||
|---|---|---|---|---|---|---|---|---|
| Total SNPs | Indels | Pairwise identity (%) | Identical sites | Total SNPs | Indels | Pairwise identity (%) | Identical sites | |
| S. viscosa | 2754 | 1479 | 98.2 | 154 351 | – | – | – | – |
| S. scabra | 314 | 178 | 99.8 | 156 277 | 2739 | 1479 | 98.3 | 154 366 |
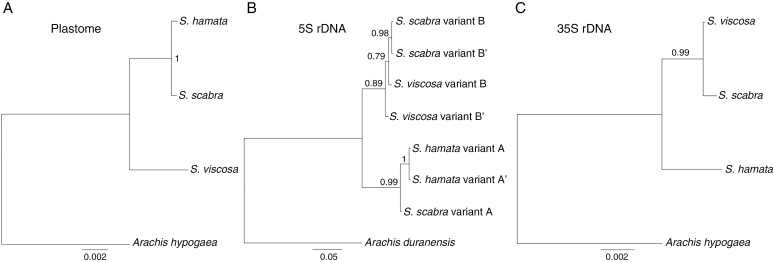
Fig. 3.
Bayesian phylogenetic comparison based on whole plastomes and rDNA units among Stylosanthes. (A) Plastome tree, (B) 5S rDNA unit variants tree and (C) 35S rDNA unit tree.
To check whether the plastomes of Stylosanthes are suitable for identification of molecular markers, we have searched for polymorphic SSRs composed of 2–6 bp units. Using the microsatellite identification tool MISA, SSRs were identified in the plastomes of S. hamata, S. viscosa and S. scabra. Afterwards we searched for polymorphic SSRs across the three plastomes. Only five polymorphic SSRs were found between S. hamata and S. scabra plastomes, three being dinucleotides and two trinucleotides (Supplementary Data Fig. S1A), which reinforces the great similarity of both plastomes. However, this number increased to 15 polymorphic SSRs when comparing S. viscosa with the other two species: ten dinucleotides, three trinucleotides and two hexanucleotides. Eighteen polymorphic SSRs were found when considering all three plastomes (Supplementary Data Fig. S1A). All dinucleotide SSRs were composed of AT motifs, while all trinucleotides and hexanucleotides were composed of AAT and AATACT motifs, respectively (Supplementary Data Fig. S1B).
rDNA sequence analyses recognize the paternal genome donor of S. scabra
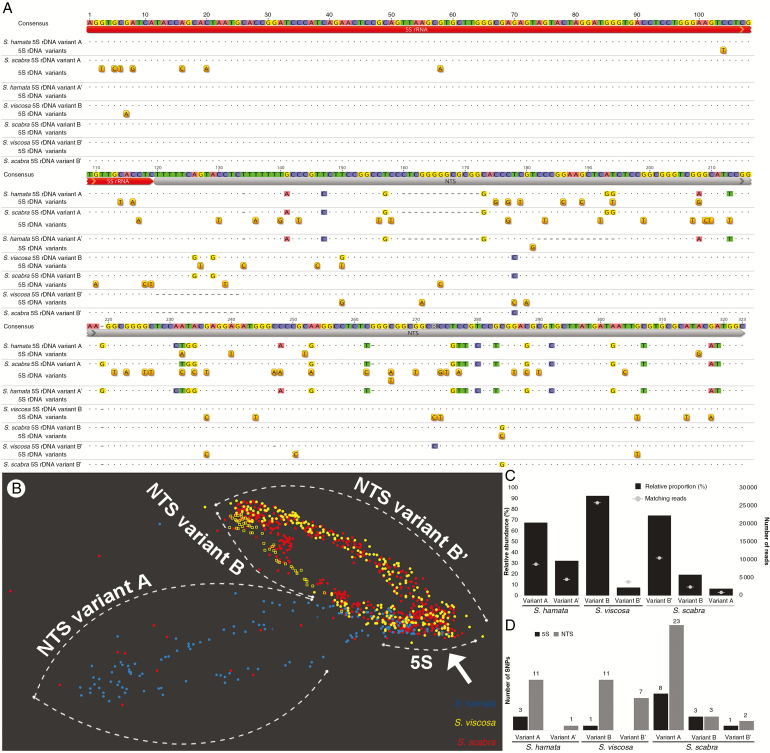
We were interested in comparing 5S rDNA sequences in S. scabra and its two putative diploid parents. In total, four 5S rDNA unit variants were found in the three species based on RepeatExplorer assembly output. Stylosanthes hamata showed two variants named A (308 bp) and A’ (295 bp), which differed mainly by an INDEL of 13 nucleotides in the NTS region (Fig. 4A). Stylosanthes viscosa also had two variants named B (322 bp) and B’ (307 bp), which differed mainly by an INDEL of 15 nucleotides at the beginning of the NTS region (Fig. 4A). Stylosanthes scabra showed three variants: A, B and B’, with close similarities to the variants identified in S. hamata and S. viscosa. By mapping the genomic reads against each variant contig with highly strict read mapping parameters (see the Materials and Methods), we were able to quantify reads matching each variant (Fig. 4C). Variant A was the most abundant in S. hamata, comprising about 68 % (8702) of 5S rDNA reads found. In S. viscosa, variant B was the most abundant, comprising 92.5 % (25 800). In S. scabra, variant B’ (similar to B’ in S. viscosa) was the most abundant, comprising 74.4 % (10 495) of matching reads, with variants B from S. viscosa (19.3 %) and A from S. hamata (6.3 %) being less abundant (Fig. 4C). We inferred a length of 119 nucleotides for the coding 5S rDNA region in Stylosanthes starting with 5’ AGG and ending with 3’ CTC instead of 120 nucleotides as found in most organisms. A shorter length for the 5S coding region has already been reported for other legume species (Hemleben and Werts, 1988; Gottlob-McHugh et al., 1990). However, we cannot be sure of this length since we annotated this region based on BLAST comparison, and further detailed analysis needs to be carried out to confirm the length of the 5S coding region of Stylosanthes. The NTS varied in length and sequence identity amongst all variants found. The lowest sequence similarity was found between the NTS from S. hamata variant A and S. viscosa variant B (77.1 %, Fig. 4A). Furthermore, the NTS regions of variants A and A’ were also characterized by a duplication of a 13 nucleotide region (Supplementary Data Fig. S2). In summary, these results reveal the predominance of the S. viscosa 5S rDNA variant B’ in the genome of S. scabra.
Fig. 4.
5S rDNA sequence characterization in Stylosanthes. (A) Alignment of 5S (red) rDNA variants including the NTS region (grey) of S. hamata, S. viscosa and S. scabra. Positions of SNPs detected are drawn under each respective sequence. (B) SeqGrapheR visualization of RepeatExplorer 5S rDNA cluster including reads of all three species. Note the close grouping of S. scabra and S. viscosa reads along the NTS region, while the coding 5S region groups reads from the three species (arrow). A few reads (red dots) from S. scabra are seen along NTS variant A, which is in agreement with the low abundance of this variant in the allopolyploid genome. (C) Relative abundance and number of reads matching each 5S rDNA variant found in Stylosanthes. (D) Number of SNPs found in each variant sorted by the 5S coding and NTS regions. Note the high level of SNPs found in variant A of S. scabra.
To provide further evidence that the 5S rDNA sequences found in S. scabra were mostly of the S. viscosa type, we performed a comparative graph-based clustering of rDNA reads generated by RepeatExplorer. Figure 4B shows that 5S rDNA reads of S. scabra and S. viscosa overlap along the length of the NTS region, indicating that their sequences are very similar. Additionally, a few hits of S. scabra grouped with S. hamata NTS hits (Fig. 4B), which is consistent with the occurrence of a low number of copies of variant A in the S. scabra genome. Bayesian phylogenetic analysis based on consensus sequences of 5S rDNA variants confirmed the close relationship between B and B’ variants from S. viscosa and S. scabra and A variants from S. hamata and S. scabra (Fig. 3B).
Because we observed several 5S rDNA variants in Stylosanthes, we were interested in checking their intragenomic SNP frequency. In S. hamata, variant A showed the highest number of intragenomic SNPs (14 SNPs), whilst variant A’ showed only one SNP along the NTS region. In S. viscosa, variant B showed 12 and variant B’ showed seven SNPs. In S. scabra, variant A showed the highest number of SNPs (31 SNPs), whilst variant B showed six and variant B’ showed three SNPs (Fig. 4D). Variant A of S. scabra showed a high number of SNPs within the 5S coding region (eight SNPs) (Fig. 4A, D).
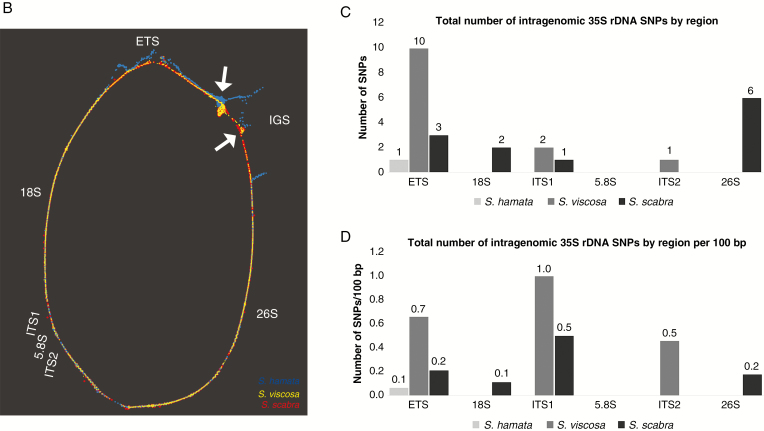
Given that there is a single 35S rDNA locus in the karyotype of S. scabra, when two sites would be expected given the number of sites in the presumed diploid progenitors, we were also interested in comparing their sequences. From HiSeq Illumina reads, we were able to assemble the genic component, ITS, ETS and IGS sequences of 35S rDNA in each species (Fig. 5A). 18S–ITS1–5.8S–ITS2–26S rDNA lengths were 5783 bp in S. hamata, 5785 bp in S. viscosa and 5785 bp in S. scabra. Coding regions were the same length in the three species. ITS1 and ITS2 lengths were the same in S. viscosa and S. scabra, but different in S. hamata. The ETS and IGS were the most divergent regions of rDNA, as expected, and differed amongst the three species (Fig. 5A; Table 3). ETS + IGS lengths were at least 3096 bp in S. hamata, 3383 bp in S. viscosa and 3061 bp in S. scabra, which include a probable repetitive domain evidenced by the ‘knot’ seen on the graphs (Fig. 5B; Supplementary Data Fig. S3C, D). Transcription initiation site (TIS) and transcription termination site (TTS) sequences were the same in all three species (TATTATAGGG and CCCTCCCC, respectively). Furthermore, alignment of the ETS–18S–ITS1–5.8S–ITS2–26S region revealed high pairwise sequence identity (98.5 %) between S. scabra and S. viscosa. In contrast, S. hamata showed less pairwise sequence identity (94.4 %) to S. scabra. Alignment of S. hamata and S. viscosa revealed a pairwise identity of 93.4 % (Fig. 5A; Table 4). These results indicate the predominance of both 5S and 35S rDNA from the S. viscosa type in the S. scabra genome.
Fig. 5.
35S rDNA sequence characterization in Stylosanthes. (A) Alignment of the 35S rDNA unit including the transcribed and non-transcribed spacers of S. hamata, S. viscosa and S. scabra. (B) SeqGrapheR visualization of the entire 35S rDNA unit including reads of all three species. Note the close overlap of reads from S. scabra and S. viscosa, while S. hamata reads do not overlap along the ETS and IGS. Reads are coloured by species. Note the ‘knot’ (arrowed) which probably reflects repeats in the IGS. (C) Number of SNPs found across the entire 35S rDNA unit sorted by region. (D) Number of SNPs found across the entire 35S rDNA unit sorted by region per 100 bp.
Table 3.
Comparative analysis of the length of 35S rDNA regions among the three species of Stylosanthes
| Species | Length of 35S rDNA regions (bp) | ||||||
|---|---|---|---|---|---|---|---|
| ETS | 18S | ITS1 | 5.8S | ITS2 | 26S | IGS | |
| S. hamata | 1594 | 1809 | 197 | 164 | 219 | 3394 | ≥1502 |
| S. viscosa | 1518 | 1809 | 200 | 164 | 218 | 3394 | ≥1865 |
| S. scabra | 1430 | 1809 | 200 | 164 | 218 | 3394 | ≥1631 |
Table 4.
18S–ITS1–5.8S–ITS2–26S pairwise identity (%) among the three species of Stylosanthes
| Species | S. hamata | S. viscosa |
|---|---|---|
| Pairwise identity (%) | Pairwise identity (%) | |
| S. viscosa | 93.4 | – |
| S. scabra | 94.4 | 98.5 |
SeqGrapheR visualization for the 35S rDNA unit showed a situation comparable with 5S rDNA analysis (Fig. 5B; Supplementary Data Fig. S3). Stylosanthes viscosa and S. scabra sequence reads were strongly overlapped along the 35S rDNA unit, while S. hamata reads overlapped only along coding regions. Transcribed and non-transcribed spacers of S. hamata show a distinct distribution pattern (Fig. 5B; Supplementary Data Fig. S3). Moreover, Bayesian phylogenetic analysis based on the consensus sequences of 35S rDNA revealed a close relationship between sequences in S. viscosa and S. scabra (Fig. 3C). These results indicate that most of the 35S rDNA units in S. scabra were similar to a progenitor with S. viscosa 35S rDNA-like sequences.
The IGS regions of all three species showed a nucleotide tandem repeat array ranging from at least 507 bp in S. scabra to 743 bp in S. viscosa. The S. hamata repeat showed a consensus monomer size of 84 nucleotides, while S. viscosa and S. scabra showed a consensus of 86 nucleotides. Furthermore, direct repeats were also found in the ETS and IGS regions of all species (Supplementary Data Fig. S4). The exact length of the IGS could not be determined due to limitation of the repetitive sequence assembly. Furthermore, overall intragenomic SNP calling across 35S rDNA units from Stylosanthes revealed on average a low number of polymorphisms along the ETS–18S–ITS1–5.8S–ITS2–26S region (Fig. 5C, D). As expected non-conding regions showed the highest level of SNPs per 100 bp (Fig. 5D). SNPs along IGS regions were not evaluated because of their highly repetitive nature, as shown above.
Genome-wide SNV sharing between S. scabra and its progenitors
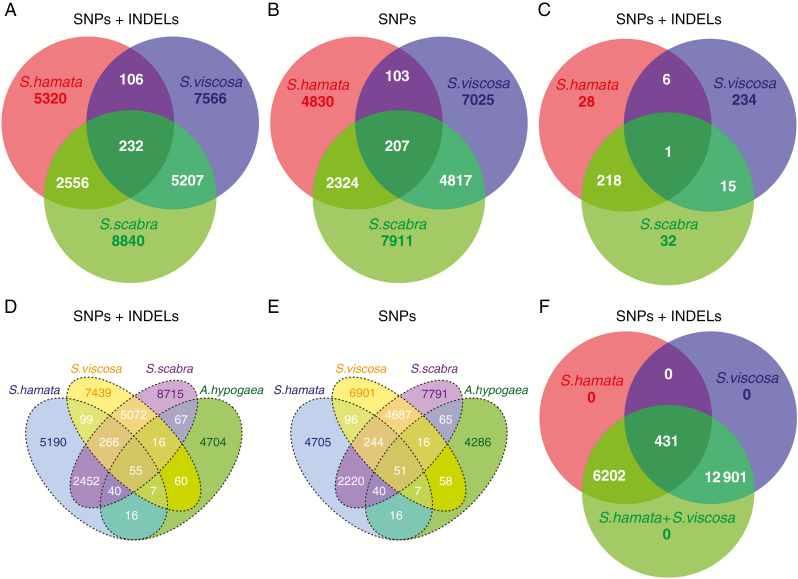
Genome-wide reference-free SNV detection with discoSnp++ has revealed that S. scabra shares a high content of isolated SNPs and INDELs with its most likely progenitors S. hamata and S. viscosa. We observed that S. hamata showed the lowest content of SNVs (8214), of which 106 were shared with S. viscosa and 2556 with S. scabra. Stylosanthes viscosa showed a higher content of SNVs (13 111), of which 5207 were shared with S. scabra. The highest level of identified variants was found in S. scabra (16 835). Only 232 variants were shared among all three samples (Fig. 6A). Despite both S. viscosa and S. hamata being diploids, the former genome showed a higher level of intragenomic SNVs (Fig. 6A). Analysis counting only SNPs, but excluding INDELs, showed similar results and revealed that most variations found are composed of SNPs (Fig. 6B; Supplementary Data Table S3). Additionally, we have also searched for plastome-wide SNVs among the three species. As expected, S. scabra shared a large quantity of SNVs (219; >80 %) with S. hamata, with both showing very few unique SNVs, while S. viscosa shared only very few SNVs with S. scabra (16; 6 %) (Fig. 6C), but showed a large number of unique SNVs (234). These results confirm our above conclusion that S. hamata and S. scabra plastomes are very similar.
Fig. 6.
Venn diagram showing the amount of genome-wide reference-free shared and unique SNVs detected in each Stylosanthes genome. (A) Genome-wide analysis of SNPs + INDELs with S. hamata, S. viscosa and S. scabra. (B) SNP only analysis with S. hamata, S. viscosa and S. scabra. (C) Plastome-wide analysis of SNPs + INDELs with S. hamata, S. viscosa and S. scabra. (D) Analysis of SNPs + INDELs with S. hamata, S. viscosa, S. scabra and A. hypogaea. (E) SNP only analysis with S. hamata, S. viscosa, S. scabra and A. hypogaea. (F) Genome-wide analysis of SNPs + INDELs with S. hamata, S. viscosa and a virtual hybrid (S. hamata + S.viscosa) as control.
To check whether SNVs were evaluated properly among the samples, we performed two controls. (1) The same analysis was carried out but also including A. hypogaea as a close outgroup. The results obtained confirmed our previous analysis and showed that even the genome of A. hypogaea, which is closely related to Stylosanthes, shared very few SNVs with the three Stylosanthes species used in the present work (Fig. 6D, E). Thus, most of the identified SNVs are indeed specific to Stylosanthes genomes and not an artefact. (2) As an internal control, we have mixed an equal number of reads from S. hamata and S. viscosa to simulate a virtual hybrid condition and performed the SNV analysis as above. Indeed, as expected, all SNVs were mapped back to the parental data sets while none of the SNVs was ‘unique’ for the virtual hybrid (Fig. 6F).
Estimating the age of allotetraploid S. scabra
Because we were interested in having an estimation of the age of S. scabra, we performed molecular dating with BEAST using ITS and plastid trnL–trnF regions. We observed that the ITS regions from S. viscosa and S. scabra were closely related and grouped together in the phylogenetic tree, with an estimated time of divergence at approx. 0.63 Mya, while S. hamata sequences were grouped in another clade (Fig. 7A). Indeed, the consensus ITS sequence obtained in the present work for S. hamata grouped together with a sample identified as S. seabrana (GenBank accession no. AJ320384), while other S. hamata sequences were grouped in a sister clade. In contrast, the plastid trnL–trnF assembled region for S. hamata and S. scabra grouped together, with an estimated time of divergence at 0.52 Mya, while other sequences from GenBank for S. hamata and S. scabra grouped in a sister clade. Sequences of S. viscosa were placed in a more distant clade (Fig. 7B). These results support an origin of S. scabra during the Middle Pleistocene (0.78–0.13 Mya).
DISCUSSION
Allotetraploid ancestry of S. scabra
The results confirm that S. scabra is an allotetraploid involving progenitor diploids with A (maternal) and B (paternal) genomes as previously proposed (Liu et al., 1999; Liu and Musial, 2001; Vander Stappen et al., 2002). Rates of divergence in plastid DNA and ITS sequences indicate that S. scabra formed in the Middle Pleistocene – which spans from 0.78 to 0.13 Mya – (with estimates being 0.63 or 0.52 Mya, respectively). In fact, this age is in agreement with the diversification of some plant lineages from neotropical savannas and seasonally dry forests in Central and North-eastern Brazil (Queiroz et al., 2017), the main centre of Stylosanthes diversification. Furthermore, the phylogenetic analyses based on our assembled sequences and GenBank accessions confirmed the close relationship of S. hamata and samples identified as S. seabrana, which grouped together in both ITS and plastid trees. This result reinforces the need for a better taxonomic review based on molecular studies for the genus. Nevertheless, because a considerable intraspecific genetic diversity has been described in Stylosanthes (Stace and Cameron, 1987; Liu et al., 1999), as we can also observe in the phylogeny (Fig. 7), we do not believe that intraspecific variation could affect the main conclusions of the study. In fact, the A and B genomes of Stylosanthes studied here were characterized as being fairly distinguishable from each other, since we were able to differentiate them by GISH, plastome, rDNA and genome-wide SNV analysis.
Since chloroplasts are considered to be maternally inherited in Stylosanthes (Liu and Musial, 2001), our combined plastome assembly and phylogenetic analysis revealed that the maternal genome donor of S. scabra is most probably closely related to a S. hamata-like genome due to their highly similar plastomes, which have even conserved the same length (156 502 bp). Because chloroplast genomes are non-recombining and uniparentally inherited, they are a valuable source of information for improving phylogenetic resolution and species delimitations (Dong et al., 2015; Biswal et al., 2017; Yin et al., 2017). Here we report for the first time the complete chloroplast genome sequence for three Stylosanthes species. The plastomes of Stylosanthes shared high similarity to the Arachis hypogaea plastome, showing >95 % pairwise identity. The similarity of these plastomes confirms a close phylogenetic relationship between the genera, as suggested previously (Cardoso et al., 2013). Furthermore, the fact that we were able to identify interspecific polymorphic SSRs in the plastomes of Stylosanthes shows that these data could serve as potential barcode markers for species discrimination, breeding programmes, genetic diversity of germplasm collections and for analyses of the genetic structure of natural populations (Santos et al., 2009a, b, c; Chandra et al., 2011; Santos-Garcia et al., 2012). Thus, these findings may contribute to the improvement of Stylosanthes.
Whilst the number of chromosomes in S. scabra (2n = 40) is additive of the parental, diploid dosage (both 2n = 20), the number of 35S rDNA units indicates the loss of a locus, most probably from the A genome origin, given that 35S rDNA units are most similar to the S. viscosa parent and that no 35S rDNA locus was found on S. hamata-labelled chromosomes. Similarly, for 5S rDNA, one locus is apparently much reduced in size. This small locus was also characterized to be of S. hamata origin based on GISH and given that most of the rDNA units identified in S. scabra are most similar to those of S. viscosa. Moreover, GISH works effectively on the chromosomes of S. scabra. However, some chromosomes of the complement are not entirely labelled with one or other of the genomic probes. GISH using both genomes as probes in the concentration ratio of 1:1 showed several chromosomes with large blocks of label from both parents, perhaps indicating intergenomic translocations or cross-hybridization of probes. Other chromosomes which were predominantly labelled with one genomic probe are heavily labelled around their (peri)centromeric regions with the other genomic probe. GISH using the genome of S. hamata as probe and blocking DNA of S. viscosa in the concentration ratio of 1:20 has increased genome differentiation. Thus, most probably cross-hybridization at (peri)centromeric regions may have occurred because of the similarity of repetitive sequences at these regions in the genomes of S. hamata and S. viscosa, which are also present in S. scabra. Indeed, similar centromeric repeats were found differentially to occupy the centromeres of soybean chromosomes, leading the authors to suggest a recent allopolyploidization event (Gill et al., 2009). A similar situation might be the case in S. scabra. Alternatively, these data might indicate intergenomic mobility of large blocks of repeats around the pericentromeric region, given that an interspersed arrangement of the different parental repeats would be expected to give a signal from both genomic probes. Such mobility of repeats between parental genomes (Zhao et al., 1998; Lim et al., 2007) has been reported previously for other allopolyploids, and may arise through little understood sequence homogenization processes.
Genome size results showed a reduction of 0.15 pg (approx. 9 %) in the genome of S. scabra of the expected sum of both S. hamata and S. viscosa genomes. Although we did not measure the genome size of other accessions of S. hamata and S. viscosa, the relatively recent origin of S. scabra (0.63–0.52 Mya) might be a reason why no great genome size reduction was observed and perhaps genome downsizing is still an ongoing process. Indeed, the loss/reduction of rDNA sites in S. scabra may have contributed to the observed genome downsizing. Genome size reduction in allopolyploids is well reported in the literature (Leitch and Bennett, 2004). Nicotiana young allopolyploids (<0.2 million years old) have shown similar genome downsizing to that found in S. scabra (Leitch et al., 2008), with loss of paternally inherited repeats and rDNA in Nicotiana tabacum (Renny-Byfield et al., 2011). In contrast, older allopolyploids (>4.5 million years old) may show a relative increase in DNA content (>16 %) associated with a replacement of ancestral repeats with new repeats and/or an expansion of ancestral repeats over time (Leitch et al., 2008; Renny-Byfield et al., 2013). Whether a similar situation is present in Stylosanthes will be a field to be explored in future work.
rDNA homogenization in S. scabra
In S. scabra, all 35S rDNAs are closely similar to those of S. viscosa and it is likely that the locus of S. hamata origin has been deleted. Uniparental inheritance, as a result of differential elimination of one of the parental rDNAs, can be accompanied by structural rearrangements of the rDNA to form new rDNA variants (Cronn et al., 1999; Volkov et al., 2007). Whilst the 35S rDNA units are similar between S. viscosa and S. scabra, they are not identical, and it is possible that small-scale mutations have amplified across multiple rDNA units at the remaining rDNA locus in S. scabra. Alternatively, the true parent of S. scabra may have had a slightly different 35S rDNA unit structure to that of the plant we sequenced. Similarly, for 5S rDNA, most units in S. scabra show high levels of similarity to units of S. viscosa origin, and the few units of S. hamata origin that remain may occur on a locus of reduced size. However, in S. viscosa, the B variant of 5S rDNA is most common, with the B’ variant occurring at a lower frequency (7.5 % of all variants). In contrast, in S. scabra, it is the B’ variant that is most common (74.4 % of all variants). As with 35S rDNA, the different abundances of variant types may be a consequence of either homogenization or variation between individuals in a population.
Distinguishing between the inheritance of variant types and homogenization, amplification and deletion processes after the polyploidy formed may prove difficult, and all these processes have been described previously in the context of allopolyploidy. In the natural allotetraploid Spartina anglica, which is of recent (within the last 120 years) origin and probably derived from a single allopolyploid event, different abundances of rDNA variants observed between individuals in a population are most likely to be due to homogenization, amplification and deletion processes across the rDNA array (Huska et al., 2016). In contrast, the natural, recent (within the last approx. 80 years) allopolyploidy events that gave rise to Tragopogon mirus and T. miscellus occurred on multiple occasions from different diploid individuals of the same species that had different 35S rDNA variants (Malinska et al., 2011; Dobesova et al., 2015). To better understand rDNA dynamics in Stylosanthes, it will be necessary to conduct similar population genetics studies and to investigate rDNA variants in multiple individuals in diploid and derived allopolyploid populations.
Intragenomic rDNA variation was found to be higher at non-coding spacer regions of both 5S and 35S rDNA in Stylosanthes, as has been shown in other plants (Matyasek et al., 2012; Lunerová et al., 2017). However, variant A of 5S rDNA, which has been found in the genomes of S. hamata and S. scabra, showed a relatively high number of intragenomic SNPs in the coding region in S. scabra, compared with the other variants in the species studied. The high level of SNPs found in 5S rDNA variant A in S. scabra could indicate that the variant is transcriptionally silent, and without function, or with reduced function, so that there is reduced selective pressure, leading to its pseudogenization (Wang et al., 2016; Volkov et al., 2017). The 5S variant that is probably generating the weak 5S rDNA signal on one of the chromosome pairs in S. scabra is probably this A variant. However, without analysis of the transcriptome, and detailed mapping of the variants at each rDNA locus, it is not possible to know where the active variants are residing. Indeed, in T. mirus, one individual had lost the majority of a variant type, yet the remaining copies were those that were most transcriptionally active (Dobesova et al., 2015).
Genomic heterogeneity of S. scabra
Generally, SNV detection methods use a reference genome. However, Stylosanthes as a non-model organism lacks a reference genome. Thus, we applied the discoSnp++ reference-free method (Uricaru et al., 2015; Peterlongo et al., 2017) to estimate genome-wide and plastome occurrence of shared and unique SNVs. This approach allowed us to compare SNV profiles among S. scabra and its putative progenitors S. hamata and S. viscosa. Overall evaluation showed that most SNVs were made up of SNPs, while a minor fraction comprised INDELs. SNPs are known to be of considerable importance because they have a much higher abundance in the genome and are often used to determine the population structure (Jain et al., 2014; Hu et al., 2015; Shen et al., 2017). Furthermore, INDELs may also be used for fine mapping and marker-assisted selection (Wang et al., 2012; Das et al., 2015).
The allotetraploid genome of S. scabra shared a high quantity of genome-wide isolated SNVs with its progenitors (almost 50 %), wherein around 16 % was shared with S. hamata and around 32 % with S. viscosa, while only about 1.3 % was shared among the three. In contrast, the diploid progenitors S. hamata and S. viscosa shared very few SNVs. Indeed, we observed in the three samples that a high number of SNVs were species specific, which in the case of S. scabra may account for its genome heterogeneity accumulated since its origin. As discussed above, the origin of S. scabra is something around 0.6–0.3 Mya. Thus, it is surprising to see so many S. scabra-specific SNVs (8840). Furthermore, the total amount of SNVs found in S. scabra accounts for >80 % of the total SNVs found in both diploids together. This could be due to either a fast-evolving genome or a ‘genomic shock’ after the allopolyploidy event (McClintock, 1984). It is known that changes can occur at the DNA sequence, epigenetic, karyotypic and transcription levels (Wendel, 2000; Renny-Byfield and Wendel, 2014). Thus, factors such as accumulation of repeats and/or pseudogenization of duplicated genes in the S. scabra genome would increase its nucleotide diversity, contributing the high level of specific intragenomic SNVs found. Moreover, the fact that S. hamata showed a much lower content of intragenomic SNVs compared with S. viscosa, despite the fact that both are diploids, indicates that the latter genome shows a higher rate of intragenomic heterogeneity. One possible explanation for such differences in the rate of intragenomic SNVs found in the diploid progenitors could have arisen from differential accumulation of repetitive sequences. When considering only plastome-matching reads, discoSnp++ showed, as expected, that most SNVs from S. hamata (86 %) and S. scabra (82 %) were shared, while S. viscosa shared only 6 % and 3 % of its plastome SNVs with S. scabra and S. hamata, respectively. These results reinforce the similarity of S. hamata and S. scabra plastomes as discussed above. The present approach used in our analysis may be a powerful tool in future works for assisting Stylosanthes breeding programmes and for rapid assessment of Stylosanthes genome heterogeneity.
CONCLUSION
This work presented for the first time conclusive evidence for the origin and genome characterization of the allotetraploid S. scabra. By combining cytogenetic and bioinformatic tools, we were able to characterize maternal and paternal genome donors of S. scabra and confirmed its previously suggested AABB genome composition. However, the precise parent of any established allopolyploid will always be an approximation, since both the diploid and the polyploid species have diverged since the allopolyploidy event. Furthermore, we report for the first time whole-plastome sequences for three Stylosanthes species, which are important sequence resources for future systematics and barcode marker studies in the genus. rDNA analysis has shown that S. scabra has undergone genome downsizing by eliminating rDNA copies from the A genome, while paternally inherited B genome rDNA copies were maintained and accumulated species-specific mutations. Finally, we show that the methodological approach used may help in elucidating the evolution and complex systematics of Stylosanthes, being extrapolatable to the study of the origin of other allopolyploid species in the genus.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: the distribution, type and presence of polymorphic SSRs among the whole plastomes of Stylosanthes. Figure S2: dotplot of 5S rDNA variants. Figure S3: SeqGrapheR visualization of RepeatExplorer 35S rDNA clusters fragmented in four sub-regions. Figure S4: repeat characterization of 35S rDNA in Stylosanthes. Table S1: number of reads, amount of data, ploidy level, genome size and coverage. Table S2: gene annotation found in the plastomes of Stylosanthes. Table S3: genome-wide reference-free SNV calling with discoSnp++.
ACKNOWLEDGEMENTS
This work was supported by the Alagoas State Research Support Foundation (FAPEAL) and by the Pernambuco Science and Technology Support Foundation (FACEPE) [grant no. APQ-0970-2.03/15]. A DCR fellowship was granted to A.M. by the National Council for Scientific and Technological Development (CNPq). The authors declare no conflict of interest.
LITERATURE CITED
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- Biswal D, Konhar R, Debnath M, Parameswaran S, Sundar D, Tandon P. 2017. Chloroplast genome sequence annotation of Dendrobium nobile (Asparagales: Orchidaceae), an endangered medicinal orchid from northeast India. PLoS Currents Tree of Life Edition 1. doi: 10.1371/currents.tol.cf1709613759c2223eb582c0fa694cc7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DF, Chakraborty S. 2004. Forage potential of Stylosanthes in different production systems. In: Chakraborty C, ed. High-yielding anthracnose-resistant Stylosanthes for agricultural systems. Canberra, Australia: Australian Centre for International Agricultural Research, 27–38. [Google Scholar]
- Cardoso D, Pennington RT, de Queiroz LP, et al. 2013. Reconstructing the deep-branching relationships of the papilionoid legumes. South African Journal of Botany 89: 58–75. [Google Scholar]
- Chandra A. 2013. Biotechnology of Stylosanthes. In: Jain S, Dutta Gupta S, eds. Biotechnology of neglected and underutilized crops. Dordrecht: Springer, 217–241. [Google Scholar]
- Chandra A, Kaushal P. 2009. Identification of diploid Stylosanthes seabrana accessions from existing germplasm of S. scabra utilizing genome-specific STS markers and flow cytometry, and their molecular characterization. Molecular Biotechnology 42: 282–291. [DOI] [PubMed] [Google Scholar]
- Chandra A, Tiwari KK, Nagaich D, Dubey N, Kumar S, Roy AK. 2011. Development and characterization of microsatellite markers from tropical forage Stylosanthes species and analysis of genetic variability and cross-species transferability. Genome 54: 1016–1028. [DOI] [PubMed] [Google Scholar]
- Chen H, Boutros PC. 2011. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Leitch AR, Soltis PS, Soltis DE. 2010. Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (Polyploidy/Hybridisation). Genes 1: 166–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cires E, Baltisberger M, Cuesta C, Vargas P, Prieto JAF. 2014. Allopolyploid origin of the Balkan endemic Ranunculus wettsteinii (Ranunculaceae) inferred from nuclear and plastid DNA sequences. Organisms, Diversity and Evolution 14: 1–10. [Google Scholar]
- Cronn RC, Small RL, Wendel JF. 1999. Duplicated genes evolve independently after polyploid formation in cotton. Proceedings of the National Academy of Sciences, USA 96: 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa LC, Valls JFM. 2010. Stylosanthes Sw. In: Forzza RC, et al, eds. Catálogo de plantas e fungos do Brasil vol. 2. Rio de Janeiro: Andrea Jakobsson Estúdio: Instituto de Pesquisas Jardim Botânico do Rio de Janeiro, 1090–1091. [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Upadhyaya HD, Srivastava R, et al. 2015. Genome-wide insertion–deletion (InDel) marker discovery and genotyping for genomics-assisted breeding applications in chickpea. DNA Research 22: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research 45: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobesova E, Malinska H, Matyasek R, et al. 2015. Silenced rRNA genes are activated and substitute for partially eliminated active homeologs in the recently formed allotetraploid, Tragopogon mirus (Asteraceae). Heredity 114: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Sgorbati S, Lucretti S. 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiologia Plantarum 85: 625–631. [Google Scholar]
- Dolezel J, Greilhuber J, Suda J. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols 2: 2233–2244. [DOI] [PubMed] [Google Scholar]
- Dong W, Xu C, Li C, et al. 2015. ycf1, the most promising plastid DNA barcode of land plants. Scientific Reports 5: 8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology & Evolution 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J, Pedrosa-Harand A. 2014. Lotus cytogenetics. In: Tabata S, Stougaard J, eds. The Lotus japonicus genome. Berlin Heidelberg: Springer-Verlag, 9–20. [Google Scholar]
- Ganley AR, Kobayashi T. 2007. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Research 17: 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastony GJ, Yatskievych G. 1992. Maternal inheritance of the chloroplast and mitochondrial genomes in cheilanthoid ferns. American Journal of Botany 79: 716–722. [Google Scholar]
- Gernhard T. 2008. The conditioned reconstructed process. Journal of Theoretical Biology 253: 769–778. [DOI] [PubMed] [Google Scholar]
- Gill N, Findley S, Walling JG, et al. 2009. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiology 151: 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlob-McHugh SG, Levesque M, MacKenzie K, Olson M, Yarosh O, Johnson DA. 1990. Organization of the 5S rRNA genes in the soybean Glycine max (L.) Merrill and conservation of the 5S rDNA repeat structure in higher plants. Genome 33: 486–494. [DOI] [PubMed] [Google Scholar]
- Greiner S, Sobanski J, Bock R. 2014. Why are most organelle genomes transmitted maternally?Bioessays 37: 80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemleben V, Werts D. 1988. Sequence organization and putative regulatory elements in the 5S rRNA genes of two higher plants (Vigna radiata and Matthiola incana). Gene 62: 165–169. [DOI] [PubMed] [Google Scholar]
- Hu J, Gui S, Zhu Z, Wang X, Ke W, Ding Y. 2015. Genome-wide identification of SSR and SNP markers based on whole-genome re-sequencing of a Thailand wild sacred Lotus (Nelumbo nucifera). PLoS One 10: e0143765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huska D, Leitch IJ, de Carvalho JF, et al. 2016. Persistence, dispersal and genetic evolution of recently formed Spartina homoploid hybrids and allopolyploids in Southern England. Biological Invasions 18: 2137–2151. [Google Scholar]
- Jain M, Moharana KC, Shankar R, Kumari R, Garg R. 2014. Genomewide discovery of DNA polymorphisms in rice cultivars with contrasting drought and salinity stress response and their functional relevance. Plant Biotechnology Journal 12: 253–64. [DOI] [PubMed] [Google Scholar]
- Jankowiak K, Rybarczyk A, Wyatt R, Odrzykoski I, Pacak A, Szweykowska-Kulinska Z. 2005. Organellar inheritance in the allopolyploid moss Rhizomnium pseudopunctatum. Taxon 54: 383–388. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology & Evolution 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A, Dadejova M, Lim YK, et al. 2008. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Annals of Botany 101: 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. 2004. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society 82: 651–663. [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, et al. 2008. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Annals of Botany, 101: 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GP, Schrire B, Mckinder B, Lock JM. 2005. The legumes of the world. Kew: Royal Botanic Gardens. [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, et al. 2007. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytologist 175: 756–763. [DOI] [PubMed] [Google Scholar]
- Liu CJ. 1997. Geographical distribution of genetic variation in Stylosanthes scabra revealed by RAPD analysis. Euphytica 98: 21–27. [Google Scholar]
- Liu CJ, Musial JM. 2001. The application of chloroplast DNA clones in identifying maternal donors for polyploid species of Stylosanthes. Theoretical and Applied Genetics 102: 73–77. [Google Scholar]
- Liu CJ, Musial JM, Thomas BD. 1999. Genetic relationships among Stylosanthes species revealed by RFLP and STS analyses. Theoretical and Applied Genetics 99: 1179–1186. [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research 41: W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Rodriguez E, Dolezel J, Santos C. 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Annals of Botany 100: 875–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunerová J, Renny‑Byfeld S, Matyášek R, Leitch A, Kovařík A. 2017. Concerted evolution rapidly eliminates sequence variation in rDNA coding regions but not in intergenic spacers in Nicotiana tabacum allotetraploid. Plant Systematics and Evolution 303: 1043–1060. [Google Scholar]
- Ma ZY, Chandra A, Musial JM, Liu CJ. 2004. Molecular evidence that Stylosanthes angustifolia is the third putative diploid progenitor of the hexaploid S. erecta (Fabaceae). Plant Systematics and Evolution 248: 171–176. [Google Scholar]
- Maass BL, Mannetje Lt. 2002. Stylosanthes seabrana (Leguminosae: Papilionoideae), a new species from Bahia, Brazil. Novon 12: 497–500. [Google Scholar]
- Maass BL, Sawkins M. 2004. History, relationships and diversity among Stylosanthes species of commercial significance. In: Chakraborty C, ed. High-yielding anthracnose-resistant Stylosanthes for agricultural systems. Canberra, Australia: Australian Centre for International Agricultural Research, 9–26. [Google Scholar]
- Malinska H, Tate JA, Mavrodiev E, et al. 2011. Ribosomal RNA genes evolution in Tragopogon: a story of New and Old World allotetraploids and the synthetic lines. Taxon 60: 348–354. [Google Scholar]
- Marques A, Ribeiro T, Neumann P, et al. 2015. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proceedings of the National Academy of Sciences, USA 112: 13633–13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyasek R, Renny-Byfield S, Fulnecek J, et al. 2012. Next generation sequencing analysis reveals a relationship between rDNA unit diversity and locus number in Nicotiana diploids. BMC Genomics 13: 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1984. The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- Nascimento MPSCB, Nascimento HTS, de Oliveira ME, Fernandes CD, Leal JA, Chakraborty S. 2001. Avaliação de acessos de Stylosanthes scabra Vog no Piauí. 38ª Reuniao Anual da Sociedade Brasileira de Zootecnia. Piracicaba: FEALQ, 184–185. [Google Scholar]
- Novak P, Neumann P, Macas J. 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Neumann P, Pech J, Steinhaisl J, Macas J. 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793. [DOI] [PubMed] [Google Scholar]
- Pathak PS, Ramesh CR, Bhatt RK. 2004. Stylosanthes in the reclamation and development of degraded soils in India. In: Chakraborty C, ed. High-yielding anthracnose-resistant Stylosanthes for agricultural systems. Canberra, Australia: Australian Centre for International Agricultural Research, 85–96. [Google Scholar]
- Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A. 2002. Chromosomal map of the model legume Lotus japonicus. Genetics 161: 1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlongo P, Riou C, Drezen E, Lemaitre C. 2017. DiscoSnp ++: de novo detection of small variants from raw unassembled read set(s). BioRxiv doi: 10.1101/209965 [DOI] [Google Scholar]
- Queiroz LP, Cardoso D, Fernandes MF, Moro MF. 2017. Diversity and evolution of the flowering plants of the Caatinga Domain. In: Silva JMC, Leal I, Tabarelli M, eds. Caatinga: The largest Tropical Dry Forest Region in South America. Cham: Springer, 23–63. [Google Scholar]
- Rambaut A. 2014. FigTree. v 1.4.2. [Google Scholar]
- Rambaut A, Suchard MA, Xie W, Drummond AJ. 2014. TRACER. v. 1.6. [Google Scholar]
- Reboud X, Zeyl C. 1994. Organelle inheritance in plants. Heredity 72: 132–140. [Google Scholar]
- Renny-Byfield S, Wendel JF. 2014. Doubling down on genomes: polyploidy and crop plants. American Journal of Botany 101: 1711–1725. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Chester M, Kovarik A, et al. 2011. Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Molecular Biology & Evolution 28: 2843–2854. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Kovarik A, Kelly LJ, et al. 2013. Diploidization and genome size change in allopolyploids is associated with differential dynamics of low- and high-copy sequences. The Plant Journal 74: 829–839. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MO, Karia CT, Resende RMS, et al. 2009. a Isolation and characterization of microsatellite loci in the tropical forage legume Stylosanthes guianensis (Aubl.) Sw. Conservation Genetics Research 1: 43–46. [Google Scholar]
- Santos MO, Sassaki RP, Chiari L, Resende RM, Souza AP. 2009b Isolation and characterization of microsatellite loci in tropical forage Stylosanthes capitata Vogel. Molecular Ecology Resources 9: 192–4. [DOI] [PubMed] [Google Scholar]
- Santos MO, Sassaki RP, Ferreira THS, et al. 2009. c Polymorphic microsatellite loci for Stylosanthes macrocephala Ferr. et Costa, a tropical forage legume. Conservation Genetics Research 1: 481–485. [Google Scholar]
- Santos-Garcia MO, Karia CT, Resende RMS, et al. 2012. Identification of Stylosanthes guianensis varieties using molecular genetic analysis. AoB Plants 2012: pls001. doi: 10.1093/aobpla/pls001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Särkinen T, Pennington RT, Lavin M, Simon MF, Hughes CE. 2012. Evolutionary islands in the Andes: persistence and isolation explain high endemism in Andean dry tropical forests. Journal of Biogeography 39: 884–900. [Google Scholar]
- Shen C, Jin X, Zhu, Lin Z. 2017. Uncovering SNP and indel variations of tetraploid cottons by SLAF-seq. BMC Genomics 18: 247. doi: 10.1186/s12864-017-3643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Mavrodiev EV, Doyle JJ, Rauscher J, Soltis PS. 2008. ITS and ETS sequence data and phylogeny reconstruction in allopolyploids and hybrids. Systematic Botany 33: 7–20. [Google Scholar]
- Song J, Shi L, Li D, et al. 2012. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS One 7: e43971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D. 1991. Structure and organization of ribosomal DNA. Biochimie 73: 631–638. [DOI] [PubMed] [Google Scholar]
- Stace HM, Cameron DF. 1984. Cytogenetics and the evolution of Stylosanthes. In: Stace MH, Edye LA, eds. The biology and agronomy of Stylosanthes. Sydney, Australia: Academic Press, 49–72. [Google Scholar]
- Stace HM, Edye LA, eds 1984. The biology and agronomy of Stylosanthes. Sydney, Australia: Academic Press. [Google Scholar]
- Stace HM, Cameron DF. 1987. Cytogenetic review of taxa in Stylosanthes hamata sensu lato. Tropical Grasslands 21: 182–188. [Google Scholar]
- Thiel T, Michalek W, Varshney RK, Graner A. 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 106: 411–22. [DOI] [PubMed] [Google Scholar]
- Uricaru R, Rizk G, Lacroix V, et al. 2015. Reference-free detection of isolated SNPs. Nucleic Acids Research 43: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Stappen J, Weltjens I, Munaut F, Volckaert G. 1999. Interspecific and progeny relationships in the genus Stylosanthes inferred from chloroplast DNA sequence variation. Comptes Rendus De L Academie Des Sciences Serie Iii-Sciences De La Vie-Life Sciences 322: 481–490. [DOI] [PubMed] [Google Scholar]
- Vander Stappen J, De Laet J, Gama-Lopez S, Van Campenhout S, Volckaert G. 2002. Phylogenetic analysis of Stylosanthes (Fabaceae) based on the internal transcribed spacer region (ITS) of nuclear ribosomal DNA. Plant Systematics and Evolution 234: 27–51. [Google Scholar]
- Vanni RO. 2017. The genus Stylosanthes (Fabaceae, Papilionoideae, Dalbergieae) in South America. Boletín de la Sociedad Argentina de Botánica 52: 549–585. [Google Scholar]
- Vanni RO, Fernandez A. 2011. The true identity of Stylosanthes seabrana BL Maass & L. ‘t Mannetje (Leguminosae Papilionoideae). Caryologia 64: 247–250. [Google Scholar]
- Volkov RA, Komarova NY, Hemleben V. 2007. Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression. Systematics and Biodiversity 5: 261–276. [Google Scholar]
- Volkov RA, Panchuk II, Borisjuk NV, Hosiawa-Baranska M, Maluszynska J, Hemleben V. 2017. Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna. BMC Plant Biology 17: 21. doi: 10.1186/s12870-017-0978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ma L, Becher H, et al. 2016. Astonishing 35S rDNA diversity in the gymnosperm species Cycas revoluta Thunb. Chromosoma 125: 683–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ren GF, Li XX, Tu JL, Lin ZX, Zhang XL. 2012. Development and evaluation of intron and insertion-deletion markers for Gossypium barbadense. Plant Molecular Biology Reporter 30: 605–613. [Google Scholar]
- Wendel JF. 2000. Genome evolution in polyploids. Plant Molecular Biology Reporter 42: 225–249. [PubMed] [Google Scholar]
- Williams RJ, Reid R, Schultze-Kraft R, Costa NMS, Thomas BD. 1984. Natural distribution of Stylosanthes. In: Stace HM, Edye LA, eds. The biology and agronomy of Stylosanthes. Sydney, Australia: Academic Press; 73–101. [Google Scholar]
- Yin D, Wang Y, Zhang X, Ma X, He X, Zhang J. 2017. Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Scientific Reports 7: 11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XP, Si Y, Hanson RE, et al. 1998. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Research 8: 479–492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.