Abstract
Histidine kinases (HKs) are primary sensor proteins that act in cell signaling pathways generically referred to as “two-component systems” (TCSs). TCSs are among the most widely distributed transduction systems used by both prokaryotic and eukaryotic organisms to detect and respond to a broad range of environmental cues. The structure and distribution of HK proteins are now well documented in prokaryotes, but information is still fragmentary for eukaryotes. Here, we have taken advantage of recent genomic resources to explore the structural diversity and the phylogenetic distribution of HKs in the prominent eukaryotic supergroups. Searches of the genomes of 67 eukaryotic species spread evenly throughout the phylogenetic tree of life identified 748 predicted HK proteins. Independent phylogenetic analyses of predicted HK proteins were carried out for each of the major eukaryotic supergroups. This allowed most of the compiled sequences to be categorized into previously described HK groups. Beyond the phylogenetic analysis of eukaryotic HKs, this study revealed some interesting findings: 1) characterization of some previously undescribed eukaryotic HK groups with predicted functions putatively related to physiological traits; 2) discovery of HK groups that were previously believed to be restricted to a single kingdom in additional supergroups, and 3) indications that some evolutionary paths have led to the appearance, transfer, duplication, and loss of HK genes in some phylogenetic lineages. This study provides an unprecedented overview of the structure and distribution of HKs in the Eukaryota and represents a first step toward deciphering the evolution of TCS signaling in living organisms.
Keywords: histidine kinases, cell signaling, two-component systems, eukaryotes, phylogenetic analysis
Introduction
Protein phosphorylation is one of the most widespread processes in living organisms and is commonly used to regulate numerous cell signaling pathways. Protein phosphorylation is mediated by enzymes belonging to the kinase superfamily. Kinases catalyze the transfer of a phosphate from ATP to various amino acids including serine/threonine (Ser/Thr kinases), tyrosine (Tyr kinases), and histidine (His kinases). To date, histidine kinases (HKs) have been described in archaea, bacteria, amoebae, plants, fungi, viruses, and bacteriophages (Delaroque et al. 2001; Mascher et al. 2006; Osakabe et al. 2013; Hargreaves et al. 2014; Hérivaux et al. 2016; Schaap et al. 2016; Galpérin et al. 2018). HK activity has also been detected in mammals (Attwood 2013), but mammalian genomes do not encode any typical bacterial or eukaryotic HK-like proteins. Consequently, approaches aiming at pharmacologically targeting HK proteins represent a promising strategy for the future development of antimicrobials in the context of human disease (Bem et al. 2015; Shor and Chauhan 2015).
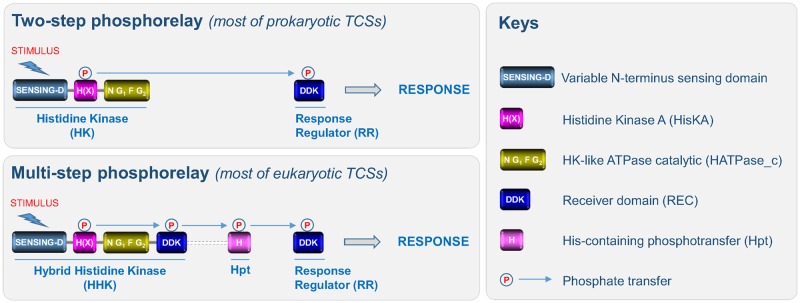
The canonical structure of HKs is composed of various domains (fig. 1). The first region corresponds to a highly variable N-terminal sequence that determines which stimulus is perceived by the HK. This region is referred to as the “sensing domain.” The central region is the transmitter domain consisting of both histidine kinase A (dimerization/phosphoacceptor) (HisKA) and cognate histidine kinase-like ATPase catalytic (HATPase_c) subdomains. The HisKA domain includes a H-box, usually containing a phosphorylatable histidine, and an X-box (Grebe and Stock 1999). The HATPase_c subdomain includes four distinct boxes: the N-, G1-, F-, and G2-boxes (Grebe and Stock 1999). In contrast to prokaryotic HKs, most eukaryotic HKs harbor an additional C-terminal receiver domain (REC) characterized by the presence of a three amino-acid signature (DDK) that includes a phosphorylatable aspartate residue. Thus, eukaryotic HKs are generally called hybrid HKs (Appleby et al. 1996).
Fig. 1.
—Domain organization and TCSs signaling. The canonical structure of HKs is composed of: 1) a highly variable N-terminal sequence that determines which stimulus is perceived by the HK (sensing-D); 2) and both histidine kinase A (H(X)) (including a phosphorylatable histidine residue in the H-box and an X-box) and histidine kinase-like ATPase catalytic (including four distinct boxes: the N-, G1-, F-, and G2-boxes) subdomains. In prokaryotes, TCSs usually consist of a two-step phosphorelay between an HK and an RR. Perception of a stimulus induces autophosphorylation of the HK which thus acts as a primary sensor, and the phosphate is then transferred to an RR which acts as a transcription factor to directly regulate a series of genes required for an adapted response. Most eukaryotic HKs harbor an additional C-terminal receiver domain (REC) characterized by the presence of a three amino acid signature (DDK). Thus, eukaryotic HKs are generally called hybrid HKs (HHKs). In eukaryotes, TCSs are composed of additional modules, such as the Hpt domain and the signaling routes that mediate the perception of stimuli correspond to a multistep phosphorelay between three families of proteins (HHK, Hpt, and RR). In most eukaryotic TCSs, the RR either acts directly as a transcription factor or governs downstream elements (MAPK cascades, the cAMP pathway, or Raf-like kinases). Note that in a few HKs, the Hpt domain is fused to the REC domain.
HKs are primary sensor proteins involved in cell signaling pathways generically referred to as “two-component systems” (TCSs) (fig. 1). TCSs are among the most widely distributed transduction systems used by both prokaryotic and eukaryotic organisms to detect and respond to a wide range of environmental cues. In prokaryotes, TCSs usually involve communication between two functional modules, that is, HKs and response regulators (RRs), via phosphorylation events (fig. 1). The perception of a stimulus induces autophosphorylation of the HK, and the phosphate is then transferred to an RR which acts as a transcription factor to directly regulate a series of target genes required for an adapted response (Appleby et al. 1996). In eukaryotes, TCSs are composed of additional modules, such as the histidine-containing phosphotransfer (Hpt) domain and the signaling routes that mediate the perception of stimuli involve a multistep phosphorelay between three families of proteins: HK → Hpt → RR (fig. 1). In most eukaryotic TCSs, the RR either acts directly as a transcription factor or regulates downstream elements such as mitogen-activated protein kinase (MAPK) cascades or cyclic adenosine monophosphate (cAMP) signaling systems (Hérivaux et al. 2016). In plants, some HKs are known to directly regulate other types of signaling proteins such as Raf-like kinases (Clark et al. 1998). Finally, few other variations in the architecture of TCSs have been described in some models (Jung et al. 2012).
It is now well documented that prokaryotic HKs govern a wide range of crucial processes, including nutrient acquisition, various metabolic pathways (nitrogen, glucose, cell wall biosynthesis), adaptation to changes in the environment (osmolarity, light, chemotaxis, temperature, oxygen), developmental pathways (sporulation, biofilm formation, competence), virulence, antibiotic resistance, and many others (e.g., flagellar assembly, secretion systems) (Mascher et al. 2006). In amoebae, HKs regulate a large panel of fundamental processes during multicellular development such as motility, morphogenesis, osmoregulation, as well as spore differentiation, encapsulation and dormancy (Du et al. 2015). In plants, HKs participate in the regulation of female gametophyte development and vegetative growth, as osmosensors by regulating drought, salt-stress and stomatal closure responses, and are involved in light and phytohormones (cytokinin and ethylene) perception and signal transduction (Osakabe et al. 2013). Finally, HKs are also widespread in fungi and to date have been reported to be involved in stress adaptation (high osmolarity and oxidant conditions), red-light perception, morphogenesis, and virulence (Hérivaux et al. 2016)
While the structure and distribution of these sensing proteins are now well documented in prokaryotes (Kim and Forst 2001; Mascher et al. 2006; Galpérin et al. 2018), information is still fragmentary for eukaryotes. Plant and fungal HKs are currently categorized into 7 and 16 groups, respectively (Schaller et al. 2008; Defosse et al. 2015). Some studies exploring the structure of HKs in Dictyosteliaceae, Physaraceae, and Protosteliaceae initiated the establishment of a classification of these sensing proteins in amoebae (Eichinger et al. 2005; Heidel et al. 2011; Schaap et al. 2016; Hillmann et al. 2018). Thus, although recent advances have provided a preliminary insight into the distribution of HKs in fungi, plants, and amoebae, the underlying mechanisms that drive the marked diversity among eukaryotic HKs, the number of genes, the structure, and the functional variations remain largely obscure. More precisely, the evolutionary paths that led to the appearance, transfer, duplication, and loss of HK genes have not been addressed broadly across the eukaryotes. In the present work, we have taken advantage of recent genomic resources to explore the structural diversity and the phylogenetic distribution of HKs in the six prominent eukaryotic clades: Archaeplastida (and sister clades Cryptophyta and Haptophyta), Opisthokonta, Amoebozoa, Apusozoa, Excavata, and SAR (Stramenopiles, Alveolates, and Rhizaria) (Grant and Katz 2014). This study represents an essential step toward deciphering the evolution of TCS cell signaling pathways in eukaryotes.
Materials and Methods
HK Sequence Search, Analysis, and Annotation
BLAST (Altschul et al. 1990) searches were performed against the NCBI and JGI genome sequence databases. The protein domains HisKA (PFAM00512) and HATPase_c (PFAM02518) were used as queries against both databases without threshold constraints. In order to eliminate redundant protein sequences, a multiple alignment was performed using the Clustal Omega algorithm (Sievers et al. 2014). After annotation, domain organization was predicted for each unique protein using the SMART (Schultz et al. 1998) and the Conserved Domain Database (Marchler-Bauer et al. 2017) algorithms. The TMHMM (Krogh et al. 2001) algorithm was used to predict transmembrane helices in the predicted proteins. Importantly, following the domain analysis, we only retained putative HK sequences that possessed at least two of the three following domains: HisKA (PFAM00512), HATPase_c (PFAM02518), and REC (PFAM00072). HK protein sequences were annotated using the first letter of the genus followed by the first four letters of the species, HK, and the number of the sequence (e.g., KniteHK1 for Klebsormidium nitens Histidine Kinase 1) except for Arabidopsis thaliana, Candida albicans, Cryptococcus neoformans, Oryza sativa, and Dictyostelium discoideum HKs for which the previously published nomenclature was conserved.
Phylogenetic Analysis
Alignments were built with Cobalt 2.0.2 (Papadopoulos and Agarwala 2007), applying an additional constraint on conserved domains that were compared with the CDD domain bank v0.75. The alphabet chosen was of the regular type and a maximum distance between two sequences of 0.7 was chosen (fraction of the number of words in common to all the words). The resulting alignments were curated using Gblocks 0.91b (Talavera and Castresana 2007) to remove nonconserved positions that could prevent correct reconstruction of the phylogeny. Several Gblocks parameter values were tested to retain between 11% and 13% of the positions in each initial alignment. For each alignment, the minimum length of a block was set to four positions and all positions corresponding to a gap in one block were retained. The parameters related to 1) the number of sequences required to consider a position as being conserved, 2) the number of sequences required to consider a position as being flanking, and 3) the maximum number of contiguous nonconserved positions that were optimized to define each block (Opisthokonta: 102, 102, 100; Archaeplastida and sister clades: 116, 116, 100; Excavata: 16, 26, 30; Amoebozoa: 64, 64, 175; SAR: 74, 79, 115; Apusozoa: 6, 6, 50; respectively). A phylogeny was constructed using the curated alignments with the package “Phangorn” (Schliep 2011) in R (R Development Core Team, 2013). A Neighbor-Joining tree was used as a primer tree, and distances between sequences were fitted using the maximum likelihood approach and the Le and Gascuel (2008) substitution matrix after several substitution models had been tested using the “Modeltest” function with both invariant sites and the gamma distribution. We retained the LG model (the model of Le and Gascuel 2008). which was always the most appropriate as determined by its Bayesian information criterion. Branch position was optimized with a nearest neighbor interchange algorithm. Branch robustness was evaluated using parametric resampling (bootstrap of 100) carried out in multicore computing machines of the CCSC (CNRS, Orléans, France).
Results
Exploring the Structure and Distribution of HKs in Eukaryota
In order to acquire a representative sampling to study the structure and distribution of HKs in the Eukaryota, we browsed the genomes of 67 species (table 1) spread throughout the eukaryotic tree of life (Grant and Katz 2014). Most eukaryotic HKs are hybrid, containing a C-terminal REC domain downstream of HisKA and HATPase_c domains, but there are some examples of eukaryotic HKs that lack the REC domain. This is the case, for example, of some diverged groups of HKs, such as plant phytochromes, ERS2-type ethylene receptors, and chloroplast TCS sensors. These proteins progressively acquired serine threonine kinase activity and non-RR functional partners during evolution (Cashmore 1998; Puthiyaveetil and Allen 2009; Schaller et al. 2011; Gallie 2015). We therefore searched all 67 genomes with the Pfam domains HisKA (PFAM00512) and HATPase_c (PFAM02518) and only retained putative HK sequences that had at least two out of the three following domains: HisKA (PFAM00512), HATPase_c (PFAM02518), and REC (PFAM00072). This strategy not only helped to remove a number of non-HK sequences that contain just a single HATPase_c domain, such as, for instance, pyruvate dehydrogenases, heat shock proteins 90, and DNA mismatch repair proteins MutL, but also provided the opportunity to capture divergent proteins related to the HK family. This allowed the compilation of 748 predicted proteins (table 1 and supplementary table S1, Supplementary Material online). The HK family is known to be highly divergent, even within a single kingdom (Mascher et al. 2006; He et al. 2016; Hérivaux et al. 2017). Thus, independent phylogenetic analyses of HK predicted proteins were carried out for each of the six major eukaryotic supergroups: Archaeplastida, Opisthokonta, Amoebozoa, Apusozoa, Excavata, and SAR (Grant and Katz 2014).
Table 1.
List of Eukaryotic Species in Which the Presence of Predicted HK Sequences Was Investigated for This Study
| Clades | Simplified Classification | Species Name | Strain Name | No. of HKs | ||
|---|---|---|---|---|---|---|
| Archaeplastida | Glaucocystophyta | Cyanophora paradoxa | CCMP329 | 1 | ||
| Rhodophyta | Cyanidioschizon merolae | 10D | 1 | |||
| Viridiplantae | Chlorophyta | Prasinophytes | Micromonas pusilla | CCMP1545 | 3 | |
| Chlorophyceae | Volvox carteri | Eve | 7 | |||
| Streptophyta | Charophytes | Klebsormidium nitens | NIES-2285 | 67 | ||
| Bryophyta | Physcomitrella patens | – | 54 | |||
| Marchantiophytes | Marchantia polymorpha | – | 20 | |||
| Lycopodiopsida | Selaginella moellendorffii | – | 16 | |||
| Acrogymnospermae | Pinus taeda | – | 10 | |||
| Magnoliophyta | Amborella trichopoda | – | 9 | |||
| Magnoliophyta | Arabidopsis thaliana | – | 16 | |||
| Magnoliophyta | Oryza sativa | P0709F06 | 13 | |||
| Cryptophyta | Guillardia theta | CCMP2712 | 10 | |||
| Haptophyta | Emiliana huxleyi | CCMP1516 | 3 | |||
| Opisthokonta | Metazoa | Eumetazoa | Cnidaria | Nematostella vectensis | CH2xCH6 | 0 |
| Arthropoda | Drosophila melanogaster | – | 0 | |||
| Platyhelminthes | Schistosoma mansoni | Puerto Rico | 0 | |||
| Chordata | Homo sapiens | – | 0 | |||
| Choanoflagellates | Monosiga brevicollis | MX1 | 1 | |||
| Mesomycetozoa | Capsaspora owczarzaki | ATCC30864 | 11 | |||
| Dikarya | Basidiomycota | Pucciniomycotina | Puccinia graminis | CDL75–36-700–3 | 5 | |
| Ustilaginomycotina | Ustilago maydis | 521 | 6 | |||
| Agaricomycotina | Cryptococcus neoformans | JC21 | 7 | |||
| Ascomycota | Pezizomycetes | Tuber melanosporum | Mel28 | 8 | ||
| Orbiliomycetes | Arthrobotrys oligospora | ATCC24927 | 17 | |||
| Eurotiomycetes | Aspergillus fumigatus | Af293 | 13 | |||
| Lecanoromycetes | Usnea florida | ATCC18376 | 10 | |||
| Leotiomycetes | Botrytis cinerea | T4 | 20 | |||
| Sordariomycetes | Fusarium verticillioides | ATCC38932 | 16 | |||
| Xylonomycetes | Xylona heveae | TC161 | 10 | |||
| Saccharomycotina | Candida albicans | SC5314 | 3 | |||
| Taphrinomycotina | Taphrina deformans | PYCC5710 | 8 | |||
| EDF | Mucoromycota | Glomeromycotina | Rhizophagus irregularis | DAOM181602 | 8 | |
| Mortierellomycotina | Mortierella elongata | AG-77 | 13 | |||
| Mucoromycotina | Umbelopsis ramanniana | AG | 12 | |||
| Zoopagomycota | Zoopagomycotina | Syncephalis plumigaleata | NRRL S24 | 3 | ||
| Entomophthoromycotina | Conidiobolus coronatus | NRRL28638 | 6 | |||
| Kickxellomycotina | Ramicandelaber brevisporus | CBS 109374 | 2 | |||
| Blastocladiomycota | Catenaria anguillulae | PL171 | 7 | |||
| Chytridiomycota | Gonapodya prolifera | JEL478 | 12 | |||
| Neocallimastigomycota | Piromyces finnis | E2 | 0 | |||
| Microsporidia | Enterocytozoon bieneusi | H348 | 0 | |||
| Cryptomycota | Rozella allomyces | CSF55 | 5 | |||
| Amoebozoa | Discosea | Longamoebia | Centramoebida | Acanthamoeba castellani | Neff | 48 |
| Mycetozoa | Myxogastria | Myxogastromycetidae | Physarum polycephalum | LU352 | 51 | |
| Dictyostelids | Dictyosteliales | Dictyostelium discoideum | AX4 | 14 | ||
| Acytosteliales | Polysphondylium pallidum | PN500 | 13 | |||
| Archamoebae | Entamoeba histolytica | HM-1: IMSS | 0 | |||
| Apusozoa | Thecamonas trahens | ATCC50062 | 11 | |||
| Excavata | Heterolobosea | Naegleria gruberi | NEG-M | 24 | ||
| Euglenozoa | Leishmania infantum | JPCM5 | 0 | |||
| Trypanosoma brucei | DAL972 | 0 | ||||
| Fornicata | Diplomonadida | Spironucleus salmonicida | ATCC 50377 | 0 | ||
| Giardia intestinalis | ATCC 50803 | 0 | ||||
| Parabasalia | Trichomonas vaginalis | G3 | 7 | |||
| SAR | Stramenopiles | Pelagophyceae | Pelagomonadales | Aureococcus anophagefferens | CCMP1984 | 2 |
| Bacillariophyta | Bacillariophyceae | Fistulifera solaris | DA0580 | 34 | ||
| PX clade | Phaeophyceae | Ectocarpus siliculosus | Ec32 | 13 | ||
| Oomycetes | Albuginaceae | Albugo laibachii | Nc14 | 2 | ||
| Alveolata | Apicomplexa | Haemosporida | Plasmodium falciparum | 3D7 | 0 | |
| Piroplasmida | Babesia bovis | T2Bo | 0 | |||
| Coccidia | Toxoplasma gondii | ARI | 0 | |||
| Coccidia | Cryptosporidium hominis | TU502 | 0 | |||
| Dinophyceae | Symbiodinium microadriaticum | CCMP2467 | 0 | |||
| Chromerida | Vitrella brassicaformis | CCMP3155 | 7 | |||
| Perkinsea | Perkinsus marinus | CB5D4 | 3 | |||
| Ciliophora | Tetrahymena thermophila | SB210 | 70 | |||
| Rhizaria | Cercozoa | Plasmodiophora brassicae | e3 | 16 | ||
| Foraminifera | Reticulomyxa filosa | — | 0 | |||
Note.—EDF, early diverging fungi.
Improving the Categorization of HK Proteins in Archaeplastida, Opisthokonta, and Amoebozoa
Archaeplastida and Sister Clades (Cryptophyta and Haptophyta)
Several recent studies have provided important advances in our understanding of the structure and distribution of land plant HKs (Schaller et al. 2008; Yasumura et al. 2012; Gruhn et al. 2014; Liu et al. 2014). However, there is a need to extend this analysis to the whole green lineage (Archaeplastida), which encompasses glaucophytes (Glaucocystophyta), red algae (Rhodophyta), green algae (Chlorophyta), and land plants (Streptophyta), together with the sister clades Cryptophyta and Haptophyta in some phylogenies (Grant and Katz 2014). Genome searches revealed that HKs are broadly represented throughout all these clades since we identified sequences in the Glaucocystophyta (Cyanophora paradoxa, one HK) (Price et al. 2012) and the Rhodophyta (Cyanidioschyzon merolae, one HK) (Matsuzaki et al. 2004). We also identified three and seven HK sequences from the genomes of the aquatic green algae Micromonas pusilla (Prasinophytes) (Worden et al. 2009) and Volvox carteri (Chlorophyceae) (Prochnik et al. 2010), respectively, 67 HKs in the genome of K.nitens (Charophyta, terrestrial green alga) (Hori et al. 2014), 54 HKs in the genome of Physcomitrella patens (Bryophytes) (Rensing et al. 2008), 20 HKs in the genome of Marchantia polymorpha (Liverworts) (Bowman et al. 2017), and 16 HKs in the genome of Selaginella moellendorffii (Lycophytes) (Banks et al. 2011). The Acrogymnospermae clade was represented by the loblolly pine genome (Pinus taeda) (Neale et al. 2014) in which we annotated ten HKs. Concerning Magnoliophyta (flowering plants), we identified nine HKs in Amborella trichopoda (Amborella Genome Project 2013), a model early-diverging species from the Magnoliophyta, to which we added the 16 previously described HKs from A.thaliana (a Eudicot model species) (Hwang et al. 2002) and the 13 HKs from O.sativa (a Monocot model species) (Pareek et al. 2006). Finally, we identified ten and three HKs in the genomes of representative species from the sister clades Cryptophyta (Guillardia theta) (Curtis et al. 2012) and Haptophyta (Emiliania huxleyi) (Read et al. 2013), respectively (table 1).
To gain insight into the phylogenetic relationships between HKs from glaucophytes (Glaucocystophyta), red algae (Rhodophyta), green algae (Chlorophyta), land plants (Streptophyta), and the sister clades Cryptophyta and Haptophyta, we generated a robust phylogenetic tree after multiple alignment of 230 predicted sequences (table 1 and supplementary table S1, Supplementary Material online). This analysis revealed that most of the deduced HK sequences tended to cluster in the previously described major plant HK groups (fig. 2) (Schaller et al. 2008; Ishida et al. 2010). These included the AHK1 group, known to act as osmosensors in Eudicots (Urao 1999; Chefdor et al. 2006), the CKI1 group (Kakimoto 1996), which constitutes key regulators of central cell specification in the Arabidopsis female gametophyte (Pischke et al. 2002), the CKI2 group (Kakimoto 1996), which was reported to be involved in stress-induced stomatal closure (Mira-Rodado et al. 2012), the phytochrome group (Schaller et al. 2008), which consists in photoswitchable red/far-red photoreceptors regulating competition with neighboring plants for photosynthetically active red light (Rockwell and Lagarias 2017), the ethylene receptor group (ETR), which has been divided into two subfamilies (Gallie 2015) that display the three predicted transmembrane helices containing the crucial residues involved in ethylene perception (Wang et al. 2006), the cytokinin receptor group (CKR), which consists of proteins displaying a cyclases/histidine kinase-associated sensing extracellular (CHASE) domain in their N-terminal region (Anantharaman and Aravind 2001; Mougel and Zhulin 2001) known to be essential for the binding of the hormones (Gruhn et al. 2014), and the more recently described membrane-associated sensor 1 (MASE1) domain-containing HKs of unknown function (Ishida et al. 2010). Most of these HK groups, including AHK1, CKR, and ETR, were found exclusively in the Streptophyta (K. nitens, P. patens, Mar. polymorpha, S. moellendorffii, Pi. taeda, Am. trichopoda, A. thaliana, and O. sativa) (fig. 2). It is noteworthy that most of the CHASE domain-containing HKs predicted from the K. nitens and P. patens genomes tended to cluster in the subfamily 2 of plant CKRs (Gruhn et al. 2014) whereas K. nitens KniteHK9 and KniteHK48 did not appear to cluster with the two previously described subfamilies, thus defining a new subfamily of putative CKRs in Archaeplastida (fig. 2). The MASE1 domain-containing HK group appeared to be restricted to early diverging land plants (P. patens, Mar. polymorpha, and S. moellendorffii). Finally, this phylogenetic analysis reveals that the CKI1 and CKI2 groups are restricted to land plants and that, interestingly, the CKI1 group has likely emerged from a structural specialization of AHK1-like ancestors (fig. 2).
Fig. 2.
—Phylogeny estimation of HK predicted protein sequences in Archaeplastida. A representative structure is provided for each defined HK group. Athal, Arabidopsis thaliana; Atric, Amborella trichopoda; Cmero, Cyanidioschyzon merolae; Cpara, Cyanophora paradoxa; Ehuxl, Emiliania huxleyi; Gthet, Guillardia theta; Knite, Klebsormidium nitens; Mpusi, Micromonas pusilla; Osati, Oryza sativa; Ppate, Physcomitrella patens; Ptaed, Pinus taeda; Smoel, Selaginella moellendorffii; Vcart, Volvox carteri.
In addition to these observations, several interesting advances were provided by the phylogenetic analysis. First, the genome of the red alga Cy. merolae encodes a unique HK (CmeroHK1) that belongs to the chloroplast sensor kinase (CSK) group (Puthiyaveetil et al. 2008) (fig. 2). The first nuclear-encoded CSK was described in A. thaliana. CSK genes are believed to have originated from cyanobacterial histidine kinase 2 (Hik2). CSK homologs are present in all major lineages of photosynthetic eukaryotes, but they were not included in the present study because they lack a typical HisKA Pfam domain. A unique aspect of CSK evolution is that these proteins occur as a canonical HKs along with an RR partner in nongreen algae but in green algae and plants they are modified HKs that have lost the conserved histidine residue and its cognate RR partner. As is also the case for land plant ethylene receptors, plant CSK has been rewired during evolution to work with a non-RR functional partner, in the case of CSKs to link chloroplast gene expression with photosynthetic electron transport (Puthiyaveetil and Allen 2009; Puthiyaveetil et al. 2010, 2013). Glaucocystophyta CparaHK1 shares high similarity with several cyanophytochromes (not shown) and tended to cluster in the tree with Streptophyta phytochromes (fig. 2). This is in accordance with previous analyses demonstrating that despite the lack of detectable sequence similarity, Archaeplastida phytochromes originated from cyanophytochromes (Kooß and Lamparter 2017). CparaHK1 was previously shown to act as a phytochrome, detecting light broadly throughout the visible spectrum (blue, green, orange, red, and far-red) in contrast to land plant phytochromes that only use the ratio of red to far-red light to detect shading by neighboring plants and to control photomorphogenesis (Rockwell et al. 2014). Few HKs were found in green algae (e.g., M. pusilla, three HKs), including notably a single phytochrome (MpusiHK1) (Duanmu et al. 2014), which appears to be distantly related to land plant phytochromes (fig. 2). The two other M. pusilla HKs (MpusiHK2 and MpusiHK3) are predicted to have multiple transmembrane regions within their N-terminus and therefore correspond to a previously undescribed HK group that displays similarity to sequences from the Cryptophyta (G. theta, GthetHK3, GthetHK5, GthetHK8, and GthetHK10) (fig. 2).
Some HKs from the Archaeplastida sister clades Cryptophyta (G. theta) and Haptophyta (E. huxleyi) were also of interest (fig. 2). These included not only G. theta GthetHK6, which clustered with the Archaeplastida phytochrome group, and GthetHK2, GthetHK7, and GtethHK9 that harbor a combination of PAS/PAC domains (Per-period circadian protein; Arnt-Ah receptor nuclear translocator protein; Sim-single minded protein) (Möglich et al. 2009) within their N-termini (referred here as multi-PAS HKs). We also detected GthetHK1 and EhuxlHK3, which are members of the Hik33/ycf26 group of HKs. This class of HKs is known to participate in the chloroplast TCS (Suzuki et al. 2001; Puthiyaveetil and Allen 2009).
One important finding was that both K. nitens, a model species representing the early transition step from aquatic algae to land plants, and, to a lesser extent, basal Embryophyta species (P. patens, Mar. polymorpha, and S. moellendorffii) possessed a broad range of HKs, some of which had features that have not been described for land plant HKs. For example, KniteHK26 and KniteHK43 share the same domain topology and cluster with the bacteriorhodopsins VcartHK1, VcartHK3, VcartHK4, VcartHK5, and VcartHK6 from the green alga V. carteri (Kianianmomeni and Hallmann 2014) (fig. 2 and supplementary fig. S1, Supplementary Material online). Homologs of these proteins, which are located in the eyespot of Chlamydomonas (another green alga), play a crucial role in the adaptation of behavioral responses in the presence of UV-A light (Kateriya et al. 2004; Troein et al. 2011; Luck et al. 2012). Five other HKs, KniteHK15, KniteHK19, KniteHK23, KniteHK28, and KniteHK34, possess an N-terminal sensing region that shares domain topology and significant similarity with the light, oxygen, or voltage (LOV) domain found in blue-light photoreceptors from cyanobacteria and chlorophyta (supplementary fig. S2, Supplementary Material online, and fig. 2) (Crosson et al. 2003; Glantz et al. 2016). This similarity suggests that green algae retained this type of light-sensing HK protein during the first steps of terrestrialization but that it was later lost in land plants and replaced by cryptochromes and phototropins, allowing them to perceive and to adapt to UV-A and blue light (Sullivan and Deng 2003). Another interesting finding was that KniteHK29, KniteHK44, SmoelHK12, SmoelHK15, and PpateHK38 possess an N-terminal sensing region that shares domain topology and significant similarity with bacterial NarX nitrate/nitrite sensors (Mascher et al. 2006) (fig. 2 and supplementary fig. S3, Supplementary Material online). It is thus possible that such transduction systems were acquired via horizontal gene transfer during the evolution of green algae and allowed adaptation to the particular form of nitrogen source offered by soils during the terrestrialization process. It is also interesting to note that KniteHK1 and KniteHK2 correspond to dual HKs, which have hitherto only been reported in fungi and the Amoebozoa (fig. 2) (Defosse et al. 2015; Schaap et al. 2016). Finally, it was also surprising to observe, for the first time in the green lineage, members belonging to the prominent fungal group X HK (KniteHK20, KniteHK56, and KniteHK59) (fig. 2). A previous study suggested that these HKs were also acquired by fungi during the land colonization process allowing their adaptation to the particular oxidant conditions imposed by terrestrial habitats (Hérivaux et al. 2017).
Opisthokonta
Opisthokonta represents a supergroup of morphologically highly diversified species, including animals (Metazoa), choanoflagellates, Mesomycetozoa, and fungi. Although some previous studies have suggested the presence of HKs in some animals (Putnam et al. 2007; Hughes and Friedman 2011; Simakov et al. 2013; You et al. 2013; Zhang et al. 2013; Zhou et al. 2013; Gulia-Nuss et al. 2016; Neave et al. 2017), our analysis demonstrated that all metazoan HK candidates (about 150 sequences) found in the NCBI database corresponded either to an inappropriate annotation or to contamination by bacterial sequences (not shown). Thus, our analysis confirms that animals do not possess any typical eukaryotic HKs.
However, HKs appear to occur broadly in the remaining Opisthokonta clades because one predicted protein was identified in a representative species of choanoflagellate (Monosiga brevicollis) (King et al. 2008) and 11 HKs were found in the mesomycetozoan model species Capsaspora owczarzaki (Suga et al. 2013). We also included HK sequences from 23 broadly distributed species from the kingdom Fungi in this phylogenetic analysis (Grigoriev et al. 2014). These included the Basidiomycota species Puccinia graminis (Pucciniomycotina) (Duplessis et al. 2011), Ustilago maydis (Ustilaginomycotina) (Kamper et al. 2006), and Cr.neoformans (Agaricomycotina) (Loftus et al. 2005); the Ascomycota species Tuber melanosporum (Pezizomycetes) (Martin et al. 2010), Arthrobotrys oligospora (Orbiliomycetes) (Yang et al. 2011), Aspergillus fumigatus (Eurotiomycetes) (Nierman et al. 2005), Usnea florida (Lecanoromycetes), Botrytis cinerea (Leotiomycetes) (Staats and van Kan 2012), Fusarium verticillioides (Sordariomycetes) (Cuomo et al. 2007), Xylona heveae (Xylonomycetes) (Gazis et al. 2016), C.albicans (Saccharomycotina) (Jones et al. 2004), and Taphrina deformans (Taphrinomycotina) (Cisse et al. 2013); and the early diverging fungi Rhizophagus irregularis Glomeromycotina) (Tisserant et al. 2013), Mortierella elongata (Mortierellomycotina) (Uehling et al. 2017), Umbelopsis ramanniana (Mucoromycotina) (Hérivaux et al. 2017), Syncephalis plumigaleata (Zoopagomycotina), Conidiobolus coronatus (Entomophthoromycotina) (Chang et al. 2015), Ramicandelaber brevisporus (Kickxellomycotina) (Hérivaux et al. 2017), Catenaria anguillulae (Blastocladiomycota) (Mondo et al. 2017), Gonapodya prolifera (Chytridiomycota) (Chang et al. 2015), Piromyces finnis (Neocallimastigomycota) (Haitjema et al. 2017), Enterocytozoon bieneusi (Microsporidia) (Akiyoshi et al. 2009), Rozella allomyces (Cryptomycota) (James et al. 2013). A total of 203 predicted fungal HK proteins was thus included in our analysis (table 1 and supplementary table S1, Supplementary Material online).
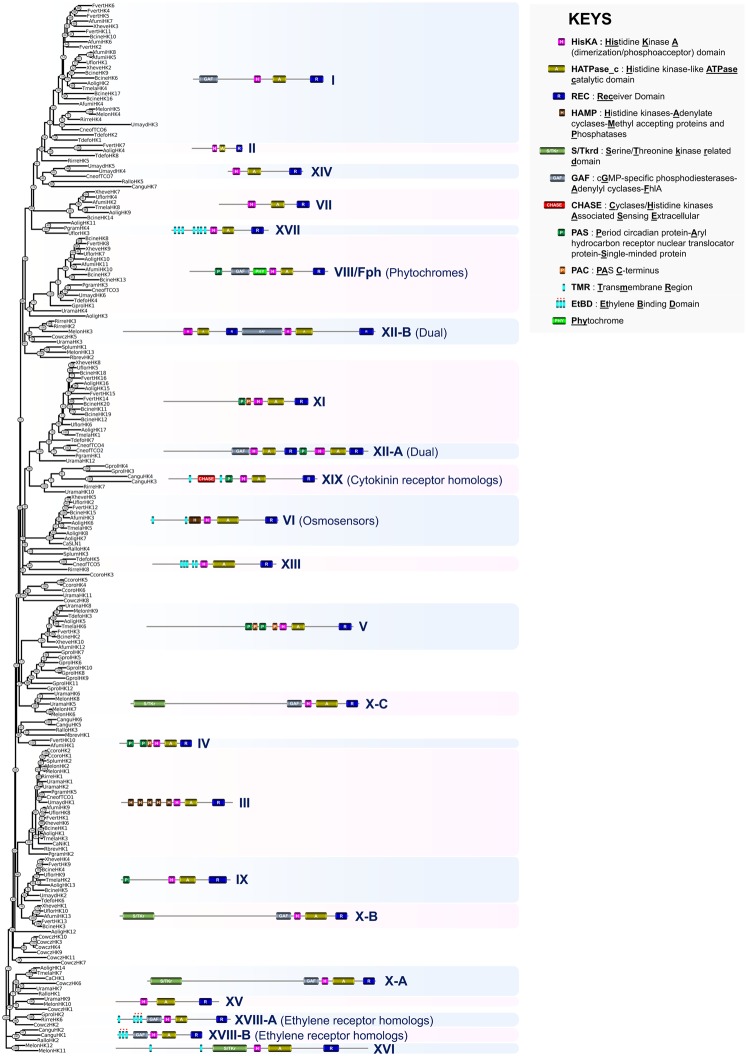
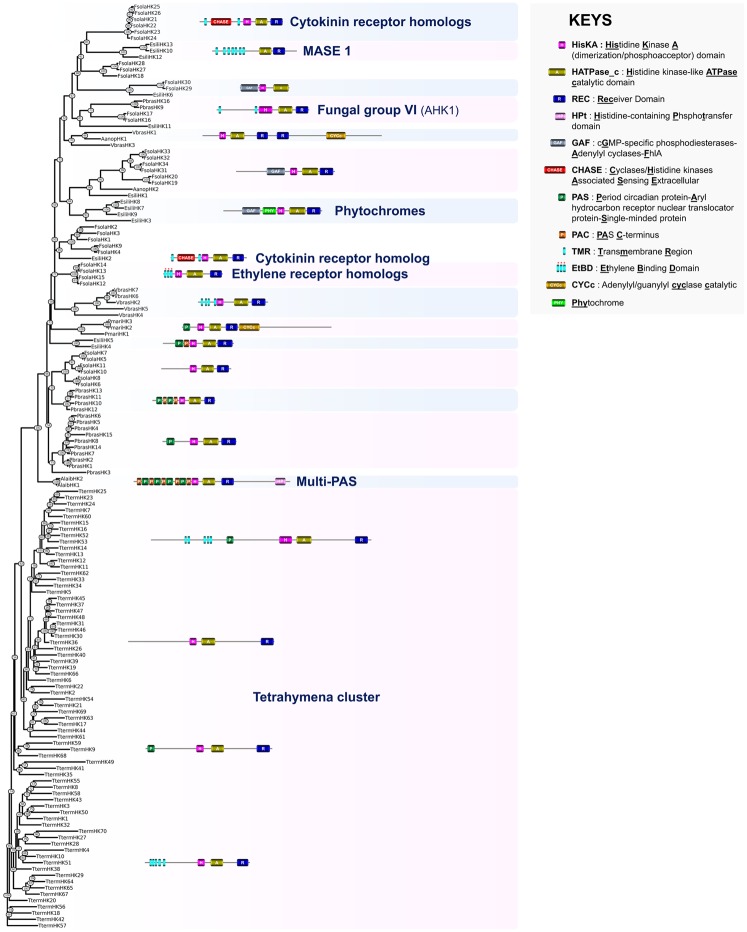
The opisthokont phylogenetic analysis allowed the 11 prominent fungal HK groups (I―XI) originally categorized by Catlett et al. (2003) to be recovered from the Ascomycota along with the five additional groups described by Defosse et al. (2015) (fig. 3). Although the function of most of these latter groups remains unknown, some fungal HK groups have been shown to be involved in the regulation of various physiological processes (Defosse et al. 2015; Hérivaux et al. 2016).
Fig. 3.
—Phylogeny estimation of HK predicted protein sequences in Opisthokonta. A representative structure is provided for each defined HK group. Afumi, Aspergillus fumigatus; Aolig, Arthrobotrys oligospora; Bcine, Botrytis cinerea; Calbi, Candida albicans; Cangu, Catenaria anguillulae; Ccoro, Conidiobolus coronatus; Cneof, Cryptococcus neoformans; Cowcz, Capsaspora owczarzaki; Fvert, Fusarium verticillioides; Gprol, Gonapodya prolifera; Mbrev, Monosiga brevicollis; Melon, Mortierella elongata; Pgram, Puccinia graminis; Rallo, Rozella allomycis; Rbrev, Ramicandelaber brevisporus; Rirre, Rhizophagus irregularis; Splum, Syncephalis plumigaleata; Tdefo, Taphrina deformans; Tmela, Tuber melanosporum; Uflor, Usnea florida; Umayd, Ustilago maydis; Urama, Umbelopsis ramanniana; Xheve, Xylona heveae.
Group I HKs are known to be highly divergent within the group (Catlett et al. 2003). These proteins are present not only in various fungal clades including the Ascomycota and Basidiomycota but also in early diverging fungi and appear to have been expanded in the phytopathogenic Ascomycota. A group I HK mutant in the rice blast fungus Magnaporthe oryzae (Ascomycota) displayed a slight decline in conidiation and an attenuated virulence (Jacob et al. 2014), but there was no apparent altered phenotype for an equivalent mutant in Cr. neoformans (Basidiomycota) (Bahn et al. 2006). Group II fungal HKs are present in the Sordariomycetes, the Orbiliomycetes, and the Leotiomycetes (Catlett et al. 2003). The function of these proteins is unknown. Most available data concerning the roles of fungal HKs were gained from functional characterization of proteins belonging to group III. Fungal group III HKs proteins contain a variable number of HAMP (histidine kinases–adenylate cyclases–methyl accepting proteins and phosphatases) domain repeats (Aravind and Ponting 1999) within the sensing domain and have been primarily reported to be implicated in virulence in human, plant, and insect pathogenic fungi but also in osmosensing, morphogenesis, conidiation, and cell wall integrity (Defosse et al. 2015; Hérivaux et al. 2016). Fungal group III HKs were only found in terrestrial species (fig. 3) suggesting that these sensing proteins were acquired for/during the land colonization process. Group IV fungal HKs were exclusively present in the Eurotiomycetes and the Sordariomycetes (fig. 3), where they have been shown to be involved in the virulence of As.fumigatus (Clemons et al. 2002). Group V fungal HKs are large proteins with PAS/PAC repeats within the sensing domain. Group V fungal HKs were found not only in several clades of the Ascomycota and Basidiomycota but also in early diverging fungi (fig. 3) and have been shown to be implicated in vegetative growth, conidiation, stress adaptation, and virulence in Mag. oryzae (Jacob et al. 2014). Group VI HKs appeared to be restricted to the Ascomycota (fig. 3) and are known to mainly act as transmembrane osmosensors by governing the high-osmolarity glycerol MAPK pathway (Defosse et al. 2015; Hérivaux et al. 2016). The canonical N-terminus of these transmembrane proteins includes two hydrophobic helices flanking an osmosensing extracellular loop, as found in Archaeplastida AHK1-related HKs (fig. 2). Group VII fungal HKs were present in the Ascomycota and Basidimycota clades (fig. 3), but the function of these proteins is unknown. Phytochromes (group VIII or Fph) are light-perceiving receptors, broadly represented in nearly all fungal clades (fig. 3). These proteins mediate red light sensing in fungi, while other types of photoreceptors are involved in sensing light of shorter wavelengths (Bayram et al. 2010). Group IX fungal HKs were found in several clades of the Ascomycota and Basidiomycota but not in early diverging fungi. Their function is unknown. Group X fungal HKs are large proteins with a canonical N-terminus that includes a serine/threonine kinase-related (S/TKr) domain and a GAF domain (Aravind and Ponting 1997). Like fungal group III HKs, group X HKs have been extensively studied and have been shown to govern virulence, morphogenesis, and peroxide adaptation (Defosse et al. 2015; Hérivaux et al. 2016). Group X HKs are broadly represented across the fungi. The phylogenetic tree indicates that this group is polyphyletic, defining fungal groups X-A, X-B, and X-C (fig. 3). Finally, group XI fungal HKs are known to be highly divergent within the group (Catlett et al. 2003) and appeared to be broadly spread in the Ascomycota and Basidiomycota but not in early diverging fungi (fig. 3). Magnaportheoryzae group XI fungal HKs have been shown to be implicated in vegetative growth, conidiation, stress adaptation, and virulence (Jacob et al. 2014).
Importantly, the phylogenetic tree shed light on additional clusters that allowed us to refine the categorization of fungal HKs by defining new fungal HK groups (fig. 3), a process that was recently initiated in Defosse et al. (2015) and Hérivaux et al. (2016). These new groups include XII-A in Basidiomycota (PgramHK1, CneoTCO2, and CneoTCO4) and XII-B in early diverging fungi (RirreHK2, RirreHK3, MelonHK3, and UramaHK3), which correspond to the polyphyletic groups of dual-HKs (Lavín et al. 2014). The dual-HK CneoTCO2 in Cr. neoformans has been shown to be involved in heavy metal and antifungal susceptibility as well as in oxidant condition adaptation (Bahn et al. 2006). Group XIII (formerly known as IB or CneoTCO5) HKs were found in Cr. neoformans, T. deformans and R. irregularis whereas group XIV, which appeared to be restricted to the Basidiomycota, includes predicted proteins in U. maydis and Cr. neoformans (UmaydHK4, UmaydHK5, and CneoTCO7) (fig. 3). Group XV HKs (MelonHK10 and UramaHK9), also referred to as MS-HKI, as well as group XVI HKs (MelonHK11 and MelonHK12), formerly called MS-HKII, were exclusively found in the Mucoromycotina (fig. 3) according to previous studies (Defosse et al. 2015; Hérivaux et al. 2016). Predicted HK proteins from the Lecanoromycetes (UflorHK3), the Orbiliomycetes (AoligHK11), and the Pucciniomycotina (PgramHK4) now define the new group XVII of fungal HKs. It is noteworthy that the recently described ethylene receptor homologs in early diverging fungi (Hérivaux et al. 2017) tended to cluster into two different groups (groups XVIII-A and XVIII-B) (fig. 3) suggesting a polyphyletic origin for these fungal phytohormone-sensing proteins. CHASE domain-containing HKs (here categorized as group XIX) appeared particularly abundant in early diverging fungi (fig. 3) and tended to cluster together suggesting a monophyletic origin for these cytokinin receptor homologs. The remaining predicted sequences that do not cluster with the above mentioned HK groups, and which were either found in a single species, were positioned in branches supported by low bootstrap values or did not have a common N-terminal domain arrangement, have not been incorporated into this refined classification of fungal HKs.
Most of the Cap. owczarzaki (Mesomycetozoa) HKs possessed a sensing region with multiple predicted transmembrane helices that could indicate that Cap. owczarzaki contains few intracellular HKs and mostly membrane bound HKs. One of these HKs, CowczHK2, clustered with ethylene receptors (group XVIII-A) from early diverging fungi (fig. 3) (Hérivaux et al. 2017). As revealed by an alignment with ethylene receptors from plants, green algae, and cyanobacteria, CowczHK2 displays the three predicted transmembrane helices and contained all of the conserved, essential residues involved in ethylene perception (supplementary fig. S4, Supplementary Material online) (Wang et al. 2006). Another remarkable finding was the occurrence of a dual-HK in Cap. owczarzaki (CowcHK5) because this group of HKs was long believed to be restricted to the Basidiomycota within the opisthokonts (Lavín et al. 2014). None of the remaining Cap. owczarzaki HKs displayed significant phylogenetic relationships with other fungal HK groups. They thus represent potential founder members of novel Opisthokonta HK groups.
Amoebozoa
More than 2,000 species of amoebozoid protists have been described. These unicellular organisms, which often possess pseudopods, are known to behave either as free-living cells in water and soils or as parasites of humans and other organisms. We incorporated the predicted HK sequences from four species broadly distributed across the phylogenetic tree of the Amoebozoa (Shadwick et al. 2009) into the present phylogenetic analysis. These included the 51 recently described Physarum polycephalum HKs (Schaap et al. 2016) and the 14 HKs of D.discoideum (Eichinger et al. 2005; Sucgang et al. 2011), and we annotated 48 and 13 predicted sequences from Acanthamoeba castellanii (Clarke et al. 2013) and Polysphondylium pallidum (Heidel et al. 2011), respectively. No HK sequences were found in the genome of the obligate human parasite Entamoeba histolytica (Loftus et al. 2005) (table 1).
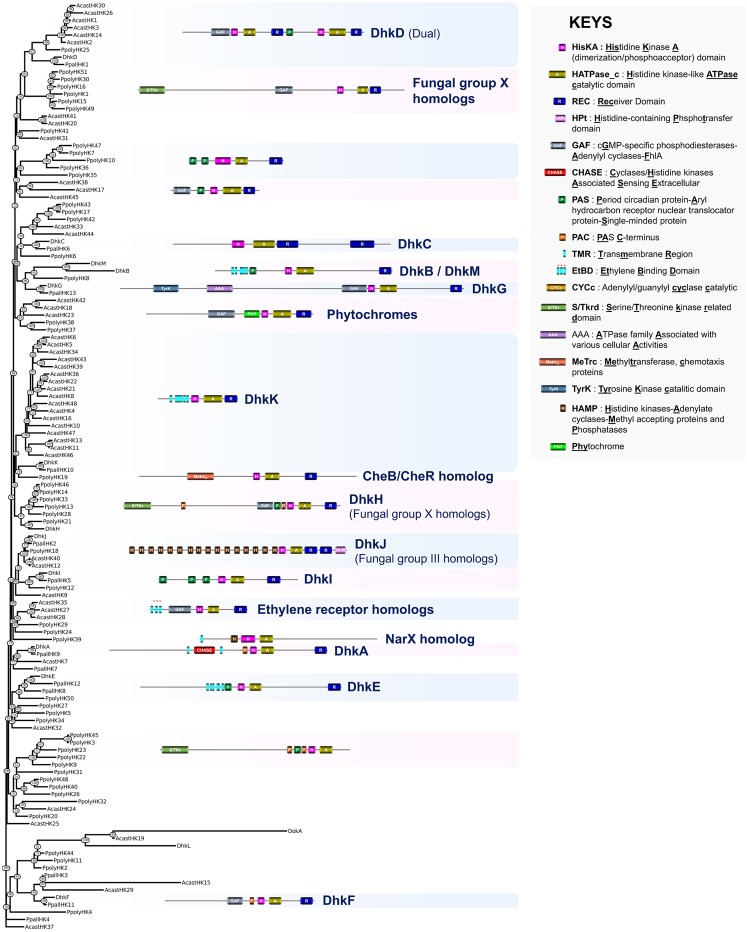
Although a recent study reported a phylogeny of HKs for Physa. polycephalum (Schaap et al. 2016), little is known about the conservation of these sensing proteins within the various clades of the Amoebozoa. We therefore constructed a phylogenetic tree based on an alignment of the 126 predicted HK sequences from the four above-mentioned species (table 1 and supplementary table S1, Supplementary Material online). To date, very little data are available about the function of HKs in Amoebozoa, and what exists is derived from studies on D. discoideum (Du et al. 2015). A homolog of D. discoideum DhkA shares a CHASE domain-containing sensing domain with plant cytokinin receptors (supplementary fig. S5, Supplementary Material online). However, in D. discoideum this domain detects a peptide, SDF-2, which functions both in the regulation of prestalk gene expression and in the control of the terminal differentiation of prespore cells (Wang et al. 1996, 1999). DhkA was also identified in P. pallidum (PpallHK9) but was not found in two other taxon-group-representative species (fig. 4). Homologs of D. discoideum DhkB, which regulates spore germination and maintenance of spore dormancy (Zinda and Singleton 1998), and DhkC, which is required for the initiation of fruiting body formation (Singleton et al. 1998) and also affects chemotaxis (Kuwayama and Kubohara 2016), were also found in P. pallidum (fig. 4). Finally, homologs of D. discoideum DhkF and DhkG, for which no functions are known, were identified in P. pallidum but were absent from the genomes of Physa. polycephalum and Ac. castellanii (fig. 4). This indicated that some of the D. discoideum HKs are likely restricted to the Dictyostelia, with possible roles in their multicellular development to fruiting bodies, which does not occur in the other Amoebozoa with sequenced genomes (Schaap 2011).
Fig. 4.
—Phylogeny estimation of HK predicted protein sequences in Amoebozoa. A representative structure is provided for each defined HK group. Acast, Acanthamoeba castellanii; Ddisc, Dictyostelium discoideum; Ppall, Polysphondylium pallidum; Ppoly, Physarum polycephalum.
Compared with the explored supergroups, the domain topology of amoebozoan HKs was highly diversified and many domains found in plant and fungal HKs, such as PAS, GAF, S/TKrd, were also found in the sensing regions of these proteins (fig. 4). Some proteins contained domains that have not been described previously in eukaryotic HKs. These included the TyrK (tyrosine kinase) domain found in DhkG and PpallHK13 and the MeTrc (methyltransferase, chemotaxis proteins) domain found in PpolyHK19. PpolyHK19 is homologous to the prokaryotic CheB/CheR HK, which plays a central role in bacterial chemotaxis (Bischoff and Ordal 1992). Interestingly, some Physa. polycephalum HK proteins contain an Hpt domain fused to the C-terminal REC domain (i.e., PpolyHK5, PpolyHK18). To date, this particular feature has been observed previously only in facultative aerobic bacteria (notably the oxygen-sensing HK ArcB, Malpica et al. 2006) and to our knowledge, this hybrid HK structure is unique to Amoebozoa within the Eukaryota.
We also detected the presence of some amoebozoan HK groups displaying topologies found in other eukaryotic supergroups. Two Physa. polycephalum HK sequences (PpolyHK37 and PpolyHK38) that contain an N-terminal GAF domain correspond to light-sensing phytochromes (Schaap et al. 2016) (fig. 4). Most of the amoebozoan Dual HKs cluster with the DhkD branch. Dual HKs were believed to be restricted to Basidiomycota, where the function of these large HKs remains obscure. In the future, it will be important to understand how these sensing proteins orchestrate their phosphorelay events during signaling and how their particular topologies dovetailed the currently accepted canonical TCS (fig. 1). The predicted amoebozoan HKs with domain topologies similar to those of fungal group X exhibited marked polyphyly, while the HKs related to fungal group III tended to cluster together in the tree (DhkJ group). We also found that PpolyHK39 displays an N-terminal sensing region that shares domain topology and significant similarity with K. nitens (terrestrial green algae) KniteHK29 and KniteHK44, and bacterial NarX nitrate/nitrite sensors (fig. 4 and supplementary fig. S3, Supplementary Material online) (Mascher et al. 2006). Finally, as revealed by alignment with ethylene receptors from plants, green algae, cyanobacteria, and early diverging fungi, three Ac. castellanii HKs (AcastHK27, AcastHK28, and AcastHK35) contained three predicted transmembrane helices containing all of the conserved residues required for ethylene perception within their N-termini (fig. 4 and supplementary fig. S4, Supplementary Material online) (Wang et al. 2006). Thus, this represents the first description of phytohormone receptor homologs in Amoebozoa.
Exploring the Structure and the Phylogeny of HK Proteins in Apusozoa, Excavata, and SAR
Apusozoa
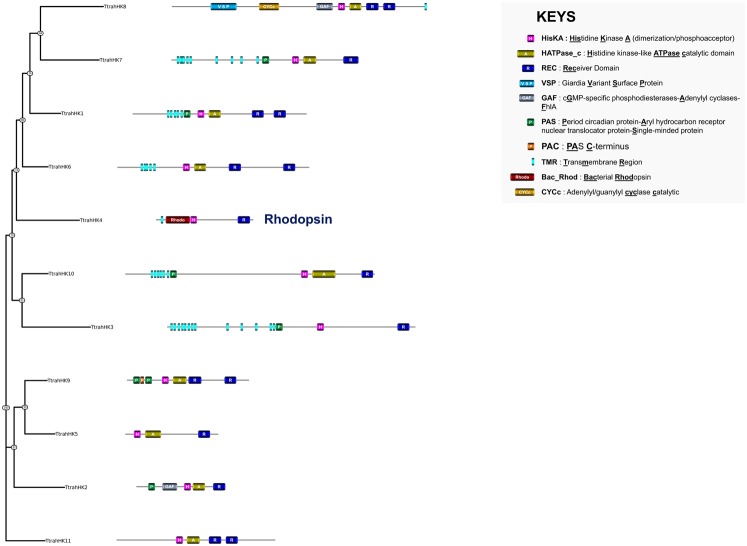
Apusozoa are unicellular flagellated organisms occurring in soils and aquatic habitats where they feed essentially on bacteria. To date, the genome of only one species has been sequenced, namely Thecamonas trahens (Russ et al. unpublished). We identified 11 predicted proteins that display the canonical HK feature in the Th. trahens genome (table 1 and supplementary table S1, Supplementary Material online). The structures of Th. trahens HKs are relatively diverse (fig. 5). Half of these HKs are likely to be located at the membrane because multiple transmembrane regions were found within their N-termini. Several domains usually associated with HK sensing, such as PAS or GAF, were observed in some Th. trahens HKs. Two particular structures stood out from the identified series of predicted proteins. These included the topology VSP-CYCc-GAF-HisKA-HATPase_c-RE-REC of TtrahHK8 (fig. 5). Although the CYCc domain (adenylyl/guanylyl cyclase catalytic) has been found in a small number of eukaryotic HKs (from the Archeaplastida and Amoebozoa), VSP (Giardia variant-specific surface protein) domains mediating antigenic variation at the surface of Giardiaintestinalis (Gargantini et al. 2016) have not been described previously in HKs. Another interesting finding in Th. trahens was the occurrence of a bacteriorhodopsin HK, that is, TtrahHK4 (previously observed in the green algae) (fig. 5 and supplementary fig. S1, Supplementary Material online). Thus, bacteriorhodopsin HKs appear to be used for UVA sensing in several distantly related lineages but sharing similar environments: some halophilic prokaryotes (Haloarchaea, in which bacteriorhodopsins were originally found) and marine unicellular eukaryotes (green algae and Th. trahens).
Fig. 5.
—Phylogeny estimation of HK predicted protein sequences in Apusozoa. The structure of each HK is provided. Ttrah, Thecamonas trahens.
Excavata
Excavata corresponds to unicellular, heterotrophic, and commonly flagellated species. Most are free-living organisms in the environment, whereas others are commensals, symbionts or parasites of animals. Seven and 24 HK sequences were predicted in the genomes of Trichomonas vaginalis (Metamonada) (Carlton et al. 2007) and Naegleria gruberi (Discoba) (Fritz-Laylin et al. 2010), respectively, but no HKs were found in the genomes of the other representative Excavata species (Spironucleus salmonicida, Gi.intestinalis, Leishmania infantum, and Trypanosoma brucei) (Morrison et al. 2007; Peacock et al. 2007; Jackson et al. 2010; Xu et al. 2014) (table 1 and supplementary table S1, Supplementary Material online).
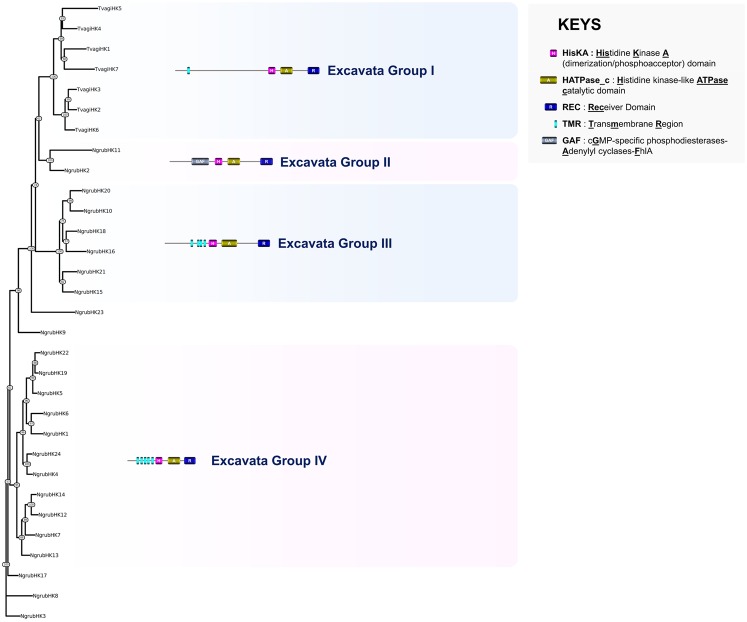
To gain insight into the phylogenetic relationships between Tr. vaginalis and N. gruberi HKs, the domain organization of each protein was investigated and a phylogenetic tree was built using a multiple alignment of all the predicted sequences from both species. Based on this analysis and, in particular, sequence similarities between HisKA domains and N-terminal sensing regions, four groups of Excavata HK are proposed (fig. 6).
Fig. 6.
—Phylogeny estimation of HK predicted protein sequences in Excavata. A representative structure is provided for each defined HK group. Ngrub, Naegleria gruberi; Tvagi, Trichomonas vaginalis.
All seven Tr. vaginalis HKs are hybrid and tended to cluster together in the phylogenetic tree (Excavata group I, fig. 6) independently of the 24 HKs from N. gruberi. Most of the Tr. vaginalis HKs possessed a sensing region with no similarity to known domains, but the presence of one or two predicted transmembrane helices in some of these proteins could indicate that Tr. vaginalis contains both intracellular and membrane-bound HKs (fig. 6). Although the structure of the predicted N. gruberi HK proteins seems relatively well conserved at a first glance, the sequences were distributed into three major clades (Excavata groups II, III, and IV, fig. 6). Most of these HKs harbor multiple predicted hydrophobic helices indicating that the majority of N. gruberi HKs are transmembrane proteins. Two N. gruberi HKs possessed domains that are often associated with the eukaryotic HK sensing region: PAS in NgrubHK23 and GAF in NgrubHK11. In contrast, five of the 24 HKs from N. gruberi did not contain any detectable N-terminal sensing region, as previously observed in some fungal HKs (Defosse et al. 2015). None of the N. gruberi and Tr. vaginalis predicted HK sequences were found to share any homology with other previously described prokaryotic or eukaryotic HK groups.
Stramenopiles, Alveolates, and Rhizaria
The SAR is a highly diversified supergroup encompassing not only various unicellular and multicellular algal and planktonic species but also ciliates and oomycetes.
HKs appear to be broadly represented in Stramenopiles because 1―34 predicted proteins were identified in pelagophytes (Aureococcus anophagefferens, two HKs) (Gobler et al. 2011), diatoms (Fistulifera solaris, 34 HKs) (Tanaka et al. 2015), brown algae (Ectocarpus sp., 13 HKs) (Cock et al. 2010; Cormier et al. 2017), and oomycetes (Albugo laibachii, two HKs) (Kemen et al. 2011). However, the distribution of HKs in Alveolata is highly heterogeneous because no HK sequence was detected in the apicomplexan genomes (e.g., Plasmodium falciparum, Babesia bovis, Toxoplasma gondii, and Cryptosporidium hominis) (Gardner et al. 2002; Xu et al. 2004; Brayton et al. 2007; Lorenzi et al. 2016) or in the Dinophyceae (Symbiodinium microadriaticum) (Aranda et al. 2016), whereas a few members were identified in the Chromerida (Vitrella brassicaformis, seven HKs) (Woo et al. 2015) and the Perkinsae (Perkinsus marinus, three HKs) (Joseph et al. 2010). Interestingly, we also observed a marked increase in the number of HK predicted sequences in the Alveolata lineage Ciliophora (70 HKs in Tetrahymena thermophila) (Eisen et al. 2006). For Rhizaria, HKs were found in the Cercozoa (Plasmodiophora brassicae, 16 HKs) (Schwelm et al. 2015) but were absent from a Foraminifera genome (Reticulomyxa filosa) (Glöckner et al. 2014) (table 1 and supplementary table S1, Supplementary Material online). Although it is well documented that the SAR includes a large number of genetically distant species with strongly divergent genomes, which could possibly complicate phylogenomic approaches (Grant and Katz 2014), we attempted to provide insight into the evolutionary relationship of HKs in this supergroup by carrying out a robust phylogenetic analysis (fig. 7).
Fig. 7.
—Phylogeny estimation of HK predicted protein sequences in SAR. A representative structure is provided for each defined HK group. Aanop, Aureococcus anophagefferens; Alaib, Albugo laibachii; Esili, Ectocarpus siliculosus; Fsola, Fistulifera solaris; Pbras, Plasmodiophora brassicae; Pmar, Perkinsus marinus; Tterm, Tetrahymena termophila; Vbras, Vitrella brassicaformis.
Surprisingly, Te. thermophila HKs tended to cluster in a single clade, consistent with the current hypothesis that paramecia have evolved rapidly and independently and therefore differ markedly from other Alveolata clades (Eisen et al. 2006; Aury et al. 2006). All except one of the Te. thermophila HKs are hybrid HKs and most of these harbor multiple predicted hydrophobic helices within the N-terminal sensing region that presumably allow these proteins to be anchored to the cell membrane. Most of these proteins do not have any known domains in the N-terminal region; exceptions include 12 HKs that have a PAS domain (supplementary table S1, Supplementary Material online, and fig. 7). Despite the large number of Te. thermophila HKs, there is a limited structural diversity suggesting that Te. thermophila HK genes, as previously reported for many other kinase families in paramecia, are the result of a rapid gene family expansion process (Bemm et al. 2009).
Concerning the phylogeny of HKs from the other selected SAR species, we identified a set of three HKs from the Alveolata group Perkinsae (Pe. marinus) that tend to cluster together (fig. 7). Among these, PmariHK1 has a CYCc domain in its C-terminus. Five out of the seven Chromerida Vi. brassicaformis HKs possess multiple hydrophobic helices (fig. 7). The genome of this species encodes another hybrid HK structure, VbrasHK1, with a domain topology (HisKA-HATPase_C-REC-REC-CYCc) that has never been observed previously in the Eukaryota. Thus, as observed in this study for green algae, some Alveolata TCS are coupled with adenylyl/guanylyl cyclase activity.
In the Rhizaria, most of the HKs found in the representative species Pl. brassicae (Cercozoa) were predicted to be cytoplasmic proteins. Some possess PAS/PAC domains and these tended to cluster within two clades in the tree. Interestingly, two Cercozoa HKs are predicted to be transmembrane proteins with two hydrophobic helices that border an osmosensing extracellular loop, as found in Archaeplastida AHK1-related HKs (figs. 2 and 7).
In the stramenopiles, the oomycete Al. laibachii possesses two HKs that differ drastically from those of other SAR clades because their N-termini contain multiple PAS domain repeats and because they display an Hpt domain fused to a C-terminal REC domain (fig. 7). We identified similar multi-PAS HKs in the Archaeplastida sister clades Cryptophyta (G. theta) and Haptophyta (E. huxleyi) (figs. 2 and 7). Aureococcusanophagefferens (pelagophyte), F. solaris (diatom), and Ectocarpus sp. (brown alga) share some common HK structures including proteins with PAS or GAF domains within their N-terminal sensing domains (fig. 7). The Ectocarpus predicted proteins EsiliHK3, EsiliHK7, EsiliHK8, and EsiliHK9 correspond to previously described light-sensing phytochromes (Rockwell and Lagarias 2017). Further analyses revealed some interesting findings concerning the HK genes detected in the genome of the brown alga Ectocarpus sp.: 1) Six of these genes (EsiliHK3, EsiliHK4, EsiliHK5, EsiliHK10, EsiliHK11, and EsiliHK13, fig. 7) are located either within a large, integrated, transcriptionally silent viral genome (Delaroque and Boland 2008; Cock et al. 2010) similar to Ectocarpus siliculosus virus 1 (Delaroque et al. 2001) located on chromosome 6 or within another inserted viral fragment on scaffold 490, and should therefore be considered as viral rather than algal genes; 2) of the remaining genes, all except EsiliHK6 share one or more features with viral HK genes including high sequence similarity (EsiliHK1, EsiliHK6, EsiliHK7, EsiliHK8, and EsiliHK12; 25―86% identity), phylogenetic proximity (EsiliHK12 clusters with EsiliHK10 and EsiliHK13, and EsiliHK7, EsiliHK8 and EsiliHK9 cluster with EsiliHK3 in fig. 7), and/or only a small number (zero or one) of introns (EsiliHK2, EsiliHK7, EsiliHK8, EsiliHK9, and EsiliHK12), suggesting possible evolutionary relationships between the viral and algal genes; 3) the N-terminal sensing regions of two of the inserted viral genes, EsiliHK10 and EsiliHK12, encode proteins with typical MASE1 domains (fig. 7) that were herein described in early diverging Archaeplastida; and 4) the Ectocarpus sp. genome possesses non-hybrid HK genes (e.g., EsiliHK6) (fig. 7) that appear to have a cyanobacterial origin. The most interesting finding of the SAR analysis was the identification of HKs displaying a CHASE domain within their N-terminus in the brown alga Ectocarpus sp. (EsiliHK2) and the diatom F. solaris (FsolaHK22 to FsolaHK26) (fig. 7 and supplementary fig. S5, Supplementary Material online). The CHASE domain is a characteristic feature of plant cytokinin receptors. Surprisingly, alignment of four F. solaris HKs (FsolaHK12―FsolaHK15) with ethylene receptors from plants, green algae, cyanobacteria, Amoebozoa, and early diverging fungi allowed the identification, within their N-termini, of three predicted transmembrane helices with all of the conserved residues necessary for ethylene perception (fig. 7 and supplementary fig. S4, Supplementary Material online) (Wang et al. 2006). Thus, this represents the first description of cytokinin and ethylene receptor homologs in SAR.
Discussion
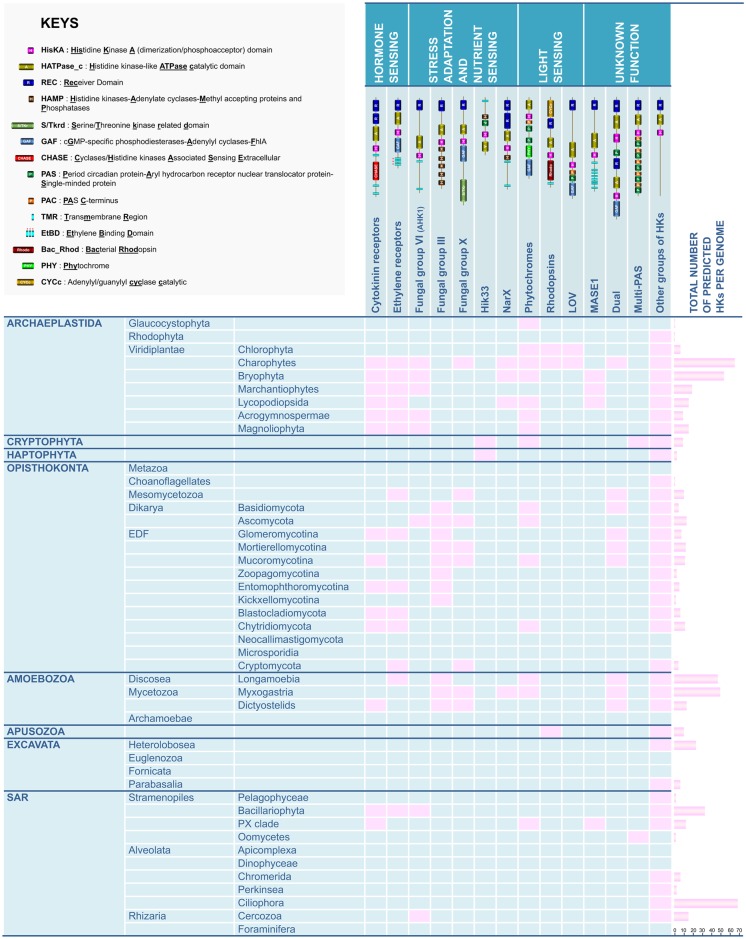
Significant advances in knowledge concerning the structure, classification, and distribution of HKs have been made within the past decade in plants, fungi, and to a lesser extent in amoebae, but these analyses have not been more broadly addressed in Eukaryota. The present study reports for the first time a preliminary but global phylogenetic analysis of this prominent family of sensing proteins in all the eukaryotic supergroups including Archaeplastida, Opisthokonta, Amoebozoa, Apusozoa, Excavata, and SAR (fig. 8). This analysis allowed most of the compiled sequences to be categorized into a previously described HK group, but we also propose to refine the HK classification for some supergroups, notably the Archaeplastida and Opisthokonta. Unfortunately, the large genetic distances between the representative species in each supergroup and the limited quantity of genomic resources for some clades have precluded the categorization of some highly divergent HKs (notably from the Amoebozoa). Beyond this phylogenetic analysis and classification of eukaryotic HKs, our study has revealed several interesting findings: 1) a description of some previously overlooked eukaryotic HK groups with predicted putative physiological functions; 2) the observation that some HK groups, previously believed to be restricted to one kingdom (plants or fungi), occur in additional supergroups; and 3) the suggestion of evolutionary paths that have led to the appearance, transfer, duplication, and loss of HK genes in some phylogenetic lineages.
Fig. 8.
—Occurrence of prominent HK groups in eukaryotic clades. For each eukaryotic clade, the presence of main HK groups is indicated with a pink box. The total number of predicted HK sequences is indicated in the right column.
HKs in Archaeplastida: Unprecedented Structures and Evolution
It is now generally admitted that a common ancestor of the Archaeplastida acquired many genes from cyanobacteria through the primary endosymbiotic event that produced their plastids approximately 1.5 Ga (Brodie et al. 2017). Therefore, it has been suggested that the initial pool of HK genes were acquired from cyanobacteria (Lacey and Binder 2016). In Archaeplastida, we observed first that unicellular aquatic species possess few HK genes (1―7 HKs in Glaucocystophyta, Rhodophyta, and Chlorophyta). The number of HKs is significantly greater in the Charophyta lineage (67 HKs in the model terrestrial algae), but these genes are less abundant in land plants (54 HKs in the model moss, and 9―16 in the remaining studied species) (fig. 8). Thus, it could be hypothesized that Archaeplastida algae progressively extended and diversified their HK battery prior to the conquest of terrestrial habitats, notably acquiring the ability to sense new nutrient sources offered by terrestrial niches (nitrate/nitrite sensor homologs in K. nitens, P. patens, and S. moellendorffii) and by developing capacity to respond to various constraints related to terrestrial habitats such as oxidant stresses (homologues of fungal group X oxidant sensors in K. nitens), osmotic stresses (proteins related to the osmosensor AthalHK1 in K. nitens) or direct sunlight exposure (homologues of bacteriorodopsins and LOV blue-light photoreceptors in K. nitens) (fig. 8). CKI2, which is involved in stress-induced stomatal closure (Mira-Rodado et al. 2012), was detected in species that have colonized terrestrial habitats. Domain-based phylogenetic analysis suggested that CKI1, which directs specification of the endosperm precursor central cell resulting in seeds containing an embryo and an endosperm, appeared within the seed-forming flowering plants (Angiosperms) (Yuan et al. 2016).
An important feature of the diversification of the structure of green algal HKs was the apparition of ethylene and cytokinin receptors from the terrestrial alga K. nitens (fig. 8). Both ethylene and cytokinin are largely documented as prominent phytohormones essential for land plant developmental programs (Zdarska et al. 2015). The apparition of ethylene and cytokinin-sensing HKs in large numbers in the terrestrial algae (K. nitens) is likely to have played an essential role in progressively shaping the first multicellular Archaeplastida for growth and nutrition in various land ecotopes.
Strengthening the Classification of HKs in Opisthokonta
For the Opisthokonta, we first confirmed the absence of typical prokaryotic or eukaryotic HKs in animals. This is of course an essential prerequisite for approaches aimed at developing antimicrobials that target HKs in the context of human disease, for example (Bem et al. 2015; Shor and Chauhan 2015).
Fungal HKs were initially categorized into 11 groups (Catlett et al. 2003) and then subsequently into 16 groups (Defosse et al. 2015). Our analysis of a large panel of Dikarya (Ascomycota and Basidiomycota) and early diverging fungi indicated that classification of fungal HKs needs to be extended because previously undescribed topologies were identified and, consequently, some new groups have been proposed.
Exploration of the structure of HKs in the mesomycetozoan model species Cap. owczarzaki revealed the unexpected presence of an HK that shared a high degree of similarity with plant ethylene receptors (fig. 8). However, we failed to detect any plant cytokinin receptor homologs in this species. Importantly, phytohormone receptor homologs (ethylene and cytokinin receptors) were recently found not only in early diverging fungi that are plant root symbionts or endophytes but also in species that colonize decaying plant material. It was thus speculated that these particular sensing proteins have promoted the interaction of early diverging fungi with plants leading to the conquest of land by these ancestral fungi (Hérivaux et al. 2017). The presence of such phytohormone receptor homologs in the Mesomycetozoa suggests that the acquisition of these sensing proteins occurred in a common ancestor of fungi and Mesomycetozoa. In cyanobacteria, ethylene receptors are bifunctional sensing proteins that mediate responses to both light and ethylene (Lacey and Binder 2016). It would be thus interesting to investigate the precise origin of ethylene receptors and their roles in the Mesomycetozoa and early diverging fungi.
A Marked Structural Diversity in Amoebozoan HKs
In contrast to plant and fungal HKs, knowledge about the function and physiological roles of HKs in amoebae remains fragmentary. The domain topologies of amoebozoan HKs were found to be highly diversified compared with other eukaryotic supergroups, indicating that they are likely to have a broad range of biochemical outputs and functions. Phylogenetic distribution provided some clues about the evolutionary paths driving the amoebozoan HKs diversity. For example, HKs with N-terminal CHASE domains (DhkA) were found exclusively in Dictyostelia, that is, amoebae that form true multicellular fruiting bodies (sorocarps) to produce spores. We also found some domain topologies in the Amoebozoa that had been observed previously only in fungi, such as group III HKs, which are reported to act as osmosensors, group X HKs, known to mediate oxidant stress adaptation (Defosse et al. 2015) and light-sensing phytochromes (fig. 8). A similar situation was observed for some domain topologies that had been observed previously only in bacteria and plants, such as nitrate/nitrite sensors (fig. 8). Most importantly, we described, for the first time, the occurrence of ethylene receptor homologs in the Amoebozoa, especially in the free-living amoeba Ac. castellanii (fig. 8). However, the physiological role of homologs of fungal group III, group X, phytochromes, and ethylene receptors remains to be established.
Globally, the number of HKs differed sharply between amoebozoan species (fig. 8). The size of an organism HKs complement appeared to be inversely related to its level of developmental complexity (D. discoideum and P. pallidum vs. Physa. polycephalum) or to its dependency on a host species (the free living Ac. castellanii vs. the obligate parasite En. histolytica). However, these trends will need to be validated using larger scale phylogenomic analyses when more amoebozoan genomes are available.
First Description of HKs in Apusozoa
In this study, we also took the opportunity to advance the description of HKs in Eukaryota by investigating the structure and distribution of these proteins in unexplored eukaryotic supergroups. We describe here for the first time predicted HK structures for the Apusozoa. The phylogenetic position of this clade is still controversial, but recent molecular evidence suggests that it is a paraphyletic phylum related to the Opisthokonta (Cavalier-Smith et al. 2015). We identified 11 predicted proteins in the Th. trahens genome that display canonical HK features and these proteins were structurally diversified. Some interesting topologies were observed, notably HKs with a VSP domain in their N-terminal sensing region and bacteriorhodopsins. However, we have not been able to characterize this protein family further due to a lack of additional genomic resources and functional genomic data for the Apusozoa, and the lack of similarity of Apusozoa HKs with other bacterial and eukaryotic HK sequences.
HKs Are Restricted to Some Clades within the Excavata Supergroup
Searches of genomic resources for Excavata identified seven and 24 predicted HK sequences in the genomes of Tr. vaginalis (Metamonada) and N. gruberi (Discoba), respectively, but no HKs were found in the genomes of other prominent genera from the Excavata (Giardia sp., Leishmania sp., and Trypanosoma sp.). This distribution seems surprising but it is important to highlight the fact that, the genetic distance between Tr. vaginalis and Gi. intestinalis is large, as that between N. gruberi and other species within the Discoba (Grant and Katz 2014). However, it was interesting to note the previously unreported occurrence of HKs in two prominent species Tr. vaginalis (human parasite) and N. gruberi (closely related to the human parasite Naegleriafowleri). Three main observations can be highlighted concerning the predicted Excavata HKs: 1) Metamonada and Discoba HKs appear to be highly diverged, reflecting the large genetic distance between these two major clades of the Excavata; 2) although a small number of the Excavata HKs are likely to act as intracellular sensing proteins, most are predicted to be anchored in the cell membrane; and 3) very few of the Excavata HKs have known domains in their N-termini that have been implicated in sensing in other prokaryotic and eukaryotic HKs. Thus, the HK family appears to have evolved structurally in a specific manner in the Excavata supergroup.
First Description of Cytokinin and Ethylene Receptor Homologs in SAR
Finally, we report here the first description of HK structures in the SAR supergroup. The distribution of these predicted proteins was highly heterogeneous within the SAR clades, and we failed to identify any homologs in the genomes of Apicomplexa. Hence, therapeutic developments based on the inhibition of HK activity will unfortunately not be useful in combatting malaria, toxoplasmosis, or other coccidian diseases. The high level of genetic divergence between the genomes of the representative SAR species selected for this analysis complicated the phylogenomic approaches, and it was not possible to draw any general conclusions about the evolutionary relationships of HKs in this supergroup. However, interesting findings were made. Notably the presence in the genomes of photosynthetic diatoms and brown algae of genes encoding HKs that bear within their N-termini a CHASE domain, the characteristic feature of plant cytokinin receptors (fig. 8). In addition, we described here for the first time the occurrence of ethylene receptor homologs in photosynthetic diatoms (fig. 8). It is currently admitted that the diatom/brown algal lineage underwent an ancient secondary endosymbiosis events involving engulfment of a red alga by a stramenopile protist cell (Brodie et al. 2017). It is thus possible that phytohormone-like receptors were acquired by some Stramenopiles lineages via this secondary endosymbiosis. This is in line with the current hypothesis that the acquisition of HKs in eukaryotic lineages occurred via both the early mitochondrial endosymbiosis and the later endosymbiosis involving a cyanobacterium-like prokaryote that gave rise to plastids (Schaller et al. 2011). We also observed indications that brown algal HK gene complements may have been enriched by exchanges with large DNA viruses, mediated by insertions of the genomes of these viruses into algal genomes. Inserted viral DNA containing HK genes was detected in Ectocarpus sp., for example. Moreover, not only did most of the bona fide algal HK genes share a high level of similarity with viral genes or cluster with them in phylogenetic trees but they also tended to be intron-poor, which is unusual for brown algal genes but is a typical feature of viral genomes. Taken together, these observations suggest that both endosymbiotic and other horizontal gene transfer mechanisms may have played an important role during the evolutionary history of stramenopile HKs.
Concluding Remarks
This study provides a preliminary overview of the structure and distribution of HKs in Eukaryota (fig. 8). Although HKs had been previously identified in plants, fungi, and Amoebozoa, we present here the first description of this family of HK proteins in unexplored clades such as Apusozoa, Excavata, and SAR. We observed conservation of some HK groups that are putatively involved in crucial physiological processes, including light perception, hormone sensing, development, and stress adaptation within eukaryotic supergroups (fig. 8). We show that data concerning the function of HK groups can be combined with robust phylogenetic analysis to decipher the evolutionary history and currently known functions of HKs, notably by correlating the presence of specific domain topologies with physiological traits. This approach was applied for the Archaeplastida and, to a lesser extent, for the Opisthokonta (mainly fungi). Critically, phylogenetic analysis suggested that cytokinin and/or ethylene signaling systems are not only found in plants but may also occur in various other eukaryotic (brown algae, diatoms, amoebae, fungi, and Mesomycetozoa) (this study and Hérivaux et al. 2017) and prokaryotic (cyanobacteria and eubacteria) clades (Lacey and Binder 2016; Wang et al. 2017; Kabbara et al. 2018) where they probably regulate cellular processes in a functional context with chemotaxis, developmental processes, and interkingdom communication.
In conclusion, this analysis extends our knowledge of the dissemination of TCS signaling pathways in eukaryotes. From an evolutionary perspective, it would be interesting in the near future to use phylogenetic methods to explore the evolutionary origins of eukaryotic HKs to predict whether they are derived from the archetypal ancestors of chloroplasts, mitochondria, or the archaebacterial host. In the same way, it would be highly relevant to identify eukaryotic HKs that were acquired thanks to more or less recent lateral gene transfer events from prokaryotes or eukaryotes. From a functional point of view, it remains also essential to accelerate the acquisition of data concerning the roles of eukaryotic HKs, notably for pathogenic fungi, Metamonada (Tr. vaginalis), Discoba (N. gruberi), and oomycetes. This could shed light on their potential as new targets for antimicrobial development, as suggested over the past few years for bacteria (Bem et al. 2015).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the U.S. Department of Energy Joint Genome Institute and the PIs of the 1000 Fungal Genomes Project (J. Spatafora, A. Gryganskyi, T. James, M.E. Smith, and J. Magnuson) for permission to use the Usnea florida, Syncephalis plumigaleata, Umbelopsis ramanniana, and Ramicandelaber brevisporus genome data before publication. We also acknowledge the Broad Institute Genome Sequencing Platform for making the Thecamonas trahens genome publicly available. We thank S. Gribaldo (Institut Pasteur, Paris) for constructive conversations about this manuscript. S.K. was supported by a fellowship from association AZM. A.H. was supported by a fellowship from the University of Angers. N.P. was funded by the Agence Nationale de la Recherche (program ANR-PRCE Mycormones).
Literature Cited
- Akiyoshi DE, et al. 2009. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog. 5(1):e1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project. 2013. The Amborella genome and the evolution of flowering plants. Science 342(6165):1241089. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L.. 2001. The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem Sci. 26(10):579–582. [DOI] [PubMed] [Google Scholar]
- Appleby JL, Parkinson JS, Bourret RB.. 1996. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86(6):845–848. [DOI] [PubMed] [Google Scholar]
- Aranda M, et al. 2016. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci Rep. 6:39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Ponting CP.. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 22(12):458–459. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP.. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 176(1):111–116. [DOI] [PubMed] [Google Scholar]
- Attwood PV. 2013. Histidine kinases from bacteria to humans. Biochem Soc Trans. 41(4):1023–1028. [DOI] [PubMed] [Google Scholar]
- Aury JM, et al. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444(7116):171–178. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Kojima K, Cox GM, Heitman J.. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell. 17(7):3122–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332(6032):960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O, Braus GH, Fischer R, Rodriguez-Romero J.. 2010. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet Biol. 47(11):900–908. [DOI] [PubMed] [Google Scholar]
- Bem AE, et al. 2015. Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem Biol. 10(1):213–224. [DOI] [PubMed] [Google Scholar]
- Bemm F, Schwarz R, Förster F, Schultz J.. 2009. A kinome of 2600 in the ciliate Paramecium tetraurelia. FEBS Lett. 583(22):3589–3592. [DOI] [PubMed] [Google Scholar]
- Bischoff DS, Ordal GW.. 1992. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol Microbiol. 6(1):23–28. [DOI] [PubMed] [Google Scholar]
- Bowman JL, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171(2):287–304.e15. [DOI] [PubMed] [Google Scholar]
- Brayton KA, et al. 2007. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 3(10):1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie J, et al. 2017. Biotic interactions as drivers of algal origin and evolution. New Phytol. 216(3):670–681. [DOI] [PubMed] [Google Scholar]
- Carlton JM, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315(5809):207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR. 1998. Higher-plant phytochrome: “I used to date histidine, but now I prefer serine.” Proc Natl Acad Sci U S A. 95(23):13358–13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett NL, Yoder OC, Turgeon BG.. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell. 2(6):1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, et al. 2015. Multigene phylogeny resolves deep branching of Amoebozoa. Mol Phylogenet Evol. 83:293–304. [DOI] [PubMed] [Google Scholar]
- Chang Y, et al. 2015. Phylogenomic analyses indicate that early fungi evolved digesting cell walls of algal ancestors of land plants. Genome Biol Evol. 7(6):1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chefdor F, et al. 2006. Osmotic stress sensing in Populus: components identification of a phosphorelay system. FEBS Lett. 580(1):77–81. [DOI] [PubMed] [Google Scholar]
- Cisse OH, et al. 2013. Genome sequencing of the plant pathogen Taphrina deformans, the causal agent of peach leaf curl. Mbio 4(3):e00055–e00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C.. 1998. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci U S A. 95(9):5401–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, et al. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 14(2):R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons KV, Miller TK, Selitrennikoff CP, Stevens DA.. 2002. Fos-1, a putative histidine kinase as a virulence factor for systemic aspergillosis. Med Mycol. 40(3):259–262. [DOI] [PubMed] [Google Scholar]
- Cock JM, et al. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465(7298):617–621. [DOI] [PubMed] [Google Scholar]
- Cormier A, et al. 2017. Re-annotation, improved large-scale assembly and establishment of a catalogue of noncoding loci for the genome of the model brown alga Ectocarpus. New Phytol. 214(1):219–232. [DOI] [PubMed] [Google Scholar]
- Crosson S, Rajagopal S, Moffat K.. 2003. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 42(1):2–10. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, et al. 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317(5843):1400–1402. [DOI] [PubMed] [Google Scholar]
- Curtis BA, et al. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492(7427):59–65. [DOI] [PubMed] [Google Scholar]
- Defosse TA, et al. 2015. Hybrid histidine kinases in pathogenic fungi. Mol Microbiol. 95(6):914–924. [DOI] [PubMed] [Google Scholar]
- Delaroque N, Boland W.. 2008. The genome of the brown alga Ectocarpus siliculosus contains a series of viral DNA pieces, suggesting an ancient association with large dsDNA viruses. BMC Evol Biol. 8(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaroque N, et al. 2001. The complete DNA sequence of the Ectocarpus siliculosus Virus EsV-1 genome. Virology 287(1):112–132. [DOI] [PubMed] [Google Scholar]
- Du Q, Kawabe Y, Schilde C, Chen Z-H, Schaap P.. 2015. The evolution of aggregative multicellularity and cell-cell communication in the Dictyostelia. J Mol Biol. 427(23):3722–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, et al. 2014. Marine algae and land plants share conserved phytochrome signaling systems. Proc Natl Acad Sci U S A. 111(44):15827–15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis S, et al. 2011. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A. 108(22):9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435(7038):43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, et al. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4(9):e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140(5):631–642. [DOI] [PubMed] [Google Scholar]
- Gallie DR. 2015. Appearance and elaboration of the ethylene receptor family during land plant evolution. Plant Mol Biol. 87(4―5):521–539. [DOI] [PubMed] [Google Scholar]
- Galpérin MY, Makarova KS, Wolf YI, Koonin EV.. 2018. Phyletic distribution and lineage-specific domain architectures of archaeal two-component signal transduction systems. J Bacteriol. 200(7):pii: e00681–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419(6906):498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargantini PR, et al. 2016. Antigenic variation in the intestinal parasite Giardia lamblia. Curr Opin Microbiol. 32:52–58. [DOI] [PubMed] [Google Scholar]
- Gazis R, et al. 2016. The genome of Xylona heveae provides a window into fungal endophytism. Fungal Biol. 120(1):26–42. [DOI] [PubMed] [Google Scholar]
- Glantz ST, et al. 2016. Functional and topological diversity of LOV domain photoreceptors. Proc Natl Acad Sci U S A. 113(11):E1442–E1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner G, et al. 2014. The genome of the foraminiferan Reticulomyxa filosa. Curr Biol. 24(1):11–18. [DOI] [PubMed] [Google Scholar]
- Gobler CJ, et al. 2011. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci U S A. 108(11):4352–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JR, Katz LA.. 2014. Building a phylogenomic pipeline for the eukaryotic tree of life—addressing deep phylogenies with genome-scale data. PloS Curr. 6:pii: ecurrents.tol.e089df47766ed1e9dabac39c76bae266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe TW, Stock JB.. 1999. The histidine protein kinase superfamily. Adv Microb Physiol. 41:139–227. [DOI] [PubMed] [Google Scholar]
- Grigoriev IV, et al. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42(Database issue):D699–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn N, Halawa M, Snel B, Seidl MF, Heyl A.. 2014. A subfamily of putative cytokinin receptors is revealed by an analysis of the evolution of the two-component signaling system of plants. Plant Physiol. 165(1):227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulia-Nuss M, et al. 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 7:10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitjema CH, et al. 2017. A parts list for fungal cellulosomes revealed by comparative genomics. Nat Microbiol. 2:17087. [DOI] [PubMed] [Google Scholar]
- Hargreaves KR, Kropinski AM, Clokie MR.. 2014. What does the talking? Quorum sensing signalling genes discovered in a bacteriophage genome. PLoS One 9(1):e85131.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, et al. 2016. Genome-wide identification and expression analysis of two-component system genes in tomato. Int J Mol Sci. 17(8):pii: E1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, et al. 2011. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 21(11):1882–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérivaux A, et al. 2016. Major sensing proteins in pathogenic fungi: the hybrid histidine kinase family. PLoS Pathog. 12(7):e1005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérivaux A, et al. 2017. The identification of phytohormone receptor homologs in early diverging fungi suggests a role for plant sensing in land colonization by fungi. mBio 31(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmann F, et al. 2018. Multiple roots of fruiting body formation in Amoebozoa. Genome Biol Evol. 10(2):591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, et al. 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 5:3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R.. 2011. A survey of schistosome protein domain types: insights into unique biological properties. Mol Biochem Parasitol. 177(2):100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen H-C, Sheen J.. 2002. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129(2):500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Nakanishi H, Mizuno T.. 2010. Classification of the genes involved in the two-component system of the moss Physcomitrella patens. Biosci Biotechnol Biochem. 74(12):2542–2545. [DOI] [PubMed] [Google Scholar]
- Jackson AP, et al. 2010. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African trypanosomiasis. PloS Negl Trop Dis. 4(4):e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S, Foster AJ, Yemelin A, Thines E.. 2014. Histidine kinases mediate differentiation, stress response, and pathogenicity in Magnaporthe oryzae. Microbiologyopen 3(5):668–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, et al. 2013. Shared signatures of parasitism and phylogenomics unite Cryptomycota and Microsporidia. Curr Biol. 23(16):1548–1553. [DOI] [PubMed] [Google Scholar]
- Jones T, et al. 2004. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 101(19):7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, et al. 2010. The alveolate Perkinsus marinus: biological insights from EST gene discovery. BMC Genomics 11:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Fried L, Behr S, Heermann R.. 2012. Histidine kinases and response regulators in networks. Curr Opin Microbiol. 15(2):118–124. [DOI] [PubMed] [Google Scholar]
- Kabbara S, Schmülling T, Papon N.. 2018. CHASEing cytokinin receptors in plants, bacteria, fungi, and beyond. Trends Plant Sci. 23(3):179–181. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. 1996. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274(5289):982–985. [DOI] [PubMed] [Google Scholar]
- Kamper J, et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444(7115):97–101. [DOI] [PubMed] [Google Scholar]
- Kateriya S, Nagel G, Bamberg E, Hegemann P.. 2004. “Vision” in single-celled algae. News Physiol Sci. 19:133–137. [DOI] [PubMed] [Google Scholar]
- Kemen E, et al. 2011. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 9(7):e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianianmomeni A, Hallmann A.. 2014. Algal photoreceptors: in vivo functions and potential applications. Planta 239(1):1–26. [DOI] [PubMed] [Google Scholar]
- Kim D, Forst S.. 2001. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiol Read Engl. 147(Pt 5):1197–1212. [DOI] [PubMed] [Google Scholar]
- King N, et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451(7180):783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooß S, Lamparter T.. 2017. Cyanobacterial origin of plant phytochromes. Protoplasma 254(1):603–607. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kuwayama H, Kubohara Y.. 2016. Differentiation-inducing factor 2 modulates chemotaxis via the histidine kinase DhkC-dependent pathway in Dictyostelium discoideum. FEBS Lett. 590(6):760–768. [DOI] [PubMed] [Google Scholar]
- Lacey RF, Binder BM.. 2016. Ethylene regulates the physiology of the Cyanobacterium synechocystis sp. PCC 6803 via an ethylene receptor. Plant Physiol. 171(4):2798–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavín JL, Sarasola-Puente V, Ramírez L, Pisabarro AG, Oguiza JA.. 2014. Dual-histidine kinases in basidiomycete fungi. C R Biol. 337(2):111–116. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel OLG.. 2008. An improved, general amino-acid replacement matrix. Mol Biol Evol. 25(7):1307–1320. [DOI] [PubMed] [Google Scholar]
- Liu Z, et al. 2014. Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. Pekinensis). DNA Res. 21(4):379–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus B, et al. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433(7028):865–868. [DOI] [PubMed] [Google Scholar]
- Loftus BJ. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307(5713):1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi H, et al. 2016. Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat Commun. 7: 10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck M, et al. 2012. A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J Biol Chem. 287(47):40083–40090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica R, Sandoval GRP, Rodríguez C, Franco B, Georgellis D.. 2006. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid Redox Signal. 8(5―6):781–795. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45(D1):D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, et al. 2010. Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464(7291):1033–1038. [DOI] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G.. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 70(4):910–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, et al. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature ; 428(6983):653–657. [DOI] [PubMed] [Google Scholar]
- Mira-Rodado V, et al. 2012. Identification of two-component system elements downstream of AHK5 in the stomatal closure response of Arabidopsis thaliana. Plant Signal Behav. 7(11):1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Ayers RA, Moffat K.. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17(10):1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondo SJ, et al. 2017. Widespread adenine N6-methylation of active genes in fungi. Nat Genet. 49(6):964–968. [DOI] [PubMed] [Google Scholar]
- Morrison HG, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317(5846):1921–1926. [DOI] [PubMed] [Google Scholar]
- Mougel C, Zhulin IB.. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem Sci. 26(10):582–584. [DOI] [PubMed] [Google Scholar]
- Neale DB, et al. 2014. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15(3):R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave MJ, Michell CT, Apprill A, Voolstra CR.. 2017. Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci Rep. 7:40579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438(7071):1151–1156. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS.. 2013. Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot. 64(2):445–458. [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R.. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23(9):1073–1079. [DOI] [PubMed] [Google Scholar]
- Pareek A, et al. 2006. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 142(2):380–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock CS, et al. 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 39(7):839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischke MS, et al. 2002. An Arabidopsis histidine kinase is essential for megagametogenesis. Proc Natl Acad Sci U S A. 99(24):15800–15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DC, et al. 2012. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335(6070):843–847. [DOI] [PubMed] [Google Scholar]
- Prochnik SE, et al. 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329(5988):223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Allen JF.. 2009. Chloroplast two-component systems: evolution of the link between photosynthesis and gene expression. Proc Biol Sci. 276(1665):2133–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, et al. 2008. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc Natl Acad Sci U S A. 105(29):10061–10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, et al. 2010. Transcriptional control of photosynthesis genes: the evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biol Evol. 2:888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Ibrahim IM, Allen JF.. 2013. Evolutionary rewiring: a modified prokaryotic gene-regulatory pathway in chloroplasts. Philos Trans R Soc Lond B Biol Sci. 368(1622):20120260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317(5834):86–94. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2013. R: a language and environment for statistical computing. Vienna (Austria) : R Foundation for Statistical Computing. Available from: http://www.R-project.org/.
- Read BA, et al. 2013. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499(7457):209–213. [DOI] [PubMed] [Google Scholar]
- Rensing SA, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319(5859):64–69. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, et al. 2014. Eukaryotic algal phytochromes span the visible spectrum. Proc Natl Acad Sci U S A. 111(10):3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC.. 2017. Phytochrome diversification in cyanobacteria and eukaryotic algae. Curr Opin Plant Biol. 37:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap P. 2011. Evolutionary crossroads in developmental biology: Dictyostelium discoideum. Development 138(3):387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap P, et al. 2016. The Physarum polycephalum genome reveals extensive use of prokaryotic two-component and metazoan-type tyrosine kinase signaling. Genome Biol Evol. 8(1):109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Kieber JJ, Shiu S-H.. 2008. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 6:e0112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Shiu SH, Armitage JP.. 2011. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 21(9):R320–R330. [DOI] [PubMed] [Google Scholar]
- Schliep KP. 2011. Phangorn: phylogenetic analysis in R. Bioinformatics 27(4):592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP.. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 95(11):5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwelm A, et al. 2015. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci Rep. 5:11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadwick LL, Spiegel FW, Shadwick JDL, Brown MW, Silberman JD.. 2009. Eumycetozoa = Amoebozoa? SSUrDNA phylogeny of protosteloid slime molds and its significance for the amoebozoan supergroup. PLoS One 4(8):e6754.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Chauhan N.. 2015. A case for two-component signaling systems as antifungal drug targets. PLoS Pathog. 11(2):e1004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2014. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O, et al. 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 493(7433):526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton CK, Zinda MJ, Mykytka B, Yang P.. 1998. The histidine kinase dhkC regulates the choice between migrating slugs and terminal differentiation in Dictyostelium discoideum. Dev Biol. 203(2):345–357. [DOI] [PubMed] [Google Scholar]
- Staats M, van Kan JA.. 2012. Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot Cell. 11(11):1413–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucgang R, et al. 2011. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol. 12(2):R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, et al. 2013. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 4:2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW.. 2003. From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol. 260(2):289–297. [DOI] [PubMed] [Google Scholar]
- Suzuki I, Kanesaki Y, Mikami K, Kanehisa M, Murata N.. 2001. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol. 40(1):235–244. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Tanaka T, et al. 2015. Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 27(1):162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserant E, et al. 2013. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci U S A. 110(50):20117–20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troein C, et al. 2011. Multiple light inputs to a simple clock circuit allow complex biological rhythms. Plant J Cell Mol Biol. 66(2):375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehling J, et al. 2017. Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ Microbiol. 19(8):2964–2983. [DOI] [PubMed] [Google Scholar]
- Urao T. 1999. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11(9):1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FF, Cheng ST, Wu Y, Ren BZ, Qian W.. 2017. A bacterial receptor PcrK senses the plant hormone cytokinin to promote adaptation to oxidative stress. Cell Rep. 21(10):2940–2951. [DOI] [PubMed] [Google Scholar]
- Wang N, Shaulsky G, Escalante R, Loomis WF.. 1996. A two-component histidine kinase gene that functions in Dictyostelium development. EMBO J. 15(15):3890–3898. [PMC free article] [PubMed] [Google Scholar]
- Wang N, Söderbom F, Anjard C, Shaulsky G, Loomis WF.. 1999. SDF-2 induction of terminal differentiation in Dictyostelium discoideum is mediated by the membrane-spanning sensor kinase DhkA. Mol Cell Biol. 19(7):4750–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, et al. 2006. Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18(12):3429–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YH, et al. 2015. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. eLife 4:e06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, et al. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324(5924):268–272. [DOI] [PubMed] [Google Scholar]
- Xu F, et al. 2014. The genome of Spironucleus salmonicida highlights a fish pathogen adapted to fluctuating environments. PLoS Genet. 10(2):e1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, et al. 2004. The genome of Cryptosporidium hominis. Nature 431(7012):1107–1112. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2011. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 7(9):e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y, Pierik R, Fricker MD, Voesenek LACJ, Harberd NP.. 2012. Studies of Physcomitrella patens reveal that ethylene-mediated submergence responses arose relatively early in land-plant evolution. Plant J Cell Mol Biol. 72(6):947–959. [DOI] [PubMed] [Google Scholar]
- You M, et al. 2013. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 45(2):220–225. [DOI] [PubMed] [Google Scholar]
- Yuan L, et al. 2016. The CKI1 histidine kinase specifies the female gametic precursor of the endosperm. Dev Cell. 37(1):34–46. [DOI] [PubMed] [Google Scholar]
- Zdarska M, et al. 2015. Illuminating light, cytokinin, and ethylene signalling crosstalk in plant development. J Exp Bot. 66(16):4913–4931. [DOI] [PubMed] [Google Scholar]
- Zhang F, Jiang Z, Xu A, Zeng Y, Li C.. 2013. Recent geological events and intrinsic behavior influence the population genetic structure of the Chiru and Tibetan gazelle on the Tibetan Plateau. PLoS One 8(4):e60712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2013. Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat Commun. 4:2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinda MJ, Singleton CK.. 1998. The hybrid histidine kinase dhkB regulates spore germination in Dictyostelium discoideum. Dev Biol. 196(2):171–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.