Abstract
Background:
A number of cohort studies have collected Scope mouthwash samples by mail which are being used for microbiota measurements. We evaluated the stability of Scope mouthwash samples at ambient temperature and determined the comparability of Scope mouthwash with saliva collection using the OMNIgene ORAL kit.
Methods:
Fifty-three healthy volunteers from Mayo Clinic and fifty cohort members from Bangladesh provided oral samples. One aliquot of the OMNIgene ORAL and Scope mouthwash were frozen immediately and one aliquot of the Scope mouthwash remained at ambient temperature for four days and then was frozen. DNA was extracted and the V4 region of the 16S rRNA gene was PCR amplified and sequenced using the HiSeq. Intraclass correlation coefficients (ICC) were calculated.
Results:
The overall stability of the Scope mouthwash samples was relatively high for alpha and beta diversity. For example, the meta-analyzed ICC for the Shannon Index was 0.86 (95% CI: 0.76, 0.96). Similarly, the ICCs for the relative abundance of the top 25 genera were generally high. The comparability of the two sample types was relatively low when measured using ICCs, but were increased by using a Spearman correlation coefficient (SCC) to compare the rank order of individuals.
Conclusions:
Overall, the Scope mouthwash samples appear to be stable at ambient temperature which suggests that oral rinse samples received by the mail can be used for microbial analyses. However, Scope mouthwash samples were distinct compared to OMNIgene ORAL samples.
Impact:
Studies should try to compare oral microbial metrics within one sample collection type.
Keywords: Oral microbiota, sampling methods, stability, concordance
INTRODUCTION
Oral microbiota has been hypothesized to be related to human health and several diseases. In cancer research, oral health has been found to be associated with cancer of the esophagus (1), stomach (2), pancreas (3), and head and neck (4). Oral health has also been found to be associated with oral microbiota (5), particularly plaque samples (6,7), which lends to the hypothesis that oral microbiota directly affects diseases such as cancer (8).
A number of prospective cohort studies have collected oral wash specimens using Scope mouthwash and these samples are being used for nested case-control studies within these cohorts to study cancer outcomes (9). Many of these cohort studies received the oral wash specimens by mail where the sample remained at ambient temperature over the course of a few days prior to processing and freezing. The impact of ambient temperature on human DNA from the oral wash sample has been considered (10–13), but the impact of ambient temperature on microbial DNA from an oral wash sample is not well understood.
Ongoing studies of oral microbiota are using other collection methods for oral samples. One available method, the OMNIgene ORAL kit, advertises stability of saliva samples at ambient temperature for up to three weeks. The comparability of an oral wash collection and saliva collected using the OMNIgene ORAL kit has not been determined.
Therefore, we evaluated the stability of Scope mouthwash samples at ambient temperature and determined the comparability of Scope mouthwash with the OMNIgene ORAL kit within two distinct populations: healthy volunteers from the Mayo Clinic and cohort members of the Health Effects of Arsenic Longitudinal Study (HEALS) in Bangladesh.
MATERIALS AND METHODS
Mayo Clinic study participants
A description of this population has previously been described in detail (14). In brief, 53 healthy volunteers were recruited from Mayo Clinic employees. Participants had to be 18 years or older, not used antibiotics or probiotics within the past two weeks, had no history of pelvic radiation, and not currently undergoing chemotherapy. All participants provided written informed consent and the study was approved by the Mayo Clinic Studies Institutional Review Board and the NCI Office of Human Subjects Research.
HEALS study participants
The HEALS study (15) and the recruitment of participants for the microbiome component of this study (16) have been previously described in detail. In brief, HEALS is a prospective cohort study which recruited participants from Araihazar, Bangladesh from October 2000 to May 2002. For the microbiome collection, HEALS participants living in the 6 nearby villages surrounding the clinic were recruited by trained village health workers to visit the study clinic. In total, 50 participants visited the clinic and completed all of the study procedures. All participants provided written informed consent and the study was approved by the University of Chicago Institutional Review Board and the NCI Office of Human Subjects Research.
Oral specimen collection
For both the Mayo Clinic and HEALS studies, participants were asked to refrain from eating or smoking at least 20 minutes prior to the oral specimen collections. First, the participant provided a saliva sample using the OMNIgene ORAL OM-505 collection device (DNAGenotek, Ontario, Canada). Next, 10 mL of Scope mouthwash was aliquoted into a sterile measuring cup from an individual sized bottle of Scope. The participant was asked to swish the sample for 5 seconds, followed by gargling for 5 seconds, and repeated the swish and gargle for a total of 30 seconds. At the end of 30 seconds, the participant spit the mouthwash back into the collection cup. Then, the participant filled out a short questionnaire regarding tobacco use, alcohol consumption, oral health habits, recent antibiotic exposure, and demographics.
Once the oral samples were collected, the OMNIgene tube was shaken and then incubated at 50°C for 1 hour in a water bath as indicated in the DNAGenotek aliquoting protocol (https://www.dnagenotek.com/us/pdf/PD-PR-00214.pdf). After incubation, one aliquot was created and frozen immediately at −80°C (Day 0). Two aliquots were created from the Scope mouthwash sample. One of the aliquots were frozen immediately at −80°C (Day 0) and the other remained at room temperature for 96 hours (Day 4). At the end of the four days, the remaining aliquot of Scope was frozen at −80°C.
DNA extraction and sequencing
The samples were shipped on dry ice to the University of California, San Diego, thawed at 4˚C, and kept on ice during plating. A wooden swab (Puritan Cotton Tipped Applicators; Puritan Medical Products) was dipped into each aliquot from the OMNIgene kit and Scope mouthwash and then the swab was used for DNA extraction.
DNA extraction, polymerase chain reaction (PCR) amplification and amplicon preparation for sequencing were performed as described previously (14,16). In brief, DNA was extracted using the MO-BIO PowerMag Soil DNA Isolation Kit. Barcoded 515F/806R primers were used to PCR amplify the V4 region of the 16S rRNA gene and barcoded amplicons were pooled with equal concentrations. DNA sequencing was conducted using the Illumina HiSeq. For the samples from Mayo, on average, the OMNIgene ORAL samples had 90,837 reads (SD 26,278 reads) and the Scope mouthwash samples had 77,153 reads (SD 29,662 reads). For the samples from Bangladesh, the OMNIgene ORAL samples had an average of 115,689 reads (SD 52,442 reads) and the Scope mouthwash samples had 115,340 reads (SD 46,941 reads).
Bioinformatic processing
Bioinformatic processing of the data was conducted as described previously (14,16). In brief, reads were demultiplexed and quality filtered using QIIME 1.9 (17). Sub-operational taxonomic units (sOTU) were obtained using the default parameters of Deblur (18). The cleaned read files were joined to make a single biom table, with each sOTU representing a unique 150 base pair sequence. Taxonomy was assigned using QIIME with both Greengenes database version 13.8 (19) and RDP classifier 2.2 (20). A phylogenetic tree for the samples was built using QIIME.
Alpha and beta diversity measures were calculated after rarefaction to 10,000 reads per sample. After rarefaction, from the Mayo Clinic samples, 46 OMNIgene ORAL samples, 50 Scope mouthwash Day 0, and 47 Scope mouthwash Day 4 samples remained. From the Bangladesh samples, 43 OMNIgene ORAL samples, 44 Scope mouthwash Day 0, and 45 Scope mouthwash Day 4 samples remained. Alpha diversity measures (observed sOTUs and the Shannon Diversity index) were calculated using the R phyloseq package (21). The Bray-Curtis distance and Jaccard index were calculated using the R vegan package and unweighted UniFrac, generalized UniFrac, and weighted UniFrac were calculated using the R GUniFrac package (22).
Statistical analysis
Descriptive characteristics of the population were determined from the questionnaire data provided by the participants. We presented the relative abundances at the phylum, family, and genus level for the two collection methods and two populations and tested for a statistical difference between populations for the same sampling method using the PERMANOVA test for the Bray-Curtis distance. Then a distance-based coefficient of determination R2 was calculated to quantify the percentage of microbiota variability explained by subject, collection method, and freezing timepoint using the ‘adonis’ function in the R vegan package using a previously described statistical model with adjustment due to the large degrees of freedom (23). Unweighted, generalized, and weighted UniFrac and the Bray-Curtis distance were used to summarize the overall variability of the microbiota and reflect the shared diversity between bacterial populations in terms of ecological distance.
The stability of the Scope mouthwash samples (Day 0 versus Day 4) and the comparability of the OMNIgene ORAL to the Scope mouthwash were calculated using an intraclass correlation coefficient (ICC) for ten representative microbial community metrics as described previously (24). These metrics included the relative abundance of the top four phyla (Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria), two alpha diversity metrics (observed sOTUs and the Shannon Diversity index), and the five beta diversity matrices (Bray-Curtis, Jaccard, and unweighted, generalized, and weighted UniFrac distances). To further investigate the effects on lower taxonomic levels, selected genera detected in at least 90% of the population in both datasets were included for analysis. The ICCs were calculated using a linear mixed effects model. For the relative abundances at the phylum and genus levels, the ICCs were calculated based on the square-root transformed abundances to reduce the influence of extremely high abundances. The transformation also made the data roughly meet the normality assumption under the mixed effects model. For the four beta-diversity matrices, we used a distance-based ICC, for which the within-subject squared distances and the between-subject squared distances were used to calculate the biological and technical variance (16). Spearman correlation coefficients (SCC) in place of ICCs were used to determine whether the rank order of samples was similar between the two collection methods. For the beta-diversity matrices, SCCs were calculated using all pairwise distances, reflecting the preservation of the inter-sample relationships. For ICC values, we calculated 95% confidence intervals (95% CI) using the R ICC package (CI=‘Smith’) with the exception of the distance-based ICCs and the SCCs which used 1,000 bootstrap samples to calculate 95% CIs.
We also conducted a differential abundance analysis using the Wilcoxon signed-rank test to identify the bacterial taxa at the phylum, family, and genus level which were differentially abundant between Day 0 and Day 4 Scope mouthwash samples or differentially abundance between the OMNIgene ORAL and the Scope mouthwash samples. Taxa read counts were normalized into proportions before analysis and taxa with a prevalence less than 10% or maximum proportion less than 0.2% were excluded from testing. False discovery rate (FDR) control using the Benjamini-Hochberg procedure was used to correct for multiple testing.
RESULTS
Population comparison and overall microbial variability
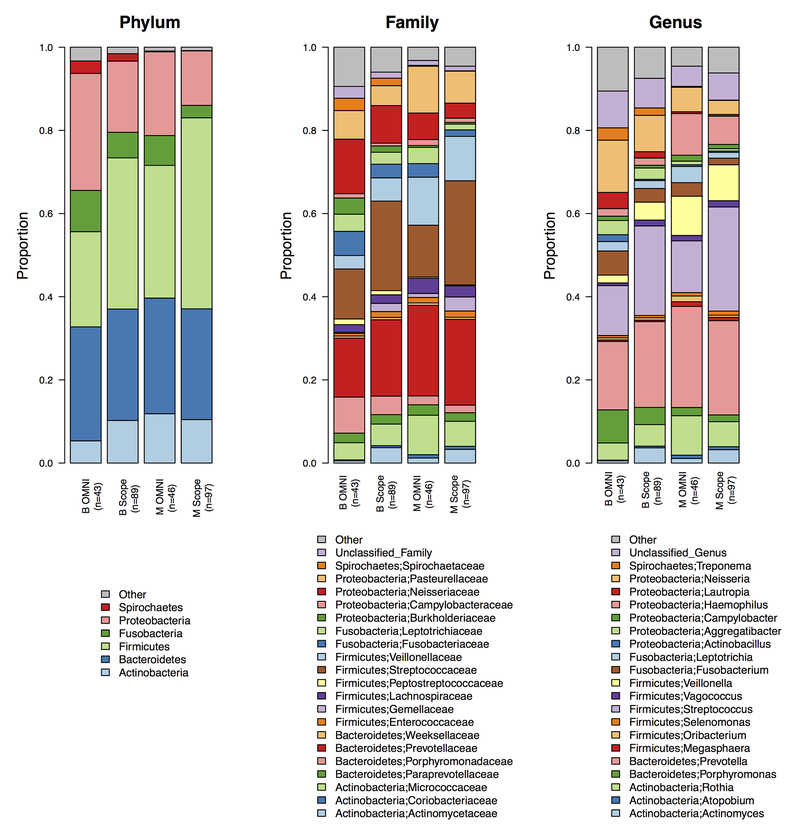
Comparing the relative abundances at the phylum, family, and genus level of the Mayo Clinic and Bangladesh samples showed some differences between populations, but also between sample collection methods. For example, the relative abundance of the phylum Spirochaetes was greater in the Bangladesh samples compared to the Mayo Clinic samples, for both the OMNIgene ORAL and Scope mouthwash. While in the OMNIgene ORAL samples, the relative abundances of the phylum Actinobacteria were greater in the Mayo Clinic samples compared to the Bangladesh samples, but when comparing Scope mouthwash, the relative abundances were similar. Overall, the taxonomic profiles for the two populations were significantly different for both sampling methods (P < 0.001 for all taxonomic ranks using PERMANOVA from the Bray-Curtis distance) (Figure 1).
FIGURE 1.
Stacked barplot of the relative abundances at the phylum, family, and genus level for OMNIgene ORAL (OMNI) and Scope mouthwash samples (both Day 0 and Day 4) from Mayo Clinic (M) and Bangladesh (B). Using the PERMANOVA test for the Bray-Curtis difference, the taxonomic profiles for the two populations were statistically different for both the OMNIgene ORAL and Scope mouthwash collections (P < 0.001).
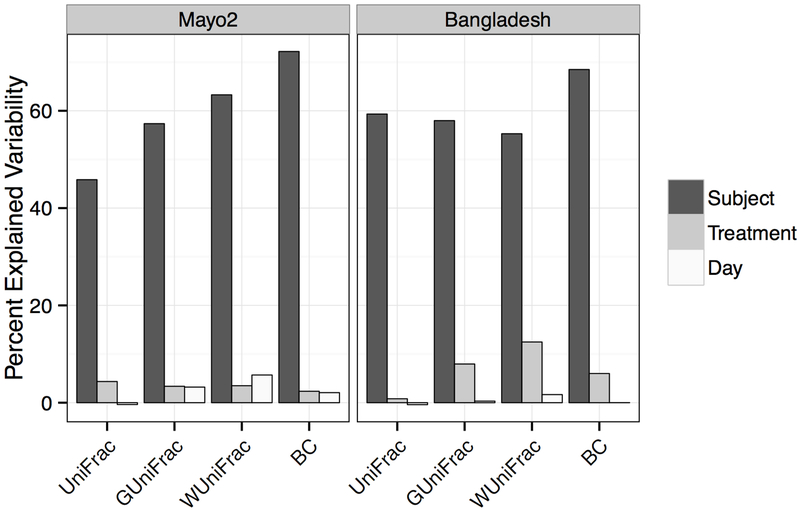
When considering the percent of microbial variability explained by inter-subject, treatment (i.e., Scope mouthwash or OMNIgene ORAL), and day of freezing (i.e., immediately or after 96 hours at ambient temperature), inter-subject variability explained the highest proportion of microbial variability for all measures of beta diversity in both study populations. Some variability was also explained in the Bangladesh samples by the collection method, particularly for weighted UniFrac (Figure 2).
FIGURE 2.
Percent of microbial variability explained by subject (black), sample collection method (grey), and day of freezing (white) was calculated using an adjusted distance-based coefficient of determination R2 for beta-diversity estimates from unweighted UniFrac, generalized UniFrac, weighted UniFrac, and Bray-Curtis (BC) distance for Mayo Clinic and Bangladesh samples.
Stability of Scope mouthwash at ambient temperature
The ICCs for stability of Scope mouthwash samples after four days at ambient temperature measured by the relative abundance of four phyla, two alpha diversity metrics, and four beta diversity matrices were relatively high. For example, the meta-analyzed ICC for the relative abundance of Actinobacteria was 0.78 (95% CI: 0.56, 1.00) and for the Shannon Index the meta-analyzed ICC was 0.86 (95% CI: 0.76, 0.96). The ICCs for the relative abundance of Firmicutes and the unweighted UniFrac matrix were lower (Figure 3; Supplemental Table 1).
FIGURE 3.
Stability of Scope mouthwash samples incubated at ambient temperature for four days (Day 4) compared to samples frozen immediately (Day 0) for the relative abundance of four phyla, two alpha-diversity metrics, and five beta-diversity matrices using intraclass correlation coefficients for Mayo Clinic and Bangladesh samples.
For the relative abundance of the top 25 genera, the ICCs were generally high overall. The meta-analyzed ICCs for the relative abundances of Atopobium, Corynebacterium, Rothia, Capnocytophaga, Porphyromonas, Prevotella, Bulleidia, Catonella, Dialister, Megasphaera, Peptostreptococcus, Selemonas, Veillonella, Fusobacterium, Aggregatibacter, Lautropia, and Neisseria were all greater than 0.75 (Supplemental Figure 1; Supplemental Table 2).
Some of the relative abundances at the phylum, family, and genus level were significantly different at a FDR less than 0.01 after four days at ambient temperature. For example, at the phylum level, an increase of Firmicutes and a decrease of Bacteroidetes, Proteobacteria, and Fusobacteria were detected in both the Mayo Clinic and Bangladesh samples. The increase in Firmicutes appeared to be related specifically to an increase in the Streptococcus genus, while the decrease in Bacteroidetes included decreases in Prevotella, Porphyromonas, and Capnocytophaga (Supplemental Figures 2A and 2B).
Comparability of Scope mouthwash with the OMNIgene ORAL kit
The ICCs for the comparability of Scope mouthwash with the OMNIgene ORAL kit were generally lower, but a few ICCs were acceptable including the relative abundance of Bacteroidetes (ICC 0.77; 95% CI: 0.58, 0.95) and the observed sOTUs (ICC 0.77; 95% CI: 0.61, 0.94) (Figure 4A; Supplemental Table 3). The SCC values for the comparability of Scope mouthwash with the OMNIgene ORAL kit were higher. The highest meta-analyzed SCC was observed for the relative abundance of Proteobacteria with an SCC of 0.81 (95% CI: 0.72, 0.90) (Figure 4B; Supplemental Table 4).
FIGURE 4.
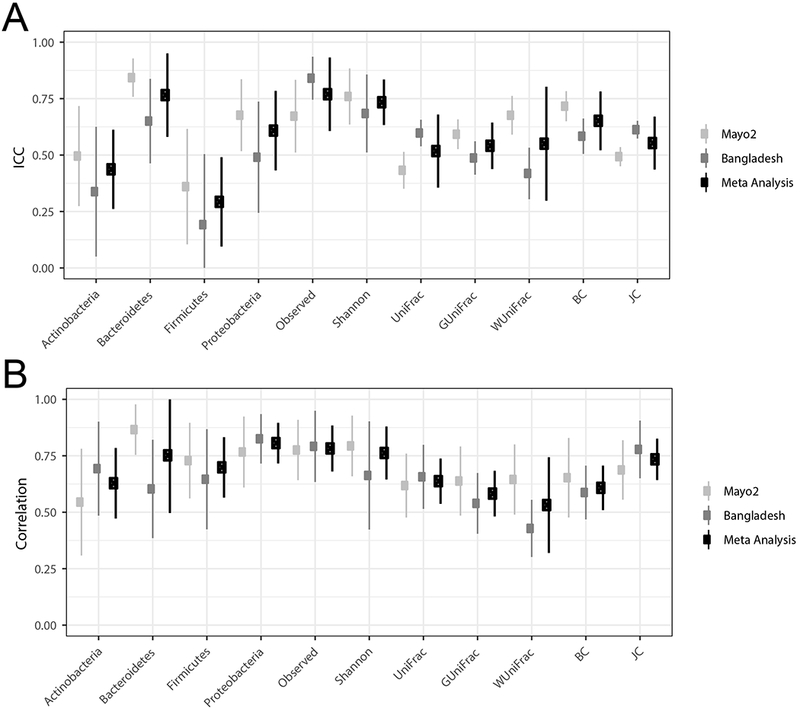
Comparability of the immediately frozen Scope mouthwash to OMNIgene ORAL kit samples for the relative abundance of four phyla, two alpha-diversity metrics, and five beta-diversity matrices using intraclass correlation coefficients (3A) and Spearman correlations (3B) for Mayo Clinic and Bangladesh samples.
For the relative abundance of the top 25 genera, the ICCs were variable, but were greater than 0.75 for Atopobium, Megasphaera, Aggregatibacter, and Lautropia (Supplemental Figure 3A; Supplemental Table 5). The SCCs overall were higher than the ICCs for the relative abundance of the top 25 genera with Porphyromonas, Catonella, Megasphaera, Oribacterium, Peptostreptococcus, Streptococcus, Fusobacterium, Aggregatibacter, Campylobacter, Lautropia, and Neisseria all with SCCs 0.75 or greater (Supplemental Figure 3B; Supplemental Table 6).
Some of the relative abundances at the phylum, family, and genus level were significantly different at a FDR less than 0.01 when comparing the Scope mouthwash with the OMNIgene ORAL samples. Compared to the OMNIgene ORAL samples, the samples collected in Scope mouthwash had higher levels of the phylum Firmicutes in both the Mayo Clinic and Bangladesh samples. The Bangladesh Scope mouthwash samples also had higher levels of the phylum Actinobacteria, but the Mayo Clinic Scope mouthwash samples had lower levels of Actinobacteria. There were consistently lower levels of the phyla Proteobacteria, Fusobacteria, and Spirochaetes in the Scope mouthwash samples compared to the OMNIgene ORAL samples for both Mayo Clinic and Bangladesh samples (Supplemental Figures 4A and 4B).
DISCUSSION
In this study of 53 healthy volunteers from Mayo Clinic and 50 individuals in the HEALS cohort in Bangladesh, microbial variability was primarily explained by between subject differences, although in the Bangladesh samples, some variability was explained by collection method. The stability of Scope mouthwash samples after four days at ambient temperature was high for the relative abundance of four phyla, two alpha diversity metrics, four beta diversity matrices, and the relative abundances of many of the top 25 genera. The relative abundances of some taxa were significantly altered in Scope mouthwash samples after four days at ambient temperature including an increased in the phylum Firmicutes and decreases in Bacteroidetes, Proteobacteria, and Fusobacteria. The comparability of the Scope mouthwash samples to the OMNIgene ORAL samples were relatively low when assessed using ICCs, but the SCC values were generally higher for the relative abundance of four phyla, two alpha diversity metrics, four beta diversity matrices, and the relative abundances of many of the top 25 genera. Specifically, there were significantly higher relative abundances of the phylum Firmicutes and lower levels of Proteobacteria, Fusobacteria, and Spirochaetes in the Scope mouthwash samples compared to the OMNIgene ORAL samples which suggests that studies should make comparisons within a single collection method.
Some previous studies have evaluated the stability of oral samples for microbial analyses. For cheek swabs collected from three individuals, room temperature storage for up to 10 days had no significant effect on microbial diversity or composition (25). Saliva samples from four adults stored in liquid dental transport medium or in an OMNIgene kit had similar bacterial diversity after room temperature storage for 2 to 7 days (26). For oral wash samples, human DNA appeared stable at room temperature for variable lengths of time (10–13) and as seen in this study, a number of microbial metrics were relatively stable at room temperature, but there were some significant differences for the relative abundances of taxa between the samples frozen immediately and those left at room temperature for four days.
Data from the Human Microbiome Project (HMP) gave evidence for distinct community types within the oral cavity (27), however an oral wash specimen was not included in the HMP. In another study which included oral sampling similar to the HMP, but also collected an oral wash sample with Scope mouthwash, found that the buccal cells derived from the oral wash samples were distinct from the other oral samples, although the buccal cells were most similar to the saliva sample (28). When a saliva sample without preservative was compared to a saliva sample collected in an OMNIgene kit, there were no significant differences in the quantity or quality of the extracted DNA. When the saliva sample without preservative was compared to an oral wash sample collected in saline solution, the oral wash sample tended to have increased alpha diversity compared to the saliva, but the difference was not statistically significant. And in general, the beta diversity plots did not show clustering by collection method (29). Overall, we did detect differences between the Scope mouthwash sample and the OMNIgene ORAL sample, but similar to previous findings, the between subject variability tended to outweigh the collection method differences.
This study has some limitations. For the stability calculations, we did not test whether the OMNIgene ORAL sample was stable at room temperature for four days since it is advertised as a kit that is stable for up to three weeks at room temperature. However, it would be important to test this claim. We also did not include an immediately extracted sample since all samples were sent to a central laboratory for DNA extraction, PCR amplification, and sequencing. However, any large epidemiological study would likely not be able to immediately extract all collected samples, so this process represents a more realistic process for sample collection and processing. We did not calculate assay-to-assay laboratory measurement error, so the stability and comparability calculations incorporate laboratory measurement error and temporal or sample collection differences. In addition, we were unable to test stability or comparability differences for rare taxa due to small sample size. Finally, we only assessed the stability and comparability of samples using 16S rRNA gene sequencing and it will be important to understand how these methods may affect other technologies, such as whole genome shotgun metagenomics.
This study also has a number of important strengths. We conducted this study in two distinct populations with unique diets and exposures with similar results for stability and comparability of the collection methods. In addition, the samples collected in Bangladesh were within a larger cohort study and this demonstrates the feasibility of collecting oral samples in a field study. Finally, we used novel statistical methods to evaluate the changes in the relative abundance of specific taxa for the stability of the Scope mouthwash samples and the comparability of the Scope mouthwash to the OMNIgene ORAL samples.
Currently, oral wash samples from a number of prospective cohort studies are being used to evaluate associations between the oral microbiota and adverse health outcomes. Though we found the room temperature storage of Scope mouthwash over four days did not affect the overall oral microbiota as much as different collection methods, we did detect growth or decline of specific taxa over four days at room temperature and the change was relatively consistent between the two studies. Thus, we suggest recording the time at room temperature which then could be adjusted for in the statistical analysis, especially when the time is correlated with the primary variable of interest. Finally, due to the differences between the two oral sample collection methods, we suggest that any new study of the oral microbiota should make comparisons within one collection method.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge Dr. Xianfeng Chen (Department of Health Sciences Research, Mayo Clinic), Dr. Adam Robbins-Pianka, Yoshiki Vazquez Baeza, Grant Gogul, James Gaffney, Greg Humphrey, and Tara Schwartz (Department of Pediatrics, University of California San Diego) for their technical assistance in this study. This work was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health; the Gerstner Family Career Development Awards and Mayo Clinic Center for Individualized Medicine (to JC); grants from the National Institutes of Health (1R01CA179243 to NC and P42ES10349 and R01CA107431 to HA); and the Howard Hughes Medical Institute and the Sloan Foundation awards (to RK).
Abbreviation list:
- CI
Confidence interval
- FDR
False discovery rate
- ICC
Intraclass correlation coefficient
- SCC
Spearman correlation coefficient
- sOTU
Sub-operational taxonomic unit
Footnotes
Conflict of interest: The authors report no conflicts of interest.
Data availability: The datasets generated for the current study are available in the European Nucleotide Archive [ERP015481 and ERP105068].
REFERENCES
- 1.Abnet CC, Kamangar F, Islami F, Nasrollahzadeh D, Brennan P, Aghcheli K, et al. Tooth loss and lack of regular oral hygiene are associated with higher risk of esophageal squamous cell carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2008;17(11):3062–8 doi 10.1158/1055-9965.epi-08-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abnet CC, Qiao YL, Mark SD, Dong ZW, Taylor PR, Dawsey SM. Prospective study of tooth loss and incident esophageal and gastric cancers in China. Cancer Causes Control 2001;12(9):847–54. [DOI] [PubMed] [Google Scholar]
- 3.Stolzenberg-Solomon RZ, Dodd KW, Blaser MJ, Virtamo J, Taylor PR, Albanes D. Tooth loss, pancreatic cancer, and Helicobacter pylori. Am J Clin Nutr 2003;78(1):176–81. [DOI] [PubMed] [Google Scholar]
- 4.Michaud DS, Liu Y, Meyer M, Giovannucci E, Joshipura K. Periodontal disease, tooth loss, and cancer risk in male health professionals: a prospective cohort study. The lancet oncology 2008;9(6):550–8 doi 10.1016/s1470-2045(08)70106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang F, Zeng X, Ning K, Liu KL, Lo CC, Wang W, et al. Saliva microbiomes distinguish caries-active from healthy human populations. The ISME journal 2012;6(1):1–10 doi 10.1038/ismej.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang S, Yang F, Zeng X, Chen J, Li R, Wen T, et al. Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC oral health 2011;11:33 doi 10.1186/1472-6831-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PloS one 2012;7(6):e37919 doi 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. British journal of cancer 2016. doi 10.1038/bjc.2015.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2016. doi 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrisin TE, Humma LM, Johnson JA. Collection of genomic DNA by the noninvasive mouthwash method for use in pharmacogenetic studies. Pharmacotherapy 2002;22(8):954–60. [DOI] [PubMed] [Google Scholar]
- 11.Heath EM, Morken NW, Campbell KA, Tkach D, Boyd EA, Strom DA. Use of buccal cells collected in mouthwash as a source of DNA for clinical testing. Archives of pathology & laboratory medicine 2001;125(1):127–33 doi . [DOI] [PubMed] [Google Scholar]
- 12.Lum A, Le Marchand L. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 1998;7(8):719–24. [PubMed] [Google Scholar]
- 13.Feigelson HS, Rodriguez C, Robertson AS, Jacobs EJ, Calle EE, Reid YA, et al. Determinants of DNA yield and quality from buccal cell samples collected with mouthwash. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2001;10(9):1005–8. [PubMed] [Google Scholar]
- 14.Vogtmann E, Chen J, Amir A, Shi J, Abnet CC, Nelson H, et al. Comparison of Collection Methods for Fecal Samples in Microbiome Studies. Am J Epidemiol 2017;185(2):115–23 doi 10.1093/aje/kww177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahsan H, Chen Y, Parvez F, Argos M, Hussain AI, Momotaj H, et al. Health Effects of Arsenic Longitudinal Study (HEALS): description of a multidisciplinary epidemiologic investigation. Journal of exposure science & environmental epidemiology 2006;16(2):191–205 doi 10.1038/sj.jea.7500449. [DOI] [PubMed] [Google Scholar]
- 16.Vogtmann E, Chen J, Kibriya MG, Chen Y, Islam T, Eunes M, et al. Comparison of fecal collection methods for microbiota studies in Bangladesh. Applied and environmental microbiology 2017. doi 10.1128/aem.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods 2010;7(5):335–6 doi 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017;2(2) doi 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology 2006;72(7):5069–72 doi 10.1128/aem.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic acids research 2009;37(Database issue):D141–5 doi 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one 2013;8(4):e61217 doi 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics (Oxford, England) 2012;28(16):2106–13 doi 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Ryu E, Hathcock M, Ballman K, Chia N, Olson JE, et al. Impact of demographics on human gut microbial diversity in a US Midwest population. PeerJ 2016;4:e1514 doi 10.7717/peerj.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha R, Chen J, Amir A, Vogtmann E, Shi J, Inman KS, et al. Collecting Fecal Samples for Microbiome Analyses in Epidemiology Studies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2016;25(2):407–16 doi 10.1158/1055-9965.epi-15-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017;5(1):52 doi 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo T, Srinivasan U, Ramadugu K, Shedden KA, Neiswanger K, Trumble E, et al. Effects of Specimen Collection Methodologies and Storage Conditions on the Short-Term Stability of Oral Microbiome Taxonomy. Applied and environmental microbiology 2016;82(18):5519–29 doi 10.1128/aem.01132-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome biology 2012;13(6):R42 doi 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G, Phillips S, Gail MH, Goedert JJ, Humphrys M, Ravel J, et al. Evaluation of Buccal Cell Samples for Studies of Oral Microbiota. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2017;26(2):249–53 doi 10.1158/1055-9965.epi-16-0538. [DOI] [PubMed] [Google Scholar]
- 29.Lim Y, Totsika M, Morrison M, Punyadeera C. The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Scientific reports 2017;7(1):8523 doi 10.1038/s41598-017-07885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.