Summary
Seven neuropeptides are expressed within the Drosophila brain circadian network. Our previous mRNA profiling suggested that Allatostatin-C (AstC) is an eighth neuropeptide and specifically expressed in dorsal clock neurons (DN1s). Our results here show that AstC is indeed expressed in DN1s, where it oscillates. AstC is also expressed in two less well-characterized circadian neuronal clusters, the DN3s and lateral posterior neurons (LPNs). Behavioral experiments indicate that clock neuron-derived AstC is required to mediate evening locomotor activity under short (winter-like) and long (summer-like) photoperiods. The AstC-Receptor 2 (AstC-R2) is expressed in LNds, the clock neurons that drive evening locomotor activity, and AstC-R2 is required in these neurons to modulate the same short photoperiod evening phenotype. Ex vivo calcium imaging indicates that AstC directly inhibits a single LNd neuron. The results suggest that a novel AstC/AstC-R2 signaling pathway, from dorsal circadian neurons to an LNd, regulates the evening phase in Drosophila.
Keywords: Neuropeptide, Circadian rhythms, Photoperiod, Behavior, Locomotion, Circadian circuitry, Drosophila
Graphical Abstract
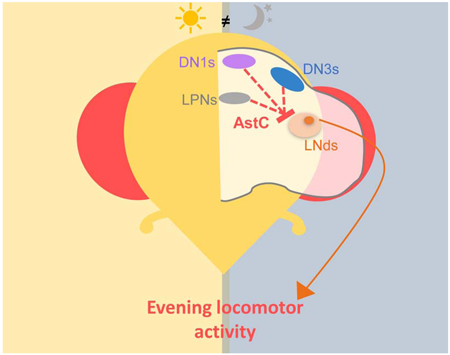
Diaz et al. characterize AstC as a new neuropeptide involved in the Drosophila circadian neural network. AstC released from a subset of the clock neurons inhibits a single LNd to modulate evening activity under short and long photoperiods.
Organisms ranging from cyanobacteria to mammals exhibit circadian rhythms or behavioral and physiological processes that occur in a near 24-hour cycle. The molecular clocks that drive these rhythms are sensitive to environmental cues, which provide time-of-day information. Indeed, one important feature of circadian clocks is their plasticity or their ability to re-entrain to different environmental conditions. Clocks can detect fluctuations in light and temperature and adjust features accordingly [1]. This allows adaptation to changing environments, such as seasonal photoperiods and temperatures.
The fruit fly Drosophila melanogaster is no exception. Under standard 12:12 light/dark (LD) cycles, it manifests a bimodal locomotor activity pattern with prominent and characteristic morning (M) and evening (E) peaks near the lights-on and lights-off transitions, respectively. Importantly, these peaks show anticipatory behaviors that precede these transitions. Flies also adjust these two locomotor activity events to photoperiod length; this presumably reflects an adaptation to different seasonal conditions. For instance, the M- and E-peaks are further apart under long, summer-like days and closer together under short, winter-like days [2].
These behavioral rhythms are driven by ~150 clock neurons within the fly brain. They are subdivided into distinct neuronal clusters: the small and large lateral neurons (s-LNvs and l-LNvs, respectively), the dorsal lateral neurons (LNds), the lateral posterior neurons (LPNs) and the dorsal neurons (DNs). The DNs are the largest group and are further subdivided into two anterior DN1s (DN1as), sixteen posterior DN1s (DN1ps), two DN2s, and approximately 30-40 DN3s. The s-LNvs are also known as morning cells (M-cells) as they are primarily responsible for the timing of the M-peak, whereas the LNds (along with the 5th s-LNv) are also known as the evening cells (E-cells) as they drive the E-peak [3, 4].
The different clock neuron subgroups must communicate within the circadian network, for example to maintain synchrony of their molecular clocks, especially in the face of varying environmental conditions. Although neuronal communication can occur via gap junctions and neurotransmitters as well as neuropeptides, we have focused here on neuropeptides. To date, only seven different neuropeptides have been identified within the circadian neurons. The neuropeptide pigment-dispersing factor (PDF) is expressed in the LNvs, acts as the synchronizing signal to most of the other circadian neurons, and is necessary to maintain rhythmicity in constant darkness [5] [6] [7]. The l-LNvs also express neuropeptide F (NPF) [8], and the s-LNvs co-express short NPF (sNPF) [9]. The activity-promoting E-cells express several different neuropeptides: the 5th s-LNv and five of the six LNds express a combination of NPF, sNPF, and ion transport peptide (ITP) [8] [9] [10]. Among the DN1s, the two DN1as are anatomically and functionally distinct from the DN1ps, partly due to expression of the neuropeptides IPNamide [11] and CCHamide1 [12] in the DN1as. The neuropeptide diuretic hormone 31 (Dh31) is synthesized in five of the DN1ps to influence sleep [13] and possibly temperature preference [14]. In contrast, there are no neuropeptides associated with any of the remaining circadian clusters and neurons: the DN2s, DN3s, the LPNs, and most of the DN1ps.
To identify additional circadian neuropeptides, we exploited RNA-seq data generated from purified LNvs, LNds, and DN1ps to identify known neuropeptide transcripts that are strongly expressed in circadian neurons [15]. We prioritized the DN1p cluster for the possibility of identifying additional neuropeptides, especially within those neurons with uncharacterized signaling mechanisms. Allatostatin-C (AstC) transcripts were highly expressed in the DN1ps and found at much lower levels in the LNvs and the LNds [15]. Intriguingly, transcripts encoding the AstC receptor 2 (AstC-R2) were also detected in the LNds [15]. Thus, we predicted that AstC/AstC-R2 would be a novel intra-clock signaling pathway.
Indeed, AstC is expressed in the DN1s as predicted; it is also expressed in the DN3s and the LPNs. Knockdown experiments indicate that AstC-expressing clock neurons contribute to the evening activity phase that occurs under altered photoperiod. Importantly, knock-down of AstC-R2 within the LNds also exhibits the same phenotype as the AstC knockdown, indicating that dorsal neurons communicate with the LNds. Ex vivo calcium imaging indicates that AstC directly inhibits a single LNd neuron. These experiments therefore identify a novel AstC/AstC-R2 pathway that regulates evening phase under varying environmental conditions.
Results
Identifying AstC as a novel circadian neuropeptide
To characterize gene expression within subpopulations of the Drosophila circadian neural network, our lab previously conducted mRNA profiling on the LNvs, LNds, and a subset of the DN1ps (Figure 1A), and we focused on neuropeptide transcripts [15]. The sequencing data indicated high levels of the Allatostatin-C (AstC) transcript in DN1ps, whereas there are lower levels in the LNds (three times lower; p < 0.0001) and no detectable AstC transcript in the LNvs (Figure 1B).
Figure 1. mRNA sequencing data suggests that Allatostatin-C (AstC) mRNA is expressed in the circadian neurons of Drosophila.
A.. One hemisphere of the clock neurons in an adult Drosophila brain are depicted schematically. The core clock consists of about 150 lateral and dorsal neurons (LNs and DNs, respectively). The ventral LNs are subdivided in the small (s-LNvs) and large neurons (l-LNvs), shown in light and dark green, respectively. The dorsal LNs (LNds) consists of six neurons and the 5th s-LNv, shown in orange. The DNs are subclassified into the approximate 16 DN1s (shown in purple), two DN2s (shown in yellow), and approximately 30-40 DN3s (shown in blue). The three lateral posterior neurons (LPNs) form the last cluster (shown in gray). B. Amount of AstC mRNA transcripts in the three neuronal clusters profiled with deep sequencing: the LNvs, LNds (including the 5th s-LNv), and the posterior DN1s. The DN1s have significantly more AstC transcripts compared to the LNvs and LNds. Values were averaged from 12 sequencing data sets across different timepoints and biological replicates. Boxplot whiskers show 10th-90th percentile. “+” denotes the mean. *** p < 0.0001, one-way ANOVA
AstC is known to bind to two G-protein coupled receptors: AstC-R1 (star1) and AstC-R2 (AlCR2) [16]. AstC-R1 expression was not detectable in the three circadian neuronal groups, but AstC-R2 transcripts was present in the LNds [15]. Because the transcripts for the peptide and one of its receptors were expressed within circadian neurons, we predicted that AstC plays a role in the clock network and pursued these initial findings.
AstC is expressed in the DN1ps, DN3s, and LPNs
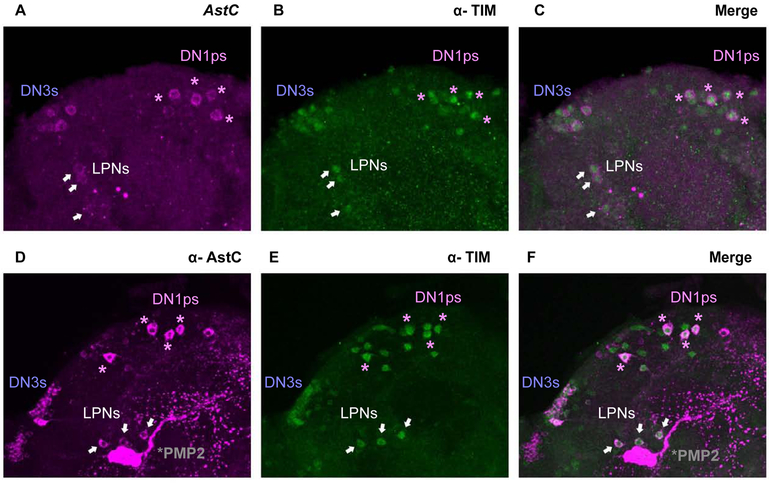
To confirm that AstC transcripts are indeed present in the clock neurons, we took advantage of a recently published fluorescent in situ hybridization (FISH) protocol to visualize AstC mRNA in whole-mount adult Drosophila brains (Figure 2A; see materials and methods). The transcripts were detectable in approximately six neurons in the dorsal protocerebrum, a distribution that is quite similar to previously schematized diagrams of AstC distribution in adult fly brains [17]. To confirm that these are indeed dorsal circadian neurons, we co-stained with antibody against the core clock transcription factor TIMELESS (TIM). The brains were stained at ZT24, a time when TIM protein is abundant (Figure 2B) [18] [19]. Of the five or six AstC-positive neurons in the dorsal brain, only four co-stain with TIM protein (Figure 2C), suggesting that this cluster contains four DN1ps, as well as one or two immediately adjacent non-circadian neurons. Surprisingly, AstC is also expressed in two additional circadian clusters, the DN3s and the LPNs (Figure 2C); these two groups were not profiled [15]. We did not observe AstC transcripts in the location of the LNds (data not shown), indicating that the low AstC transcript levels in the LNds indicated by the sequencing is not detectable by FISH or perhaps reflects non-specific background from the RNA sequencing (Figure 1B; see Methods).
Figure 2. Visualizing AstC in the clock neurons of the adult Drosophila brain.
A. Fluorescent in situ hybridization (FISH) for AstC mRNA transcripts at ZT24. B. TIM antibody staining showing the dorsal clock neurons. C. FISH coupled with immunostaining shows the co-localization of AstC mRNA transcripts (magenta) and TIM antibody (green), revealing AstC transcripts in four DN1ps (purple asterisks), three LPNs (white arrows), and the DN3s. D. Immunostaining of the AstC neuropeptide at ZT20.The posterior medial protocerebral 2 (PMP2, gray asterisk) are previously known to contain AstC. E. TIM antibody staining showing the dorsal clock neurons. F. Co-localization of AstC neuropeptide (magenta) is indeed expressed in a subset of the dorsal clock neurons (anti-TIM, green): four DN1ps (purple asterisks), three LPNs (white arrows), and the DN3s. Images are from maximum projections. Only the posterior, dorsal region of one hemisphere is shown here.
To test whether the AstC mRNA is translated into functional peptide within the circadian neurons, we performed co-immunostaining on adult brains during the late night (ZT20) using antibodies against AstC (Figure 2D) and TIM (Figure 2E). The expression pattern of AstC is very similar to that observed by FISH. The co-localization of AstC and TIM confirms that the AstC peptide is expressed in four DN1ps, approximately 20 of the ~30-40 DN3s neurons, and the three LPNs (Figure 2F). AstC is also present within elaborate arborizations in the dorsal regions. Notably, there is a pair of massive non-circadian bi-lateral neurons near the LPNs, which are AstC-positive with likely extensive processes in nearly the entire brain; these cells were previously annotated as posterior medial protocerebral 2 neurons [17, 20] (PMP2; Figure 2D and 2F, asterisk).
AstC is the first identified neuropeptide in the enigmatic DN3 and LPN circadian clusters. Because previous work from our lab identified the neurotransmitter glutamate within the subset of the DN1ps known to express Dh31 [21], we asked whether AstC is also expressed within this subset of glutamatergic DN1ps. To this end, we expressed GFP with a specific DN1p driver (R51H05-GAL4) known to include Dh31 [13] and glutamate [21]. The co-immunostaining with antibodies against AstC and GFP indicated that the four AstC-positive DN1ps are indeed included within this subset that also produces Dh31 and glutamate (Figure S1A).
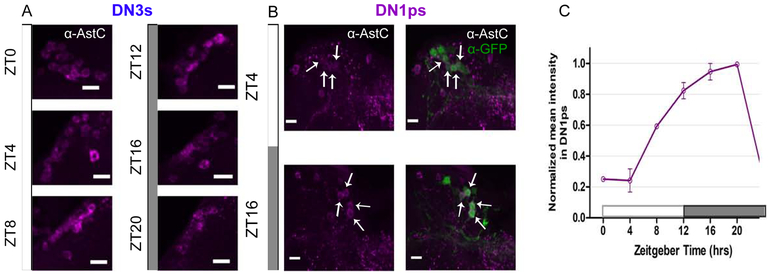
We next asked whether AstC protein levels changes (cycles) in these dorsal clock neurons as a function of time-of-day. We expressed GFP in a subset of the DN1ps and conducted immunohistochemistry co-labeling with anti-GFP and anti-AstC antibodies at six timepoints throughout the 12:12 light:dark (LD) day: ZT0, 4, 8, 12, 16, and 20 (Figure 3).
Figure 3. AstC cycles in the DN1ps, and not in the DN3s.
Young males flies expressing GFP in a subset of the DN1ps (clk4.1-GAL4) were entrained to six timepoints throughout the day under a 12:12 light:dark (LD) cycle. A. The DN3s express AstC throughout all the timepoints. The DN3s were identified by their anatomical location. B. Two representative timepoint images of the DN1ps immunolabeled with anti-AstC (magenta) and GFP (green). The arrows indicate which four AstC-expressing neurons co-localize as the DN1ps. C. Normalized quantification of AstC cycling in the DN1ps under 12:12 LD cycle. AstC is more abundant during the dark phase in comparison to the light phase. Two biological replicates are averaged. Error bars are SEM. Images are from maximum projections. n ≥5 brains per condition. Scale bar= 10 μm. See also Figure S1.
Indeed, AstC cycles throughout the day in the DN1ps: AstC is dramatically reduced during the light phase (between ZT0 and 4), and it accumulates throughout the dark phase, reaching a maximum near ZT20 (Figure 3C). To address whether this cycling pattern in the DN1ps is light-driven, we conducted the same immunolabeling experiment in constant darkness (DD) and observed the same DN1p cycling pattern in DD (Figure S1B), indicating that this is light-independent and therefore almost certainly clock-driven.
The DN3s were identified by their anatomical location. Because AstC can be easily visualized with immunohistochemistry at all time points in LD (Figure 3A), we suspect that there is little synchronous cycling in this large circadian cluster. However, it is impossible to rule out potential AstC cycling in a subset of the DN3s, because these neurons are difficult to quantify due to their number, size variation and dense arrangement. The LPNs are also difficult to visualize across timepoints but for a different reason: they are often obscured by the enormous PMP2 neuron that can completely eclipse this clock cluster.
AstC in the clock neurons contributes to proper timing of the evening phase under short and long photoperiod conditions
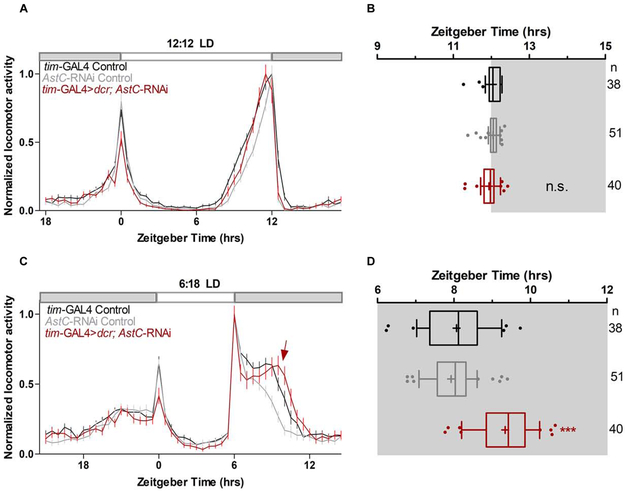
To explore the function of AstC within the clock network, we knocked-down AstC mRNA levels in all clock cells via tim-GAL4 driver-mediated expression of an RNAi and then examined the locomotor behavior of these flies under standard 12:12 LD conditions (see materials and methods). We hypothesized that AstC produced in the dorsal neurons may bind to the AstC-R2 in the LNds and affect the evening activity peak (E-peak). However, the behavior of the AstC knockdown and control flies was indistinguishable under these standard conditions (Figure 4A). The amplitude and timing of the E-peak were also not significantly different among the genotypes (p > 0.05; Figure 4B). The negative result was probably not due to inefficient knockdown as indicated by immunostaining of the AstC RNAi strain (Figure S2). To address whether AstC influences the free-running period, flies were shifted to constant darkness (dark:dark, DD) after entraining under standard 12:12 LD conditions. AstC is not required to maintain normal behavioral rhythms (Table S1), and the free-running period was only marginally lengthened compared to the genetic controls (one-way ANOVA; not statistically significant, p > 0.05).
Figure 4. AstC in clock neurons regulates evening phase in short photoperiod days.
A. Normalized averaged actograms of the tim-GAL4 control (black, n= 38), AstC-RNAi control (gray, n= 51), and the AstC RNAi knock-down in all clock cells mediated by tim-GAL4 (red, n= 40) under standard 12:12 light:dark (LD) conditions at 27°C. White and dark boxes indicates the respective light and dark phases. The error bars represent SEM. B. Boxplot distribution showing the evening peak phase from individual flies. There are no significant differences (p ≥ 0.05). C. Normalized averaged actograms of the tim-GAL4 control, AstC-RNAi control, and the AstC RNAi knock-down in all clock cells mediated by tim-GAL4 under short photoperiods of 6:18 LD at 27°C. The red arrow denotes the delay of the evening peak when AstC is knocked-down compared to the two genetic controls. D. Boxplot distribution showing the evening peak phase from individual flies. The tim-GAL4 mediated AstC-RNAi knock-down (red) is significantly delayed compared to both the tim-GAL4 control (black) and AstC-RNAi control (gray; p < 0.0001). These data are combined from two independent biological replicates. *** p < 0.0001. n.s. no significant difference (p ≥ 0.05). “+” indicates the mean and the whiskers denotes the 10th/90th percentiles. At least two biological replicates are averaged here. See also Figures S2, S3, S4, S5, and Table S1.
Hyperactivity generated by the lights-off event at ZT12 in standard 12:12 LD conditions can override or “mask” more subtle effects on the evening peak [22] [23]. We therefore sought to uncouple the “true” E-peak from this photo-entrainment effect by assaying flies under a short photoperiod. To this end, flies were then shifted to 6:18 LD (6 hours light and 18 hours dark) after one week of entrainment under more standard 12:12 LD conditions (see materials and methods). Under these conditions, control flies exhibit an interruption of their day-time siesta by the lights-off transition at ZT6 with the maximum E-peak occurring approximately two hours later at ZT8 (Figure 4C, D). Flies lacking AstC show a very similar locomotor behavior pattern; however, the E-peak is delayed ~ 1 hour (Figure 4C; arrow). Quantification of the maximum evening peak in individual flies (Figure S3) showed that there was a significant delay in its timing when AstC was knocked-down in clock neurons (9.34 ± 0.11 hrs) in comparison to the controls (GAL4: 8.07 ± 0.15 hrs; UAS: 7.92 ± 0.09 hrs; p < 0.0001; Figure 4D; see materials and methods for details).
To verify this effect, we also used long photoperiod conditions (18:6 LD), under which flies extend their day-time siesta and broaden their evening activity peak. The maximum E-peak is dissociated from the lights-off transition at ZT18 and peaks earlier, at approximately ZT16. Interestingly, AstC knockdown flies exhibited a significant one-hour delay in the E-peak (16.48 ± 0.07 hrs) compared to the controls (GAL4: 15.80 ± 0.10 hrs; UAS: 15.77 ± 0.08 hrs; p < 0.0001) (Figure S5A, B), indicating that AstC can modulate evening activity under long as well as short photoperiods days
To further dissect the role of AstC in the circadian circuit, we focused on short photoperiods. Because tim-GAL4 also drives expression in several non-circadian cell types, we did several experiments to identify from where the knock-down of AstC was affecting the E-peak phenotype. First, AstC immunostaining was performed in the tim-GAL4> UAS-4stC-RNAi flies alongside the parental controls (Figure S2). AstC staining was eliminated in the DN1s, DN3s and LPNs with no changes detected in AstC-expressing neurons outside the circadian network. Second, we knocked-down AstC only in the eye with GMR-GAL4 driver; there were no significant behavioral differences (p > 0.05; Figure S4), consistent with the notion that clock neuron-derived AstC is responsible for mediating the phenotype.
To determine which AstC-positive clock neurons are responsible for this effect, we utilized well-characterized circadian drivers to limit the RNAi expression to more restricted sub-populations. clk856-GAL4 targets nearly the entire clock network [24], including the four AstC-positive DN1ps and three LPNS (Figure S4). Nonetheless, AstC knock down with clk856-GAL4 did not change the evening peak compared to the controls (p > 0.05; Figure S4). There were also no significant differences when AstC was downregulated in the DN1ps with clk4.1M-GAL4 (p > 0.05; Figure S4), also suggesting that DN1 AstC alone is not responsible for the phenotype. These negative data suggest that the remaining AstC-expressing DN3s, unlabeled with the clk856-GAL4 and clk4.1M-GAL4, may be principally responsible for mediating the short photoperiod evening peak phenotype (Figure S4).
AstC acts upon AstC-R2 in the LNds
Given the role of the LNds in generating the evening locomotor activity peak [3] [4], we predicted that AstC acts upon the LNds to affect the evening phase short photoperiod phenotype. Of the two receptors, only AstC-R2 mRNA was detected in the LNds by RNA-sequencing [15]. We therefore knocked-down AstC-R2 in the LNds using dvPDF-GAL4, PDF-GAL80 mediated RNAi expression (Figure 5). Although the AstC-R2 knock-down was nearly identical to the controls in standard 12:12 LD conditions (Figure 5A, B), there was a significant delay in the timing of the E-peak in the AstC-R2 knock-down (9.67 ± 0.11 hrs) compared to the genetic controls (GAL4: 8.32 ± 0.11 hrs; UAS: 7.92 ± 0.11 hrs; p < 0.0001, one-way ANOVA) (Figure 5C, D) under 6:18 LD conditions. Similar effects were seen using a second and novel split GAL4 driver line (MB122B-sGAL4) [25, 26], which also targets the LNds (Figure S6), i.e., there was a significant delay (p < 0.001) in the timing of the E-peak in the knock-down strain (9.05 ± 0.26 hrs) compared to the genetic controls (sGAL4: 7.87 ± 0.10 hrs; UAS: 8.18 ± 0.23 hrs). These phenotypes are essentially identical to those from the AstC knockdowns (Figure 4) and strongly suggest that AstC from dorsal circadian neurons binds to AstC-R2 in the LNds to regulate the timing of the evening peak.
Figure 5. AstC-R2 in LNds is required to regulate evening phase in short photoperiod days.
A. Normalized averaged actograms of the dvPDF-GAL4, PDF-GAL80 control (gray, n= 26), AstC R2-RNAi control (black, n= 15), and the AstC R2-RNAi knock-down in the LNds mediated by dvPDF-GAL4, PDF-GAL80 (orange, n= 21) under standard 12:12 LD condition at 27°C. White and dark boxes indicate the respective light and dark phases. The error bars represent SEM. B. Boxplot distribution showing the evening peak phase from individual flies. There are no significant differences (p ≥ 0.05). C. Normalized averaged actograms of the dvPDF-GAL4, PDF-GAL80 control (gray, n= 40), AstC R2-RNAi control (black, n= 45), and the AstC R2 RNAi knock-down in the LNds mediated by dvPDF-GAL4, PDF-GAL80 (orange, n= 46) under short photoperiod of 6:18 LD at 27°C. The orange arrow denotes the delay of the evening peak when AstC R2 is knocked-down compared to the two genetic controls. D. Boxplot distribution showing the evening peak phase from individual flies. The dvPDF-GAL4, PDF-GAL80 mediated AstC R2-RNAi knock-down (orange) is significantly delayed compared to both the dvPDF-GAL4, PDF-GAL80 control (gray) and AstC R2-RNAi control (black; p < 0.001). ** p < 0.001. n.s. no significant difference (p ≥ 0.05). “+” indicates the mean and the whiskers denotes the 10th/90th percentiles. At least two biological replicates are averaged here. See also Figure S6.
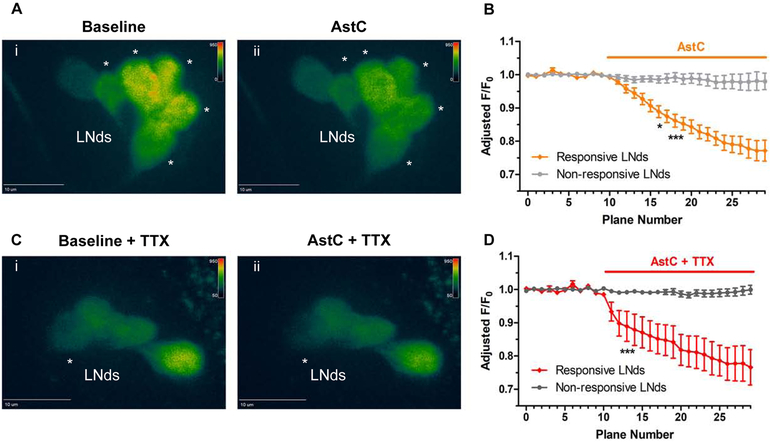
To address how AstC may modulate the LNds, we conducted functional imaging with the calcium sensor GCaMP6f. Young adult fly brains were collected in the evening between ZT9-12, when LNd calcium is expected to be highest [26, 27]. A baseline period was recorded before exposing the explanted brains to 10μM of synthetic AstC peptide (see methods). We observed significant decreases in fluorescence in a subset of the LNds upon AstC application, indicating that these neurons were inhibited (Figure 6A, B).
Figure 6. Functional calcium imaging of the LNds responding to AstC (10 μM).
Flies expressing the calcium sensor GCaMP6 in most clock neurons (clk856-GAL4> UAS-GCaMP6f) were collected in the evening, ZT9-12, and brains were explanted. A. A baseline recording of the LNds was obtained for 10 planes (Ai). When the synthetic AstC peptide (10 μM) was added into the bath, a substantial decrease in fluorescence was observed in multiple LNd neurons (Aii, denoted by asterisks). B. Quantification of the fluorescence relative to the initial baseline recording (F/F0) over time. Values were adjusted to remove trending bleach lines. The LNd neurons showed either a significant decrease in calcium (orange, ‘Responsive LNds’) or were unaffected (light gray, ‘Non-responsive LNds’, less than 10% change in fluorescence). C. To address whether AstC directly inhibited the LNds, the same experiment was conducted with the sodium channel blocker tetrodotoxin (TTX) added to the bath. Similar to (A), a baseline recording was obtained for the LNds (Ci) before exposing the brains to AstC (Cii). The asterisk denotes the single LNd neuron that showed a significant decrease in calcium in response to the AstC treatment. D. the quantification of the direct inhibition of AstC onto a single LNd neuron (red, ‘Responsive LNds’). Most LNd neurons were considered unaffected by having a less than 10% change in fluorescence. n = 6 brains per condition. * p < 0.05, *** p <0.001, two-way ANOVA.
Because the number of inhibited LNds neurons varied widely from only one to five, we were concerned about indirect network interference. To address the extent to which AstC directly inhibits the LNds, the same functional imaging experiments were conducted in the presence of tetrodotoxin (TTX), a voltage-gated sodium channel blocker. Similar decreases of LNd fluorescence signal were observed (Figure 6C, D), indicating that AstC indeed directly inhibits the LNds. However, there was much less variability in the number of responsive LNds in TTX. Although most of the LNds showed no significant changes compared to baseline recording upon AstC application (Figure 6D, gray), a single LNd neuron was consistently responsive with a dramatically decreased calcium signal (Figure 6Cii, asterisk; 6D, red). These data indicate that AstC directly inhibits a single LNd neuron, indicating striking specificity within a substantially heterogeneous clock neuronal cluster.
Discussion
To learn more about how the ~150 clock neurons within the adult fly brain communicate, we examined RNA sequencing data from the LNds, LNvs and DN1s for neuropeptides not yet associated with this circuitry. AstC was a promising candidate because mRNAs encoding both the peptide and one of its receptors (AstC-R2) were identified within the three clock neuron clusters; these data suggested a novel intra-clock circuitry signaling pathway. AstC transcripts as well as the neuropeptide are indeed well-expressed in the DN1s, and the neuropeptide signal undergoes strong cycling in DD as well as LD conditions. Moreover, AstC is also expressed in two other circadian neuron subgroups, the DN3s and the LPNs, and it is the first neuropeptide identified in these circadian clusters. Behavioral data after RNAi knockdown experiments indicate that the AstC binds to AstC-R2 expressed in E-cells to modulate the timing of evening locomotor activity. Ex vivo calcium imaging indicates that AstC directly inhibits a single LNd neuron.
Functional role of AstC and AstC-R2 in clock neurons
AstC is required in the clock neurons to regulate the evening locomotor activity phase in short and long photoperiods, suggesting that “masking” effects in 12:12 LD obscured a phenotypic effect. This shift in the timing of the evening peak occurs when AstC is reduced in all circadian neurons (tim-GAL4 driver).
AstC cycles in the DN1s not only under standard 12:12 LD conditions (Fig 3B, C), but also under DD (Figure S1B) and the short photoperiod condition of 6:18 LD (Figure S1C). In all cases, AstC staining intensity in the DN1 soma is lowest during the early day. We hypothesize that the AstC staining is reduced at this time because the peptide is being transported from the soma to the dendritic arbors for secretion. Indeed, this early day timing correlates with DN1 firing, as DN calcium and firing frequency are highest at this time [25] [28] and thus may be associated with activity-dependent peptide release [29]. This temporal regulation could coincide with the timing of the AstC effect on the phase of the evening locomotor peak. If this is the case however, residual AstC remaining after knock-down in the DN1s with the clk4.1M-GAL4 driver must still be sufficient to promote a wild-type phenotype (Figure S4).
In addition, the LPNs are probably not a key circadian source of AstC: their AstC levels were also dramatically reduced in the clk856-GAL4 mediated knock-down without any phenotypic effect (Figure S4). All AstC-expressing DN3s are targeted by the tim-GAL4 driver, yet most of these DN3s are not included in the clk856-GAL4 driver (Figure S4). Although we cannot exclude a tim-GAL4 source from outside the circadian network, these results suggest that the DN3s are the key source of AstC. We note that this tentative conclusion is based on negative data, and the lack of a DN3-specific GAL4 driver makes it impossible to test this model directly. Therefore, we propose three possible models: 1) the DN3s are the primary source of AstC within the circadian circuit (Figure 7A); 2) the DN1s, DN3s, and LPNs, or some combination, are functionally redundant (Figure 7B); 3) A small amount of residual AstC within DN1s is sufficient for its behavioral role in the evening activity peak assay. Although neuron-specific deletion of AstC might contribute to distinguishing between these three possibilities, we do not have an accurate and efficient CRISPR-based strategy for achieving temporal and spatial specificity.
Figure 7. Proposed model for AstC/AstC-R2 signaling in the circadian circuitry of Drosophila.
A. The DN3s are the integrator for light/dark duration to sense changes in photoperiod conditions. AstC solely in the DN3s inhibits a single LNd neuron via AstC-R2 to regulate the evening phase, as revealed under short photoperiod. B. It is unclear which group of clock neurons serves as the critical integrator of the different photoperiod conditions. Due to lack of a DN3-specific driver, we propose this second model: a combination of the AstC-containing DN1ps, DN3s, and LPNs, together inhibit an LNd neuron to regulate the evening peak, indicating a functional redundancy in neuropeptide signaling. C-D. The functional role of AstC in the circadian circuitry is to precisely regulate evening phase, as seen under short (C) and long (D) photoperiod condition. Because the reduction of AstC leads to a delay in the evening phase, AstC must normally act to regulate the locomotor evening offset.
Once released by dorsal circadian neurons, AstC signals to the LNds via binding to its receptor, AstC-R2. This is because AstC-R2 knock-down in the LNds and knockdown of AstC in the entire circadian circuit give rise to the identical delayed evening peak phenotype (Figure 4, Figure 5, Figure S6). Moreover, the DNs and the LNds both extend projections to the dorsal protocerebrum region (near the pars intercerebralis), where they come in close proximity [30]. In summary, we add a new player to the neuronal circuitry governing E-peak modulation. We suggest that the LNd neuronal activity is not only modulated by signaling from M-cells [31] but also by AstC signaling from the dorsal region of the brain.
Our functional imaging strongly indicates that AstC binding to LNd-localized AstC-R2 leads to neuronal inhibition. The effect is consistent with several previous electrophysiology experiments [16, 32, 33] and contributes to an emerging theme of LNd inhibition [21, 26, 27, 34]. It is also one of the first pieces of evidence indicating that the dorsal neurons can be a source of inhibition onto the LNds [21]. Interestingly, only a single LNd neuron is directly AstC-sensitive, further attesting to LNd heterogeneity [9] [8] [10] [35] and suggesting that behavioral regulation of the evening phase arises from this signaling to a single LNd neuron. We suggest that this response is communicated directly to the rest of the LNds, for example via gap junctions, but we cannot exclude a more indirect and circuitous route of communication.
We note that there are several caveats to the current model. The AstC and AstC-R2 knockdown experiments were done with only a single RNAi line each, due to the lack of additional functional RNAi lines. However, the two experiments show essentially identical phenotypes suggesting that off-target effects are unlikely to pose a big problem. We have also been unable to rule out the possibility that the AstC knockdown phenotypes are not due to a developmental requirement. Experiments to address this point are challenging because when the temperature is raised to reduce tubgal80ts repression and allow for adult-only knockdown, the heat itself dramatically changes the evening peak timing (data not shown). Lastly, lack of LPN and DN3-specific drivers precluded addressing whether DN3-derived AstC is required for this evening activity peak modulation, and whether the DN3s can directly inhibit the LNds.
A conserved role of Allatostatin-C/ mammalian somatostatin in photoperiodism
Although the AstC peptide sequence is highly conserved among insect species [36] [37] [38], only the AstC-R2 receptor has a mammalian homolog, the somatostatin/galanin/opioid receptor family [16, 32, 33]. Inhibitory somatostatin (SST) interneurons are present in the mammalian equivalent of a central core clock, the suprachiasmatic nucleus (SCN) [39] [40] [41]. SST interneurons are also known to affect sleep [42] and circadian behaviors [40]. Interestingly, SST is associated with proper adaptation under photoperiod conditions for both diurnal and nocturnal mammals [43-45], suggesting a highly conserved function with AstC/AstC-R2 for adaptation under different equinox environments. It will be interesting to see whether the SCN-resident SST interneurons are important for this adaptation, like the AstC-containing clock neurons described in this study.
Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Rosbash (rosbash@brandeis.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster strains were reared on standard cornmeal/agar medium supplemented with yeast under 12:12 LD cycles at 25 °C. The following transgenic flies were used for behavior: Tim-GAL4 (yw; Tim-GAL4/CyO) [46], Clk4.1M-GAL4 [47], Clk856-GAL4 [24], GMR-GAL4 [48], dvPDF-GAL4 [49], PDF-GAL80 [4], and UAS-Dicer2. The LNd split-GAL4 (JRC_MB122B)) was provided by Gerald M. Rubin. UAS-AstC RNAi and UAS-AstC-R2 RNAi lines (nos. 102735KK and 106146KK, respectively) were from the Vienna Drosophila RNAi Center. The GAL4 controls were crossed into the empty background vector of the RNAi (60100KK). Microscopy experiments involved w1118 for wild-type flies, R51H05-GAL4 [25], and UAS-EGFP. Young, male flies were used for all experiments.
METHOD DETAILS
Fluorescent in situ hybridization (FISH)
FISH was performed as described previously [51] onto wild-type flies (w1118) at ZT24 with the following exceptions: custom oligo probes were ordered against the entire AstC mRNA sequence, including the 5’ and 3’ untranslated regions, and conjugated with Quasar 670 dye (Stellaris Probes, Biosearch Technologies) (Table S2). Probes were aliquoted and frozen at −20°C with a stock concentration of 25 μM. For the hybridization reaction of the probes onto the brains, the oligo probes were diluted to a final concentration of 0.75 μM. Immediately following the FISH protocol, the brains were blocked with 10% normal goat serum for two hours at room temperature before incubating in primary antibody overnight at 4°C (α- TIM 1:200). The brains were then fluorescently labeled with Alexa Flour 488 conjugated anti-rat at 1:500 for four hours at room temperature. Lastly, the brains were washed before mounting onto slides with Vectashield Mounting Medium (Vector Laboratories). The slides were immediately viewed on a Zeiss 880 series confocal microscope with a 25x oil objective. The z-stack was sequentially imaged in 0.8-μm sections.
Immunostaining
Wild-type flies (w1118) were entrained for three days before collecting at their respective timepoints. Fly heads were fixed in PBS with 4% paraformaldehyde and 0.008% Triton X-100 for 60-65 min at room temperature while rotating. Fixed heads were washed in PBS with 0.5% Triton X-100 (PBS-T) and then dissected in PBS-T. The brains were blocked in 10% normal goat serum (NGS; Jackson Immunoresearch) for an hour at room temperature. The brains were later incubated with primary antibodies at 4 °C for three nights. For TIM and AstC co-staining, rat anti-TIM antibody (1:200) and rabbit anti-Manduca AstC antibody (1:250, gift from Dr. Jan Veenstra) [52] were used as primary antibodies. For GFP and AstC co-staining, mouse anti-GFP antibody (1:1000) and rabbit anti-Manduca AstC antibody (1:250, gift from Dr. Jan Veenstra) were used as primary antibodies. After three washes with PBST, the brains were incubated with either Alexa Fluor 488-conjugated anti-rat or anti-mouse, and Alexa Fluor 635-conjugated anti-rabbit (Molecular Probes) at 1:500 dilutions in 10% NGS. After the brains were washed three more times, the brains were mounted in Vectashield Mounting Medium (Vector Laboratories). The slides were immediately viewed on a Leica SP5 confocal microscope with a 20x objective and sequentially imaged in 0.8-μm sections. For experiments showing the validation of the RNAi efficiency, the laser intensity and other settings were set the same across the different genotypes for each experiment.
Locomotor behavior assay
We used the Drosophila Activity Monitoring system (Trikinetics, Waltham, MA, USA) to record the number of beam crosses caused by the fly in one-min intervals. Young, male flies were individually placed in glass tubes containing agar-sucrose media. The temperature was constant at 27°C for increased knock-down efficiency in all experiments [53]. Flies were allowed one day of habituation before six days of entrainment under 12:12 LD. For the determination of the evening peak timing under short and long days, the flies were then transferred to either 6:18 LD or 18:6 LD for six additional days. Only the final four days of locomotor activity were used in our analysis (i.e., days 3-6 of short or long photoperiod).
Functional calcium imaging
The calcium sensor GCaMP6f was expressed in most clock neurons using clk856-GAL4. Young male flies were entrained to a standard 12:12 LD to be collected at evening time between ZT9-12. Adult male fly brains were dissected in room temperature adult hemolymph-like saline (AHL) (108 mM NaCl, 5 mM KCl, 2 mM CaCl2, 8.2 mM MgCl2, 4 mM NaHCO3, 1 mM NaH2PO4 -H2O, 5 mM trehalose, 10 mM sucrose, 5 mM HEPES, pH 7.5) [55]. Brains were then pinned to a layer of Sylgard (Dow Corning, Midland, MI) silicone under a small bath of AHL contained within a recording/perfusion chamber (Warner Instruments, Hamden, CT, RC-26G) and bathed with room temperature AHL. Brains expressing GCaMP6f were exposed to the fluorescent light for approximately one to two minutes before imaging to allow for baseline fluorescence stabilization of the LNds. Perfusion flow was established over the brain with a gravity-fed ValveLink perfusion system (Automate Scientific, Berkeley, CA). A spinning disk confocal (Intelligent Imaging Innovations, Inc., Denver, CO) was used to visualize all the LNds possible by recording a z-stack with a step size ranging from 2-2.5 μm. After 10 planes of baseline recording with AHL, 10 μM AstC (GenScript, Piscataway, New Jersey) was delivered by switching the perfusion flow until the end of the recording for a total of 30 planes.
For tetrodotoxin (TTX) experiments, brains were dissected with regular AHL before being pre-incubated in AHL containing 1 uM TTX for at least 5 minutes. The brains were then recorded in the same parameters as above, with AHL+TTX (1 uM) as a baseline recording before exposing the brains to AstC (10 uM) + TTX (1 uM).
QUANTIFICATION AND STATISTICAL ANALYSIS
For the distribution of the differential sequencing of AstC, the mRNA transcripts were averaged across the entire AstC gene for each of the twelve data sets previously described in [15, 50]. The twelve independent data sets originate from two biological replicates of each of the six time-of-day collections. Statistical analysis was performed on the Prism 5 software (GraphPad) using a one-way ANOVA with a Tukey's multiple-comparisons test. For the behavioral analyses, average actograms for either short or long photoperiod days 3–6 were plotted for each fly (Figure S3). To quantify the timing of the evening peak, actograms from individual flies (average of days 3-6) were analyzed. The highest activity bout after the lights-off startle response was scored as the short-day evening peak. Under long days, the highest activity bout preceding the lights-off transition was scored as the evening peak as described previously [54]. It is important to note that the evening peak phase of different groups can be quite similar even though the average actogram looks quite different (Figure 4C and D). Small changes in activity from fly-to-fly can lead to differences in evening peak timing that are not apparent in the averaged activity data. See the sample distribution Figure 4D and individual fly examples in Figure S3. Statistical analyses were performed on the Prism 5 software (GraphPad) using a one-way ANOVA with a Tukey’s multiple-comparisons test. For the functional calcium imaging, the SlideBook Reader software (Intelligent Imaging Innovations, Inc., Denver, CO) was used to subtract the background from the signal intensities of the regions of interest. After the background subtraction, the initial F/F0 values were exported. Based on the 10 planes of baseline recording, a bleach trend line was calculated for each sample (MATLAB 2017, MathWorks, Natick, MA), and subtracted from the F/F0. Therefore, the adjusted F/F0 reflects any true fluorescent signal changes over time. Individual LNd neurons were considered ‘responsive’ if there was at least a 10% change in fluorescence signal (Figure 6). Statistical analyses were performed on the Prism 5 software (GraphPad) using a two-way ANOVA. For each dataset, details of n-values can be found in the corresponding Figure Legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Manduca sexta anti-AstC | Laboratory of Jan Veenstra, France [52] | N/A |
| Mouse monoclonal anti-GFP | Sigma-Aldrich | Cat#G6539 |
| Rat monoclonal anti-TIM | Laboratory of Michael Rosbash, USA [19] | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AstC synthetic peptide | This paper | N/A |
| Tetrodotoxin | Abcam | Cat#ab120054 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: Tim-GAL4 | Laboratory of Jeff Hall, USA | RRID: BDSC_7126 |
| D. melanogaster: Clk856-GAL4 | Laboratory of Orie Shafer, USA | N/A |
| D. melanogaster: Clk4.1M-GAL4 | Laboratory of Paul Hardin, USA | RRID: BDSC_36316 |
| D. melanogaster: GMR-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_1104 |
| D. melanogaster: R51H05-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_69036 |
| D. melanogaster: dvPDF-GAL4 | Laboratory of Jae Park, USA | N/A |
| D. melanogaster: JRC_MB122B- split GAL4 | Laboratory of Gerald Rubin, USA | N/A |
| D. melanogaster: PDF-GAL80 | Laboratory of Michael Rosbash, USA | N/A |
| D. melanogaster: UAS-EGFP | Bloomington Drosophila Stock Center | RRID: BDSC_5428 |
| D. melanogaster: UAS-GCaMP6f | Bloomington Drosophila Stock Center | RRID: BDRC_42747 |
| D. melanogaster: UAS- AstC RNAi | Vienna Drosophila Resource Center | VDRC ID: 102735 KK; RRID: FlyBase FBst0478291 |
| D. melanogaster: UAS- AstC-R2 RNAi | Vienna Drosophila Resource Center | VDRC ID: 106146 KK |
| D. melanogaster: Control KK | Vienna Drosophila Resource Center | VDRC ID: 60100 KK |
| Oligonucleotides | ||
| AstC FISH probes, see Table S2 | This paper | N/A |
| Software and Algorithms | ||
| Stellaris Probe Designer version 4.2 | LGC Biosearch Technologies | https://www.biosearchtech.com/stellaris-designer |
| Leica Application Suite X | Leica Application Suite X | RRID:SCR_013673 |
| Zeiss LSM 800 | Zeiss LSM 800 | RRID:SCR_015963 |
| SlideBook | Intelligent Imaging Innovations | RRID: SCR_014300 |
| ImageJ | ImageJ | RRID:SCR_003070 |
| MATLAB R2017b | MathWorks | RRID: SCR_001622 |
| Microsoft Excel | Microsoft | RRID: SCR_016137 |
| Prism5 | GraphPad | RRID:SCR_002798 |
| Other | ||
| RNA-sequencing of isolated clock neurons | [50] [15] | N/A |
| Original fluorescent in situ hybridization method | [51] | N/A |
| Evening phase analysis in varying photoperiod | [54] | N/A |
Highlights.
AstC is a novel signaling neuropeptide in the Drosophila clock circuit.
AstC is expressed in the DN1, DN3, and LPN clock neuron clusters.
AstC inhibits a single LNd clock cell to modulate seasonal evening activity.
Acknowledgements
Many thanks to members of the Rosbash lab for several thoughtful discussions, and especially Dr. Fang Guo, Meghana Holla, Patrick Weidner, and José Carlos González for comments. Also, Muibat Yussuf for assistance and fly maintenance. Much appreciation to members of the Griffith Lab at Brandeis University, especially Johanna “Joey” Adams and Dr. Timothy Wiggin for post-hoc analysis of the functional imaging. Thanks to Dr. Orie Shafer for guidance. Thank you to Drs. Robert Singer and Timothee Lionnet for technical assistance with the FISH protocol and to Ed Dougherty for microscopy support. The AstC antibody was a generous gift from Dr. Jan Veenstra. We thank the Bloomington Stock Center and Vienna Drosophila RNAi Center for flies. M.D. was sponsored by NIH Neuroscience Training Grant #T32MH019929. M.S. was sponsored by a German Research Foundation (DFG) fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- 1.Majercak J, Sidote D, Hardin PE, and Edery I (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230. [DOI] [PubMed] [Google Scholar]
- 2.Yoshii T, Rieger D, and Helfrich-Forster C (2012). Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila. Prog Brain Res 199, 59–82. [DOI] [PubMed] [Google Scholar]
- 3.Grima B, Chelot E, Xia R, and Rouyer F (2004). Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873. [DOI] [PubMed] [Google Scholar]
- 4.Stoleru D, Peng Y, Agosto J, and Rosbash M (2004). Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868. [DOI] [PubMed] [Google Scholar]
- 5.Renn SC, Park JH, Rosbash M, Hall JC, and Taghert PH (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. [DOI] [PubMed] [Google Scholar]
- 6.Peng Y, Stoleru D, Levine JD, Hall JC, and Rosbash M (2003). Drosophila Free-Running Rhythms Require Intercellular Communication. PLoS Biol 1, E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Stormo GD, and Taghert PH (2004). The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. The Journal of neuroscience : the official journal of the Society for Neuroscience 24, 7951–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermann C, Yoshii T, Dusik V, and Helfrich-Forster C (2012). Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. The Journal of comparative neurology 520, 970–987. [DOI] [PubMed] [Google Scholar]
- 9.Johard HA, Yoishii T, Dircksen H, Cusumano P, and Rouyer F (2009). Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. The Journal of comparative neurology 516, 59–73. [DOI] [PubMed] [Google Scholar]
- 10.Hermann-Luibl C, Yoshii T, Senthilan PR, Dircksen H, and Helfrich-Forster C (2014). The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 9522–9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shafer OT, Helfrich-Forster C, Renn SC, and Taghert PH (2006). Reevaluation of Drosophila melanogaster's neuronal circadian pacemakers reveals new neuronal classes. The Journal of comparative neurology 498, 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara Y, Hermann-Luibl C, Katsura M, Sekiguchi M, Ida T, Helfrich-Förster C, and Yoshii T (2018). The CCHamide1 Neuropeptide Expressed in the Anterior Dorsal Neuron 1 Conveys a Circadian Signal to the Ventral Lateral Neurons in Drosophila melanogaster. Frontiers in Physiology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, and Nitabach MN (2014). Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol 24, 2652–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goda T, Tang X, Umezaki Y, Chu ML, and Hamada FN (2016). Drosophila DH31 Neuropeptide and PDF Receptor Regulate Night-Onset Temperature Preference. The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 11739–11754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, Shafer O, and Rosbash M (2017). RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet 13, e1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreienkamp HJ, Larusson HJ, Witte I, Roeder T, Birgul N, Honck HH, Harder S, Ellinghausen G, Buck F, and Richter D (2002). Functional annotation of two orphan G-protein-coupled receptors, Drostar1 and −2, from Drosophila melanogaster and their ligands by reverse pharmacology. The Journal of biological chemistry 277, 39937–39943. [DOI] [PubMed] [Google Scholar]
- 17.Price MD, Merte J, Nichols R, Koladich PM, Tobe SS, and Bendena WG (2002). Drosophila melanogaster flatline encodes a myotropin orthologue to Manduca sexta allatostatin. Peptides 23, 787–794. [DOI] [PubMed] [Google Scholar]
- 18.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, and Young MW (1996). Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 271, 1736–1740. [DOI] [PubMed] [Google Scholar]
- 19.Zeng H, Qian Z, Myers MP, and Rosbash M (1996). A light-entrainment mechanism for the Drosophila circadian clock. Nature 380, 129–135. [DOI] [PubMed] [Google Scholar]
- 20.Zitnan D, Sehnal F, and Bryant PJ (1993). Neurons producing specific neuropeptides in the central nervous system of normal and pupariation-delayed Drosophila. Developmental biology 156, 117–135. [DOI] [PubMed] [Google Scholar]
- 21.Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, and Rosbash M (2016). Circadian neuron feedback controls the Drosophila sleep--activity profile. Nature 536, 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieger D, Stanewsky R, and Helfrich-Forster C (2003). Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. Journal of biological rhythms 18, 377–391. [DOI] [PubMed] [Google Scholar]
- 23.Lu B, Liu W, Guo F, and Guo A (2008). Circadian modulation of light-induced locomotion responses in Drosophila melanogaster. Genes Brain Behav 7, 730–739. [DOI] [PubMed] [Google Scholar]
- 24.Gummadova JO, Coutts GA, and Glossop NR (2009). Analysis of the Drosophila Clock promoter reveals heterogeneity in expression between subgroups of central oscillator cells and identifies a novel enhancer region. Journal of biological rhythms 24, 353–367. [DOI] [PubMed] [Google Scholar]
- 25.Guo F, Chen X, and Rosbash M (2017). Temporal calcium profiling of specific circadian neurons in freely moving flies. Proc Natl Acad Sci U S A 114, E8780–E8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang X, Holy TE, and Taghert PH (2017). A Series of Suppressive Signals within the Drosophila Circadian Neural Circuit Generates Sequential Daily Outputs. Neuron 94, 1173–1189 e1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang X, Holy TE, and Taghert PH (2016). Synchronous Drosophila circadian pacemakers display nonsynchronous Ca(2)(+) rhythms in vivo. Science 351, 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flourakis M, Kula-Eversole E, Hutchison AL, Han TH, Aranda K, Moose DL, White KP, Dinner AR, Lear BC, Ren D, et al. (2015). A Conserved Bicycle Model for Circadian Clock Control of Membrane Excitability. Cell 162, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richhariya S, Jayakumar S, Kumar Sukumar S, and Hasan G (2018). dSTIM and Ral/Exocyst Mediated Synaptic Release from Pupal Dopaminergic Neurons Sustains Drosophila Flight. eneuro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helfrich-Forster C (2003). The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Tech 62, 94–102. [DOI] [PubMed] [Google Scholar]
- 31.Stoleru D, Nawathean P, Fernandez MP, Menet JS, Ceriani MF, and Rosbash M (2007). The Drosophila circadian network is a seasonal timer. Cell 129, 207–219. [DOI] [PubMed] [Google Scholar]
- 32.Birgul N, Weise C, Kreienkamp HJ, and Richter D (1999). Reverse physiology in drosophila: identification of a novel allatostatin-like neuropeptide and its cognate receptor structurally related to the mammalian somatostatin/galanin/opioid receptor family. Embo j 18, 5892–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenz C, Williamson M, and Grimmelikhuijzen CJ (2000). Molecular cloning and genomic organization of a second probable allatostatin receptor from Drosophila melanogaster. Biochem Biophys Res Commun 273, 571–577. [DOI] [PubMed] [Google Scholar]
- 34.Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, and Taghert PH (2008). Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron 58, 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao Z, and Shafer OT (2014). The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343, 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson M, Lenz C, Winther AM, Nassel DR, and Grimmelikhuijzen CJ (2001). Molecular cloning, genomic organization, and expression of a C-type (Manduca sexta-type) allatostatin preprohormone from Drosophila melanogaster. Biochem Biophys Res Commun 282, 124–130. [DOI] [PubMed] [Google Scholar]
- 37.Veenstra JA (2009). Allatostatin C and its paralog allatostatin double C: The arthropod somatostatins. Insect biochemistry and molecular biology 39, 161–170. [DOI] [PubMed] [Google Scholar]
- 38.Veenstra JA (2016). Allatostatins C, double C and triple C, the result of a local gene triplication in an ancestral arthropod. General and Comparative Endocrinology 230-231, 153–157. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka M, Okamura H, Matsuda T, Shigeyoshi Y, Hisa Y, Chihara K, and Ibata Y (1996). Somatostatin neurons form a distinct peptidergic neuronal group in the rat suprachiasmatic nucleus: a double labeling in situ hybridization study. Neurosci Lett 215, 119–122. [PubMed] [Google Scholar]
- 40.Fukuhara C, Hamada T, Shibata S, Watanabe S, Aoki K, and Inouye SI (1994). Phase advances of circadian rhythms in somatostatin depleted rats: effects of cysteamine on rhythms of locomotor activity and electrical discharge of the suprachiasmatic nucleus. J Comp Physiol A 175, 677–685. [DOI] [PubMed] [Google Scholar]
- 41.Biemans BA, Gerkema MP, and Van der Zee EA (2002). Increase in somatostatin immunoreactivity in the suprachiasmatic nucleus of aged Wistar rats. Brain Res 958, 463–467. [DOI] [PubMed] [Google Scholar]
- 42.Funk CM, Peelman K, Bellesi M, Marshall W, Cirelli C, and Tononi G (2017). Role of Somatostatin-Positive Cortical Interneurons in the Generation of Sleep Slow Waves. The Journal of neuroscience : the official journal of the Society for Neuroscience 37, 9132–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dulcis D, Jamshidi P, Leutgeb S, and Spitzer NC (2013). Neurotransmitter switching in the adult brain regulates behavior. Science 340, 449–453. [DOI] [PubMed] [Google Scholar]
- 44.Deats SP, Adidharma W, and Yan L (2015). Hypothalamic dopaminergic neurons in an animal model of seasonal affective disorder. Neurosci Lett 602, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumbell RA, Scherbarth F, Diedrich V, Schmid HA, Steinlechner S, and Barrett P (2015). Somatostatin Agonist Pasireotide Promotes a Physiological State Resembling Short-Day Acclimation in the Photoperiodic Male Siberian Hamster (Phodopus sungorus). J Neuroendocrinol 27, 588–599. [DOI] [PubMed] [Google Scholar]
- 46.Kaneko M, and Hall JC (2000). Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. The Journal of comparative neurology 422, 66–94. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, and Emery P (2010). Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol 20, 600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freeman M (1996). Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87, 651–660. [DOI] [PubMed] [Google Scholar]
- 49.Bahn JH, Lee G, and Park JH (2009). Comparative analysis of Pdf-mediated circadian behaviors between Drosophila melanogaster and D. virilis. Genetics 181, 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abruzzi K, Chen X, Nagoshi E, Zadina A, and Rosbash M (2015). RNA-seq Profiling of Small Numbers of Drosophila Neurons. Methods in enzymology 551, 369–386. [DOI] [PubMed] [Google Scholar]
- 51.Long X, Colonell J, Wong AM, Singer RH, and Lionnet T (2017). Quantitative mRNA imaging throughout the entire Drosophila brain. Nature methods 14, 703–706. [DOI] [PubMed] [Google Scholar]
- 52.Veenstra JA, Agricola H-J, and Sellami A (2008). Regulatory peptides in fruit fly midgut. Cell and Tissue Research 334, 499–516. [DOI] [PubMed] [Google Scholar]
- 53.Duffy JB (2002). GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34, 1–15. [DOI] [PubMed] [Google Scholar]
- 54.Schlichting M, and Helfrich-Forster C (2015). Photic entrainment in Drosophila assessed by locomotor activity recordings. Methods in enzymology 552, 105–123. [DOI] [PubMed] [Google Scholar]
- 55.Wang JW, Wong AM, Flores J, Vosshall LB, and Axel R (2003). Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112, 271–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.