Fig. 1.
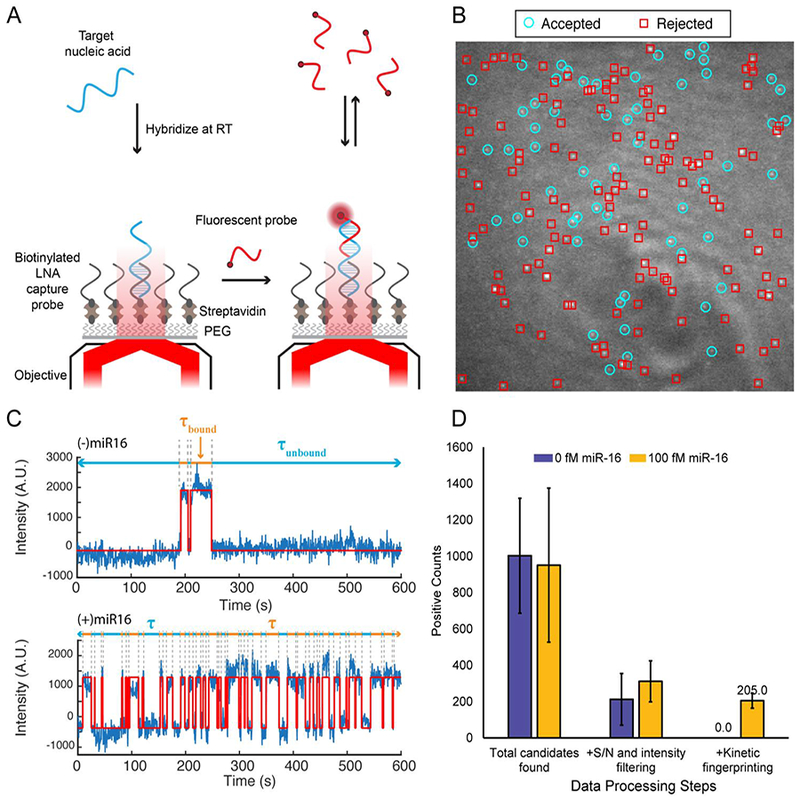
Overview of the SiMREPS technique for low-background, high-specificity detection of single molecules. (A) Schematic illustrating the experimental principles of SiMREPS. A target analyte is captured at the surface of a coverslip via a biotinylated capture probe. Then, using TIRF microscopy, each copy of surface-bound analyte is detecting by monitoring the repeated transient binding of a fluorescent probe, which yields a distinctive kinetic fingerprint. (B) Single movie frame from a representative field of view from SiMREPS using objective-type TIRF microscopy. Red squares indicate positions of binding events that were rejected as likely background binding by kinetic fingerprinting, and the cyan circles indicate positions of repeated binding events with kinetics that suggest the presence of the analyte (C) Representative fluorescence-versus-time traces observed in the presence and absence of a miRNA target, hsa-miR-16. The kinetics of transitions between FP-bound and FP-unbound states are analyzed to distinguish between true and false positives at the single-molecule level. (D) Number of spots counted in positive and negative control experiments for miR-16 before (‘total counts’) and after (‘accepted counts’) kinetic filtering. While filtering based on intensity and signal-to-noise (S/N) alone does not yield a significant difference between positive and negative controls (due to background binding of the probe), the application of kinetic filtering criteria (see section 2.7.4) reduces accepted counts in the negative control to essentially zero.
