Abstract
Adipose inflammation is an important cause for obesity-associated metabolic disorders, including insulin resistance and hypertension. Here we investigated that a circular RNA (circRNA), which we termed circARF3 (ADP-ribosylation factor 3), acts as an endogenous miR-103 sponge to alleviate adipose inflammation by promoting mitophagy. On the other hand, miR-103 aggravated inflammation by inhibiting mitophagy, revealing that miR-103 acts as a positive regulator of adipose inflammation. Furthermore, we found that tumor necrosis factor receptor-associated factor 3 (TRAF3), as a miR-103 downstream target, mediates the functions of miR-103 in adipose inflammation. Overexpressing TRAF3 attenuated miR-103-induced inflammation by accelerating mitophagy. Moreover, we identified that circARF3 blocked miR-103 effects, which resulted in an increase in TRAF3 expression. TRAF3 restrained the nuclear factor κB (NF-κB)-signaling pathway, heightened mitophagy, and suppressed NLRP3 inflammasome activation ultimately. Our data showed that circARF3 acts as an endogenous miR-103 sponge to inhibit mitophagy-mediated adipose inflammation both in vitro and in vivo. These findings disclose a new regulatory pathway for adipose inflammation, which consists of circARF3, miR-103, and TRAF3. This study can be a useful addition to our knowledge, as it provides a new strategy for the prevention of adipose inflammation in obesity disorder.
Keywords: circRNA, miR-103, TRAF3, mitophagy, inflammation, adipose tissue
Introduction
Obesity has become a global health problem, which is often accompanied by a variety of metabolic diseases, including insulin resistance, type 2 diabetes (T2D), inflammation, nonalcoholic fatty liver disease (NAFLD), hypertension, and atherosclerosis.1 In obesity, adipocytes become hypertrophic, resulting in increased pro-inflammatory adipokines such as tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, and monocyte chemotactic protein (MCP)-1 and decreased anti-inflammatory adipokines such as adiponectin and IL-10, thus promoting the occurrence of inflammation.2 It is now clear that inflammation is an underlying cause of T2D as well as many other obesity-induced diseases.3 Inflammatory pathways interfere with insulin signaling4 and contribute to impaired glucose metabolism in adipocytes, hepatocytes, and muscle cells.5 Therefore, it is an important task to investigate molecules regulating inflammation to prevent metabolic syndrome.
MicroRNAs (miRNAs) can directly regulate about 30% of human and mouse genes, and they play an important role in multiple metabolic tissues.6, 7 A large number of published studies have shown that miRNAs can be used as a biomarker of multiple diseases, including metabolic diseases, making them play a crucial role in the diagnosis and treatment of diseases.8, 9 In obesity, miRNAs have been recognized as powerful regulators of numerous genes and the pathogenesis of inflammation.10 For example, miR-145 promotes TNF-α production, and it induces inflammation by activating nuclear factor kappa B (NF-κB) in human adipose tissue.11 miR-132 induces NF-κB activation and IL-8 and MCP-1 production in human adipocytes.12 Adipose tissue shows obvious inflammatory status, accompanied by IL-6, IL-1β, TNF-α, and other inflammatory factors, after silencing miR-223.13 It had been reported that miR-103 was upregulated in the adipose tissue of obesity mice.14 Besides, miR-103 was found to suppress Krüppel-like factor 4 and promote endothelial inflammation to increase monocyte adhesion to endothelial cells.15 However, whether miR-103 regulates adipose inflammation has never been studied.
Circular RNAs (circRNAs) are a group of non-coding RNAs (ncRNAs) characterized by the presence of a covalent bond linking 3′ and 5′ ends produced by backsplicing.16 circRNAs are widely spread with stable structure, conserved sequences, and cell- or tissue-specific expression.17, 18 Studies have shown that circRNAs can function in multiple ways.19, 20 However, the function of circRNAs is only now beginning to be understood. ciRS-7/CDR1as has been identified as a miR-7 sponge to inhibit miR-7 activity.21 Luan et al.22 found that circHLA-C acted as a sponge for miR-150 and played an important role in the pathogenesis of lupus nephritis. These studies strongly supported the idea that circRNAs could be used as natural miRNA sponges to affect disease progressions. A large number of circRNAs have been found in different organisms.23 However, it is not reported whether there are circRNAs as miR-103 sponges in adipocytes.
Autophagy is a highly conserved physiological process in eukaryotes that maintains cell homeostasis.24 The process of autophagy is very complex and sophisticated, and it is mainly regulated by autophagy-related protein (ATG). Especially, LC3, an autophagic substrate, can be used to monitor the number of autophagosomes as well as autophagic activity.25 Mitochondria are critical in metabolism: they not only provide energy for metabolic activity but also produce reactive oxygen species (ROS) to transmit signals and regulate gene expressions.26 The dysfunction and damage of mitochondria cause serious consequences and even lead to cell death.27 Damaged mitochondria usually degrade through a complex physiological process called mitophagy.28 Mitophagy is an autophagic response that specifically targets mitochondria.29 Mitophagy participates in various physiological processes, including inflammation. Studies have shown that inflammasome activation can be restrained by mitophagy.30, 31 Therefore, we attempted to explore the roles of mitophagy in adipose inflammation.
In this study, we presented that circARF3 (ADP-ribosylation factor 3) functions as an endogenous miR-103 sponge to inhibit miR-103 activity, resulting in an increase of TNF receptor-associated factor 3 (TRAF3) expression, a target of miR-103. We further demonstrated that circARF3 alleviated inflammation in mouse adipose tissue by enhancing mitophagy.
Results
miR-103 Promotes Inflammation in Mouse Adipose
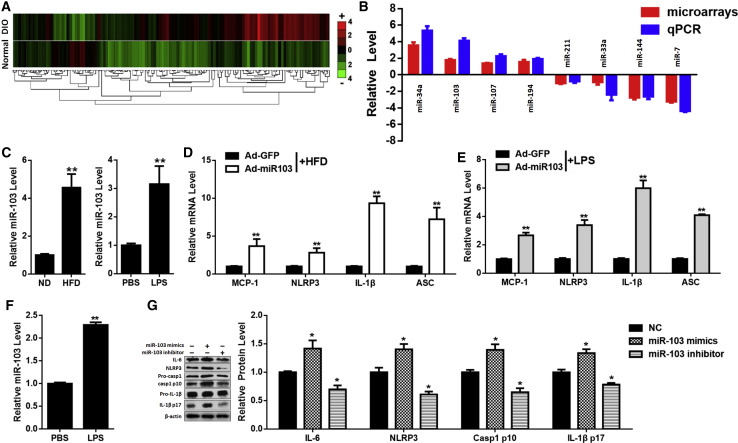
It has been reported that miR-103 is much more abundant in adipose tissue than in liver and muscle, and adipose-specific miR-103 silencing leads to improved insulin sensitivity.14 miR-103 is one of the most upregulated miRNAs in the adipose tissue of diet-induced-obesity (DIO) mice (Figures 1A and 1B). To detect the relationship between miR-103 and adipose inflammation, two inflammatory models were established: high-fat-diet (HFD) mice or normal-diet (ND) mice injected with lipopolysaccharide (LPS).
Figure 1.
miR-103 Promotes Inflammation in Mouse Adipose
(A) Heatmaps of miRNAs in adipose tissue of normal or DIO mice (GEO: GSE85101). (B) Relative levels of the most upregulated or downregulated miRNA in DIO mouse adipose tissue. (C) The relative level of miR-103 in adipose tissue of ND or HFD mice or ND mice injected with PBS or LPS. (D) Relative levels of MCP-1, NLRP3, IL-1β, and ASC mRNAs in HFD mouse adipose after treatment with Ad-miR103 or Ad-GFP. (E) Relative levels of MCP-1, NLRP3, IL-1β, and ASC mRNAs in Ad-miR103/Ad-GFP- and LPS-treated ND mouse adipose. (F) The relative miR-103 level in adipocytes after treatment with PBS or LPS. (G) Relative IL-6, NLRP3, Caspase 1, and IL-1β protein levels in adipocytes after transfection with miR-103 mimics, inhibitor, or NC and treatment with LPS. Error bar, SE. All data are represented as the mean ± SEM (*p < 0.05 and **p < 0.01; n ≥ 3).
Noticeably, miR-103 was dramatically elevated in adipose tissues of the two inflammatory models (Figure 1C). To study the effects of miR-103 in adipose inflammation, mice were intraperitoneally (i.p.) injected with miR-103 or control adenoviruses. We found that, in chronic and acute inflammation models, Ad-miR103 upregulated mRNA levels of MCP-1, IL-1β, NLR family, pyrin domain containing (NLRP) 3, and apoptosis-associated speck-like protein containing CARD (ASC) (Figures 1D and 1E). Besides, Ad-miR103 increased protein levels of NLRP3, cleavage Caspase 1, and IL-1β (Figures S1B and S1D).
Furthermore, adipocytes were incubated with LPS (500 ng/mL) for 8 hr to study the roles of miR-103 in adipose inflammation in vitro. miR-103 was also increased after LPS treatment in adipocytes (Figure 1F). It was found that overexpressing miR-103 by miR-103 mimics increased the transcription levels of IL-6, IL-1β, and MCP-1, while silencing miR-103 by miR-103 inhibitor decreased the mRNA levels of these pro-inflammatory adipokines (Figure S1F). miR-103 mimics also enhanced the protein levels of IL-6 and NLRP3 and cleavage levels of Caspase 1 and IL-1β, while miR-103 inhibitor downregulated them (Figure 1G). These data indicated that miR-103 promoted adipose inflammation both in vitro and in vivo.
miR-103 Inhibits Mitophagy in Mouse Adipose
As autophagy is altered in LPS treatment and obesity,32, 33 we tested whether miR-103 regulated autophagy as well. We found that overexpressing miR-103 in mouse adipose led to a significant increase in SQSTM1/p62 mRNA and protein levels and a decrease in ATG7 and Beclin1 mRNA levels and LC3II protein level (Figures 2A and 2B; Figures S1A–S1D). Moreover, miR-103 also inhibited autophagy in adipocytes (Figure 2C; Figure S1G), which was consistent with our results.
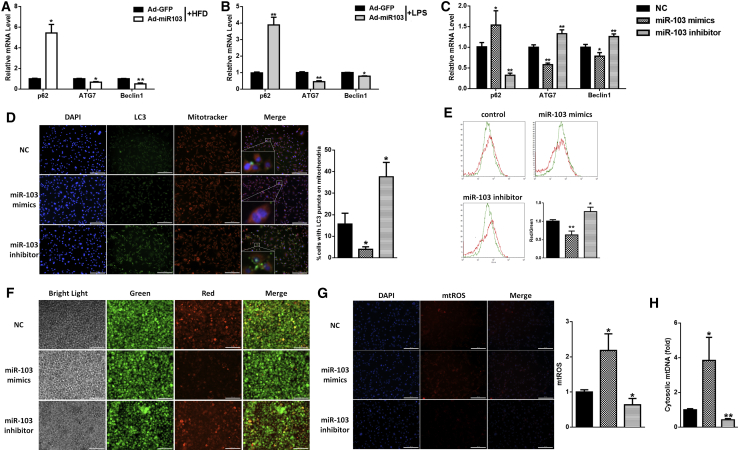
Figure 2.
miR-103 Inhibits Mitophagy in Mouse Adipose
(A and B) Relative mRNA levels of p62, ATG7, and Beclin1 in adipose tissue of ND or HFD mice (A) or ND mice injected with PBS or LPS (B). (C) Relative p62, ATG7, and Beclin1 mRNA levels in adipocytes after transfection with miR-103 mimics, inhibitor, or NC and treatment with LPS in vitro. (D) Intracellular distribution of LC3 (LC3-GFP) and mitochondria (Mitotracker). (E) Mitochondrial membrane potentials were detected by flow cytometry after JC-1 staining. (F) Red and green fluorescence distributions were observed by fluorescence microscopy after JC-1 staining. (G) Relative mtROS amounts were determined by MitoSOX staining. (H) Relative concentrations of mtDNA in adipocytes. Scale bar, 200 μm. Error bar, SE. All data are represented as the mean ± SEM (*p < 0.05 and **p < 0.01; n ≥ 3).
LC3-GFP vector was used to observe autophagy. We found that the LC3 puncta were either around or overlapping with mitochondria (Figure 2D). We suspected that the autophagy that miR-103 inhibited mainly occurred in mitochondria, namely mitophagy. As mitophagy is a process of clearing damaged mitochondria and mitochondrial depolarization is an important event of mitophagy,34 we measured the effect of miR-103 on mitochondrial membrane potential (Δψm) after JC-1 staining. Flow cytometry analysis showed that miR-103 mimics decreased red fluorescence intensity and enhanced green fluorescence intensity, which were contrary to miR-103 inhibitor (Figures 2E and 2F), meaning that miR-103 induced the loss of Δψm. Moreover, we found that miR-103 mimics heightened mitochondrial ROS (mtROS) production (Figure 2G) and raised mtDNA release (Figure 2H), which are correlated with the accumulation of damaged mitochondria. Cell fractionation confirmed these results, which suggested that miR-103 mimic treatment resulted in much more p62 and Parkin being recruited into the mitochondrial fraction, while it possessed less lipidated LC3 (Figure 3A). In summary, these findings indicated that miR-103 inhibited mitophagy in mouse adipose.
Figure 3.
miR-103 Promotes Inflammation by Blocking Mitophagy in Adipocytes
Adipocytes were transfected with miR-103 mimics, inhibitor, or NC and treated with LPS. (A) Subcellular distributions of p62, LC3, and Parkin in adipocytes. (B and C) Relative mRNA levels of MCP-1, NLRP3 (B), Beclin1, and ATG7 (C). (D) The protein levels of NLRP3, Caspase 1, IL-1β, p62, and LC3 were determined after CsA treatment. Error bar, SE. Data are represented as the mean ± SEM (*p < 0.05 and **p < 0.01; n ≥ 3).
miR-103 Promotes Inflammation by Blocking Mitophagy in Adipocytes
It has been reported that NLRP3 inflammasome activation can be restrained by autophagy.30, 31 We tested whether miR-103 promoted adipose inflammation by inhibiting mitophagy. A low dose (100 ng/mL) of ethidium bromide (EtBr) was used to eliminate mitochondrial signals.35 We found that EtBr treatment reduced inflammation and induced autophagy sharply and miR-103 mimics attenuated the effect of EtBr, as the mRNA levels of MCP-1 and NLRP3 were increased (Figure 3B) and Beclin1 and ATG7 mRNA levels were decreased compared with those in the relative control group (Figure 3C). On the contrary, miR-103 inhibitor strengthened EtBr effects, for MCP-1 and NLRP3 mRNA levels were decreased while those of Beclin1 and ATG7 were increased (Figures 3B and 3C). Moreover, miR-103 reinforced the effects of cyclosporin A (CsA), an inhibitor of mitophagy.36 Western blotting suggested that miR-103 mimics increased CsA-induced NLRP3, p62, cleavage Caspase 1, and IL-1β levels and decreased the CsA-blocked LC3II level, which were contrary to miR-103 inhibitor (Figure 3D). These data suggested that miR-103 promoted inflammation by blocking mitophagy in adipocytes.
TRAF3 Is a Downstream Target of miR-103
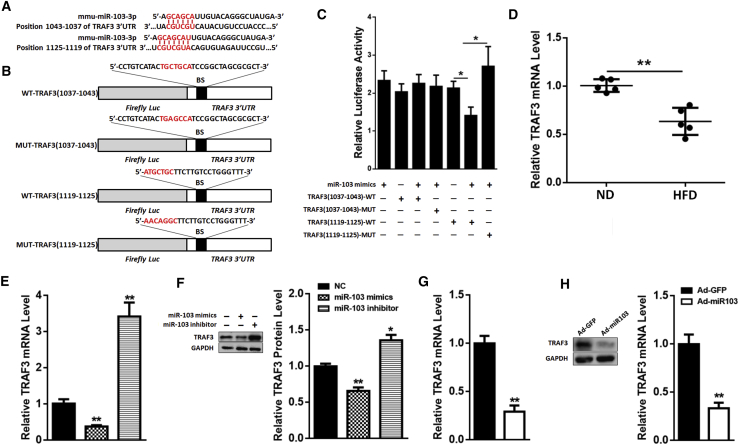
Bioinformatic analysis was carried out to search for the candidate miR-103 target genes, and we found there are two assumed binding sites on the TRAF3 3′ UTR for miR-103 (Figure 4A). We constructed wild-type (WT) and mutant (MUT) luciferase vectors of TRAF3 (Figure 4B), and we measured the relative luciferase activities after transfecting them with miR-103 mimics. We found that the luciferase activity of TRAF3(1,119–1,125)-WT was dramatically decreased compared to the control group or MUT group, while there was no difference between TRAF3(1,037–1,043)-WT group and control group or MUT group (Figure 4C), indicating that the TRAF3(1,119–1,125) site may be the potential binding site for miR-103. Besides, the mRNA level of TRAF3 was lower in the adipose tissue of HFD mice than that in ND mice (Figure 4D), which was opposite to miR-103. Moreover, in adipocytes, miR-103 mimics downregulated the mRNA and protein levels of TRAF3, while miR-103 inhibitor upregulated them (Figures 4E and 4F). In the adipose tissue of HFD mice, Ad-miR103 reduced TRAF3 transcriptional and post-transcriptional levels (Figures 4G and 4H). These data demonstrated that TRAF3 is a downstream target of miR-103.
Figure 4.
TRAF3 Is a Downstream Target of miR-103
(A) Scheme of the potential binding sites of miR-103 in the TRAF3 3′ UTR. (B) miR-103-binding sites in TRAF3 wild-type 3′ UTR (3′ UTR-WT) and mutant 3′ UTR (3′ UTR-MUT) are shown. (C) Double luciferase assay was performed in 293T cells 48 hr after miR-103 mimic transfection with TRAF3-WT 3′ UTR, TRAF3-MUT 3′ UTR, or control. (D) The level of TRAF3 mRNA in ND or HFD mouse adipose tissue. (E and F) Adipocytes were transfected with miR-103 mimics, inhibitor, or NC, and TRAF3 mRNA (E) and protein (F) levels were measured. (G and H) HFD mice were i.p. injected with Ad-miR103 or Ad-GFP, and TRAF3 mRNA (G) and protein (H) levels were detected. Error bar, SE. All data are represented as the mean ± SEM (*p < 0.05 and **p < 0.01; n ≥ 3).
miR-103 Promotes Inflammation by Targeting TRAF3 in Adipocytes
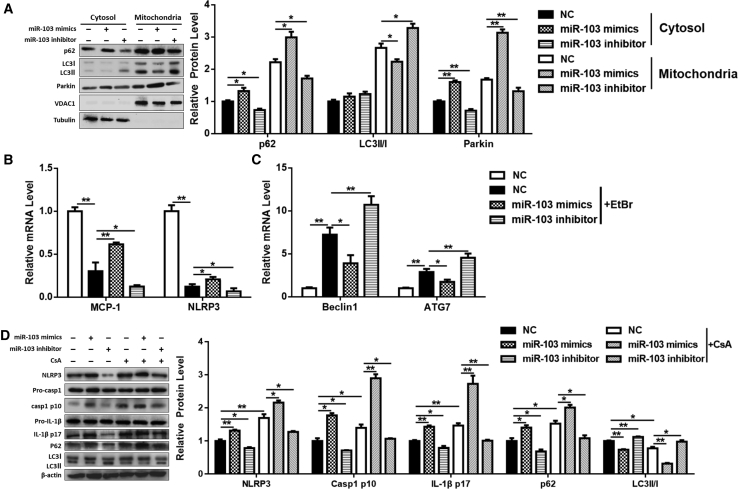
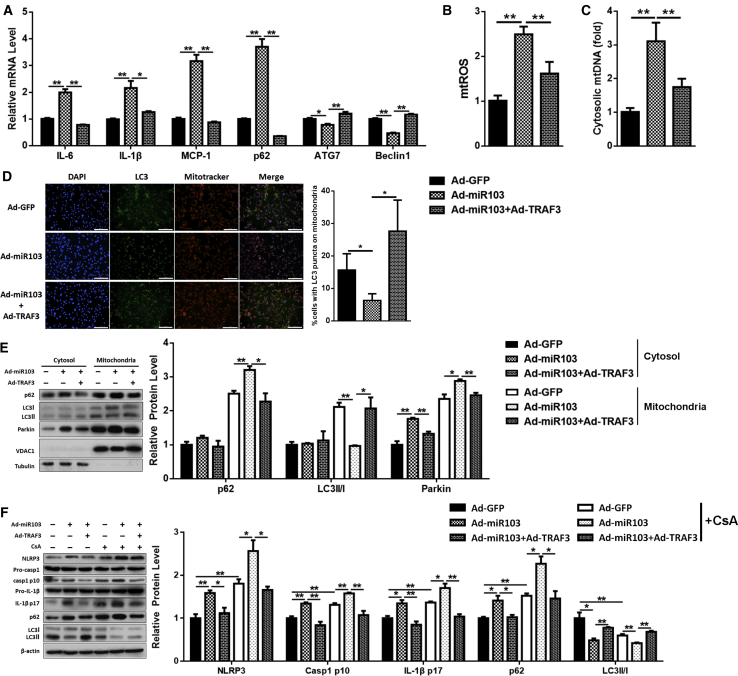
To make sure that miR-103 promoted inflammation in adipose tissue by inhibiting TRAF3, we detected the effects of miR-103 combined with TRAF3. Adipocytes were treated with miR-103 and TRAF3 adenoviruses. Compared to the miR-103 treatment group, the co-treatment of miR-103 and TRAF3 caused a decrease of IL-6, IL-1β, MCP-1, and p62 mRNA, led to a rise in ATG7 and Beclin1 mRNA levels (Figure 5A), suppressed the activation of Caspase 1 and IL-1β (Figure S2A), and cut down mtROS production and mtDNA release (Figures 5B and 5C; Figure S2B). Besides, the addition of TRAF3 promoted the formation of autophagosomes on mitochondria (Figure 5D). The mitochondrial fraction, which also showed that TRAF3 addition reduced p62 and Parkin levels and enhanced LC3II level (Figure 5E), was consistent with our results.
Figure 5.
miR-103 Promotes Inflammation by Targeting TRAF3 in Adipocytes
Adipocytes were treated with Ad-GFP, Ad-miR103, or Ad-miR103 + Ad-TRAF3. (A) Relative levels of IL-6, IL-1β, MCP-1, p62, ATG7, and Beclin1 mRNAs. (B) Relative mtROS amounts. (C) Relative concentrations of mtDNA. (D) Intracellular distribution of LC3 and mitochondria. (E) Subcellular distribution of p62, LC3II, and Parkin. (F) Western blot analysis of NLRP3, Caspase 1, IL-1β, p62, and LC3II protein levels with or without CsA treatment. Scale bar, 200 μm. Error bar, SE. Data are represented as the mean ± SEM (*p < 0.05 and **p < 0.01; n ≥ 3).
Moreover, compared to miR-103 treatment, miR-103 and TRAF3 co-treatment strengthened the effect of EtBr, as MCP-1 and NLRP3 mRNA levels were decreased (Figure S2C) and mRNA levels of ATG7 and Beclin1 were increased (Figure S2D). Furthermore, TRAF3 addition whittled the effects of CsA and miR-103, for the protein levels of NLRP3, p62, cleavage Caspase 1, and IL-1β were decreased and LC3II was increased (Figure 5F). These data indicated that miR-103 promoted NLRP3 inflammasome activation by inhibiting TRAF3.
circARF3 Acts as an Endogenous miR-103 Sponge to Relieve the Inhibitory Effect on TRAF3
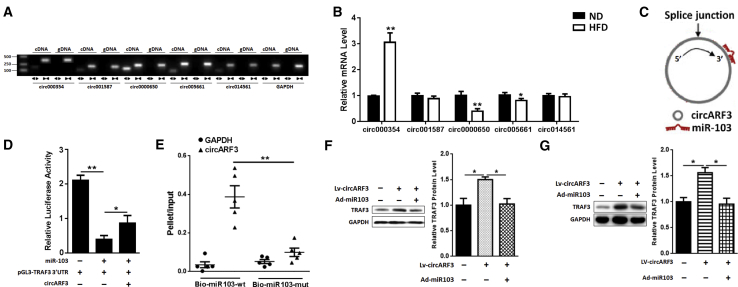
circRNAs composed of exonic sequences can act as miRNA sponges to interact with miRNAs and affect the activities of miRNAs.19, 20, 21 This inspired us to investigate whether there are circRNAs that can function as miR-103 sponges to regulate mitophagy and inflammation. Thus, we screened circRNAs that have been predicted to interact with miR-103 from the databases published online. We detected these circRNA expressions in adipose tissue of ND and HFD mice, and we found that circ0000650 was abundant in mouse adipose tissue (Figure 6A) and was less in HFD mouse adipose tissue than in that of ND mice (Figure 6B). As circ0000650 is transcribed from ARF3, we named it circARF3.
Figure 6.
circARF3 Acts as an Endogenous miR-103 Sponge to Relieve the Inhibitory Effect on TRAF3
(A) Divergent primers amplify circRNAs in cDNA, but not in genomic DNA (gDNA). (B) Relative miR-103-related circRNA expression in ND and HFD mouse adipose tissue. (C) circARF3 contains a site complementary to miR-103, as analyzed by the bioinformatic program RNAhybrid. (D) Double luciferase assay was performed in 293T cells 48 hr after miR-103 mimic transfection with pGL3-TRAF3-WT 3′ UTR and pc-circARF3. (E) qPCR was used to detect circARF3 and GAPDH levels after streptavidin capture. (F) The expression of TRAF3 in adipocytes after circARF3 lentivirus, circARF3 lentivirus + miR-103 adenovirus, or control empty virus treatment. (G) The expression of TRAF3 in adipose tissue of HFD mice after injection with LV-circARF3 or Ad-miR103. Error bar, SE. Data are represented as the mean ± SEM (*p < 0.05 and **p < 0.01; n ≥ 3).
It was predicted that there is a binding site in circARF3 for miR-103 (Figure 6C). We measured the luciferase activity to detect whether circARF3 could act as a miR-103 sponge. We found that transfection of circARF3 with miR-103 and pGL3-TRAF3 3′ UTR resulted in a remission of luciferase activity compared to the miR-103 and pGL3-TRAF3 3′ UTR group (Figure 6D). Besides, circARF3 was much more enriched in the miR-103-captured fraction compared with the corresponding mutant group (Figure 6E). In adipocytes, TRAF3 protein level was upregulated after circARF3 lentivirus treatment, and the addition of miR-103 adenoviruses reduced TRAF3 expression (Figure 6F). Furthermore, we found circARF3 lentiviruses elevated TRAF3 expression significantly, and co-treatment of circARF3 lentiviruses and miR103 adenoviruses reduced TRAF3 protein level in HFD mouse adipose tissue (Figure 6G). These data suggested that circARF3 could function as a miR-103 sponge.
circARF3 Alleviates Adipose Inflammation by Activating Mitophagy
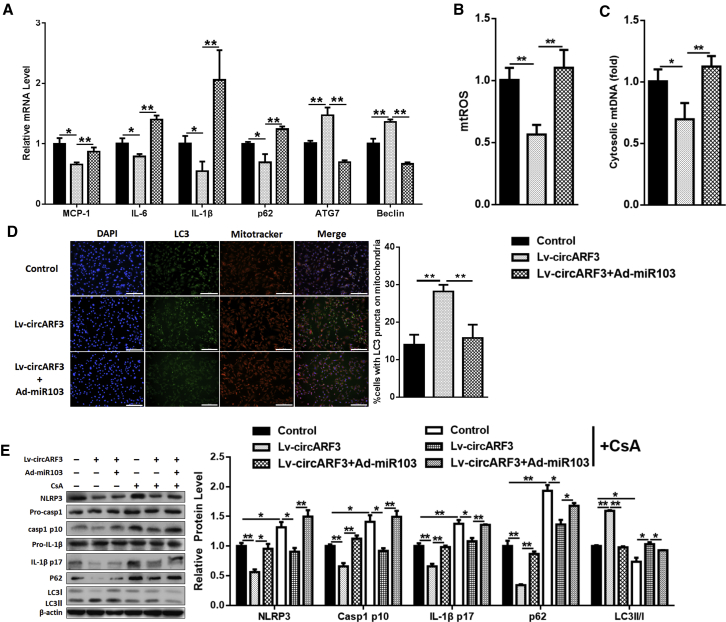
To study the effects of circARF3 in adipose inflammation and autophagy, we detected the expressions of inflammatory and autophagy factors, and we found that circARF3 reduced MCP-1, IL-6, IL-1β, and p62 mRNA levels and elevated ATG7 and Beclin1 mRNA levels (Figure 7A), suggesting that circARF3 alleviates adipose inflammation and accelerates autophagy. Western blot analysis, which indicated that circARF3 lowered the cleavage levels of Caspase 1 and IL-1β (Figure S3A), confirmed our results. circARF3 was also shown to reduce mtROS production and mtDNA release (Figures 7B and 7C; Figure S3B), while miR-103 mimic addition enhanced them. Autophagosomes on mitochondria were raised after circARF3 treatment (Figure 7D). Mitochondrial fraction results showed that circARF3 reduced p62 and Parkin while it enhanced lipidated LC3 level on mitochondria and miR-103 addition reversed the effects of circARF3 (Figure S3C). Furthermore, circARF3 enhanced the effects of EtBr on inflammation and autophagy, as circARF3 decreased the mRNA levels of MCP-1 and NLRP3 (Figure S3D) and increased Beclin1 and ATG7 mRNA levels (Figure S3E). In addition, circARF3 weakened the effects of CsA: circARF3 decreased the CsA-induced NLRP3, p62, cleavage Caspase 1, and IL-1β levels, and it increased LC3II level (Figure 7E). Also, miR-103 addition reversed the effects of circARF3 (Figures 7A–7E). These results indicated that circARF3 functions as a miR-103 sponge to alleviate adipose inflammation.
Figure 7.
circARF3 Alleviates Adipose Inflammation via Activating Mitophagy
Adipocytes were treated with circARF3 lentiviruses, circARF3 lentiviruses + miR-103 adenoviruses, or control empty viruses. (A) qPCR analysis was performed to compare relative levels of MCP-1, IL-6, IL-1β, p62, ATG7, and Beclin1 mRNAs. (B) Relative mtROS amounts. (C) Relative concentrations of mtDNA. (D) Intracellular distribution of LC3 and mitochondria. (E) Western blot analysis of NLRP3, Caspase 1, IL-1β, p62, and LC3 protein levels with CsA treatment. Scale bar, 200 μm. Error bar, SE. Data are represented as the mean ± SEM (*p < 0.05 and **p < 0.01; n ≥ 3).
circARF3 Inhibits Adipose Inflammation by Blocking NF-κB Signaling
TRAF3 is a negative regulator of the NF-κB-signaling pathway.37 We wondered whether circARF3-inhibited adipose inflammation was in relation to the NF-κB-signaling pathway. A significant decrease of p65 in the nucleus was observed after circARF3 treatment, and miR-103 addition elevated p65 content in the nucleus (Figure S4B). BAY11-7082, a specific inhibitor of NF-κB, was used to verify our results. We found that circARF3 treatment reinforced the effect of BAY11-7082. circARF3 suppressed p65 phosphorylation, p62, cleavage Caspase 1, and IL-1β levels, which were inhibited by BAY11-7082, and it raised BAY11-7082-induced LC3II level (Figure S4C). miR-103 addition reversed the effects of circARF3. These results demonstrated that circARF3 inhibited adipose inflammation by blocking the NF-κB-signaling pathway.
circARF3 Alleviates Adipose Inflammation in HFD Mouse Adipose Tissue
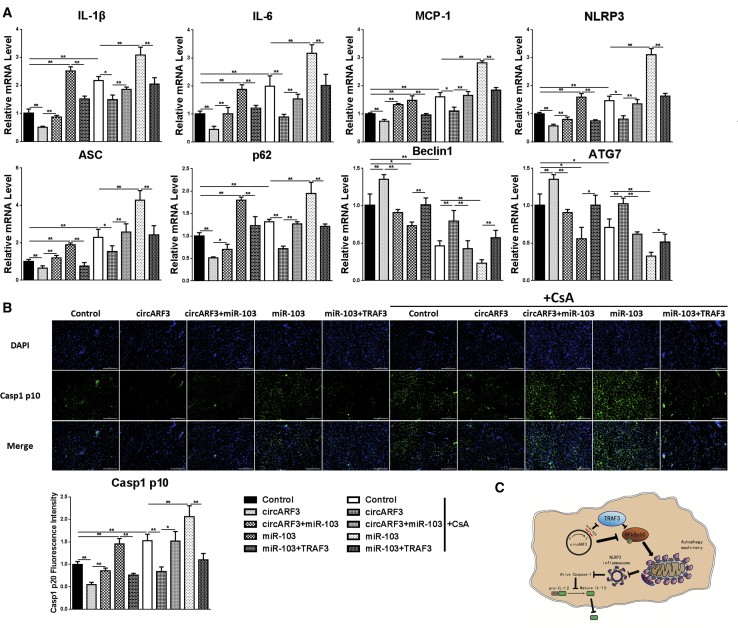
To verify the roles of circARF3 in mitophagy and adipose inflammation in vivo, HFD mice were challenged with high-titer viruses of circARF3, miR-103, and TRAF3 combined with CsA. As Figure 8A shows, we found that circARF3 decreased the mRNA levels of MCP-1, NLRP3, IL-6, IL-1β, ASC, and p62 and increased mRNA levels of Beclin1 and ATG7, which were opposite to miR-103, suggesting that circARF3 inhibited inflammation and induced autophagy while miR-103 promoted inflammation and inhibited autophagy in adipose tissue. As expected, co-treatment with circARF3 and miR-103 impaired circARF3-alleviated inflammation and suppressed circARF3-induced autophagy. miR-103 and TRAF3 co-treatment attenuated miR-103-promoted inflammation and enhanced miR-103-inhibited autophagy. Furthermore, CsA treatment further confirmed our results. Besides, Casp1 p10 immunofluorescence staining in mouse adipose tissue also showed that miR-103 augmented the expression of Casp1 p10 while circARF3 and TRAF3 reduced it (Figure 8B), supporting our notion that circARF3 functions as a miR-103 sponge to alleviate adipose tissue inflammation by promoting mitophagy.
Figure 8.
circARF3 Alleviates Adipose Inflammation In Vivo
HFD mice were i.p. injected with LV-circARF3, LV-circARF3 + Ad-miR103, Ad-miR103, Ad-miR103 + Ad-TRAF3, or control for 1 week. (A) The mRNA levels of IL-1β, IL-6, MCP-1, NLRP3, ASC, p62, Beclin1, and ATG7. (B) Casp1 p10 immunofluorescence staining was observed by fluorescence microscopy. (C) Scheme of circARF3-miR-103-TRAF3 pathway regulating adipose inflammation. circARF3 sponged miR-103, thus crippling the effect of miR-103 on TRAF3 degradation. TRAF3 blocked the NF-κB-signaling pathway and inhibited mitophagy, which induced NLRP3 inflammasome formation and inflammatory cytokine release. Scale bar, 200 μm. Error bar, SE. Data are represented as the mean ± SEM (*p < 0.05 and **p < 0.01; n ≥ 3).
Discussion
Studies have discovered that circRNAs can function as endogenous miRNA sponges, which suggests that circRNAs compete with mRNAs for miRNA binding in the cytoplasm and thus regulate gene expression. For example, ciRS-7/CDR1as binds miR-7 to sequester away miR-7, leading to miR-7 targets levels increasing.38 As the covalently closed loop structure and being widely expressed in eukaryotes, circRNAs can become a new biomarker and therapeutic target of diseases.39, 40 Consistent with these findings, our research displays that circARF3 acts as a miR-103 sponge to enhance TRAF3 level, thus alleviating inflammation in mouse adipose tissue.
Accumulating evidence indicated that chronic inflammation in obesity contributes to systemic metabolic dysfunction. Adipose inflammation regulation is a complex network. Amounts of factors, including transcription factors, adipokines, and miRNAs, regulate adipose tissue inflammation.41 miR-103 belongs to a highly conserved family of miRNA and is abundant in adipose tissue. Dozens of miR-103 target genes, such as Dicer, Cav1, Dapk, Klf4, Fadd, and Mef2d, have been demonstrated to regulate adipogenesis, insulin sensitivity, cell migration, and metastasis.42, 43, 44, 45 miR-103 has been reported to suppress Krüppel-like factor 4 and promote endothelial inflammation to increase monocyte adhesion to endothelial cells.15 In addition, miR-103 ensures proper end-stage autophagy, and its insufficiency causes dysregulation of macropinocytosis with the formation of large vacuoles in a stem cell-enriched epithelium.46 However, the effects of miR-103 on inflammation and autophagy in adipose tissue are still not determined. In this study, it has been shown that miR-103 promotes adipose tissue inflammation by inhibiting mitophagy and miR-103 functions by targeting TRAF3.
TRAF3 is a member of the TRAF family. Studies have indicated that TRAF3 regulates the homeostasis of multiple cell types through different mechanisms.47, 48 NF-κB-inducing kinase (NIK), a core signaling molecule of the NF-κB pathway, functions together with an inhibitor of NF-κB kinase α (IKKα) to induce NF-κB phosphorylation-dependent ubiquitination and process. Normally, NIK is continuously degraded by a TRAF3-dependent E3 ubiquitin ligase. In response to signals mediated by TNF receptor superfamily members, NIK becomes stabilized due to TRAF3 degradation, resulting in the activation of the noncanonical NF-κB pathway.49 To estimate whether NF-κB signaling was involved in NLRP3 inflammasome activation regulated by circARF3-miR103-TRAF3, we detected nuclear translocation of NF-κBp65 in adipocytes. We found that miR-103 facilitated NF-κBp65 delivering to nuclear and phosphorylation, which were contrary to circARF3 and TRAF3.
As a self-protection mechanism, autophagy functions to maintain the stability of the intracellular environment.50 It has been reported that various diseases like obesity, diabetes, neurodegenerative diseases, immune disorders, and cancer are associated with abnormal autophagy.51, 52 Mitophagy is a target-specific autophagy in cells, which can remove damaged mitochondria and excess ROS and block the activation of mitochondrial dysfunction-induced inflammasomes and release of inflammatory cytokines.53 Here we found that miR-103 promoted mitochondrial damage and inhibited mitophagy. Damaged mitochondria released a series of signals, including mtROS and mtDNA, which induced NLRP3 inflammasome activation.54 Parkin, an E3 ubiquitin ligase, recognizes damaged mitochondria and binds to PTEN-induced kinase 1 (PINK1) on the mitochondrial membrane. Parkin undergoes phosphorylation and is capable of ubiquitinating mitochondrial substrates, which in turn bound to LC3 to initiate mitophagy.55 In our research, we found that miR-103 promoted Parkin and p62 recruitment to damaged mitochondria but the mitophagy was inhibited. However, the mechanism involved needs to be further studied.
The present study demonstrated that circARF3-miR103-TRAF3 participate in regulating adipose inflammation through mitophagy. We observed that miR-103 promoted inflammation by inhibiting mitophagy in adipose tissue, whereas circARF3 and TRAF3 inhibited mitophagy-mediated inflammation. Adipose tissue inflammation regulation is a very complex network. Although our present research showed that circARF3 could function as a miR-103 sponge, we did not rule out other circRNAs regulating miR-103 or other miR-103 target genes in adipose tissue inflammation.
In this study, we confirmed that circARF3 repressed adipose inflammation and circARF3 functions as a miR-103 sponge to alleviate adipose inflammation by blocking mitophagy. circARF3 sponged miR-103, thus crippling the effect of miR-103 on TRAF3 degradation. TRAF3 blocked the NF-κB-signaling pathway and inhibited mitophagy, which promoted NLRP3 inflammasome activation and inflammatory cytokine release (Figure 8C). Our study provides new strategies for improving adipose inflammation in obesity.
Materials and Methods
Animal Experiment
Mice handling protocols were conducted following the guidelines and regulations approved by the Animal Ethics Committee of Northwest A&F University. C57BL/6J mice were housed at 25°C ± 1°C temperature, 55% ± 5% humidity, 12-hr-light and 12-hr-dark cycles, and they were provided ad libitum with water and diet. 8-week-old male mice were fed an HFD (fat provides 60% of total energy) for 8 weeks to obtain a chronic inflammation model. To obtain an acute inflammation model, 16-week-old male mice fed an ND (fat provides 10% of total energy) were injected with LPS (10 mg/kg, Sigma, St. Louis, MO, USA) for 6 hr.
HFD mice (n = 60) were randomly divided into ten groups (n = 6 each) using a 5 × 2 design. Mice were treated with circARF3 lentiviruses, circARF3 lentiviruses + miR-103 adenoviruses, miR-103 adenoviruses, miR-103 adenoviruses + TRAF3 adenoviruses, or control empty viruses at 1 × 109 plaque-forming units (PFUs) in 0.2 mL PBS through i.p. injection for 1 week, respectively. To inhibit mitophagy, half of the mice received a daily i.p. injection of 10 mg/kg CsA (Selleck, Shanghai, China) in 1% DMSO for 3 days, and the other half was injected with 1% DMSO.
ND mice (n = 12) were randomly divided into two group. Half of the mice were i.p. injected with miR-103 adenoviruses at 1 × 109 PFUs in 0.2 mL PBS, and the other half received control empty viruses for 1 week. For the LPS-challenged experiment in vivo, mice were injected with LPS (10 mg/kg) for 6 hr.
Primary Adipocyte Culture and Stimulation
Primary preadipocyte culture proposal was conducted as described before.56 Preadipocytes were induced to differentiation as follows:57 2 days after preadipocytes grew to 100% confluence, cells were treated with DMEM/F12 medium containing 10 μg/mL insulin (Sigma, St. Louis, MO, USA), 1 μM dexamethasone (Sigma, St. Louis, MO, USA), and 0.5 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma, St. Louis, MO, USA) for 2 days; then they were maintained in induction medium supplemented with 10 μg/mL insulin (Sigma, St. Louis, MO, USA). For stimulation, mature adipocytes were treated with 100 ng/mL EtBr (Sigma, St. Louis, MO, USA) for 3 days or 5 μM CsA (Selleck, Shanghai, China) for 30 min combined with LPS (500 ng/mL) for 8 hr.
For in vitro studies, adipocytes were transfected with miRNA negative control (NC), miR-103 mimics, miR-103 inhibitor, LC3-GFP, or empty plasmid using X-tremeGENE HP DNA Transfection Reagent (Roche, Switzerland). miR-103 (Ad-miR103), TRAF3 (Ad-TRAF3), circARF3 (Lv-circARF3), or control empty viruses were added into adipocytes at 1 × 109 PFUs/mL for 48 hr.
RNA Extraction and cDNA Synthesis
Total RNA, including miRNA, was extracted from adipose tissue or adipocytes using the TRIzol method. Then cDNA was synthesized using a PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China).
qRT-PCR Analysis
miRNA and mRNA levels were measured by qPCR.58 U6 and GAPDH were chosen as reference genes to normalize miRNA and mRNA levels, respectively.
Western Blot
Western blot was used to detect the protein levels.59 Protein samples were separated by electrophoresis on 12% and 5% SDS-PAGE gels using slab gel apparatus and then transferred to polyvinylidene fluoride (PVDF) nitrocellulose membranes (Millipore, USA), blocked with a mixture of 5% Skim Milk Powder and TBST (tris-buffered saline and Tween 20) at room temperature for 2 hr. Membranes were incubated with the primary antibodies at 4°C overnight and further incubated with the secondary antibodies for 2 hr at room temperature. Primary antibodies against IL-6 (ab208113), NLLRP3 (ab214185), Pro-IL-1β (ab106035), IL-1β p17 (ab106035), Pro-Casp1 (ab179515), Casp1 p10 (ab179515), p62 (ab109012), LC3 (ab51520), β-actin (ab8227), Parkin (ab15954), VDAC1 (ab15895), Tubulin (ab6046), GAPDH (ab9385), TRAF3 (ab36988), p65 (ab32536), and p-p65 (ab86299) were purchased from Abcam (Cambridge, UK), and secondary antibody was purchased from Baoshen (Beijing, China).
miRNA Target Gene and circRNA Prediction
miRNA target prediction was performed in TargetScan, miRBase, and PicTar, and circRNA prediction was performed in CircNet, circBase, and starBase. Their intersections were chosen to improve the accuracy of the forecast.
Biotin-Coupled miRNA Capture
Biotin-labeled WT or MUT miR-103 was transfected into adipocytes. Cells were collected and treated as described previously.38
Dual Luciferase Reporter Assay
The dual luciferase reporter assay was performed as described previously.60 A Dual Luciferase Reporter Assay kit (Promega, Madison, WI, USA) was used to measure the luciferase activity, and the procedure followed the manufacturer’s protocol.
Mitochondrial Membrane Potential Analysis
Mitochondrial membrane potential assay kit with JC-1 was purchased from Beyotime Biotechnology (China), and it was used to detect mitochondrial membrane potential following the manufacturer’s protocol.
mtROS Measurement
mtROS was measured with MitoSOX (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instructions. In brief, cells were incubated with 5 μM MitoSOX at 37°C for 10 min and washed three times with PBS. Then cells were fixed and incubated with DAPI for nuclear staining.
Immunofluorescence Staining
Immunofluorescence staining was performed by the standard method. After being fixed, permeabilized, and blocked, samples were incubated with primary antibodies and secondary fluorescent antibodies, and DAPI was used for nuclear staining.
Nuclear Protein Extraction
Nuclear and cytoplasmic fractions were prepared using Mitochondria Isolation Kit (Beyotime, China). The procedure followed the protocol provided by the manufacturer.
Data Analysis
All experiments were repeated at least three times. Data were expressed as the mean ± SEM. The statistical analysis differences were performed in SPSS using t test. *p < 0.05 was statistically significant and **p < 0.01 was very significant.
Author Contributions
All authors contributed to this article. Experimental Design, Z.Z. and C.S.; Experiments, all authors; Data Analysis, T.Z. and R.F.; Writing, Z.Z. All authors assure that they met the criteria for authorship and reviewed the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was financially supported by Major National Scientific Research Projects (2015CB943102), the National Natural Science Foundation of China (31572365), the Key Sci-tech innovation team of Shaanxi Province (2017KCT-24), and the Key Sci-tech innovation team of Northwest A&F University.
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.11.014.
Supplemental Information
References
- 1.Franks P.W., McCarthy M.I. Exposing the exposures responsible for type 2 diabetes and obesity. Science. 2016;354:69–73. doi: 10.1126/science.aaf5094. [DOI] [PubMed] [Google Scholar]
- 2.Blüher M. Adipose tissue dysfunction contributes to obesity related metabolic diseases. Best Pract. Res. Clin. Endocrinol. Metab. 2013;27:163–177. doi: 10.1016/j.beem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Gregor M.F., Hotamisligil G.S. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 4.Rehman K., Akash M.S.H. Mechanisms of inflammatory responses and development of insulin resistance: how are they interlinked? J. Biomed. Sci. 2016;23:87. doi: 10.1186/s12929-016-0303-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohlgruber A., Lynch L. Adipose tissue inflammation in the pathogenesis of type 2 diabetes. Curr. Diab. Rep. 2015;15:92. doi: 10.1007/s11892-015-0670-x. [DOI] [PubMed] [Google Scholar]
- 6.Ameres S.L., Zamore P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 7.Arner P., Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 2015;11:276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 8.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 9.Rottiers V., Näär A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebane A., Akdis C.A. MicroRNAs: Essential players in the regulation of inflammation. J. Allergy Clin. Immunol. 2013;132:15–26. doi: 10.1016/j.jaci.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Lorente-Cebrián S., Mejhert N., Kulyté A., Laurencikiene J., Åström G., Hedén P., Rydén M., Arner P. MicroRNAs regulate human adipocyte lipolysis: effects of miR-145 are linked to TNF-α. PLoS ONE. 2014;9:e86800. doi: 10.1371/journal.pone.0086800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strum J.C., Johnson J.H., Ward J., Xie H., Feild J., Hester A., Alford A., Waters K.M. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 2009;23:1876–1884. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang G., Meng C., Guo X., Cheruku P.S., Shi L., Xu H., Li H., Wang G., Evans A.R., Safe S. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125:2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 14.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann P., Zhou Z., Natarelli L., Wei Y., Nazari-Jahantigh M., Zhu M., Grommes J., Steffens S., Weber C., Schober A. Endothelial Dicer promotes atherosclerosis and vascular inflammation by miRNA-103-mediated suppression of KLF4. Nat. Commun. 2016;7:10521. doi: 10.1038/ncomms10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: Identification, biogenesis and function. Biochim. Biophys. Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 20.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 21.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 22.Luan J., Jiao C., Kong W., Fu J., Qu W., Chen Y., Zhu X., Zeng Y., Guo G., Qi H. circHLA-C Plays an Important Role in Lupus Nephritis by Sponging miR-150. Mol. Ther. Nucleic Acids. 2018;10:245–253. doi: 10.1016/j.omtn.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Jones S.A., Mills K.H., Harris J. Autophagy and inflammatory diseases. Immunol. Cell Biol. 2013;91:250–258. doi: 10.1038/icb.2012.82. [DOI] [PubMed] [Google Scholar]
- 25.Rogov V., Dötsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schapira A.H. Mitochondrial diseases. Lancet. 2012;379:1825–1834. doi: 10.1016/S0140-6736(11)61305-6. [DOI] [PubMed] [Google Scholar]
- 28.Wei H., Liu L., Chen Q. Selective removal of mitochondria via mitophagy: distinct pathways for different mitochondrial stresses. Biochim. Biophys. Acta. 2015;1853(10 Pt B):2784–2790. doi: 10.1016/j.bbamcr.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Kubli D.A., Gustafsson Å.B. Mitochondria and mitophagy: the yin and yang of cell death control. Circ. Res. 2012;111:1208–1221. doi: 10.1161/CIRCRESAHA.112.265819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong Z., Umemura A., Sanchez-Lopez E., Liang S., Shalapour S., Wong J., He F., Boassa D., Perkins G., Ali S.R. NF-κB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minton K. Inflammasome: Anti-inflammatory effect of mitophagy. Nat. Rev. Immunol. 2016;16:206. doi: 10.1038/nri.2016.33. [DOI] [PubMed] [Google Scholar]
- 32.Chen S., Yuan J., Yao S., Jin Y., Chen G., Tian W., Xi J., Xu Z., Weng D., Chen J. Lipopolysaccharides may aggravate apoptosis through accumulation of autophagosomes in alveolar macrophages of human silicosis. Autophagy. 2015;11:2346–2357. doi: 10.1080/15548627.2015.1109765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovsan J., Blüher M., Tarnovscki T., Klöting N., Kirshtein B., Madar L., Shai I., Golan R., Harman-Boehm I., Schön M.R. Altered autophagy in human adipose tissues in obesity. J. Clin. Endocrinol. Metab. 2011;96:E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Enriquez S., He L., Lemasters J.J. Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int. J. Biochem. Cell Biol. 2004;36:2463–2472. doi: 10.1016/j.biocel.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Nakahira K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basit F., van Oppen L.M., Schöckel L., Bossenbroek H.M., van Emst-de Vries S.E., Hermeling J.C., Grefte S., Kopitz C., Heroult M., Hgm Willems P., Koopman W.J. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 2017;8:e2716. doi: 10.1038/cddis.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu H., Brittain G.C., Chang J.H., Puebla-Osorio N., Jin J., Zal A., Xiao Y., Cheng X., Chang M., Fu Y.X. OTUD7B controls non-canonical NF-κB activation through deubiquitination of TRAF3. Nature. 2013;494:371–374. doi: 10.1038/nature11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 39.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 40.Hsiao K.Y., Lin Y.C., Gupta S.K., Chang N., Yen L., Sun H.S., Tsai S.J. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reilly S.M., Saltiel A.R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017;13:633–643. doi: 10.1038/nrendo.2017.90. [DOI] [PubMed] [Google Scholar]
- 42.Martello G., Rosato A., Ferrari F., Manfrin A., Cordenonsi M., Dupont S., Enzo E., Guzzardo V., Rondina M., Spruce T. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 43.Chen H.Y., Lin Y.M., Chung H.C., Lang Y.D., Lin C.J., Huang J., Wang W.C., Lin F.M., Chen Z., Huang H.D. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 44.Wang J.X., Zhang X.J., Li Q., Wang K., Wang Y., Jiao J.Q., Feng C., Teng S., Zhou L.Y., Gong Y. MicroRNA-103/107 Regulate Programmed Necrosis and Myocardial Ischemia/Reperfusion Injury Through Targeting FADD. Circ. Res. 2015;117:352–363. doi: 10.1161/CIRCRESAHA.117.305781. [DOI] [PubMed] [Google Scholar]
- 45.Li M., Liu Z., Zhang Z., Liu G., Sun S., Sun C. miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol. Chem. 2015;396:235–244. doi: 10.1515/hsz-2014-0241. [DOI] [PubMed] [Google Scholar]
- 46.Park J.K., Peng H., Katsnelson J., Yang W., Kaplan N., Dong Y., Rappoport J.Z., He C., Lavker R.M. MicroRNAs-103/107 coordinately regulate macropinocytosis and autophagy. J. Cell Biol. 2016;215:667–685. doi: 10.1083/jcb.201604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao Y., Jin J., Chang M., Chang J.H., Hu H., Zhou X., Brittain G.C., Stansberg C., Torkildsen Ø., Wang X. Peli1 promotes microglia-mediated CNS inflammation by regulating Traf3 degradation. Nat. Med. 2013;19:595–602. doi: 10.1038/nm.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panda S., Nilsson J.A., Gekara N.O. Deubiquitinase MYSM1 Regulates Innate Immunity through Inactivation of TRAF3 and TRAF6 Complexes. Immunity. 2015;43:647–659. doi: 10.1016/j.immuni.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Sun S.C. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 51.Choi A.M., Ryter S.W., Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 52.Deretic V., Saitoh T., Akira S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green D.R., Galluzzi L., Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elliott E.I., Sutterwala F.S. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 2015;265:35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eiyama A., Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr. Opin. Cell Biol. 2015;33:95–101. doi: 10.1016/j.ceb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Z., Wu S., Muhammad S., Ren Q., Sun C. miR-103/107 promote ER stress-mediated apoptosis via targeting the Wnt3a/β-catenin/ATF6 pathway in preadipocytes. J. Lipid Res. 2018;59:843–853. doi: 10.1194/jlr.M082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z., Gan L., Xu Y., Luo D., Ren Q., Wu S., Sun C. Melatonin alleviates inflammasome-induced pyroptosis through inhibiting NF-κB/GSDMD signal in mice adipose tissue. J. Pineal Res. 2017;63:e12414. doi: 10.1111/jpi.12414. [DOI] [PubMed] [Google Scholar]
- 58.Gan L., Liu Z., Luo D., Ren Q., Wu H., Li C., Sun C. Reduced endoplasmic reticulum stress-mediated autophagy is required for leptin alleviating inflammation in adipose tissue. Front. Immunol. 2017;8:1507. doi: 10.3389/fimmu.2017.01507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gan L., Liu Z., Wu T., Feng F., Sun C. αMSH promotes preadipocyte proliferation by alleviating ER stress-induced leptin resistance and by activating Notch1 signal in mice. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:231–238. doi: 10.1016/j.bbadis.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 60.Liu G., Li M., Saeed M., Xu Y., Ren Q., Sun C. αMSH inhibits adipose inflammation via reducing FoxOs transcription and blocking Akt/JNK pathway in mice. Oncotarget. 2017;8:47642–47654. doi: 10.18632/oncotarget.17465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.