The built environment contains a variety of microorganisms, some of which pose critical human health risks (e.g., hospital-acquired infection, antibiotic resistance dissemination). We uncovered a combination of complex biological functions that may play a role in bacterial survival under the presumed selective pressures in a model built environment—the International Space Station—by using an approach to compare pangenomes of bacterial strains from two clinically relevant species (B. cereus and S. aureus) isolated from both built environments and humans. Our findings suggest that the most crucial bacterial functions involved in this potential adaptive response are specific to bacterial lifestyle and do not appear to have direct impacts on human health.
KEYWORDS: International Space Station, bacterial adaptation, built environment microbiome, pangenome
ABSTRACT
Understanding underlying mechanisms involved in microbial persistence in the built environment (BE) is essential for strategically mitigating potential health risks. To test the hypothesis that BEs impose selective pressures resulting in characteristic adaptive responses, we performed a pangenomics meta-analysis leveraging 189 genomes (accessed from GenBank) of two epidemiologically important taxa, Bacillus cereus and Staphylococcus aureus, isolated from various origins: the International Space Station (ISS; a model BE), Earth-based BEs, soil, and humans. Our objectives were to (i) identify differences in the pangenomic composition of generalist and host-associated organisms, (ii) characterize genes and functions involved in BE-associated selection, and (iii) identify genomic signatures of ISS-derived strains of potential relevance for astronaut health. The pangenome of B. cereus was more expansive than that of S. aureus, which had a dominant core component. Genomic contents of both taxa significantly correlated with isolate origin, demonstrating an importance for biogeography and potential niche adaptations. ISS/BE-enriched functions were often involved in biosynthesis, catabolism, materials transport, metabolism, and stress response. Multiple origin-enriched functions also overlapped across taxa, suggesting conserved adaptive processes. We further characterized two mobile genetic elements with local neighborhood genes encoding biosynthesis and stress response functions that distinctively associated with B. cereus from the ISS. Although antibiotic resistance genes were present in ISS/BE isolates, they were also common in counterparts elsewhere. Overall, despite differences in microbial lifestyle, some functions appear common to remaining viable in the BE, and those functions are not typically associated with direct impacts on human health.
IMPORTANCE The built environment contains a variety of microorganisms, some of which pose critical human health risks (e.g., hospital-acquired infection, antibiotic resistance dissemination). We uncovered a combination of complex biological functions that may play a role in bacterial survival under the presumed selective pressures in a model built environment—the International Space Station—by using an approach to compare pangenomes of bacterial strains from two clinically relevant species (B. cereus and S. aureus) isolated from both built environments and humans. Our findings suggest that the most crucial bacterial functions involved in this potential adaptive response are specific to bacterial lifestyle and do not appear to have direct impacts on human health.
Author Video: An author video summary of this article is available.
INTRODUCTION
Indoor surfaces and dust are widely colonized by human-associated and environmental microorganisms introduced via direct contact and passive deposition from inhabitants, transported materials, and the air supply (1, 2). Numerous metagenomics and 16S rRNA gene amplicon sequencing studies have detected diverse microbial communities on interior surfaces throughout homes, schools, offices, athletic facilities, hospitals, subway stations, and cleanrooms and aboard the International Space Station (ISS) (3–13). In addition to well-characterized bacterial survival strategies (e.g., biofilm formation and sporulation), it has been suggested that complex metabolisms, biosynthetic pathways, and antibiotic resistance genes (ARGs) may also play important roles in adaptation to these built environments (BEs) (8, 12, 14). While a variety of building features (e.g., chemical cleaning frequency, human occupancy, room type, surface materials, and ventilation) have been correlated with indoor microbial diversity (1, 2), much remains unknown about species-level population genetics associated with microbial persistence under the presumed physical and chemical selective pressures (e.g., desiccation, limited resource availability, and biocide and detergent residues from cleaning products).
The indoor microbiome has important implications for human health and safety. Elevated levels of mold (e.g., Aspergillus, Cladosporium, and Penicillium) are a precursor to biodegradation of building materials and can induce human development of allergies and asthma-like symptoms (15, 16). In U.S. acute-care hospitals, approximately 4% of inpatients develop nosocomial infection (closer to 10% in less industrialized countries), which leads to estimated annual economic burdens in the range of $35 to 45 billion (17, 18). Controlling the spread of hospital-acquired infections has been challenged by the widespread emergence of antibiotic-resistant pathogens, such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (19). Dissemination of mobile ARGs or enrichment for antibiotic-resistant organisms may be intensified by the concentration of anthropogenic chemicals from cleaning/consumer products that accumulate indoors (5, 12). Testing the hypothesis that BEs impose specific selective pressures that result in characteristic adaptive responses would advance our understanding of molecular mechanisms that could be leveraged to develop novel strategies for creating and maintaining “healthier” buildings.
Insights into potential bacterial species’ adaptability and health risks are manifested in their pangenomes, i.e., the cumulative set of genes belonging to all genomes of a taxonomic group (20, 21). Population survival under constant environmental pressures is enhanced by substantial intraspecific variation generated through rapid evolution involving mobile genetic element (MGE)-mediated horizontal gene transfer (HGT), mutation to existing genes, and DNA rearrangement or loss (22). Genomic heterogeneity within a bacterial species (or any defined taxonomic group) includes nucleotide variants within the “core” component of the pangenome (i.e., essential genes conserved across all strains) and the presence/absence of genes in the “accessory” component of the pangenome (i.e., dispensable genes in one or more, but not all, strains). Enrichments in accessory genes under specific environmental conditions may represent adaptation to the particular site or host. For example, gene presence/absence has been reported to significantly differ among Enterococcus isolates from human- versus environmentally sourced samples (23), as well as among Prevotella strains across human body sites (e.g., skin, oral cavity, and gut) (24). Moreover, the size and expansiveness of pangenomes (i.e., number of new genes discovered in each new genome analyzed) more broadly reflect a taxon’s ability to adapt and evolve (20). While relatively small and predictably bound pangenomes associate with limited lifestyles (e.g., Buchnera aphidicola, an endosymbiont of aphids), having a high propensity for increasing gene repertoire supports a more versatile metabolic and potential pathogenicity range (e.g., Bacillus cereus and Escherichia coli) (25, 26). A comparative pangenomic assessment of BE strains with human-associated and environmental counterparts would be useful to discern genetic signatures for niche-specific microbial function and biogeography.
The ISS is a relevant model system for investigating microbial adaptations to the BE due to its constant human occupancy and controlled environmental conditions (e.g., temperature, humidity, and air circulation), along with routine microbial monitoring to ensure crew safety, for nearly two decades (27). Viable members of the ISS microbiome are presumably acclimated to selective pressures of the BE (e.g., low-nutrient, dry settings) as well as spaceflight (e.g., microgravity, elevated CO2, and cosmic radiation). The former is underscored by ISS microbial community composition appearing more similar to that in homes on Earth than to the human microbiome (7). The hypothesis that BE conditions may have a more selective influence on microbes than spaceflight warrants investigation.
Of the several hundred bacterial strains that have been isolated from the ISS BE, B. cereus and S. aureus are among the most prevalent species in the culture collection with sequenced genomes (28–30). These economically and epidemiologically important taxa represent model organisms with drastically different lifestyles, survival strategies, and disease implications. B. cereus is ubiquitous in nature (primarily soilborne) and forms endospores (31). It is an opportunistic pathogen involved in foodborne illness (enterotoxin production) and is sometimes associated with infectious disease in immunocompromised individuals (32). In contrast, S. aureus is a highly abundant commensal within the human microbiome, often capable of biofilm formation, and increasingly implicated in nosocomial infection (e.g., MRSA) (33, 34). Accordingly, the survival dynamics of these two taxa in the ISS BE are likely distinct. While Bacillus spores may persist in the ISS for months or even years (35), S. aureus experiences about a 4- to 5-log reduction on surfaces over the span of a month (36). Thus, strains of the latter that have been isolated from the ISS were probably deposited from whomever was aboard during the prior few weeks, and population persistence may depend on reseeding via transfers between humans and the BE. In addition to their presence in the ISS microbiome, strains of both taxa have been frequently isolated and sequenced from various environments (e.g., BEs and soil) and human clinical samples on Earth (29–31, 37–54). In the present work, whole-genome sequencing (WGS) data from these diverse studies were leveraged to characterize the pangenomes of B. cereus and S. aureus. Our objectives were to (i) distinguish key differences in the pangenomic composition of the generalist (B. cereus) and that of the host-associated (S. aureus) model organism, (ii) determine the sets of genes and functions associated with potentially adaptive responses to the BE, and (iii) identify genomic signatures of these important members of the ISS microbiome that may present potential risk to inhabitants (e.g., ARGs, MGEs, and virulence).
RESULTS
Interspecies pangenome variation.
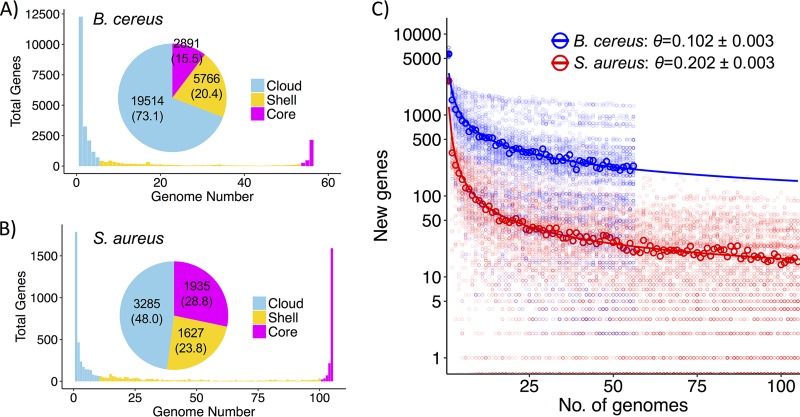
The pangenome of B. cereus contained approximately 28,171 genes, with 5,617 ± 277 genes per genome (mean ± SD) (Fig. 1; see also Fig. S1 in the supplemental material). That of S. aureus contained approximately 6,847 genes, with 2,645 ± 91 genes per genome (mean ± SD) (Fig. 1; Fig. S1). According to a power-law regression, both species pangenomes were in an “open” state (Fig. 1C). The model-predicted high likelihood of continuous discovery of new genes per genome sequenced (i.e., pangenome “openness”) indicated that populations of both species, especially B. cereus, may expand and/or alter gene repertoire over time. The predicted N50 (i.e., new genes per 50th genome analyzed) was 227.6 for B. cereus and 25.7 for S. aureus. Thus, while the two species’ average genome and pangenome sizes differed by roughly 2- and 4-fold, respectively, the numbers of new genes per genome were, disproportionately, 9 to 10 times greater for B. cereus. In summary, both pangenomes appeared boundless, though that of the generalist (B. cereus) was relatively more expansive and heterogeneous, while that of the human commensal (S. aureus) contained a more prominent core.
FIG 1.
Pangenome summary statistics. (A and B) Histogram distributions of cloud, shell, and core genes. Pie chart displays numbers of total genes with percentages in parentheses. (C) Power-law fit to the mean number of new genes per genome (bold points) after 100 pangenome permutations (i.e., background points). Θ < 1 indicates that the pangenome is in the “open” state (79).
Box plot rarefaction for total genes per genome for B. cereus (A) and S. aureus (B). Data were sourced from the total genes permutation Roary output file. Download FIG S1, TIF file, 0.6 MB (638.6KB, tif) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strain origins significantly correlate with genome contents.
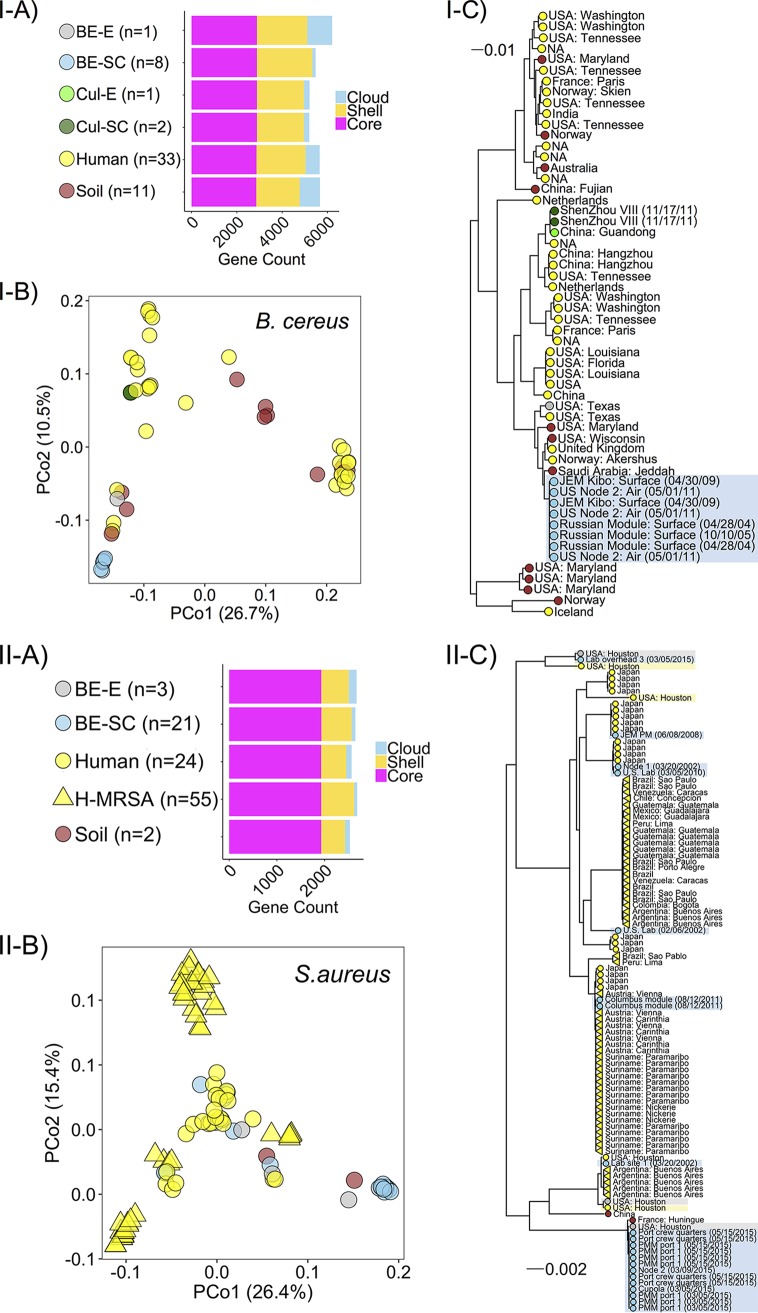
Core genome diversity significantly correlated with accessory genome diversity for both B. cereus (Mantel r = 0.881, P = 0.001) and S. aureus (Mantel r = 0.760, P = 0.001), suggesting that evolutionary trends for bacterial mutation have a relatively similar biogeography to gene gain/loss events. Despite large intraspecific variation in the ISS (Fig. 2), each set of strains exhibited similar genomic diversity (i.e., core gene variation and accessory gene presence/absence) in the relative context of counterpart Earth-based strains (Fig. 3B and C). Strain origin (e.g., BE-spacecraft, BE-Earth, soil, and human) significantly correlated with overall gene presence/absence for B. cereus (PERMANOVA R2 = 0.203, P < 0.001, n = 56) and S. aureus (PERMANOVA R2 = 0.233, P < 0.001, n = 105) (Table 1). Genomes of ISS-sourced isolates of each species clustered more closely with counterparts from Earth-based BEs and soil than with those from humans (Fig. 3). Importantly, genomes of the ISS-associated S. aureus were, on average, more similar to human-associated strains that were not reported as pathogens (J = 0.384) than to known pathogenic variants (J = 0.435), i.e., those isolated from patients with MRSA or bacteremia. These data suggest that (i) environment-based strains, regardless of being collected on Earth or in space, contain core and accessory genomic contents that are somewhat distinct from human-derived counterparts and (ii) S. aureus isolates from the ISS were more closely related to putatively commensal than pathogenic strains.
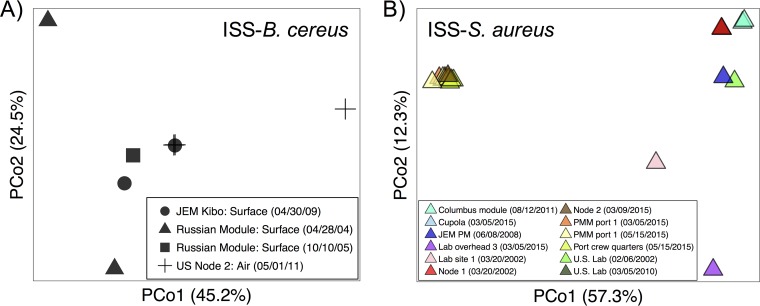
FIG 2.
Heterogeneity in gene presence/absence among ISS-associated B. cereus (A) and S. aureus (B) strains. Symbol shape or color corresponds to sample area and date.
FIG 3.
Bacterial species-level genomic diversity (i.e., gene presence/absence and core gene variants) correlates with strain origin. B. cereus and S. aureus are represented in the I and II panels, respectively. (A) Total gene counts for each fraction of the pangenome by strain origin: built environment-Earth (BE-E), -spacecraft (BE-SC), culture-Earth (Cul-E), -spacecraft (Cul-SC), human, and soil samples. BE-SC samples were taken aboard the ISS; Cul-SC samples were clonal isolates sent to space aboard the Shenzhou VIII. (B) PCoA for gene presence/absence among strains. Color/shape corresponds to sample origin. (C) Phylogenetic tree constructed from core gene codon alignment with midpoint rooting. ISS-, human-, and BE-E-sourced strains from the work of Checinska Sielaff et al. (29) and the Wallace and Voorhies data set (Table S1) are shaded in blue, yellow, and gray, respectively.
TABLE 1.
Differences in gene presence/absence for each taxon based on strain origin, sequencing technology, sequence assembler method, culture medium, and study/reference
| Taxon and factor | Variables (n) | R2a | P valuea |
|---|---|---|---|
| B. cereus | |||
| Strain origin | BE-Earth (1), BE-ISS (8), culture-Earth (1), culture-Shenzhou VIII (2), human (33), soil (11) |
0.203 | 0.001 |
| Culture mediumb |
B. cereus selective agar (8), brain heart infusion medium (2), custom medium (6), fastidious broth to blood agar (3), HEPA filter to R2A agar (3), Luria-Bertani medium (8), Trypticase soy agar (3) |
0.327 | 0.002 |
| Sequencing technology |
Illumina (32), 454 (12), combination (10) | 0.100 | 0.002 |
| Assembler | A5 (3), ABySS (10), CANU (1), Celera (4), CLC NGS Cell (2), IDBA-UD (5), combination (5), Newbler (11), SOAPdenovo (3), SPAdes (8), Velvet (1) |
0.323 | 0.001 |
| Study | 19 different studies/NCBI references (Table S1) | 0.484 | 0.001 |
| S. aureus | |||
| Strain origin | BE-Earth (3), BE-ISS (21), human (24), human-MRSA (55), soil (2) |
0.233 | 0.001 |
| Culture mediumb | Brain heart infusion agar (2), HCH-supplemented liquid medium (1), Trypticase soy agar (28), Trypticase soy broth (37) |
0.218 | 0.001 |
| Sequencing technology |
Illumina (32), PacBio (15) | 0.027 | 0.004 |
| Assembler | A5 (13), CLC Genomics Workbench (51), PacBio HGAP3 (15), SeqMan NGen (16), SOAPdenovo (1), SPAdes (8), Velvet (1) |
0.385 | 0.001 |
| Study | 9 different studies/NCBI references (Table S1) | 0.472 | 0.001 |
PERMANOVA results for genome associations with each factor.
Reflects the medium that was used in initial bacterial isolation or that which was used in isolate collection/processing, where available (i.e., not all studies provided culture method details, and some provided only information for how strains were processed rather than initial isolation).
Accession numbers, metadata, and CheckM assembly statistics for genomes utilized in the pangenomic meta-analysis. Download Table S1, XLSX file, 0.1 MB (63.9KB, xlsx) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To discern genetic signatures that may be associated with spaceflight conditions (e.g., microgravity and radiation) or from BE conditions (e.g., desiccation and chemical cleaning product residues), we focused on genomes from a study where clonal B. cereus isolates were sent to space aboard the Shenzhou VIII in containers where they were grown in Luria-Bertani medium (49). It was reported that after 16 days in spaceflight, compared to Earth-based controls that were cultivated the same way, the strains developed three polymorphic loci and experienced changes in growth rate, antibiotic resistance, and levels of metabolic expression and function (49). Despite these mutations and physiological changes that occurred in response to spaceflight conditions, the strains that were sent to space aboard the Shenzhou VIII in culture medium did not become more similar to the set of spacecraft BE strains (i.e., in Fig. 3, part I, Cul-SC does not diverge from Cul-E, and both remain distinct from BE-SC). This may be a reflection of the Cul-SC/E samples growing in a rich medium for a relatively short time, while the BE-SC samples were likely not growing for some time prior to sampling. We can still infer that spaceflight alone was probably not responsible for the drastic genomic profile differences in the ISS versus counterpart strains (Fig. 3); BE conditions may have played a role as well, with an influence from sampling date and location (i.e., ISS interior site) (Fig. 2).
To evaluate correlations between genome content and strain origin without potential biases associated with study-specific factors (e.g., sampling location and date, criteria used to select strain for further cultivation and sequencing, and factors displayed in Table 1), we focused on the subset of genomes from the Wallace and Voorhies data set (Table S1). S. aureus had been isolated from the ISS-BE (n = 8), preflight BE (n = 3; cargo bags and hardware surfaces), and preflight astronauts (n = 4; human skin swabs). Pairwise PERMANOVA indicated no differences between gene presence/absence in preflight BE and the ISS-BE isolates (R2 = 0.134, P = 0.177) or in preflight BE and preflight astronaut isolates (R2 = 0.089, P = 0.972). Alternatively, there were subtle, yet not significant, differences in gene presence/absence between preflight astronaut and ISS-BE isolates (R2 = 0.139, P = 0.097). While the BE surface strains may resemble “local” human-associated commensal strains, potential genomic differences in strains from the ISS-BE and humans may reflect site-specific factors.
Function enrichments in isolates from the ISS, soil, and humans.
Inferring potential microbial adaptive responses to a particular environment requires focusing on genome-encoded functions. A total of 2,907 and 1,729 unique functions (not counting “hypothetical proteins”) were encoded in B. cereus and S. aureus pangenomes, respectively. Consistent with the trends for gene presence/absence and core gene variation, there were significant differences in bacterial functional profiles based on strain origin (B. cereus PERMANOVA R2 = 0.209, P < 0.001, n = 56; S. aureus PERMANOVA R2 = 0.299, P < 0.001, n = 105) (Fig. S2).
Bacterial species-level genomic diversity based on gene product (encoded function) presence-absence correlates with sample origin: built environment-Earth (BE-E), -spacecraft (BE-SC), culture-Earth (Cul-E), -spacecraft (Cul-SC), human, and soil. All BE-SC samples were taken aboard the ISS, and the culture samples were clonal isolates sent to space aboard the Shenzhou VIII. Download FIG S2, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
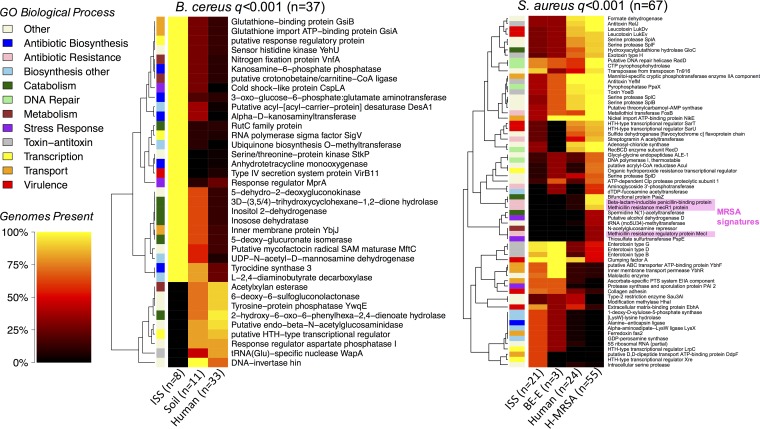
According to the generalized linear model (GLM), 262 B. cereus functions and 104 S. aureus functions were significantly associated with strain origin (P < 0.01 and FDR q < 0.1) (Table S2). The most strongly associated functional enrichments (FDR q < 0.001) are displayed in Fig. 4. For both taxa, greater proportions of ISS and Earth-based BE strains than human strains encoded key functions involved in material transport, antibiotic biosynthesis (i.e., kanosamine, tetracycline, and tyrocidine in B. cereus; bacilysin in S. aureus), and other biosynthetic processes (i.e., fatty acids and ubiquinone in B. cereus; amino acids, isoprene, and lipopolysaccharides in S. aureus) (Fig. 4; Table S2). ISS-associated B. cereus strains were also enriched with unique metabolism (i.e., carbohydrate and nitrogen), catabolism (i.e., aromatic hydrocarbon and inositol), and stress response (cold shock and starvation) processes (Fig. 4; Table S2).
FIG 4.
Strain origin-enriched gene products. The heat map displays all function enrichments with FDR q < 0.001 for B. cereus (left) and S. aureus (right). Heat color corresponds to percentage of genomes per origin type containing at least 1 gene encoding the listed product. Row colors indicate the biological process group for gene products. On the S. aureus panel, the functions associated with the staphylococcal cassette chromosome mec (IWG-SCC 2009) are shaded.
Bacterial gene product correlations with strain origin. Gene products with significant correlation (P < 0.01 and FDR q < 0.1) are shaded, and those with highest significance (q < 0.001) are in bold. For the latter, the UniProtKB pathway, gene ontology (GO) molecular function, and GO biological process are listed (i.e., manual search at http://www.uniprot.org; visited on 14 June 2018). The designated GO biological process (i.e., antibiotic biosynthesis, antibiotic resistance, biosynthesis other, catabolism, DNA repair, metabolism, ribosome, stress response, toxin-antitoxin, transcription, transport, virulence, or other) was selected in accordance with the cumulative UniProtKB information. Download Table S2, XLSX file, 0.3 MB (310.7KB, xlsx) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Regarding implications for astronaut health, there were several virulence factors (i.e., the UniProtKB biological process was “virulence” or “pathogenesis”) enriched in the ISS-derived B. cereus (i.e., type IV secretion system protein Vir11B) and S. aureus (i.e., clumping factor A, collagen adhesion, and extracellular matrix-binding protein EbhA) (Fig. 4). Additionally, several resistance functions were enriched in the genomes of ISS-derived strains compared to Earth-based counterparts (i.e., multidrug efflux pumps for B. cereus; beta-lactamase and heavy metal for S. aureus), though to a lesser extent (i.e., 0.01 < q < 0.1) than the above lifestyle-associated processes with q < 0.001 (e.g., biosynthesis, catabolism, material transport, and metabolism), as these resistances were often common in isolates from humans/soil elsewhere (Tables 2 and 3).
TABLE 2.
Antibiotic resistance gene products enriched by strain origin (P < 0.01 and FDR q < 0.1) for B. cereus for origins with n ≥ 3a
| B. cereus ARG product | Antibiotic class or other |
% of strains from origin (n): |
FDR q | ||
|---|---|---|---|---|---|
| BE-ISS (8) | Soil (11) | Human (33) | |||
| Metallothiol transferase FosB 2 | Fosfomycin | 0.0 | 45.5 | 84.8 | 0.001 |
| UDP-4-amino-4-deoxy-l-arabinose–oxoglutarate aminotransferase | Polymyxin | 0.0 | 54.5 | 0.0 | 0.005 |
| Multidrug resistance protein EbrB | Multidrug | 100.0 | 27.3 | 57.6 | 0.045 |
| Beta-lactamase 3 | Penicillin | 0.0 | 63.6 | 18.2 | 0.077 |
| Cadmium resistance transcription regulatory protein CadC | Heavy metal* | 0.0 | 9.1 | 24.2 | 0.077 |
| Multidrug resistance protein Stp | Multidrug | 100.0 | 63.6 | 100.0 | 0.080 |
Percentage of strains from the respective origins encoding each function are listed. Gene products that were more frequently present in ISS strains than soil- and/or putatively commensal human-derived strains are displayed in bold. *, not an antibiotic class, though can be associated with antibiotic resistance.
TABLE 3.
Antibiotic resistance gene products enriched by strain origin (P < 0.01 and FDR q < 0.1) for S. aureus for origins with n ≥ 3a
| S. aureus ARG product | Antibiotic class | % of strains from origin (n): |
FDR q | |||
|---|---|---|---|---|---|---|
| BE-ISS (21) | BE-Earth (3) | Human (24) | Human-MRSA (55) | |||
| Beta-lactam-inducible penicillin-binding protein | Penicillin | 0.0 | 0.0 | 8.3 | 96.4 | <0.001 |
| Methicillin resistance MecR1 protein | Penicillin | 0.0 | 0.0 | 8.3 | 90.9 | <0.001 |
| Aminoglycoside 3′-phosphotransferase | Aminoglycoside | 0.0 | 33.3 | 0.0 | 58.2 | <0.001 |
| Metallothiol transferase FosB | Fosfomycin | 38.1 | 66.7 | 62.5 | 100.0 | <0.001 |
| Streptogramin A acetyltransferase | Streptogramin | 23.8 | 0.0 | 50.0 | 87.3 | <0.001 |
| Methicillin resistance regulatory protein MecI | Penicillin | 0.0 | 0.0 | 0.0 | 34.5 | <0.001 |
| Bleomycin resistance protein | Antitumor | 0.0 | 0.0 | 4.2 | 30.9 | 0.006 |
| Cadmium resistance transcription regulatory protein | Heavy metal* | 95.2 | 66.7 | 54.2 | 92.7 | 0.007 |
| Kanamycin nucleotidyltransferase | Aminoglycoside | 0.0 | 0.0 | 8.3 | 30.9 | 0.017 |
| Macrolide export ATP-binding/permease protein MacB | Macrolide | 4.8 | 0.0 | 29.1 | 43.6 | 0.031 |
| Beta-lactamase | Penicillin | 100.0 | 100.0 | 70.8 | 96.4 | 0.036 |
Percentage of strains from the respective origins encoding each function are listed. Gene products that were more frequently present in ISS strains than soil- and/or putatively commensal human-derived strains are displayed in bold. *, not an antibiotic class, though can be associated with antibiotic resistance.
Although gene product presence/absence for S. aureus isolated from the ISS BE was correlated with year of sampling (PERMANOVA R2 = 0.692, P < 0.001, n = 21) and study/reference (PERMANOVA R2 = 0.467, P < 0.001, n = 21), several gene products enriched (or absent) in the ISS-BE were generally conserved (Fig. S3). None of the BE genomes contained MRSA signatures (i.e., beta-lactam-inducible penicillin-binding protein, MecR1 methicillin resistance protein, and methicillin resistance regulatory protein MecI) that were, conversely, present in human-associated strains. Human-associated pathogenic S. aureus happened to also be enriched with additional antibiotic resistances (e.g., macrolide, fosfomycin, and streptogramin), virulence factors, and DNA repair processes (Fig. 4; Table 3). Similarly, compared to the ISS strains of B. cereus, those that were soil- and/or human-borne encoded additional resistances more frequently (i.e., fosfomycin, polymyxin, penicillin, and heavy metal) (Table 2). Collectively, these data suggest that microbial adaptations to the ISS/BE are largely related to general lifestyle responses involving biosynthesis, material transport, metabolism, and stress tolerance. As these enriched gene products are part of broader functional pathways, and KEGG pathways encoded in the genomes that we leveraged appeared to correlate with phylogeny as measured by core gene distance (not strain origin per se, at least for S. aureus) (Fig. S4), it remains somewhat unclear whether the BE selects for overall functional potential of bacteria.
Profile of S. aureus strain origin-enriched gene products (q < 0.001; n = 67) for the ISS isolates (n = 21). Heat map displays gene product presence (yellow)/absence (black) for the gene products from Fig. 4. Reference and sampling date correspond to colors shown in the top two rows. Download FIG S3, TIF file, 1.7 MB (1.8MB, tif) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Phylogenetic tree constructed from core gene codon alignment with midpoint rooting (same as in Fig. 3, part II), from which one BE-SC isolate from each branch containing a BE-SC isolate and one counterpart isolate on the same branch were processed for KEGG pathway analysis in GhostKOALA (M. Kanehisa, Y. Sato, and K. Morishima, J Mol Biol 428:726–731, 2016, https://doi.org/10.1016/j.jmb.2015.11.006). Note that only 53.3% ± 1.3% of the open reading frames were able to be annotated with KEGG Ontology (mean ± SD; n = 14). (B) PCoA illustrates significant association between KEGG profile (collapsed to “C” level) and tree branch position (PERMANOVA R2 = 0.633; P = 0.002). Download FIG S4, TIFF file, 1.7 MB (56.7MB, tiff) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
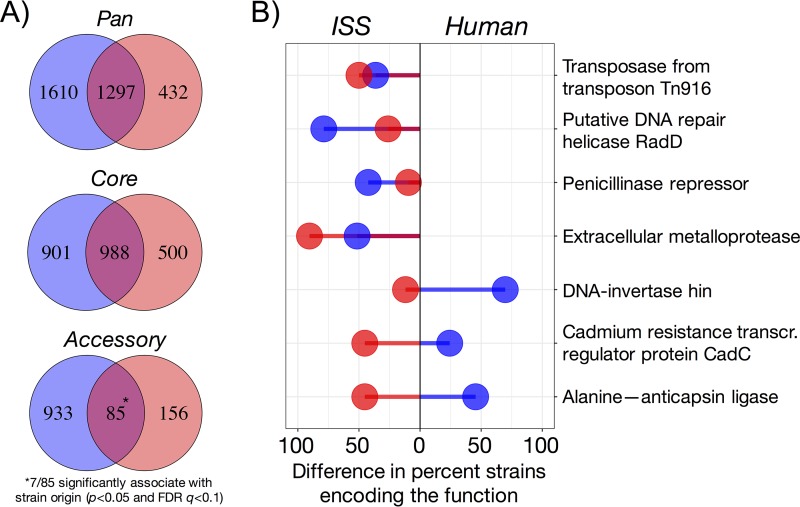
Gene products conserved across taxa enriched in a distinct environment may reflect more fundamental bacterial adaptations to said environment. Since B. cereus and S. aureus are both Gram-positive members of the same phylum, Firmicutes, we anticipated an overlap in most of their core and some of their accessory functions. Indeed, 52.3% of B. cereus core gene products were also core in S. aureus, and 66.4% of S. aureus core gene products were also core in B. cereus (Fig. 5A). Of the 85 overlapping accessory functions, 7 were significantly associated with strain origin (P < 0.01 and FDR q < 0.1). Focusing on ISS-BE and putatively commensal human-associated strains only, 4 of these functions were encoded more frequently in the former for both taxa, while the 3 others were more frequently encoded in human-associated B. cereus and ISS-associated S. aureus compared to respective counterparts (Fig. 5B). For example, an MGE-associated transposase and penicillin resistance regulatory protein were more common in the ISS strains of both taxa (Fig. 5B), suggesting a potential implication for interspecies ARG mobilization in the confined environment. In contrast, bacilysin biosynthesis (i.e., involving alanine-anticapsin ligase) and a cadmium resistance regulator were more often associated with ISS-derived S. aureus and human-derived B. cereus (Fig. 5B). Perhaps the functions with opposing sample origin associations may play a role in bacterial persistence away from traditional niches (i.e., human-derived S. aureus in the BE and environment-borne B. cereus in humans).
FIG 5.
Overlap in gene products across taxa. (A) Numbers of shared and distinct functions encoded in the pangenomes, core genomes, and accessory genomes of B. cereus (blue) and S. aureus (red). (B) Shared accessory gene products across taxa that significantly correlated (P < 0.01 and FDR q < 0.1) with strain origin for origins containing n ≥ 3 strains (i.e., B. cereus: ISS, human, soil; S. aureus: ISS, BE-Earth, human, human-MRSA). Each segment corresponds to the differences in percentage of strains (blue, B. cereus; red, S. aureus) isolated from the ISS-BE and human samples (i.e., putatively commensal S. aureus only, not MRSA) encoding the gene product. For example, functions with both segments in the same direction demonstrate association with the same origin or vice versa for segments in the opposite direction.
Potentially mobile functions unique to the ISS.
To identify potentially mobile functions that correlated with the ISS, we characterized the genes that associated with enriched MGEs. In an effort to limit potential sequencing technology and assembler method biases (Table 1), we limited the scope of this analysis to the B. cereus genomes from Illumina-based studies with raw reads available from NCBI-SRA (n = 22) and used a standardized sequence assembly method (i.e., all SRA files were processed with SPAdes whereas the original assemblies had been processed with ABYSS, Celera, IDBA-UD, CLC NGS Cell, or combinational approaches, as described in Table S1). There were 18/22 genome assemblies that passed quality control, which was similar to the proportion for the original assemblies that were processed (Table S1). Notably, there was significantly less variation in gene presence/absence of annotated genomes for the new assemblies (J = 0.338 ± 0.014; mean ± SE) compared to the original assemblies (J = 0.362 ± 0.013) (Wilcoxon P = 0.048). This finding further supports the concept of sequence assembler bias (55) and suggests that comparative genomics studies should use a standardized approach, if possible.
Scoary (56) analysis of the pangenome constructed from the new B. cereus assemblies identified significant correlations between ISS strains and pepF1 (product: oligopeptidase F, plasmid) (P = 0.001, FDR q < 0.021), bin3 (product: putative transposon Tn552 DNA-invertase bin3) (P < 0.001, FDR q = 0.004), and Int-Tn (product: transposase from transposon Tn916) (P = 0.001, FDR q = 0.021) (Table S3). Because bin3 and Int-Tn were each only present in 3 human strains and 1 soil strain, while all non-ISS strains also encoded a variant of pepF1 (Table S3), we focused on the two former for subsequent analysis.
Scoary output displaying full list of genes significantly associated with ISS-BE B. cereus genomes for the subset of B. cereus strains with raw sequencing data from Illumina-based studies processed with our standardized pipeline (n = 22). We note that this analysis was not performed for S. aureus due to limited raw sequencing data available (n = 1 as shown in Table S1). Download Table S3, XLSX file, 0.1 MB (118.6KB, xlsx) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Local neighborhood genes (i.e., flanking ±5 genes on the contig) of the two transposon-associated genes were characterized to test the hypothesis that the ISS strain-specific MGEs may (i) potentially carry different genes on the same MGE in counterpart strains and (ii) present potential risk for astronaut health (i.e., carry transmissible ARGs or virulence factors). Int-Tn was linked with lysN (product: 2-aminoadipate transaminase), ddl (product: d-Ala–d-Ala ligase), and tenA (product: aminopyrimidine aminohydrolase) in 75%, 63%, and 63% of the ISS strains, respectively (Table S4). That is, the transposase in ISS isolates consistently associated with lysine, thiamine, and peptidoglycan biosynthesis. In contrast, Int-Tn in human- and soil-derived strains appeared to associate with different genes altogether. In addition, bin3 was found to associate with rapG (product: response regulator aspartate phosphatase G) in 87.5% of ISS strains, but none of the Earth-based counterparts (Table S4). Considering the variability in B. cereus sampling time and location within the ISS and the local intraspecific genome variation (i.e., Japanese module on 30 April 2009, Russian module on 28 April 2004 and 10 October 2005, and U.S. node on 1 May 2011, as indicated in Fig. 2), these two MGEs and the mechanisms they may mobilize (i.e., biosynthesis and stress response pathways) are likely important for persistence in the ISS. Moreover, there was no indication that the ISS-enriched MGEs carried ARGs, which supports the hypothesis that the ARGs in the ISS-borne B. cereus and S. aureus were intrinsic and not acquired after deposition to the BE.
Local neighborhood genes of bin3 and Int-Tn in B. cereus. The genes (and gene products, in parentheses) that are located 1 to 5 positions upstream or downstream from each copy of the putative MGE are displayed, along with contig number and base pair range. “HP” indicates hypothetical protein, and “NA: end contig” indicates that the location at the upstream or downstream position is nonexistent due to reaching the end of the contig. Download Table S4, XLSX file, 0.05 MB (50.9KB, xlsx) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
We present the first study to use comparative pangenomics to uncover underlying genes and functions that may be involved in microbial colonization and persistence in the BE. The pangenomes of two common members of the indoor microbiome with high economic importance, B. cereus and S. aureus, were defined from WGS data of strains isolated from the ISS (i.e., a model BE), BEs on Earth, soil, and humans. Consistent with prior studies on these taxa, the numbers of new genes per genome indicated substantial intraspecific variation and overall pangenome “openness” associated with a broad niche range (25, 26, 33, 57). Indeed, B. cereus and S. aureus are versatile in host- and environmentally associated microbiomes as commensal or pathogenic variants (31, 44). Comparing the two pangenomes indicated that the host-associated taxon contains a more dominant fraction of core genes, while that of the generalist is more boundless, perhaps to a disproportionate extent (i.e., ratios of new genes per genome were exceedingly greater than ratios in average genome size and total pangenome size). This disparity may partially reflect genomic biases associated with different culture methods for the two taxa, since media and even preparation technique influence which strains will grow (58). Nevertheless, the associations we observed for both B. cereus and S. aureus genome contents with strain origins, along with the key distinctions in their pangenomes and general lifestyles, highlight the potential for characteristic microbial response to the BE.
Our findings suggest that diverse biological processes play a central role in bacterial adaptation to the BE; i.e., genetically distinct members of introduced populations may endure the local selective pressures (e.g., desiccation, limited resource availability, and biocide and detergent residues from cleaning products) (12, 59). Strains of B. cereus and S. aureus isolated from the ISS and Earth-based BEs (i.e., B. cereus from Earth-based BEs unable to be compared due to low sample size; n = 1) were enriched with functions involved in biosynthesis (e.g., fatty acids, amino acids, and antibiotics), catabolism (e.g., sugars and aromatics), material transport, metabolism (e.g., nitrogen and carbohydrate), and stress response (e.g., cold shock), compared to their respective human- and/or soil-derived counterparts. We recognize that correlations identified between strain origin and genomic content may have been influenced, at least partially, by potential biases associated with isolate genomic content across studies due to study-specific factors: e.g., methods used for isolate collection randomization, “batch effects” associated with sample processing, sampling date and location, and sequencing technology and assembler, etc. As such, the relative genomic similarity in BE strains and heterogeneity among Earth strains may reflect the scarcity of BE WGS data available (i.e., to our knowledge, we leveraged all available WGS data for B. cereus and S. aureus isolated from the BE, which are largely limited to the ISS). When evaluating correlations in genomic content and strain origin without such biases (i.e., focusing on the subset of S. aureus genomes from the Wallace and Voorhies data set; Table S1), we still found subtle differences in overall gene presence/absence between strains isolated from the ISS-BE and preflight astronauts. Of course, these differences may reflect site-specific factors (i.e., different humans were the source of S. aureus in the ISS). To confirm the hypothesis of a bacterial adaptive response to presumed selective pressures in the BE, there is an urgent need for future studies designed to control for the above limitations and expand culture/WGS data repositories for BE strains. Longitudinal sampling of clinically relevant isolates collected from the BE and human occupants in parallel (e.g., ISS or on Earth, such as in a hospital setting), across several locations, warrants investigation.
Genes that confer antibiotic resistance (e.g., β-lactamases, heavy metal resistance, and multidrug efflux) may play a significant role in BE selection as well (8, 12, 60). In fact, long-term microbial exposures to benzalkonium chloride, the primary cleaning disinfectant used on interior surfaces of the ISS (13, 61), are known to influence ARG dissemination (62) and could possibly select for intrinsic mutations that confer resistance (e.g., mdep expression, decreased porin uptake, and changes in cell wall composition) (63). However, in our analysis, the role of ARGs in ISS selection was less emphasized than that suggested from metagenomics assessments in other BEs (e.g., hospitals and athletic facilities) (8, 12). The lack of associations between BE strain origins and a larger number of ARGs may be due to (i) discrepancies between culture-independent and -dependent analyses (e.g., biases toward specific, cultivable organisms depending on the culture medium used), (ii) reduced transmission of undesirable strains (e.g., multidrug-resistant pathogens) to the ISS BE because of preflight health monitoring/screening (64), (iii) potential limitations to uncovering the absolute ARG diversity (e.g., gene annotation sensitivity yielding uncharacterized hypothetical proteins), or (iv) the spaceflight environment not necessarily selecting for the same ARGs as BEs on Earth. Perhaps Earth-based BEs may demonstrate more selection for ARGs than the ISS due to human occupancy-dependent microbial transfers and long-term evolution. While hospital staff members work at the same facility for years at a time, ISS astronauts are cycled in and out every few months. The strong correlation we found between S. aureus overall gene product presence/absence and sampling date in the ISS, along with the inactivation rates of this taxon (36), suggests that the BE isolates were probably deposited from whomever was aboard during the prior few weeks. The role of microbial reseeding and cycling between surface and host in propagation of ARGs within a population is an interesting avenue for future research.
Microbial selection in the ISS may have been influenced by selective pressures from spaceflight (e.g., microgravity, elevated CO2, and radiation) and/or BE conditions (e.g., desiccation, limited resource availability, and biocide and detergent residues from cleaning products). Importantly, physiological responses of bacteria in the BE, specifically the ISS, were partially consistent with genomic signatures we identified. The phenotypes of B. cereus were reported to be nonvirulent (i.e., non-toxin-producing and lacking toxin-encoding plasmids pXO1 and pXO2) (30). Our genomic assessment further indicated an absence of cytK and nhe, which encode other toxins commonly associated with B. cereus pathogenicity (38). Additionally, spaceflight analog culture investigations have demonstrated that S. aureus adopts a colonization phenotype with a repression of virulence characteristics (65). Culture-based resistance assays had also previously indicated that the majority of both sets of strains were resistant to penicillin and some S. aureus strains were resistant to erythromycin and rifampin as well (29, 30). In the present work, we found that only penicillin resistance was significantly enriched in the ISS-associated S. aureus genomes, which may simply reflect the fact that ARGs identified in the ISS genomes were either not conserved or sometimes common in counterpart strains elsewhere. Moreover, spaceflight conditions alone (i.e., separate from BE) undoubtedly influence genomic and physiological responses, despite being potentially less evident than adaptations to the BE. Spaceflight and microgravity simulations have been reported to enhance growth, virulence, biofilm formation, nutrient scavenging, stress tolerance, and/or antimicrobial resistance of B. cereus, Cupriavidus metallidurans, Escherichia coli, Micrococcus luteus, Pseudomonas aeruginosa, Salmonella sp., and S. aureus in vitro (49, 65–73). In the present study, not uncovering enrichments related to these functions (aside from stress tolerance) suggests that microbes in the ISS, and even in other BEs, may undergo potential physiological changes that are not necessarily reflected in their genomes as gene gains or losses. It may also likely reflect the low-humidity, well-ventilated environment of the ISS not being conducive to bacterial growth; i.e., bacteria in the BE do not need nutrients per se but only to withstand stresses associated with being stranded on surfaces or in dust. To reconcile differences in microbial physiological and genomic responses to the ISS and distinguish selective effects of spaceflight from BEs, time-series in situ experiments on genomic, transcriptomic, and proteomic dynamics of microbial isolates and communities on surfaces warrant investigation.
While mobile genetic elements (MGEs; e.g., plasmid, transposon, and phage) that mediate horizontal gene transfer (HGT) may enhance bacterial population survival under constant environmental pressures, they are also responsible for the dissemination of genes involved in antimicrobial resistance and virulence (74). In this study, we identified two transposon genes (Int-Tn and bin3) frequently associated with the same genes encoding putatively beneficial biological processes (i.e., biosynthesis and stress tolerance) in the ISS B. cereus genomes. Since acquired mobile genes are representative of the unique history of the microorganism, uncovering these similarities further supports the role of these functions in potential adaptation to the ISS BE. Biofilm formation/incorporation, which creates opportunities for gene exchange between bacteria (75), may be enhanced during spaceflight (68). However, it remains unclear whether the HGT involving Int-Tn and bin3 actually occurred in the BE, as this was probably unlikely. Barriers to HGT on surfaces/in dust may include (i) physical distance separating microbes, (ii) lack of moisture sources that may otherwise enable mobility and nutrient transport, (iii) general stresses that induce dormancy, and/or (iv) lack of compatibility between strains. Thus, an alternative explanation is that similar strains/spores have persisted in the closed system for long durations of time. Since B. cereus is a sporeformer, it is possible that the isolates were a result of bacteria that were deposited months to years before sampling or were from dust brought aboard with supplies. Regardless, our findings suggest that the few ARGs that were enriched in the ISS strains were likely intrinsic and not mobile/acquired. Testing the hypothesis that interactions in BE microbiota may mediate enhanced bacterial survival and, potentially, virulence and resistance dissemination is an interesting area for future research.
Overall, our comprehensive pangenomic analysis suggests that members of the BE microbiome, both on Earth and in the ISS, contain characteristic genomic signatures distinct from human- and/or soil-derived counterpart strains. Such signatures involve complex biological processes that may reflect local adaptations, the most crucial of which do not appear to have direct impacts on human health.
MATERIALS AND METHODS
Genome assembly processing.
GenBank genome assemblies for 83 strains of B. cereus (76 B. cereus and 7 Bacillus sp., grouped with B. cereus in this text) and 106 strains of S. aureus that were isolated from spacecraft, humans, or soil were retrieved from the NCBI Assembly Database. Accession numbers and associated metadata (e.g., strain origin, location, culture medium, sequencing technology, and assembler) are listed in Table S1 in the supplemental material. Assembly quality was evaluated with CheckM v1.0.7 (76), and genomes with less than 97% completeness or greater than 3% contamination were excluded from further analyses. The remaining genomes (B. cereus, n = 56; S. aureus, n = 105) were annotated with Prokka v1.12, referencing the respective genus (77). Output .gff files were processed in Roary v3.12.0 with minimum blastp identity of 90% to build pangenome matrices (78).
Pangenome analyses.
Statistical analyses and data visualization were performed in R v3.2.1. Genes were grouped into categories of “cloud,” “shell,” and “core” corresponding to presence in <10%, 10 to 95%, and >95% of genomes analyzed, respectively. Power-law regression was used to estimate the size and expansiveness of each pangenome based on 100 random permutations of new genes per genome: N(n) = α · n−θ, where N is the expected number of genes, n is the number of genomes sequentially added, and θ determines whether the pangenome is open (<1) or closed (>1) (79).
The associations between the presence/absence of genes and strain origin, culture medium, sequencing technology, sequence assembler, and study were evaluated with principal coordinate analysis (PCoA) and permutational analysis of variance (PERMANOVA) using Jaccard’s index with binary standardization as the beta diversity metric. To infer phylogenetic similarity, the core gene amino acid sequence alignments that were generated from Roary were processed with FastTree v2.1.10 using the Jones-Taylor-Thornton model and CAT approximation (80). The Newick trees were processed with Phangorn v2.4.0 (81) for midpoint rooting and plotted with Ape v5.1 (82). Correlation between evolutionary diversity within each taxon (i.e., core genome distance) and diversity in accessory genome content was evaluated with the Mantel test. We further assessed phylogenetic similarity associations with functional diversity via a PCoA and PERMANOVA for potential correlation between core gene alignment tree branch position and KEGG pathway gene ontology abundances, as determined with GhostKOALA (83), for select BE- and human-associated strains.
In search of microbial functions enriched by strain origin, we evaluated gene product presence/absence associations with strain origin (for origins with n ≥ 3) using a generalized linear model (GLM) with binomial error distribution. The resulting P values were adjusted to q values using the Benjamini-Hochberg false discovery rate (FDR) procedure (84), and associations with P < 0.01 and q < 0.1 were considered significant. The list of gene products with significant strain origin enrichments was screened for those that may confer antibiotic resistance via manual search for appropriate keywords (e.g., “resistance,” “lactamase,” “macrolide,” and “tetracycline”). This keyword-based approach is supported by the notion that Roary groups genes based on percent identity and assigns each group a gene/gene product name based on the most common annotation. For positive hits in the screen, UniProtKB (http://www.uniprot.org) was used to confirm antibiotic resistance as the biological process (e.g., ensure that it was not antibiotic biosynthesis).
ISS MGEs: controlling for batch effect biases.
A subset of the genome assemblies had raw sequence data available in the NCBI Sequence Read Archive (n = 35/189; see Table S1 in the supplemental material). To remove potential biases associated with sequencing and assembly protocol (Table 1), the raw sequence data for B. cereus strains from the paired-end Illumina sequencing studies (n = 22/83 B. cereus genomes) were downloaded through the SRA toolkit v.2.8.1 and processed with a standardized pipeline. We note that this analysis was not able to be performed for S. aureus due to limited raw sequencing data available (n = 1; Table S1). For the 22 B. cereus genomes, Trim Galore v0.4.4 (85) was utilized to remove residual adapter sequences and trim reads at nucleotides with a Phred score below 30. Genomes were assembled de novo with SPAdes v3.12.0 using default parameters (86). Scaffolds that passed CheckM quality assessment (n = 18/22 scaffolds; note, that since only 56/83 original assemblies had passed quality control, a limited number of raw sequence data sets yielding high-quality genomes was expected) were further processed for functional annotation and pangenome matrix construction using methods described above. The Wilcoxon test was applied to assess differences in gene presence/absence variation (i.e., Jaccard distance) in genomes annotated from the standardized assemblies compared to the original assemblies.
Scoary v1.6.16 was used to identify genes significantly associated with the ISS strains compared to all other sample types (56). MGE-related gene products (e.g., transposons) that were significantly associated with sample type (P < 0.01 and FDR q < 0.1) were further analyzed for similarities in local neighborhood genes (i.e., ±5 flanking genes before or after the MGE on the contig).
All data and bioinformatics and R scripts that may be used to reproduce our analyses are available at https://github.com/hartmann-lab/BE_ISS_pangenomes.
ACKNOWLEDGMENTS
This work was supported in part by the Searle Leadership Fund and through the computational resources and staff contributions provided by the Genomics Compute Cluster, which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg’s Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Compute Cluster is part of Quest, Northwestern University’s high-performance computing facility, with the purpose to advance research in genomics. R.A.B. is supported by a TL1 award (number TL1R001423) from the National Institutes of Health, National Center for Advancing Translational Science.
REFERENCES
- 1.Stephens B. 2016. What have we learned about the microbiomes of indoor environments? mSystems 1:e00083-16. doi: 10.1128/mSystems.00083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert JA, Stephens B. 2018. Microbiology of the built environment. Nat Rev Microbiol 16:661–670. doi: 10.1038/s41579-018-0065-5. [DOI] [PubMed] [Google Scholar]
- 3.Chase J, Fouquier J, Zare M, Sonderegger DL, Knight R, Kelley ST, Siegel J, Caporaso JG. 2016. Geography and location are the primary drivers of office microbiome composition. mSystems 1:e00022-16. doi: 10.1128/mSystems.00022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn RR, Fierer N, Henley JB, Leff JW, Menninger HL. 2013. Home life: factors structuring the bacterial diversity found within and between homes. PLoS One 8:e64133. doi: 10.1371/journal.pone.0064133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann EM, Hickey R, Hsu T, Betancourt Román CM, Chen J, Schwager R, Kline J, Brown GZ, Halden RU, Huttenhower C, Green JL. 2016. Antimicrobial chemicals are associated with elevated antibiotic resistance genes in the indoor dust microbiome. Environ Sci Technol 50:9807–9815. doi: 10.1021/acs.est.6b00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, Bohannan BJM, Brown GZ, Green JL. 2012. Architectural design influences the diversity and structure of the built environment microbiome. ISME J 6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang JM, Coil DA, Neches RY, Brown WE, Cavalier D, Severance M, Hampton-Marcell JT, Gilbert JA, Eisen JA. 2017. A microbial survey of the International Space Station (ISS). PeerJ 5:e4029. doi: 10.7717/peerj.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lax S, Sangwan N, Smith D, Larsen P, Handley KM, Richardson M, Guyton K, Krezalek M, Shogan BD, Defazio J, Flemming I, Shakhsheer B, Weber S, Landon E, Garcia-Houchins S, Siegel J, Alverdy J, Knight R, Stephens B, Gilbert JA. 2017. Bacterial colonization and succession in a newly opened hospital. Sci Transl Med 9:eaah6500. doi: 10.1126/scitranslmed.aah6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahnert A, Moissl-Eichinger C, Berg G. 2015. Microbiome interplay: plants alter microbial abundance and diversity within the built environment. Front Microbiol 6:887. doi: 10.3389/fmicb.2015.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkateswaran K, La Duc MT, Horneck G. 2014. Microbial existence in controlled habitats and their resistance to space conditions. Microbes Environ 29:243–249. doi: 10.1264/jsme2.ME14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu T, Joice R, Vallarino J, Abu-Ali G, Hartmann EM, Shafquat A, DuLong C, Baranowski C, Gevers D, Green JL, Morgan XC, Spengler JD, Huttenhower C. 2016. Urban transit system microbial communities differ by surface type and interaction with humans and the environment. mSystems 1:e00018-16. doi: 10.1128/mSystems.00018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahimipour AK, Ben Mamaar S, McFarland AG, Blaustein RA, Chen J, Glawe AJ, Kline J, Green JL, Halden RU, Van Den Wymlenberg K, Huttenhower C, Hartmann EM. 2018. Antimicrobial chemicals associate with microbial function and antibiotic resistance indoors. mSystems 3:e00200-18. doi: 10.1128/mSystems.00200-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Be NA, Avila-Herrera A, Allen JE, Singh N, Checinska Sielaff A, Jaing C, Venkateswaran K. 2017. Whole metagenome profiles of particulates collected from the International Space Station. Microbiome 5:81. doi: 10.1186/s40168-017-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mora M, Mahnert A, Koskinen K, Pausan MR, Oberauner-Wappis L, Krause R, Perras AK, Gorkiewicz G, Berg G, Moissl-Eichinger C. 2016. Microorganisms in confined habitats: microbial monitoring and control of intensive care units, operating rooms, cleanrooms and the International Space Station. Front Microbiol 7:1573. doi: 10.3389/fmicb.2016.01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oluwole O, Kirychuk SP, Lawson JA, Karunanayake C, Cockcroft DW, Willson PJ, Senthilselvan A, Rennie DC. 2017. Indoor mold levels and current asthma among school-aged children in Saskatchewan, Canada. Indoor Air 27:311–319. doi: 10.1111/ina.12304. [DOI] [PubMed] [Google Scholar]
- 16.Ryparová P, Rácová Z. 2016. The occurrence of mold in construction materials before inbuilt into new building and protection against this type of biodegradation. Key Eng Mater 714:44–50. doi: 10.4028/www.scientific.net/KEM.714.44. [DOI] [Google Scholar]
- 17.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott RD. 2009. The direct medical costs of healthcare-associated infections in U.S. hospitals and the benefits of prevention. CS200891-A. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 19.Dancer SJ. 2014. Controlling hospital-acquired infection: focus on the role of the environment and new technologies for decontamination. Clin Microbiol Rev 27:665–690. doi: 10.1128/CMR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. 2005. The microbial pan-genome. Curr Opin Genet Dev 15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 21.McInerney JO, McNally A, O’Connell MJ. 2017. Why prokaryotes have pangenomes. Nat Microbiol 2:17040. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- 22.Ziebuhr W, Ohlsen K, Karch H, Korhonen T, Hacker J. 1999. Evolution of bacterial pathogenesis. Cell Mol Life Sci 56:719–728. doi: 10.1007/s000180050018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snipen L, Liland KH. 2015. micropan: an R-package for microbial pan-genomics. BMC Bioinformatics 16:79. doi: 10.1186/s12859-015-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta VK, Chaudhari NM, Iskepalli S, Dutta C. 2015. Divergences in gene repertoire among the reference Prevotella genomes derived from distinct body sites of human. BMC Genomics 16:153. doi: 10.1186/s12864-015-1350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mira A, Martín-Cuadrado AB, D’Auria G, Rodríguez-Valera F. 2010. The bacterial pan-genome: a new paradigm in microbiology. Int Microbiol 13:45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- 26.Bazinet AL. 2017. Pan-genome and phylogeny of Bacillus cereus sensu lato. BMC Evol Biol 17:176. doi: 10.1186/s12862-017-1020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi N, Roberts M, Castro S, Oubre C, Makimura K, Leys N, Grohmann E, Sugita T, Ichijo T, Nasu M. 2014. Microbial monitoring of crewed habitats in space—current status and future perspectives. Microbes Environ 29:250–260. doi: 10.1264/jsme2.ME14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Aeronautics and Space Administration. 2018. Microbial analysis of International Space Station (ISS) air, surfaces and water (ISS_Micro_Analysis). NASA Life Sciences Data Archive. https://lsda.jsc.nasa.gov/Experiment/exper/13823. Accessed 1 July 2018.
- 29.Checinska Sielaff A, Singh NK, Allen JE, Thissen J, Jaing C, Venkateswaran K. 2016. Draft genome sequences of biosafety level 2 opportunistic pathogens isolated from the environmental surfaces of the International Space Station. Genome Announc 4:e01263-16. doi: 10.1128/genomeA.01263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkateswaran KB, Nitin K, Singh ACS, Pope RK, Bergman NH, van Tongeren SP, Patel NB, Lawson PA, Satomi M, Williamson CHD, Sahl JW, Keim P, Pierson DJP. 2017. Non-toxin-producing Bacillus cereus strains belonging to the B. anthracis clade isolated from the International Space Station. mSystems 2:e00021-17. doi: 10.1128/mSystems.00021-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang T, Rosch JW, Gu Z, Hakim H, Hewitt C, Gaur A, Wu G, Hayden RT. 2018. Whole-genome characterization of Bacillus cereus associated with specific disease manifestations. Infect Immun 86:e00574-17. doi: 10.1128/IAI.00574-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasko DA, Altherr MR, Han CS, Ravel J. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev 29:303–329. doi: 10.1016/j.femsre.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Jamrozy DM, Mohamed N, Anderson AS, Harris SR, Parkhill J, Tan CY, Peacock SJ, Holden MTG. 2016. Pan-genomic perspective on the evolution of the Staphylococcus aureus USA300 epidemic. Microb Genom 2:e000058. doi: 10.1099/mgen.0.000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindsay JA, Holden MTG. 2006. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct Integr Genomics 6:186–201. doi: 10.1007/s10142-005-0019-7. [DOI] [PubMed] [Google Scholar]
- 35.Horneck G, Moeller R, Cadet J, Douki T, Mancinelli RL, Nicholson WL, Panitz C, Rabbow E, Rettberg P, Spry A, Stackebrandt E, Vaishampayan P, Venkateswaran KJ. 2012. Resistance of bacterial endospores to outer space for planetary protection purposes—experiment PROTECT of the EXPOSE-E mission. Astrobiology 12:445–456. doi: 10.1089/ast.2011.0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otter JA, French GL. 2009. Survival of nosocomial bacteria and spores on surfaces and inactivation by hydrogen peroxide vapor. J Clin Microbiol 47:205–207. doi: 10.1128/JCM.02004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abo-Aba SEM, Sabir JSM, Baeshen MN, Sabir MJ, Mutwakil MHZ, Baeshen NA, D’Amore R, Hall N. 2015. Draft genome sequence of Bacillus species from the rhizosphere of the desert plant Rhazya stricta. Genome Announc 3:e00957-15. doi: 10.1128/genomeA.00957-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Böhm ME, Huptas C, Krey VM, Scherer S. 2015. Massive horizontal gene transfer, strictly vertical inheritance and ancient duplications differentially shape the evolution of Bacillus cereus enterotoxin operons hbl, cytK and nhe. BMC Evol Biol 15:246. doi: 10.1186/s12862-015-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egidi E, Wood JL, Mathews E, Fox E, Liu W, Franks AE. 2016. Draft genome sequence of Bacillus cereus LCR12, a plant growth-promoting rhizobacterium isolated from a heavy metal-contaminated environment. Genome Announc 4:e01041-16. doi: 10.1128/genomeA.01041-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gee JE, Marston CK, Sammons SA, Burroughs MA, Hoffmaster AR. 2014. Draft genome sequence of Bacillus cereus strain BcFL2013, a clinical isolate similar to G9241. Genome Announc 2:e00469-14. doi: 10.1128/genomeA.00469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, Maiden MCJ, Priest FG, Barker M, Jiang L, Cer RZ, Rilstone J, Peterson SN, Weyant RS, Galloway DR, Read TD, Popovic T, Fraser CM. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci U S A 101:8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson SL, Minogue TD, Teshima H, Davenport KW, Shea AA, Miner HL, Wolcott MJ, Chain PSG. 2015. Finished genome sequence of Bacillus cereus strain 03BB87, a clinical isolate with B. anthracis virulence genes. Genome Announc 3:e01446-14. doi: 10.1128/genomeA.01446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson SL, Daligault HE, Davenport KW, Jaissle J, Frey KG, Ladner JT, Broomall SM, Bishop-Lilly KA, Bruce DC, Gibbons HS, Coyne SR, Lo C-C, Meincke L, Munk AC, Koroleva GI, Rosenzweig CN, Palacios GF, Redden CL, Minogue TD, Chain PS. 2015. Complete genome sequences for 35 biothreat assay-relevant Bacillus species. Genome Announc 3:e00151-15. doi: 10.1128/genomeA.00151-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, Cookson BT, Shendure J, Salipante SJ. 2015. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet 11:e1005413. doi: 10.1371/journal.pgen.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zwick ME, Joseph SJ, Didelot X, Chen PE, Bishop-Lilly KA, Stewart AC, Willner K, Nolan N, Lentz S, Thomason MK, Sozhamannan S, Mateczun AJ, Du L, Read TD. 2012. Genomic characterization of the Bacillus cereus sensu lato species: backdrop to the evolution of Bacillus anthracis. Genome Res 22:1512–1524. doi: 10.1101/gr.134437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasc C, Richard J-Y, Peyret P. 2016. Genome sequence of Bacillus subtilis strain HUK15, isolated from hexachlorocyclohexane-contaminated soil. Genome Announc 4:e00273-16. doi: 10.1128/genomeA.00273-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pena-Gonzalez A, Marston CK, Rodriguez-R LM, Kolton CB, Garcia-Diaz J, Threppote A, Frace M, Konstantinidis KT, Hoffmaster AR. 2017. Draft genome sequence of Bacillus cereus LA2007, a human-pathogenic isolate harboring anthrax-like plasmids. Genome Announc 5:e00181-17. doi: 10.1128/genomeA.00181-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sabat AJ, Hermelijn SM, Akkerboom V, Juliana A, Degener JE, Grundmann H, Friedrich AW. 2017. Complete-genome sequencing elucidates outbreak dynamics of CA-MRSA USA300 (ST8-spa t008) in an academic hospital of Paramaribo, Republic of Suriname. Sci Rep 7:41050. doi: 10.1038/srep41050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su L, Zhou L, Liu J, Cen Z, Wu C, Wang T, Zhou T, Chang D, Guo Y, Fang X, Wang J, Li T, Yin S, Dai W, Zhou Y, Zhao J, Fang C, Yang R, Liu C. 2014. Phenotypic, genomic, transcriptomic and proteomic changes in Bacillus cereus after a short-term space flight. Adv Space Res 53:18–29. doi: 10.1016/j.asr.2013.08.001. [DOI] [Google Scholar]
- 50.Zhang G, Wang Q, Su Y, Li S, Liu H. 2017. Genome sequence of Staphylococcus aureus PX03, an acetoin-producing strain with a small-sized genome. Genome Announc 5:e00753-17. doi: 10.1128/genomeA.00753-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lepuschitz S, Huhulescu S, Hyden P, Springer B, Rattei T, Allerberger F, Mach RL, Ruppitsch W. 2018. Characterization of a community-acquired-MRSA USA300 isolate from a river sample in Austria and whole genome sequence based comparison to a diverse collection of USA300 isolates. Sci Rep 8:9467. doi: 10.1038/s41598-018-27781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vílchez JI, Tang Q, Kaushal R, Chen S, Liu R, Zhang H. 2018. Genome sequence of Bacillus cereus strain TG1-6, a plant-beneficial rhizobacterium that is highly salt tolerant. Genome Announc 6:e00351-18. doi: 10.1128/genomeA.00351-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panesso D, Planet PJ, Diaz L, Hugonnet JE, Tran TT, Narechania A, Munita JM, Rincon S, Carvajal LP, Reyes J, Londoño A, Smith H, Sebra R, Deikus G, Weinstock GM, Murray BE, Rossi F, Arthur M, Arias CA. 2015. Methicillin-susceptible, vancomycin-resistant Staphylococcus aureus, Brazil. Emerg Infect Dis 21:1844–1848. doi: 10.3201/eid2110.141914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arias CA, Reyes J, Carvajal P, Rincon S, Diaz L, Panesso D, Ibarra G, Rios R, Munita JM, Salles MJ, Alvarez-Moreno C, Labarca J, Garcia C, Luna CM, Mejia-Villatoro C, Zurita J, Guzman-Blanco M, Rodriguez-Noriega E, Narechania A, Rojas LJ, Planet PJ, Weinstock GM, Gotuzzo E, Seas C. 2017. A prospective cohort multicenter study of molecular epidemiology and phylogenomics of Staphylococcus aureus bacteremia in nine Latin American countries. Antimicrob Agents Chemother 61:e00816-17. doi: 10.1128/AAC.00816-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vollmers J, Wiegand S, Kaster AK. 2017. Comparing and evaluating metagenome assembly tools from a microbiologist’s perspective—not only size matters! PLoS One 12:e0169662. doi: 10.1371/journal.pone.0169662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bosi E, Monk JM, Aziz RK, Fondi M, Nizet V, Palsson BØ. 2016. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc Natl Acad Sci U S A 113:E3801–E3809. doi: 10.1073/pnas.1523199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka T, Kawasaki K, Daimon S, Kitagawa W, Yamamoto K, Tamaki H, Tanaka M, Nakatsu CH, Kamagata Y. 2014. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl Environ Microbiol 80:7659–7666. doi: 10.1128/AEM.02741-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gianoulis TA, Raes J, Patel PV, Bjornson R, Korbel JO, Letunic I, Yamada T, Paccanaro A, Jensen LJ, Snyder M, Bork P, Gerstein MB. 2009. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc Natl Acad Sci U S A 106:1374–1379. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schiwon K, Arends K, Rogowski KM, Fürch S, Prescha K, Sakinc T, Van Houdt R, Werner G, Grohmann E. 2013. Comparison of antibiotic resistance, biofilm formation and conjugative transfer of Staphylococcus and Enterococcus isolates from International Space Station and Antarctic Research Station Concordia. Microb Ecol 65:638–651. doi: 10.1007/s00248-013-0193-4. [DOI] [PubMed] [Google Scholar]
- 61.Ichijo T, Yamaguchi N, Tanigaki F, Shirakawa M, Nasu M. 2016. Four-year bacterial monitoring in the International Space Station—Japanese Experiment Module “Kibo” with culture-independent approach. NPJ Microgravity 2:16007. doi: 10.1038/npjmgrav.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tandukar M, Oh S, Tezel U, Konstantinidis KT, Pavlostathis SG. 2013. Long-term exposure to benzalkonium chloride disinfectants results in change of microbial community structure and increased antimicrobial resistance. Environ Sci Technol 47:9730–9738. doi: 10.1021/es401507k. [DOI] [PubMed] [Google Scholar]
- 63.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 64.Mermel LA. 2013. Infection prevention and control during prolonged human space travel. Clin Infect Dis 56:123–130. doi: 10.1093/cid/cis861. [DOI] [PubMed] [Google Scholar]
- 65.Castro SL, Nelman-Gonzalez M, Nickerson CA, Ott CM. 2011. Induction of attachment-independent biofilm formation and repression of hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl Environ Microbiol 77:6368–6378. doi: 10.1128/AEM.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leys N, Baatout S, Rosier C, Dams A, s’Heeren C, Wattiez R, Mergeay M. 2009. The response of Cupriavidus metallidurans CH34 to spaceflight in the International Space Station. Antonie Van Leeuwenhoek 96:227–245. doi: 10.1007/s10482-009-9360-5. [DOI] [PubMed] [Google Scholar]
- 67.Lynch SV, Mukundakrishnan K, Benoit MR, Ayyaswamy PS, Matin A. 2006. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl Environ Microbiol 72:7701–7710. doi: 10.1128/AEM.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zea L, Prasad N, Levy SE, Stodieck L, Jones A, Shrestha S, Klaus D. 2016. A molecular genetic basis explaining altered bacterial behavior in space. PLoS One 11:e0164359. doi: 10.1371/journal.pone.0164359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mauclaire L, Egli M. 2010. Effect of simulated microgravity on growth and production of exopolymeric substances of Micrococcus luteus space and earth isolates. FEMS Immunol Med Microbiol 59:350–356. doi: 10.1111/j.1574-695X.2010.00683.x. [DOI] [PubMed] [Google Scholar]
- 70.Crabbé A, Pycke B, Van Houdt R, Monsieurs P, Nickerson C, Leys N, Cornelis P. 2010. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ Microbiol 12:1545–1564. doi: 10.1111/j.1462-2920.2010.02184.x. [DOI] [PubMed] [Google Scholar]
- 71.Kim W, Tengra FK, Young Z, Shong J, Marchand N, Chan HK, Pangule RC, Parra M, Dordick JS, Plawsky JL, Collins CH. 2013. Spaceflight promotes biofilm formation by Pseudomonas aeruginosa. PLoS One 8:e62437. doi: 10.1371/journal.pone.0062437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson JW, Ott CM, Höner zu Bentrup K, Ramamurthy R, Quick L, Porwollik S, Cheng P, McClelland M, Tsaprailis G, Radabaugh T, Hunt A, Fernandez D, Richter E, Shah M, Kilcoyne M, Joshi L, Nelman-Gonzalez M, Hing S, Parra M, Dumars P, Norwood K, Bober R, Devich J, Ruggles A, Goulart C, Rupert M, Stodieck L, Stafford P, Catella L, Schurr MJ, Buchanan K, Morici L, McCracken J, Allen P, Baker-Coleman C, Hammond T, Vogel J, Nelson R, Pierson DL, Stefanyshyn-Piper HM, Nickerson CA. 2007. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc Natl Acad Sci U S A 104:16299–16304. doi: 10.1073/pnas.0707155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tixador R, Richoilley G, Gasset G, Templier J, Bes JC, Moatti N, Lapchine L. 1985. Study of minimal inhibitory concentration of antibiotics on bacteria cultivated in vitro in space (Cytos 2 experiment). Aviat Space Environ Med 56:748–751. [PubMed] [Google Scholar]
- 74.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 75.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 76.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 78.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 80.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paradis E, Schliep K. 2018. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics . doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 83.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 84.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 85.Andrews S, Krueger F, Segonds-Pichon A, Biggins L, Virk B, Dalle-Pezze P. 2015. Trim Galore! Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- 86.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Box plot rarefaction for total genes per genome for B. cereus (A) and S. aureus (B). Data were sourced from the total genes permutation Roary output file. Download FIG S1, TIF file, 0.6 MB (638.6KB, tif) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers, metadata, and CheckM assembly statistics for genomes utilized in the pangenomic meta-analysis. Download Table S1, XLSX file, 0.1 MB (63.9KB, xlsx) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial species-level genomic diversity based on gene product (encoded function) presence-absence correlates with sample origin: built environment-Earth (BE-E), -spacecraft (BE-SC), culture-Earth (Cul-E), -spacecraft (Cul-SC), human, and soil. All BE-SC samples were taken aboard the ISS, and the culture samples were clonal isolates sent to space aboard the Shenzhou VIII. Download FIG S2, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial gene product correlations with strain origin. Gene products with significant correlation (P < 0.01 and FDR q < 0.1) are shaded, and those with highest significance (q < 0.001) are in bold. For the latter, the UniProtKB pathway, gene ontology (GO) molecular function, and GO biological process are listed (i.e., manual search at http://www.uniprot.org; visited on 14 June 2018). The designated GO biological process (i.e., antibiotic biosynthesis, antibiotic resistance, biosynthesis other, catabolism, DNA repair, metabolism, ribosome, stress response, toxin-antitoxin, transcription, transport, virulence, or other) was selected in accordance with the cumulative UniProtKB information. Download Table S2, XLSX file, 0.3 MB (310.7KB, xlsx) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Profile of S. aureus strain origin-enriched gene products (q < 0.001; n = 67) for the ISS isolates (n = 21). Heat map displays gene product presence (yellow)/absence (black) for the gene products from Fig. 4. Reference and sampling date correspond to colors shown in the top two rows. Download FIG S3, TIF file, 1.7 MB (1.8MB, tif) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Phylogenetic tree constructed from core gene codon alignment with midpoint rooting (same as in Fig. 3, part II), from which one BE-SC isolate from each branch containing a BE-SC isolate and one counterpart isolate on the same branch were processed for KEGG pathway analysis in GhostKOALA (M. Kanehisa, Y. Sato, and K. Morishima, J Mol Biol 428:726–731, 2016, https://doi.org/10.1016/j.jmb.2015.11.006). Note that only 53.3% ± 1.3% of the open reading frames were able to be annotated with KEGG Ontology (mean ± SD; n = 14). (B) PCoA illustrates significant association between KEGG profile (collapsed to “C” level) and tree branch position (PERMANOVA R2 = 0.633; P = 0.002). Download FIG S4, TIFF file, 1.7 MB (56.7MB, tiff) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Scoary output displaying full list of genes significantly associated with ISS-BE B. cereus genomes for the subset of B. cereus strains with raw sequencing data from Illumina-based studies processed with our standardized pipeline (n = 22). We note that this analysis was not performed for S. aureus due to limited raw sequencing data available (n = 1 as shown in Table S1). Download Table S3, XLSX file, 0.1 MB (118.6KB, xlsx) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Local neighborhood genes of bin3 and Int-Tn in B. cereus. The genes (and gene products, in parentheses) that are located 1 to 5 positions upstream or downstream from each copy of the putative MGE are displayed, along with contig number and base pair range. “HP” indicates hypothetical protein, and “NA: end contig” indicates that the location at the upstream or downstream position is nonexistent due to reaching the end of the contig. Download Table S4, XLSX file, 0.05 MB (50.9KB, xlsx) .
Copyright © 2019 Blaustein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.