The dissemination of carbapenemase-producing Enterobacteriaceae (CPE) has led to the increased use of colistin, which has resulted in the emergence of colistin-resistant Enterobacteriaceae worldwide. One of the most threatening scenarios is the dissemination of colistin resistance in CPE, particularly the plasmid-encoded resistance element MCR.
KEYWORDS: MCR, polymyxin, sensitivity, specificity
ABSTRACT
The dissemination of carbapenemase-producing Enterobacteriaceae (CPE) has led to the increased use of colistin, which has resulted in the emergence of colistin-resistant Enterobacteriaceae worldwide. One of the most threatening scenarios is the dissemination of colistin resistance in CPE, particularly the plasmid-encoded resistance element MCR. Thus, it has now become mandatory to possess reliable media to screen for colistin-resistant Gram-negative bacterial isolates, especially Enterobacteriaceae. In this study, we evaluated the performances of the Superpolymyxin medium (ELITechGroup) and the ChromID Colistin R medium (bioMérieux) to screen for colistin-resistant Enterobacteriaceae from spiked rectal swabs. Stool samples were spiked with a total of 94 enterobacterial isolates (Escherichia coli, Klebsiella pneumoniae, Salmonella enterica, Enterobacter cloacae), including 53 colistin-resistant isolates. ESwabs (Copan Diagnostics) were then inoculated with those spiked fecal suspensions, and culture proceeded as recommended by both manufacturers. The sensitivity of detection of colistin-resistant Enterobacteriaceae was 86.8% (95% confidence interval [95% CI] = 74.0% to 94.0%) using both the Superpolymyxin medium and the ChromID Colistin R plates. Surprisingly, the isolates that were not detected were not the same for both media. The specificities were high for both media, at 97.9% (95% CI = 87.3% to 99.9%) for the Superpolymyxin medium and 100% (95% CI = 90.4% to 100%) for the ChromID Colistin R medium. Both commercially available media, ChromID Colistin R and Superpolymyxin, provide useful tools to screen for colistin-resistant Enterobacteriaceae from patient samples (rectal swabs) regardless of the level and mechanism of colistin resistance.
INTRODUCTION
Colistin and polymyxin B represent some of the few remaining options for the treatment of infections caused by multidrug-resistant and extremely drug-resistant Gram-negative bacteria, especially carbapenemase-producing Enterobacteriaceae (CPE) (1). Uncertainty remains over the best treatment option that should be used to manage infections caused by CPE. Treatment with carbapenem in combination with amikacin and treatment with colistin have achieved therapeutic results in some cases (2). Unfortunately, due to the dissemination of CPE, the increased use of colistin has led to the emergence of colistin-resistant Enterobacteriaceae worldwide (3). Colistin is a cationic antimicrobial peptide that interacts with the lipid A moiety of the lipopolysaccharide (LPS), disrupting the negatively charged outer membrane of Gram-negative bacteria. In Gram-negative bacteria, the main resistance mechanisms consist of LPS modification through the addition of positively charged 4-amino-4-deoxy-l-arabinose or phosphoethanolamine. In Enterobacteriaceae, the operons encoding enzymes involved in these modifications are arnBCADTEF and pmrCAB, respectively (4–6). Activation of the LPS-modifying genes is associated with chromosome-encoded resistance mechanisms, such as mutations in the PmrA/PmrB or PhoP/PhoQ two-component system, or through alterations to the master regulator MgrB (5, 6). In 2016, the expression of a plasmid-encoded phosphoethanolamine transferase, named MCR-1, was described as being involved in colistin resistance in Enterobacteriaceae (7). Since then, eight families of mcr genes (mcr-1 to mcr-8) have been assigned, and descriptions of seven were published previously (7–13). One of the most threatening scenarios is the wide dissemination of mcr in CPE isolates, again limiting therapeutic options. In addition, with (i) the rapid rise of mcr variants and (ii) the probability that an unknown number of polymyxin resistance mechanisms are as yet unidentified, the use of molecular techniques for the identification and the screening of colistin-resistant isolates is not universally possible. Accordingly, it has now become mandatory to possess reliable media to screen for colistin-resistant isolates (3).
Superpolymyxin and ChromID Colistin R are ready-to-use selective agar media designed for the screening for colistin resistance in Gram-negative bacteria. The target microorganisms are Enterobacteriaceae (mostly Escherichia coli, Klebsiella pneumoniae, Salmonella spp., and Enterobacter spp.) for both media and Acinetobacter spp. and Pseudomonas aeruginosa for the Superpolymyxin medium only. ChromID Colistin R is a chromogenic medium that distinguishes E. coli (pink), Klebsiella spp., Enterobacter spp., Serratia spp. (blue), and Salmonella spp. (colorless), while Superpolymyxin contains eosin Y and methylene blue dyes, which help to distinguish lactose-positive organisms (purple) from lactose nonfermenters (colorless). Both media are claimed to work on bacterial cultures, stool samples, rectal swabs (cecal samples from poultry, pigs, and calves might also be used). The present study aimed to compare the performance of these media with a collection of well-characterized colistin-resistant Enterobacteriaceae spiked into stool samples at different concentrations and inoculated onto swabs mimicking rectal swab samples.
RESULTS
The sensitivities for the detection of colistin-resistant Enterobacteriaceae were 86.8% (50% confidence interval [CI] = 74.0% to 94.0%) and 84.9% (95% CI = 71.8% to 92.8%) using the Superpolymyxin medium and a ChromID Colistin R plate, respectively, after 24 h of incubation. The sensitivity of both media was the same after 48 h of incubation (86.8% [95% CI = 74.0% to 94.0%]). Surprisingly, the isolates that were not detected were not the same for both media (Table 1). The specificities were high for both media, at 97.5% (95% CI = 85.6% to 99.9%) and 100% (95% CI = 89.3% to 100%) for the Superpolymyxin medium and the ChromID Colistin R medium, respectively. Overall, the ChromID Colistin R medium performed slightly better with K. pneumoniae and Salmonella enterica than the Superpolymyxin medium, with sensitivities of 100% (50% CI = 85.0% to 100%) and 96.2% (50% CI = 78.4% to 99.8%), respectively, and specificities of 100% (50% CI = 80.8% to 100%) and 87.0% (50% CI = 65.3% to 96.6%), respectively. Conversely, ChromID Colistin R did not detect 7/25 colistin-resistant E. coli isolates, while only 4 strains did not grow on Superpolymyxin (Table 1). The lack of detection was not correlated with the colistin MICs or the presence or absence of mcr-like genes (Table 1). For colistin-resistant isolates detected on both media (14 E. coli isolates, 24 K. pneumoniae isolates, and 1 S. enterica isolate), the limit of detection (LOD) was at least 1 log lower for ChromID Colistin R for 69.2% (27/39) of the isolates, equivalent for both media for 20.5% (8/39) of the isolates, and at least 1 log better for the Superpolymyxin medium for 7.7% (3/39) of the tested isolates (all E. coli). This lower LOD of the ChromID Colistin R protocol might be the result of the 4-h enrichment step in colistin-supplemented broth. In order to decipher whether such an enrichment step might increase the performance of the Superpolymyxin medium, the seven colistin-resistant isolates which did not grow on the Superpolymyxin medium were subjected to an enrichment step similar to that performed for the ChromID Colistin R protocol. This additional step did not allow them to grow on the Superpolymyxin medium, suggesting that this enrichment should not be recommended for use with this selective medium. As previously reported by Jayol et al. for the Superpolymyxin medium, prolongation of the incubation from 24 to 48 h did not modify the performance of the Superpolymyxin medium (14). Regarding ChromID Colistin R, prolongation of the incubation time to 48 h for one Mcr-1-producing E. coli isolate (strain CNR 164 A5) allowed us to identify typical pink colonies that were barely detectable at 24 h of incubation. Finally, one Enterobacter cloacae isolate positive for mcr-4.2 was not detected by either medium. As previously described for mcr-3 and mcr-4 variants of CPE isolates (15), the presence of mcr-4.2 does not confer phenotypic resistance to polymyxins in this E. cloacae isolate (colistin MIC, 0.5 mg/liter).
TABLE 1.
Limit of detection of colistin-resistant Enterobacteriaceae on ChromID Colistin R and Superpolymyxin media
| Colistin susceptibility and species | Strain name | Colistin MIC (mg/liter) |
Colistin resistance |
Lowest LOD (CFU/ml) ina: |
Reference or source |
||||
|---|---|---|---|---|---|---|---|---|---|
| Plasmid or chromosome encodedb |
Mechanism | ESwab Amies buffer |
Spiked stools |
||||||
| ChromID Colistin R |
Superpolymyxin | ChromID Colistin R |
Superpolymyxin | ||||||
| Colistin-resistant Enterobacteriaceae (n = 53) | |||||||||
| Escherichia coli | CNR 111 J7 | 16 | Chr | PmrB mutations (D14N, S71C, V83A) | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | 21 |
| CNR 20160039 | 4 | Chr | Unknown | 1 × 102 | >1 × 106 | 1 × 104 | >1 × 108 | 21 | |
| CNR 20160235 | 8 | Chr | MgrB mutation (V8A) | 1 × 105 | >1 × 106 | 1 × 107 | >1 × 108 | 21 | |
| CNR 1728 | 8 | Chr | PmrB mutation (G160E) | 1 × 106 | 1 × 104 | 1 × 108 | 1 × 106 | 21 | |
| 41489 | 4 | P | mcr-1 | 1 × 103 | 1 × 105 | 1 × 105 | 1 × 107 | 21 | |
| J53 + mcr-1f | 8 | P | mcr-1 | 1 × 106 | 1 × 104 | 1 × 108 | 1 × 106 | 21 | |
| CNR20140385 | 4 | P | mcr-1 | >1 × 106 | 1 × 104 | >1 × 108 | 1 × 106 | 21 | |
| S08-056 | 4 | P | mcr-1 | 1 × 104 | 1 × 103 | 1 × 106 | 1 × 105 | 21 | |
| CNR 117 G7 | 4 | P | mcr-1 | >1 × 106 | 1 × 104 | >1 × 108 | 1 × 106 | 22 | |
| CNR 121 G9 | 4 | P | mcr-1 | 1 × 106 | 1 × 105 | 1 × 108 | 1 × 107 | 23 | |
| R12 F5 | 4 | P | mcr-2 | 1 × 103 | >1 × 106 | 1 × 105 | >1 × 108 | 11 | |
| CNR 1745 | 4 | P | mcr-1 | >1 × 106 | 1 × 104 | >1 × 108 | 1 × 106 | 21 | |
| CNR 1604 | 4 | P | mcr-1 | 1 × 106 | 1 × 104 | 1 × 108 | 1 × 106 | 21 | |
| CNR 1790 | 4 | P | mcr-1 | 1 × 103 | 1 × 103 | 1 × 105 | 1 × 105 | 21 | |
| CNR 1859 | 4 | P | mcr-1 | 1 × 103 | 1 × 103 | 1 × 105 | 1 × 105 | 21 | |
| CNR 1886 | 4 | P | mcr-1 | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | 21 | |
| TOP10 + mcr-5f | 8 | P | mcr-5 | 1 × 103 | 1 × 105 | 1 × 105 | 1 × 107 | 21 | |
| 4222 | 4 | P | mcr-1 | 1 × 102 | 1 × 103 | 1 × 104 | 1 × 105 | 21 | |
| 4070 | 4 | P | mcr-1 | 1 × 104 | 1 × 103 | 1 × 106 | 1 × 105 | 21 | |
| 979 | 4 | P | mcr-1 | 1 × 103 | 1 × 103 | 1 × 105 | 1 × 105 | 21 | |
| 6383 | 4 | P | mcr-1.5 | 1 × 103 | 1 × 104 | 1 × 105 | 1 × 106 | 21 | |
| 1724 | 4 | P | mcr-1 | 1 × 104 | 1 × 103 | 1 × 106 | 1 × 105 | 21 | |
| 1670 | 4 | P | mcr-1.5 | 1 × 105 | 1 × 105 | 1 × 107 | 1 × 107 | 21 | |
| 36070 | 8 | P | mcr-3.2 | 1 × 105 | 1 × 105 | 1 × 107 | 1 × 107 | 24 | |
| CNR 164 A5 | 4 | P | mcr-1 | 1 × 105c | >1 × 106 | 1 × 107 | >1 × 108 | This study | |
| Klebsiella pneumoniae | CNR 20140042 | 16 | Chr | MgrB N42Y and K43I | 1 × 103 | 1 × 103 | 1 × 105 | 1 × 105 | This study |
| CNR 20140661 | 64 | Chr | MgrB Q30 stop | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | This study | |
| CNR 20151119 | 64 | Chr | MgrB L4 stop | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | This study | |
| CNR 20150622 | 64 | Chr | MgrB Y41 stop | 1 × 102 | 1 × 103 | 1 × 104 | 1 × 105 | This study | |
| CNR 20150777 | 128 | Chr | MgrB Y41 stop | 1 × 103 | 1 × 104 | 1 × 105 | 1 × 106 | This study | |
| CNR 20150944 | 64 | Chr | MgrB modified sequence starting at aad 42 | 1 × 102 | 1 × 103 | 1 × 104 | 1 × 105 | This study | |
| CNR 20150309 | 64 | Chr | MgrB modified sequence starting at aa 37 | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | This study | |
| CNR 20150675 | 64 | Chr | mgrB truncated in ORFe by IS10 | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | This study | |
| CNR 20140483 | 32 | Chr | mgrB truncated in ORF by IS1F-like | 1 × 103 | 1 × 104 | 1 × 105 | 1 × 106 | This study | |
| CNR 20140563 | 64 | Chr | mgrB truncated in ORF by IS1R | 1 × 102 | 1 × 103 | 1 × 104 | 1 × 105 | This study | |
| CNR 20150050 | 32 | Chr | mgrB truncated in promoter by IS1R | 1 × 103 | 1 × 103 | 1 × 105 | 1 × 105 | This study | |
| CNR 20140591 | 64 | Chr | mgrB truncated in ORF by IS5-like | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | This study | |
| CNR 20140550 | 32 | Chr | mgrB truncated in promoter by IS903D | 1 × 103 | 1 × 104 | 1 × 105 | 1 × 106 | This study | |
| CNR 20151285 | 32 | Chr | mgrB truncated in ORF by IS903-like | 1 × 103 | 1 × 104 | 1 × 105 | 1 × 106 | This study | |
| S14-002 | 64 | Chr | mgrB truncated in promoter by ISKpn14 | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | This study | |
| CNR 20140101 | 32 | Chr | ΔmgrB | 1 × 103 | 1 × 104 | 1 × 105 | 1 × 106 | This study | |
| CNR 2015007 | 32 | Chr | ΔmgrB | 1 × 103 | 1 × 104 | 1 × 105 | 1 × 106 | This study | |
| CNR 20150066 | 16 | Chr | ΔmgrB | 1 × 103 | >1 × 106 | 1 × 105 | >1 × 108 | This study | |
| CNR 20151223 | 32 | Chr | ΔmgrB | 1 × 102 | 1 × 103 | 1 × 104 | 1 × 105 | This study | |
| S15 | 64 | Chr | mgrB truncated in ORF by ISKpn25 | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | 25 | |
| CNR 1630 | 64/32 | Chr | mgrB truncated in ORF by IS5 | 1 × 102 | 1 × 105 | 1 × 104 | 1 × 107 | This study | |
| CNR 1861 | 16 | Chr | PmrB mutation (T157P) | 1 × 103 | 1 × 104 | 1 × 105 | 1 × 106 | This study | |
| CNR 1601 | 32 | Chr + P | mcr-1 + mgrB truncated in ORF by IS5 | 1 × 102 | 1 × 104 | 1 × 104 | 1 × 106 | This study | |
| CNR 1732 | 4 | P | mcr-1 | 1 × 103 | 1 × 103 | 1 × 105 | 1 × 105 | This study | |
| CNR 1853 | 4 | P | mcr-1 | 1 × 103 | 1 × 103 | 1 × 105 | 1 × 105 | This study | |
| Salmonella enterica | |||||||||
| Serovar Paratyphi B d-tartrate + S. enterica biotype java | 201610686 | 8 | P | mcr-1 | 1 × 103 | 1 × 105 | 1 × 105 | 1 × 107 | This study |
| Serovar Typhimurium | CNR 1776 | 8 | P | mcr-1 | 1 × 103 | >1 × 106 | 1 × 105 | >1 × 108 | This study |
| Serovar Paratyphi B d-tartrate + S. enterica biotype java | 13-SA01718 | 8 | P | mcr-5 | 1 × 103 | >1 × 106 | 1 × 105 | >1 × 108 | 8 |
| Colistin-susceptible Enterobacteriaceae (n = 41) | |||||||||
| Escherichia coli | TOP10 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | ||
| 1608071881 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1608072264 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1608073733 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1608073228 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1608078635 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1608078858 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1608062671 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1608064819 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 2H6 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| LAN 10.48 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| VER 9.39 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1F1 | 0.25 | >1 × 106 | 1 × 106 | >1 × 108 | 1 × 108 | 21 | |||
| 1A6 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 1A8 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 2A1 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 2D9 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 2C4 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| 2D5 | 0.25 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | 21 | |||
| K. pneumoniae | 1609056413 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | ||
| 1609061149 | 1 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 2 E8 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 2 I4 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 2 F1 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 2 I5 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 3 B4 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 3 B7 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 1 B6 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| CNR 173 F9 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 1 C9 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 1 E3 | 1 | >1 × 106 | 1 × 104 | >1 × 108 | 1 × 106 | This study | |||
| 2 B1 | 1 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| CNR 173 E3 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 2 C6 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| 2 D2 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | |||
| Salmonella enterica | |||||||||
| 4,12:i:− | 201604739 | 1 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | ||
| Serovar Enteritidis | 201608919 | 1 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | ||
| Serovar Typhimurium | 201606509 | 1 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | ||
| Serovar Enteritidis | 201607559 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | ||
| Serovar Veneziana | 201610299 | 0.5 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study | ||
| Enterobacter cloacae | CNR 131 G4 | 0.5 | P | mcr-4.2 | >1 × 106 | >1 × 106 | >1 × 108 | >1 × 108 | This study |
Underlined CFU counts are considered negative results. The sensitivity was 86.8% (95% CI = 74.0% to 94.0%) for both media after 48 h of incubation. The specificity was 100% (95% CI = 89.3% to 100%) and 97.5 (95% CI = 85.6% to 99.9%), respectively, for ChromID Colistin R and Superpolymyxin.
P, plasmid; Chr, chromosome.
After 48 h of incubation (no colony at 24 h).
aa, amino acid.
ORF, open reading frame.
DISCUSSION
Based on this study performed with spiked rectal swabs, ChromID Colistin R and Superpolymyxin selective media showed very similar performances. The main advantage of the Superpolymyxin medium is that it could be directly inoculated with the rectal swabs without any enrichment step (4 h) in colistin-supplemented broth, whereas ChromID Colistin R requires an enrichment step. On the other hand, the main advantage of ChromID Colistin R lies in the use of chromogenic molecules enabling the rapid presumed identification of growing colonies (pink for E. coli, blue for Klebsiella, Enterobacter, and Serratia, and white for Salmonella). Indeed, the morphological aspect of the colonies on the Superpolymyxin medium was indistinguishable between E. coli, K. pneumoniae, and Salmonella enterica (Fig. 1). As species cannot easily be differentiated on Superpolymyxin, clinical labs must then identify the growing colonies before reporting results. In our study, the selectivity of both media was good, since no Gram-positive bacteria or fungi grew on them.
FIG 1.
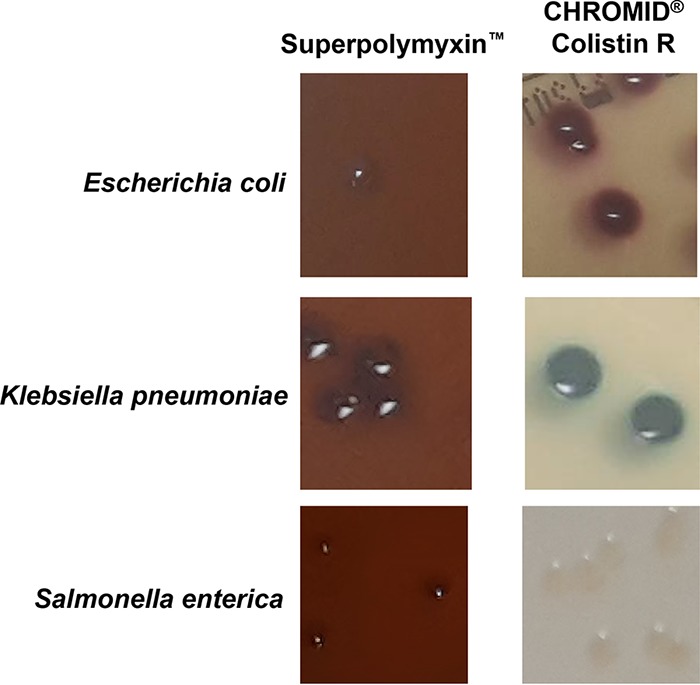
Morphological aspect of colonies of E. coli, K. pneumoniae, and Salmonella enterica grown on Superpolymyxin and ChromID Colistin R media.
Of note, unlike the ChromID Colistin R medium, which is currently limited to use with Enterobacteriaceae, the Superpolymyxin medium is also claimed to be able to detect colistin resistance in all Gram-negative bacteria, including Acinetobacter spp. and P. aeruginosa. Accordingly, we tested the Superpolymyxin medium with three colistin-resistant isolates (all producing the OXA-23 carbapenemase) and four colistin-susceptible Acinetobacter baumannii isolates. In all three colistin-resistant isolates, a mutation of PmrB (A226T, A226V, and R263H) resulted in MICs ranging from 16 to 64 mg/liter. The Superpolymyxin medium fully detected all colistin-resistant isolates, while none of the four susceptible strains grew on the medium.
As the rate of colistin resistance is likely to increase in the near future, clinical microbiology laboratories will require rapid and reliable screening media to identify carriers in hospital settings. Here, we have shown that both commercially available media, ChromID Colistin R and Superpolymyxin, are useful tools to screen for colistin-resistant Enterobacteriaceae from patient samples (rectal swabs) regardless of the level and mechanism of colistin resistance.
MATERIALS AND METHODS
Susceptibility testing.
MICs were determined by broth microdilution according to the guidelines of a CLSI and EUCAST joint subcommittee (16). Results were interpreted using EUCAST breakpoints, as updated in 2018.
Bacterial isolates.
Ninety-four enterobacterial isolates, including 53 isolates exhibiting resistance to colistin (MIC > 2 mg/liter), were tested. The colistin resistance mechanism of all these isolates has been characterized at the molecular level (Table 1). The tested isolates were as follows: colistin-resistant isolates with colistin MICs of ≥4 mg/liter, consisting of Escherichia coli (n = 25, including 20 isolates carrying mcr genes), Klebsiella pneumoniae (n = 25, including 3 isolates carrying mcr genes), and Salmonella enterica (n = 3 isolates carrying mcr genes); colistin-susceptible E. coli (n = 19), K. pneumoniae (n = 16), and Salmonella enterica (n = 5) isolates; and one mcr-4.2-positive Enterobacter cloacae isolate (Table 1). Chromosomally encoded mutations in genes responsible for colistin resistance (the pmrA, pmrB, phoP, phoQ, mgrB, and crrB genes) were also searched as described previously (17).
Spiked rectal swabs.
Suspensions of bacterial strains with an optical density of a 0.5 McFarland standard (inoculum, ∼108 CFU/ml) were serially diluted in water, and 10-fold dilutions of pure solution to 10−3 were used to spike liquid stools from healthy volunteers (1 g in 1 ml of sterile water), as previously described (18). The bacterial suspensions that were used to spike stools from healthy volunteers were verified by the concomitant inoculation of Mueller-Hinton agar with 10 µl of the suspension diluted to 10−4 in water. Ten microliters of bacterial suspension was added to 90 µl of stool. The totality (100 µl) of this spiked stool was then absorbed on the ESwab and introduced into 1 ml Amies transport medium (Copan Diagnostics, Murrieta, CA, USA) to mimic true rectal swabs. Each ESwab containing stool with each dilution of bacteria was then cultured according to the recommendations of both manufacturers (see Fig. S1 in the supplemental material). Briefly, 10 microliters of the inoculated Amies medium was transferred to the Superpolymyxin agar (ELITechGroup, Puteaux, France) and spread with a plate spreader without an enrichment step. The ChromID Colistin R agar plates (bioMérieux, La Balmes-Les-Grottes, France) were inoculated after an enrichment step, as follows: 200 µl of each inoculated Amies suspension was introduced into 10 ml of brain heart infusion (BHI) medium (bioMérieux) supplemented with one disc of colistin (10 µg) and incubated for 4 h at 37°C before seeding of 50 µl in dials.
Determination of LOD.
The lowest limit of detection (LOD) corresponds to the minimum number of bacteria that must be present in the sample to obtain growth on selective medium. In contrast to other studies that evaluated the performance of selective media with cultured bacteria (14, 19, 20), our study was performed on inoculated rectal swabs. This involves further dilution of the spiked stool sample in the ESwab Amies buffer (Fig. S1). As indicated by the manufacturer of the Superpolymyxin medium (ELITechGroup), the threshold value for susceptible strains could not be greater than 5 × 106 CFU/ml (directly from a bacterial suspension) because susceptible bacteria could benefit from an inoculum artifact to grow on the selective medium. Accordingly, the threshold for the LOD value was set at ≥1 × 106 CFU/ml in ESwab Amies buffer, corresponding to an initial concentration of 1 × 108 CFU/ml in the spiked stool (Table 1; Fig. S1). A fecal suspension without addition of bacteria was used as a negative control. In addition, 10 randomly selected strains were tested by a second experimenter to assess reproducibility. In all cases the results were identical between all experimenters.
Statistical analysis.
The sensitivity and specificity values with their respective 95% confidence intervals (CI) were calculated using the free software vassarStats (website for statistical computation, http://vassarstats.net/).
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by bioMérieux and ELITechGroup.
We thank Youri Glupczynski, Pierre Bogaerts, and Richard Bonnet for providing some well-characterized colistin-resistant E. coli and K. pneumoniae isolates.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01618-18.
REFERENCES
- 1.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 2.Stewart A, Harris P, Henderson A, Paterson D. 13 August 2018. Treatment of infections with OXA-48 producing Enterobacteriaceae. Antimicrob Agents Chemother. doi: 10.1128/AAC.01195-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerin F, Isnard C, Sinel C, Morand P, Dhalluin A, Cattoir V, Giard JC. 2016. Cluster-dependent colistin hetero-resistance in Enterobacter cloacae complex. J Antimicrob Chemother 71:3058–3061. doi: 10.1093/jac/dkw260. [DOI] [PubMed] [Google Scholar]
- 5.Jeannot K, Bolard A, Plesiat P. 2017. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 49:526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 6.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 9.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22(31):pii=30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, Zhang S, Shen J, Shen Z, Wang Y. 2018. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21(27):pii=30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 12.Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. 17 April 2018. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 13.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayol A, Poirel L, Andre C, Dubois V, Nordmann P. 2018. Detection of colistin-resistant Gram-negative rods by using the SuperPolymyxin medium. Diagn Microbiol Infect Dis 92:95–101. doi: 10.1016/j.diagmicrobio.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Teo JWP, Kalisvar M, Venkatachalam I, Ng OT, Lin RTP, Octavia S. 2018. mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J Clin Microbiol 56:e01562-17. doi: 10.1128/JCM.01562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standard Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) Joint Subcommittee. 20 June 2017, posting date Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. Clinical and Laboratory Standard Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST). [Google Scholar]
- 17.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naas T, Ergani A, Carrer A, Nordmann P. 2011. Real-time PCR for detection of NDM-1 carbapenemase genes from spiked stool samples. Antimicrob Agents Chemother 55:4038–4043. doi: 10.1128/AAC.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdul Momin MHF, Bean DC, Hendriksen RS, Haenni M, Phee LM, Wareham DW. 2017. CHROMagar COL-APSE: a selective bacterial culture medium for the isolation and differentiation of colistin-resistant Gram-negative pathogens. J Med Microbiol 66:1554–1561. doi: 10.1099/jmm.0.000602. [DOI] [PubMed] [Google Scholar]
- 20.Nordmann P, Girlich D, Poirel L. 2012. Detection of carbapenemase producers in Enterobacteriaceae by use of a novel screening medium. J Clin Microbiol 50:2761–2766. doi: 10.1128/JCM.06477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dortet L, Bonnin RA, Pennisi I, Gauthier L, Jousset AB, Dabos L, Furniss RCD, Mavridou DAI, Bogaerts P, Glupczynski Y, Potron A, Plesiat P, Beyrouthy R, Robin F, Bonnet R, Naas T, Filloux A, Larrouy-Maumus G. 1 September 2018. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: the MALDIxin test. J Antimicrob Chemother. [DOI] [PubMed] [Google Scholar]
- 22.Leroy AG, Naze F, Dortet L, Naas T, Jaubert J. 2018. Plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate in the Indian Ocean Commission. Med Mal Infect 48:426–428. doi: 10.1016/j.medmal.2018.04.388. [DOI] [PubMed] [Google Scholar]
- 23.Beyrouthy R, Robin F, Lessene A, Lacombat I, Dortet L, Naas T, Ponties V, Bonnet R. 2017. MCR-1 and OXA-48 in vivo acquisition in KPC-producing Escherichia coli after colistin treatment. Antimicrob Agents Chemother 61:e02540-16. doi: 10.1128/AAC.02540-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haenni M, Beyrouthy R, Lupo A, Chatre P, Madec JY, Bonnet R. 2018. Epidemic spread of Escherichia coli ST744 isolates carrying mcr-3 and blaCTX-M-55 in cattle in France. J Antimicrob Chemother 73:533–536. doi: 10.1093/jac/dkx418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jousset AB, Bonnin RA, Rosinski-Chupin I, Girlich D, Cuzon G, Cabanel N, Frech H, Farfour E, Dortet L, Glaser P, Naas T. 21 April 2018. 4.5 years within-patient evolution of a colistin resistant KPC-producing Klebsiella pneumoniae ST258. Clin Infect Dis. doi: 10.1093/cid/ciy293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


