Carbapenems are “last resort” β-lactam antibiotics used to treat serious and life-threatening health care-associated infections caused by multidrug-resistant Gram-negative bacteria. Unfortunately, the worldwide spread of genes coding for carbapenemases among these bacteria is threatening these life-saving drugs.
KEYWORDS: New Delhi metallo-β-lactamase, antibiotic resistance, enzyme mechanism, enzyme structure, metallo-β-lactamase
ABSTRACT
Carbapenems are “last resort” β-lactam antibiotics used to treat serious and life-threatening health care-associated infections caused by multidrug-resistant Gram-negative bacteria. Unfortunately, the worldwide spread of genes coding for carbapenemases among these bacteria is threatening these life-saving drugs. Metallo-β-lactamases (MβLs) are the largest family of carbapenemases. These are Zn(II)-dependent hydrolases that are active against almost all β-lactam antibiotics. Their catalytic mechanism and the features driving substrate specificity have been matter of intense debate. The active sites of MβLs are flanked by two loops, one of which, loop L3, was shown to adopt different conformations upon substrate or inhibitor binding, and thus are expected to play a role in substrate recognition. However, the sequence heterogeneity observed in this loop in different MβLs has limited the generalizations about its role. Here, we report the engineering of different loops within the scaffold of the clinically relevant carbapenemase NDM-1. We found that the loop sequence dictates its conformation in the unbound form of the enzyme, eliciting different degrees of active-site exposure. However, these structural changes have a minor impact on the substrate profile. Instead, we report that the loop conformation determines the protonation rate of key reaction intermediates accumulated during the hydrolysis of different β-lactams in all MβLs. This study demonstrates the existence of a direct link between the conformation of this loop and the mechanistic features of the enzyme, bringing to light an unexplored function of active-site loops on MβLs.
INTRODUCTION
β-Lactams, the most frequently prescribed class of clinically available antibiotics, are used to treat infections caused by both Gram-negative and Gram-positive bacteria. β-Lactams inhibit bacterial cell wall synthesis by targeting transpeptidases and carboxypeptidases (bacterial cell wall-synthesizing enzymes) (1). The main mechanism of bacterial resistance against β-lactams in Gram-negative bacteria is the expression of β-lactamases, enzymes that selectively hydrolyze the β-lactam ring, rendering the antibiotic ineffective (2, 3). Two distinct types of β-lactamases are currently known: serine-β-lactamases, which employ a Ser residue as the active nucleophile in catalysis, and metallo-β-lactamases (MβLs), which are metal-dependent hydrolytic enzymes. MβLs are of medical concern given their ability to hydrolyze and confer resistance to virtually all classes of β-lactam antibiotics. Notably, all MβLs show hydrolytic capacities against carbapenems, the most potent β-lactam antibiotics to date, routinely used as “last resort drugs” (1). Although some compounds have been found to be effective as MβL inhibitors (4–12), none of them are yet available to treat clinical infections, leading to a crisis in antimicrobial chemotherapy (13–18).
MβLs are subdivided in three subclasses (B1, B2, and B3) based on the identity of active-site essential residues, Zn(II) requirements, and substrate profile (15, 16, 19–22). B1 MβLs are of major clinical concern because they are broad-substrate-spectrum β-lactamases and are encoded on mobile genetic elements in pathogenic and opportunistic bacteria. The most clinically relevant B1 lactamases belong to the NDM (New Delhi metallo-β-lactamase), VIM (Verona integron-encoded MβL), and IMP (imipenemase MβL) families, with NDM-1, VIM-2, and IMP-1 being the most widespread allelic variants. In particular, NDM-1 is a membrane-anchored enzyme (23) and is one of the most widespread MβLs with a potent carbapenemase activity (16, 24, 25).
Two Zn(II) ions are required for the catalytic activity of B1 MβLs (26, 27). Both Zn(II) atoms are bound to a conserved ligand set: one Zn(II) ion is coordinated to three His residues (116, 118, and 196, after the standard BBL numbering [28]) and a hydroxide molecule (3H site), whereas the other is bound to the same (bridging) OH, an extra water molecule, and residues Asp120, Cys221, and His263 (DCH site) (29–31). The metal ion at the DCH site has been shown to be essential in stabilizing key reaction intermediates during hydrolysis of chromogenic cephalosporins (32) and carbapenems (33, 34). This active site is located in a shallow and broad groove flanked by two loops: active-site loop L10 and active-site loop L3. Amino acid substitutions in both loops are associated with changes in the substrate profile in B1 MβLs (35–40). Even though different families of B1 MβLs present a conserved active-site and global protein fold, these β-lactamases share very low sequence identity. This diversity has posed additional challenges for the development of an MβL inhibitor. Thus, the identification of common and distinct features is crucial for understanding their mechanism and substrate recognition profile.
Loop L3 has been the focus of several studies in MβLs. Crystallographic and NMR studies identified variable conformations of this loop in many B1 MβLs (38, 41–45), and its role in specific interactions with their substrates is well documented (36, 39, 44, 46). Moreover, an increase of the dynamics of this loop was hypothesized with the broadening of the substrate profile in an in vitro evolved lactamase (37). The consensus identifies loop L3 as a mobile flap able to adapt its conformation upon small molecule binding in the active site. However, the sequence heterogeneity observed in the loop L3 from different MβLs has limited generalizations about its specific role.
To explore the role of the loop L3 in the scaffold of NDM-1, we designed a series of variants in which we replaced the native loop by those of IMP-1 or VIM-2, and a third one in which a Pro residue was introduced at the C terminus of the loop. We show here that the substrate spectra and the active-site structure display minor perturbations in these chimeric proteins, despite previous expectations. Crystal structures of two of the obtained chimeras show that different loops in the same scaffold adopt quite different conformations, spanning from an open to a closed active site in the unbound form of the enzyme. However, the loop conformation cannot correlate to the substrate profile observed for the different variants. Instead, the loop conformation directly impacts on the accumulation of the anionic reaction intermediates, disclosing an auxiliary structural determinant of the mechanism of hydrolysis. These findings suggest a new role of this mobile loop in the catalytic mechanism of MβLs.
RESULTS
Loop L3 engineering gives rise to active and stable NDM variants.
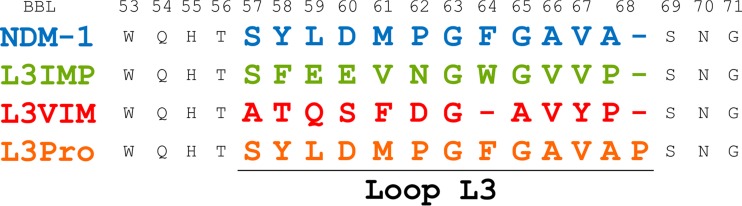
A closer look at the primary sequence of the loop L3 of IMP-1, VIM-2, and NDM-1 reveals significant differences (Fig. 1). For instance, the loop L3 of IMP-1 displays the same length as that from NDM-1, but it contains more polar residues. In contrast, the VIM-2 loop L3 shows a similar charge distribution but is one residue shorter and less hydrophobic than NDM-1 loop L3. Also, a proline residue is located at the C terminus of the loop in all cases, except in NDM-1. To study the function of loop L3 in NDM-1, we designed two chimeric proteins in which the NDM-1 loop was replaced by the loops of IMP-1 and VIM-2 (including the Pro residues). These variants were designated L3IMP and L3VIM, respectively. In order to assess how an insertion could impact in the function of loop L3, we also engineered an extra MβL in which a Pro residue was inserted at the base of the NDM-1 loop, the L3Pro variant.
FIG 1.
Engineered substitutions of loop L3 in NDM-1. Sequence alignment of the L3 variants, highlighting the differences at the loop L3 region, including the standard BBL numbering (28).
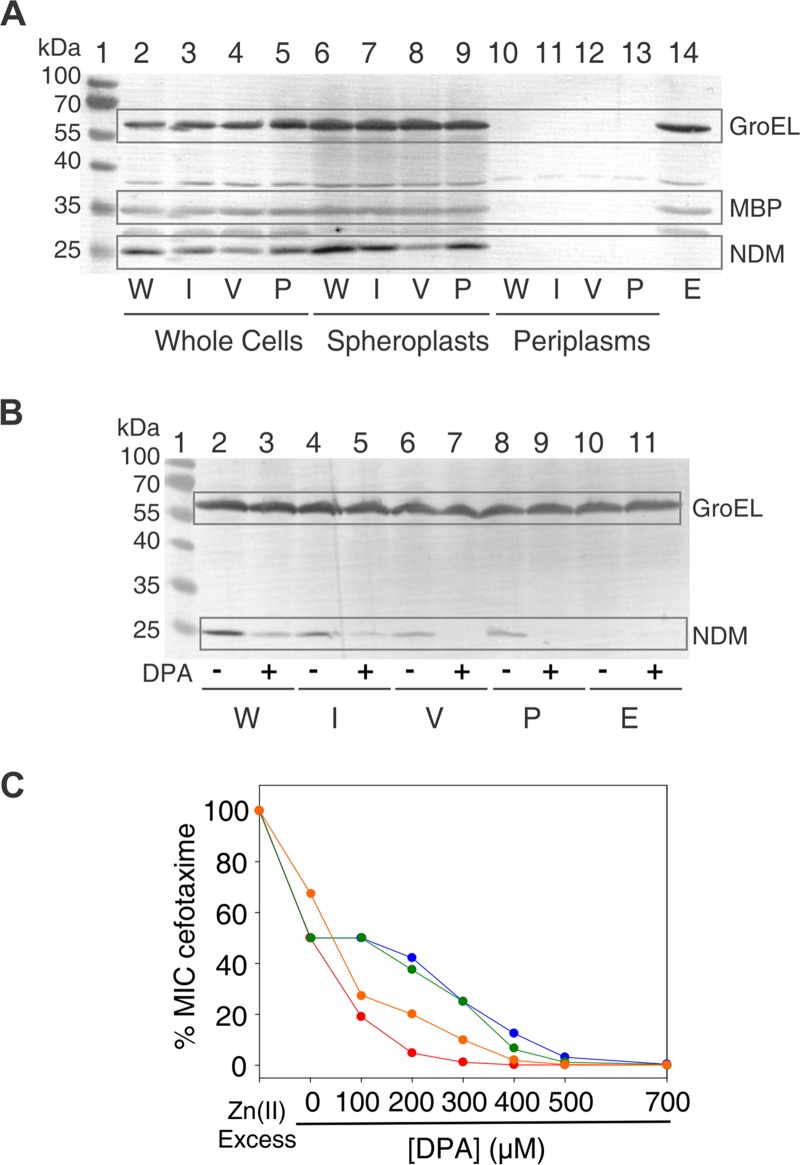
We used both in bacteria and in vitro approaches to analyze the L3 variants. Escherichia coli cells expressing the chimeric proteins were used to analyze MβLs properties in a natural background. We evaluated the expression levels in whole cells, spheroplasts, and periplasmic extracts by immunoblotting (Fig. 2A). All variants were expressed, including the native leader peptide of NDM-1, containing the canonical lipidation sequence LSGC (lipobox), which anchors the protein to the inner leaflet of the outer membrane (23, 43). None of the variants was in the periplasmic extracts, revealing an adequate processing of the leader peptide. The analysis of whole-cell extracts and spheroplasts showed that NDM-1 and variants L3IMP and L3Pro showed similar expression levels and that the L3VIM variant was expressed at lower levels, but without compromising protein stability (Fig. 2A). These experiments show that our loop L3 engineering has been successful in eliciting stable proteins.
FIG 2.
Expression levels and Zn(II) limitation susceptibility of NDM-1 and L3 variants. (A) Immunoblot demonstrating steady-state expression in E. coli DH5α. Proteins were detected from whole-cell lysates (lanes 2 to 5 and lane 14), spheroplasts (lanes 6 to 9), and periplasmic extracts (lanes 10 to 13). Wild-type NDM-1 (W) corresponds to lanes 2, 6, and 10; L3IMP (I) corresponds to lanes 3, 7, and 11; L3VIM (V) corresponds to lanes 4, 8, and 12; L3Pro (P) corresponds to lanes 5, 9, and 13. Empty plasmid (E) corresponds to lane 14. Lane 1 shows the protein ladder marker. The GroEL molecular weight is 60 kDa and that for MBP is 47 kDa. (B) Immunoblot demonstrating steady-state expression of wild-type NDM-1 and the L3 variants in E. coli DH5α treated with DPA. After induction, cells were incubated with (+) or without (–) DPA, and protein expression was detected from of whole-cell lysates. Wild-type NDM-1 (W) corresponds to lanes 2 and 3, L3IMP (I) corresponds to lanes 4 and 5, L3VIM (V) corresponds to lanes 6 and 7, L3Pro (P) corresponds to lanes 8 and 9, and empty plasmid (E) corresponds to lanes 10 and 11. Untreated cells were loaded before treated ones. Lane 1 shows the protein ladder marker. (C) Antimicrobial susceptibility profiles of E. coli DH5α/pMBLe producing β-lactamases against cefotaxime at increasing DPA concentrations. E. coli DH5α expressing NDM-1 is shown in blue, E. coli DH5α expressing L3IMP is shown in green, E. coli DH5α expressing L3VIM is shown in red, and E. coli DH5α expressing L3Pro is shown in orange.
We next tested the susceptibility of E. coli cells expressing the three NDM-1 L3 variants against a broad panel of β-lactam antibiotics. MIC analyses (Table 1) reveal that the L3 variants confer somehow lower levels of resistance against all tested substrates. In the case of cefepime, the impact of the mutations in the MIC values is larger. MβLs with reduced Zn(II) binding capabilities provide lower levels of resistance because metal binding takes place in the periplasmic space where Zn(II) availability is limited (26, 47, 48). We tested the effect of Zn(II) deprivation on protein expression levels by treating E. coli cells expressing NDM-1 variants with the metal chelator dipicolinic acid (DPA). This chelator strongly affected the expression of L3Pro and L3VIM variants, while the impact was moderate for variant L3IMP and wild-type NDM-1 (Fig. 2B). We also tested the ability of our NDM-1 variants to bind Zn(II) within the cell by determining the impact of DPA in the MIC values against cefotaxime. The sensitivity of L3IMP to Zn(II) deprivation resembled that of NDM-1, whereas it was increased for L3Pro and even more for the L3VIM variant (Fig. 2C).
TABLE 1.
Antimicrobial susceptibility profiles of E. coli DH5α pMBLe producing NDM-1 and its variants at loop L3
| Variant | MIC (mg/liter) |
|||||
|---|---|---|---|---|---|---|
| Imipenem | Meropenem | Piperacillin | Cefotaxime | Ceftazidime | Cefepime | |
| NDM-1 | 4 | 2 | 128 | 64 | 1,024 | 16 |
| L3IMP | 2 | 1 | 64 | 16 | 256 | 16 |
| L3VIM | 1 | 1 | 64 | 32 | 256 | 2 |
| L3Pro | 1 | 1 | 16 | 64 | 256 | 0.5 |
| DH5α pMBLe | 0.25 | 0.03 | 2 | 0.03 | 0.25 | 0.016 |
We then expressed and purified all variants in the soluble form, a truncated version in which the first 38 residues, including the signal peptide and the lipidation site, were removed (Δ38). All variants were obtained by expression in rich (Luria-Bertani [LB]) medium with yields similar to those observed with wild-type NDM-1, except for L3VIM, for which yields one order of magnitude lower were obtained. Also, the Zn(II) affinity of L3VIM was lower than the other variants (Table S1). This could explain the higher sensitivity of L3VIM when challenged with DPA (Fig. 2C) and the lower metal content (see Materials and Methods).
We measured the stability of the purified variants and their apo derivatives by thermal shift analysis (Table 2). The L3 variants were slightly less stable than the wild-type enzyme, with Tm values spanning a narrow range (4°C). Instead, the nonmetallated forms displayed a more pronounced destabilization effect. The L3VIM variant features the largest gap between the Tm values of metallated and nonmetallated forms, indicating that in this case the Zn(II) ions play a crucial role in stabilizing the MβLs. These results added to the differences on the expression levels under DPA addition demonstrate that the stability of the nonmetallated form determines steady-state protein expression levels and confirms that the metal uptake in the periplasm is crucial for MβL stability.
TABLE 2.
Melting temperatures of apo- and holo-enzymes measured by thermal shift assay
| Variant | Mean ± SD |
||
|---|---|---|---|
| Tm (°C) | ΔTm (°C) | ||
| NDM-1 | Holo | 56.6 ± 0.1 | 18.3 ± 0.1 |
| Apo | 38.3 ± 0.1 | ||
| L3IMP | Holo | 55.3 ± 0.1 | 24.4 ± 0.4 |
| Apo | 30.9 ± 0.3 | ||
| L3VIM | Holo | 55.2 ± 0.1 | 31.5 ± 0.1 |
| Apo | 23.7 ± 0.1 | ||
| L3Pro | Holo | 52.5 ± 0.1 | 19.0 ± 0.2 |
| Apo | 33.5 ± 0.1 | ||
The activity of the enzymes was then studied by steady-state kinetics using purified MβLs. The determined kcat/Km values were within the same range of those measured for NDM-1, and all variants showed broad substrate spectra (Table 3). In general, the L3Pro variant displayed the lowest catalytic efficiencies, and L3IMP was the only variant that was catalytically more efficient compared to NDM-1 against some penicillins and cephalosporins. We stress that L3Pro displayed both elevated Km and kcat values, with larger increases in Km resulting in overall lower catalytic performances. The kinetic parameters measured for L3VIM, in contrast, are closer to those of NDM-1. Since L3VIM presents a lower expression level than NDM-1 (Fig. 2A), it is likely that the MIC values obtained may have been lower due to a decreased expression level or Zn(II) uptake in the periplasm. The hydrolysis parameters for nitrocefin were practically unaltered upon loop replacement. On the other hand, for carbapenem hydrolysis, both Km and kcat values presented differences within the variants: L3IMP parameters were lower than the wild-type values, and for L3VIM and L3Pro these values were higher than in the wild-type enzyme. Notwithstanding these changes in the substrate preferences, a clear trend in the substrate profile elicited by the loop replacement could not be identified in the chimeric proteins.
TABLE 3.
Steady-state kinetic parameters for the NDM-1 and its L3 variantsa
| Substrate | Variant | Mean ± SD |
||
|---|---|---|---|---|
| Km (μM) | Kcat (s−1) | Kcat/Km (μM−1 s−1) | ||
| Imipenem | NDM-1 | 150 ± 30 | 570 ± 30 | 4 ± 1 |
| L3IMP | 60 ± 10 | 160 ± 6 | 2.7 ± 0.5 | |
| L3VIM | 136 ± 7 | 631 ± 9 | 4.6 ± 0.3 | |
| L3Pro | 780 ± 90 | 1,200 ± 60 | 1.5 ± 0.2 | |
| Meropenem | NDM-1 | 140 ± 20 | 960 ± 40 | 7 ± 1 |
| L3IMP | 90 ± 10 | 39 ± 1 | 0.4 ± 0.1 | |
| L3VIM | 540 ± 50 | 1,810 ± 70 | 3.4 ± 0.4 | |
| L3Pro | 1,100 ± 200 | 2,500 ± 200 | 2.3 ± 0.6 | |
| Ertapenem | NDM-1 | 25 ± 2 | 420 ± 10 | 17 ± 2 |
| L3IMP | 29 ± 6 | 29 ± 1 | 1.0 ± 0.2 | |
| L3VIM | 110 ± 20 | 750 ± 40 | 5 ± 1 | |
| L3Pro | 270 ± 20 | 710 ± 20 | 2.6 ± 0.3 | |
| Penicillin G | NDM-1 | 80 ± 10 | 690 ± 20 | 8 ± 1 |
| L3IMP | 13 ± 1 | 272 ± 4 | 21 ± 2 | |
| L3VIM | 40 ± 5 | 323 ± 9 | 8 ± 1 | |
| L3Pro | 690 ± 99 | 755 ± 49 | 1.1 ± 0.1 | |
| Piperacillin | NDM-1 | 120 ± 10 | 1,190 ± 40 | 10 ± 1 |
| L3IMP | 67 ± 8 | 232 ± 6 | 3.5 ± 0.5 | |
| L3VIM | 410 ± 30 | 1,820 ± 70 | 4.4 ± 0.5 | |
| L3Pro | 700 ± 100 | 630 ± 70 | 0.9 ± 0.2 | |
| Ceftazidime | NDM-1 | 60 ± 10 | 620 ± 20 | 10 ± 2 |
| L3IMP | 60 ± 10 | 131 ± 5 | 2.2 ± 0.4 | |
| L3VIM | 58 ± 8 | 250 ± 8 | 4.1 ± 0.7 | |
| L3Pro | 90 ± 20 | 120 ± 10 | 1.3 ± 0.4 | |
| Cefepime | NDM-1 | 50 ± 10 | 300 ± 20 | 6 ± 2 |
| L3IMP | 6.1 ± 0.6 | 178 ± 2 | 28 ± 3 | |
| L3VIM | ND | ND | 1.2 ± 0.1 | |
| L3Pro | ND | ND | 0.9 ± 0.5 | |
| Nitrocefin | NDM-1 | 1.3 ± 0.3 | 38 ± 2 | 29 ± 8 |
| L3IMP | 2.9 ± 0.9 | 67 ± 7 | 23 ± 3 | |
| L3VIM | 4.0 ± 0.9 | 48 ± 3 | 12 ± 3 | |
| L3Pro | 2.6 ± 0.5 | 85 ± 4 | 32 ± 1 | |
Experimental conditions (buffer: 10 mM HEPES [pH 7.5], 200 mM NaCl, 20 μM ZnSO4, 20 μg/ml bovine serum albumin; temperature: 30°C). ND, not determined.
Loop L3 conformation models the active-site cavity size and accessibility with minor alterations on metal ligands.
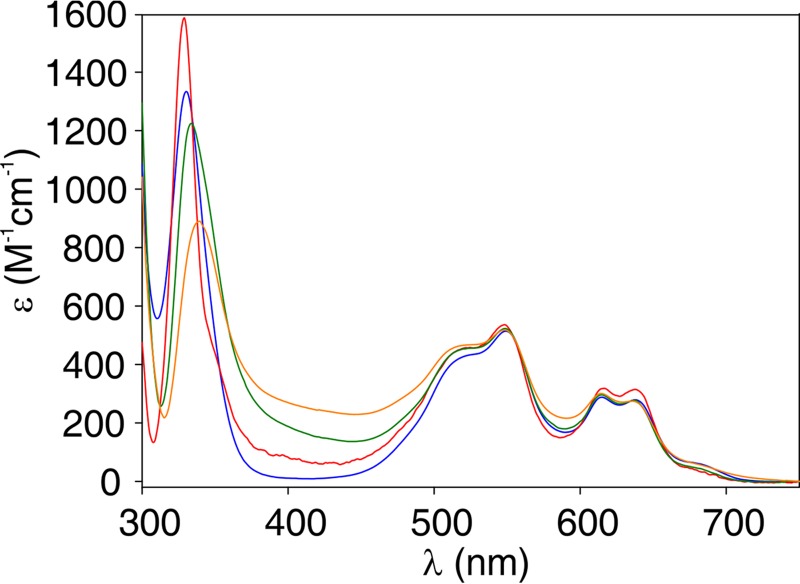
Although residues in loop L3 do not directly interact with the Zn(II) ions, the different sensitivities to Zn(II) deprivation prompted us to test whether the loop L3 replacements induced changes in the metal binding sites in the variants. For that purpose, we used Co(II) as a spectroscopic probe of the metal site coordination geometry, replacing the spectroscopically silent Zn(II) ion (49). Figure 3 shows the electronic absorption difference spectra in the UV-Vis range of all Co(II) derivatives. These spectra are characterized by distinctive features in two regions: (i) the Laporte-forbidden d-d transitions in the visible range (450 to 650 nm), which provide information on the metal site geometry (mainly of the 3H site), and (ii) a ligand-to-metal charge transfer transition (LMCT), ca. 330 nm, which reports on the Cys-Co(II) interaction at the DCH site (29). The band pattern and intensity of the d-d bands were preserved in all variants, revealing that the geometry at the 3H site was conserved (Fig. 3). The Co(II) derivatives displayed subtle changes on the position and intensity of the LMCT band, which reflects minor changes in the Co(II)-thiolate interactions at the DCH site, especially for the L3Pro and L3VIM variants. These results suggest that loop replacement did not significantly alter the coordination sphere of the metal ion in the active site.
FIG 3.
Difference spectrum of Co(II)-substituted wild-type NDM-1 and L3 variants. The difference spectra of the Co(II)-substituted MβLs were obtained by subtraction of the spectrum of the nonmetallated β-lactamase from the one corresponding to the final bi-Co(II) substituted variant. The difference spectrum of the wild-type NDM-1 is shown in blue; that for L3IMP is shown in green, that for L3VIM is shown in red, and that for L3Pro is shown in orange.
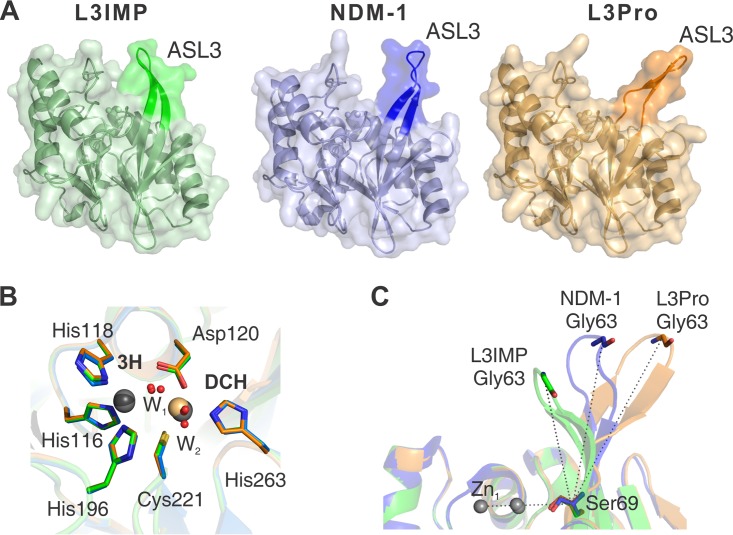
To rationalize the impact of changes in loop L3, we performed X-ray crystallographic studies. The L3IMP and L3Pro variants were crystallized, and both structures were solved at a resolution of 1.65 and 1.80 Å, respectively (Table S2). Attempts to obtain crystals of the L3VIM variant were unsuccessful. The structures of both variants are very similar to the previously reported crystal structure of native NDM-1 (PDB code 3SPU) (43). The overall structure of the enzymes and their active sites are highly conserved (Fig. 4A), as accounted for the low RMSD values of the core structure without considering loop L3 (<0.60 Å over all Cα). The active sites of L3IMP and L3Pro displayed bimetallic occupancy, i.e., with metal ions at the 3H and the DCH sites. The presence of Zn(II) was verified by anomalous diffraction. In the case of L3Pro, peaks of 40 to 50 RMSD were observed at the 3H site, confirming the presence of Zn(II), whereas no signal was observed at the DCH site (Fig. S1). Instead, the electron density could be properly accounted for by assuming the presence of Cd(II) (from the crystallization buffer) at this position. This metal substitution with a preserved geometry at the active site has already been reported for NDM-1 (3ZR9) (41).
FIG 4.
X-ray crystal structures of the L3IMP and L3Pro variants compared to NDM-1. Crystal structures are indicated by color: NDM-1 (PDB code 3SPU, chain D) in blue, L3IMP in green (PDB code 6C6I, 1.65 Å), and L3Pro in orange (PDB code 6CAC, 1.80 Å). The images were generated after the complete alignment of NDM-1 and the two L3 variants. (A) The loop L3 position is highlighted in a darker color. (B) Relevant conserved amino acids from the active sites of NDM-1 (blue), L3IMP (green), and L3Pro (orange) are represented by sticks; metal ions (Zn(II) in gray and Cd(II) in light orange) and water molecules (red) are represented as spheres. The position and orientation of the metal ligands is conserved among the three structures. The distances between the ions in the two sites are very similar (≈3.8 Å) among the different proteins. The positions of the ions in the DCH site display a slight variability among the structures, while in the 3H site the position is unchanged. (C) Angle determined by the loop L3 and the plane of the active site of each mutant. Angles were calculated between Zn1, Cα of Ser69, and Cα of Gly63. The values obtained for each loop L3 were as follows: L3IMP, 68°; NDM-1, 88°; and L3Pro, 110°.
The structures did not reveal significant changes in the metal binding sites (Fig. 4B). The position of the Zn(II) ion and the three ligands in the 3H site remains unaltered among the L3 variants. In the DCH site, despite the different identity of the metals [Zn(II) in L3IMP and Cd(II) in L3Pro], a variation of only 0.5 Å for the metal ion position was observed, with the ligand residues displaying identical conformations. This observation agrees with the spectroscopic data on the Co(II)-substituted enzymes that reveal a slightly perturbed DCH site and a conserved geometry at the 3H site.
The conformation of loop L3, instead, is considerably different among the variants (Fig. 4C). The electron density is well defined in these loops in both variants, with B-factors slightly higher than those in the protein core (around 20 and 30 Å2 higher, respectively, for L3IMP and L3Pro) (Fig. S2). In L3Pro, the interactions among the residues shaping the β-sheet at the base of loop L3 are disrupted by the insertion of a Pro residue, giving rise to a more open loop conformation. In the case of L3IMP, the loop is stabilized by hydrophobic interactions of Trp64 with residues His263, Val67, and Val61, which pull Trp64 toward the active site, inducing a more closed conformation of the loop (Fig. S3). The angle subtended by Zn1, the Cα atom of Ser69 (located on the base of loop L3) and the Cα atom of Gly63 (located on the tip of the loop), provides a bona fide description of the loop 3 conformation (the larger the angle, the more open the loop), varying from 68° (L3IMP), to 88° (NDM-1), to 110° (L3Pro) (Fig. 4C). Thus, the L3 sequence dictates the conformation of the loop in the unbound form of this enzyme within the same protein scaffold.
Loop L3 determines the accumulation of catalytic reaction intermediates.
The catalytic mechanism of MβLs takes place by accumulation of an anionic intermediate that has been characterized for the hydrolysis of the chromogenic cephalosporin, nitrocefin, and several carbapenems (33, 34, 50, 51). The rate-determining step of the reaction is, in both cases, the protonation of this intermediate, leading to the final product. Moali et al. reported that changes in loop L3 altered the accumulation of the intermediate during nitrocefin hydrolysis (39). Thus, we decided to study the hydrolysis of nitrocefin and carbapenems by our variant MβLs under pre-steady-state conditions with a photodiode array detector coupled to a stopped-flow mixing device (32, 33).
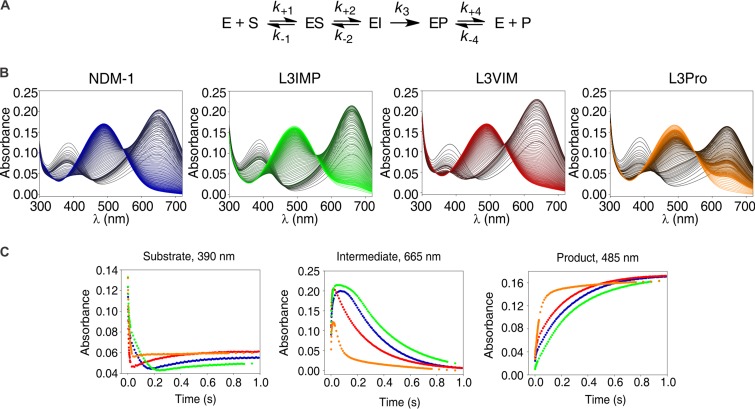
Three absorption bands are observed during nitrocefin hydrolysis at 390, 485, and 605 nm, which correspond to the substrate, product, and the anionic intermediate of the reaction, respectively (Fig. 5A) (32, 50). When we performed the hydrolysis reaction with all the L3 variants, the three bands were evidenced with similar maximum absorbance intensities (Fig. 5B). An analysis of the time course of the reaction (Fig. 5C) reveals that the time frame for accumulation and decay of the intermediate is similar for NDM-1 and the L3IMP variant, with L3IMP displaying the intermediate with the longest accumulation time. Instead, the intermediate is less stable in the L3VIM and, in particular, the L3Pro variants. Supporting this line, product formation is evidently faster in L3Pro, followed by L3VIM, NDM-1, and L3IMP, indicating that the decrease on the accumulation of the reaction intermediate is due to an increment on the protonation rate. Substrate consumption is also slower in NDM-1 and L3IMP, which could also contribute to a decrease in the rate of the reaction catalyzed by these enzymes.
FIG 5.
Photodiode array stopped-flow spectra and traces of nitrocefin hydrolysis by NDM-1 and its L3 variants. (A) Reaction mechanism for nitrocefin hydrolysis by NDM-1 (adapted from Yang et al. [32]). (B) Electronic absorption spectra upon the reaction of 10 μM nitrocefin and 10 μM enzyme in 100 mM HEPES (pH 7.5), 0.2 M NaCl, and 0.3 mM ZnSO4 at 6°C. The reaction progresses from black to color: NDM-1 in blue, L3VIM in red, L3IMP in green, and L3Pro in orange. Absorption bands with peaks at 390, 485, and 605 nm correspond to maximum absorption of the substrate, product, and the anionic intermediate of the reaction, respectively. (C) Temporary profiles of substrate (390 nm), intermediate (665 nm), and product (485 nm) during the reaction described in panel A. Traces: NDM-1, blue; L3VIM, red; L3IMP, green; L3Pro, yellow.
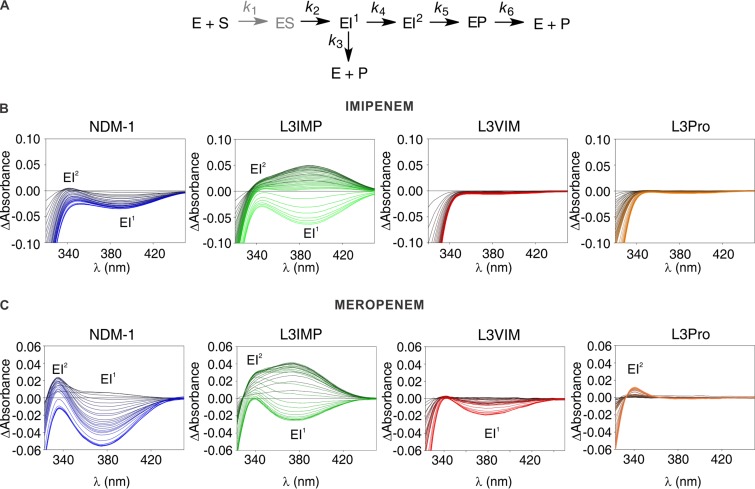
Carbapenem hydrolysis by MβLs of the three subclasses takes place by a branched mechanism with two anionic intermediate species (EI1 and EI2; Fig. 6A) (33, 34, 52). Both species are productive, but the final product and the nature of the proton donor differ. EI1 is the first intermediate produced, absorbs at 390 to 375 nm (in the case of imipenem and meropenem hydrolysis, respectively, by NDM-1), and its N protonation is produced by a water molecule bridging the Zn(II) ions (33). This species could give rise either to product or to a second intermediate species, EI2. This second intermediate species absorbs at 340 to 336 nm (for imipenem and meropenem, respectively) and is later protonated by a bulk water molecule (not bound to the metal site), leading to the formation of an EP complex lacking a metal-bound water and, later, to a product stereoselectively protonated at C-2 (33).
FIG 6.
Photodiode array stopped-flow spectra of carbapenems hydrolysis by NDM-1 and the L3 variants. (A) General reaction mechanism for carbapenems hydrolysis by MβLs (adapted from Lisa et al. [33]). The ES complex does not accumulate and is thus depicted in a lighter color (gray). (B) Sequence of difference spectra collected upon the reaction of 100 μM imipenem and 100 μM β-lactamase. The reactions progress from black to color: NDM-1, blue; L3VIM, red; L3IMP, green; and L3Pro, orange. The ionic intermediates, EI1 and EI2, were detected as absorption bands with maxima at 390 and 343 nm. The time interval spans up to 0.2 s. (C) Sequence of difference spectra upon the reaction of 100 μM meropenem and 100 μM enzyme. The reactions progress from black to color: NDM-1, blue; L3VIM, red; L3IMP, green; and L3Pro, orange. The ionic intermediates, EI1 and EI2, were detected as absorption bands with maxima at 390 and 330 nm. The time interval spans up 0.2 s.
As shown in Fig. 6B, the formation of both reaction intermediates was detected during the hydrolysis of imipenem catalyzed by NDM-1 and the L3IMP variant, being more abundant in the second case. However, in the reaction catalyzed by L3VIM or L3Pro, we could not detect the accumulation of any of those intermediates, indicating that the protonation occurred too fast for the species to accumulate to a detectable amount. Since accumulation of EI1 also depends on the formation and consumption of EI2, the decrease in the accumulation of the first intermediate species could be due only to an alteration of the protonation rate of EI2, by a bulk water molecule. For meropenem hydrolysis, the formation of both reaction intermediates was detected with NDM-1 and the L3IMP variant, being, again, more abundant in the second case (Fig. 6C). On the contrary, the hydrolysis by L3VIM and L3Pro showed accumulation of only one intermediate (EI1 in L3VIM and EI2 in L3Pro) and to a lower extent than in the wild-type enzyme (Fig. 6C). Since these results resemble the pattern observed for imipenem, we conclude that these data demonstrate a general behavior for the hydrolysis of carbapenems (33). Our observations correlate with the steady-state parameters for carbapenems, where kcat values in L3IMP were lower than the ones of wild-type NDM-1, indicating that the reaction rate is diminished, probably due to a decrease in the protonation rate. L3VIM and L3Pro present higher kcat values for carbapenems, correlating with the increased protonation rate.
DISCUSSION
A series of experimental studies of the loop L3 in B1 MβLs performed here have provided critical information on its role, mostly regarding substrate recognition and mobility. Substitutions at this loop in IMP-1, IMP-12, IMP-43, IMP-18 (36), IMP-2 (45), and VIM-31 (38) elicited changes in the substrate profile. Moali et al. reported changes in the catalytic efficiency of BcII by engineering the loop of IMP-1 (39). However, a definitive structural description of the effect of the alterations in this loop is not available. An increased mobility of loop L3 in BcII variants was correlated with a broadening in the substrate profile (37). DEER (double electron resonance) spectroscopy studies in NDM-1 have demonstrated that loop L3 closes over the active site during catalysis, returning to its original position after hydrolysis (53, 54). Theoretical calculations have predicted a correlation between the movement of loop L3 and the catalytic efficiency in IMP-1 and IMP-6 (55, 56). Nuclear magnetic resonance (NMR) studies have shown in detail the flexibility of loop L3 in different B1 enzymes (37, 57–59), while crystal structures of enzyme inhibitor or enzyme product adducts have pointed out how this loop reacts upon small molecule binding to the active site of these MβLs (5–7, 44, 45, 58–62). We show here that loop replacement in the scaffold of NDM-1 gives rise to stable, folded proteins and does not shape the substrate profile of this enzyme. Instead, loop engineering affects the catalytic mechanism, governing the accumulation of key reaction intermediates based on its conformation.
Loop replacement induced distinct levels of destabilization in the NDM-1 scaffold. Surprisingly, the largest destabilizing effects in vitro were evident in the apo (nonmetallated) variants, particularly in L3VIM. The destabilization induced in this variant also correlates with a decreased affinity of the protein toward Zn(II), a higher sensitivity to Zn(II) deprivation in bacteria, and lower expression levels. It has recently been reported that, under conditions of Zn(II) deprivation, soluble periplasmic MβLs are degraded in the apo forms (23). Our previous results account for the link between the reduced Zn(II) affinity and the phenotype observed in the case of L3VIM. Overall, these data highlight the role of Zn(II) binding for the stabilization of MβLs.
The crystal structures and spectroscopic data point to a structurally conserved metal site, with a minor perturbation in the DCH site. Although crystals for the L3VIM variant could not be obtained, the good agreement of Co(II) substitution experiments with the crystal structures allow us to extrapolate the spectroscopic results on this variant with confidence to the native Zn(II) enzyme. We therefore expect the metal site in L3VIM to be less perturbed compared to NDM-1. Under this assumption, the observed changes could be attributed to changes in loop L3, as observed for the other two variants.
Major changes in the reported structures in this analysis are related to the conformation of the engineered loops (Fig. 4). Importantly, our results show that different loops can adopt a wide range of conformations within a given MβL scaffold and reveal details on how the loop L3 sequence defines its conformation. The loop L3 in IMP-1 is closer to the one observed in the engineered L3IMP in the NDM scaffold, while inhibitor binding to IMP-1 does not alter the loop conformation as reported here. The same holds for NDM-1, for which inhibitor binding (bisthiazolidines or captopril) does not affect the loop conformation as much as loop engineering does (5–7, 44, 60). We conclude that the loop L3 sequence (regardless of the MβL scaffold) strongly determines its conformation.
The catalytic performances of the engineered variants do not reveal a substantial change in the substrate profile of NDM-1. Instead, we report a significant impact on the catalytic mechanism as witnessed by changes in the accumulation of reaction intermediates in the hydrolysis of nitrocefin, imipenem and meropenem. The stability of these reaction intermediates depends on electrostatic interactions with the metal ions, particularly with the DCH site (33, 34, 63, 64), and it has been shown that subtle changes on the MβL active site could tune the stability of these species (26, 63, 65–68). However, the structure of the catalytic Zn(II) center is not perturbed by loop replacement, indicating that other structural features modulate the half-life time of the intermediate species.
The conformation of loop L3 directly affects the stability of these intermediates: a more closed loop (as in L3IMP) leads to an enhanced accumulation of the intermediate, while a more open active site (such as in L3Pro) decreases the amount of intermediate (Fig. 6). Since the half-life of the anionic reaction intermediates depends on their protonation rates (33, 64), these results suggest that the protonation step could be modulated by the solvent accessibility in the different mutants. This also suggests that it is likely that the presence of residues in loop L3 that favor interactions with active-site residues (such as Trp64 in L3IMP) could disfavor water accessibility, making the protonation process less effective. We cannot discard the presence of specific interactions of loop L3 with the intermediate species that may increase its stability. This effect may account for the lower kcat values observed for this variant for carbapenem hydrolysis. Overall, these findings show that loop L3 plays an important role in the mechanism of β-lactam hydrolysis by MβLs by tuning the rate of the rate-limiting step and controlling the accumulation of key reaction intermediates according to its conformation.
MATERIALS AND METHODS
Bacterial strains and cloning.
Escherichia coli DH5α was used for construction and expression of plasmid pMBLe, as well as for all microbiological and biochemical studies. pET26-blaNDM-1 was kindly provided by James Spencer (University of Bristol, Bristol, UK). The construction of pMBLeblaNDM-1 has been described previously (23), and the same procedure was used for variant cloning. The full-length blaNDM-1 gene (including its native peptide leaders) was amplified by the addition of a C-terminal Strep-tag sequence (for comparative protein detection and quantification) and subcloned into the pMBLe plasmid. The expression of bla was induced by the addition of 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside). Addition of the Strep tag at the C terminus does not affect MβL ability to confer resistance (23).
Construction of the L3 variants.
blaNVIM, blaNIMP, and blaNPro containing the blaNDM-1 gene with the loop L3 of VIM-2, IMP-1, and a Pro residue introduced at the base of the loop, respectively, were custom synthesized (Celtek Genes). The region exchanged comprised residues between Ser57 and Ala68 of the NDM-1 structure.
MβL detection.
MβL expression was measured by immunoblotting of cell extracts as described previously (23). Briefly, 5-ml cultures of E. coli DH5α cells carrying the pMBLe blaNDM-1, blaNVIM, blaNIMP, and blaNPRO plasmids were grown aerobically at 37°C in LB broth with 20 μg/ml gentamicin to log phase (optical density at 600 nm [OD600] = 0.4). MβL expression was then induced with 100 μM IPTG, and cultures were left to grow to an OD600 of 1. Cultures were pelleted, and cells were washed once with 20 mM Tris–150 mM NaCl (pH 8.0). An aliquot of cell crude extract were separated on this step. The rest of the washed cells were pelleted again and resuspended in 20 mM Tris, 0.1 mM EDTA, 20% (wt/vol) sucrose, 1 mg/ml lysozyme (from chicken egg white [Sigma-Aldrich]; protein, ≥90%), and 0.5 mM phenylmethylsulfonyl fluoride (pH 8; the resuspension volume was normalized based on the OD600). The cells were incubated with agitation at 4°C for 30 min. The cells were pelleted, and the periplasmic extract was obtained from the supernatant. The pellet consisting of spheroplasts was washed in 20 mM Tris, 0.1 mM EDTA, and 20% (wt/vol) sucrose (pH 8) and resuspended in the same volume of this buffer. A total of 120 µl of the different extracts was mixed with 30 µl of loading dye, separated by SDS-PAGE (10 µl of whole-cell lysate, 25 µl of spheroplasts, and 25 µl of periplasmic extracts), and transferred to a polyvinylidene difluoride (PVDF) membrane (Novex; Life Technologies, Carlsbad, CA) by electroblotting. Strep-Tag II monoclonal antibodies (at a 1:1,000 dilution from a 200-μg/ml solution; Novagen) and immunoglobulin G-alkaline phosphatase conjugates (at a 1:3,000 dilution) were used to detect MβL expression. GroEL and MPB antibodies were added as loading controls. Protein band intensities were quantified from PVDF membranes with ImageJ software (69).
Cell-based assays.
To test the phenotypic effect of the loop L3 substitutions, the MICs of piperacillin, ceftazidime, cefotaxime, cefepime, imipenem, and meropenem were determined for each clone in LB medium using the agar macrodilution method according to CLSI guidelines (70). Protein expression was induced with 100 µM IPTG, except for ceftazidime, where induction was performed with 20 µM IPTG. In order to measure the effect of Zn(II) availability on antibiotic resistance, the growth medium was supplemented with various concentrations of the metal chelator dipicolinic acid (DPA [Merck], >98%). In all cases, plasmid expression was induced with 100 μM IPTG (71). An extra measurement was performed with 500 μM ZnSO4 to reach the maximum activity of the proteins (100%). DPA or ZnSO4 was added to the LB plate, along with gentamicin and IPTG.
For the steady-state expression of proteins in cells treated with DPA, E. coli DH5α cells expressing the NDM variants were incubated, after 1 h of induction with 100 μM IPTG, for 15 min with or without 250 μM DPA. Protein expression was detected from 10 μl of whole-cell lysates by Western blotting as previously described.
Protein purification.
For kinetic studies, mature MβLs (residues 39 to 270) were produced in E. coli BL21(DE3) and purified as previously published for NDM-1 (5), with the following two modifications. First, LB medium supplemented with 50 μg/ml kanamycin was used instead of minimal media. Second, MβL production was induced by the addition of 0.25 mM IPTG. The protein concentration was measured spectrophotometrically using ε280 = 27,960 M−1 cm−1 for NDM-1, L3VIM, and L3Pro and ε280 = 31,970 M−1 cm−1 for L3IMP. The metal content was measured using the colorimetric reagent 4-(2-pyridylazo) resorcinol (PAR) under denaturing conditions (72). The average metal contents of the variants [equivalents of Zn(II) per enzyme] were lower (1.5 for L3VIM, 1.6 for L3Pro, and 1.65 for L3IMP) compared to the wild-type protein [1.8] (33).
Determination of Zn(II) affinity constants.
Dissociation constants for Zn(II) were estimated by competition with the chromophoric chelator PAR, as previously described (26). Briefly, PAR is a metallochromic compound, whose UV-visible absorption spectrum is modified upon metal uptake, as reflected by a shift of its maximum absorption wavelength from 414 to 500 nm. Using the previously published molar absorption coefficients of free PAR (εPAR414nm = 36,868 ± 1,843 M−1 cm−1; εPAR500nm = 1,289 ± 65 M−1 cm−1), PAR-Zn(II) complex (εPAR2Zn414nm = 12,788 ± 576 M−1 cm−1; and εPAR2Zn500nm = 80,000 ± 4,000 M−1 cm−1) and the disassociation constant (Kd) for the PAR-Zn(II) complex (2.6 ± 0.2 10−12 M), we were able to quantify the amount of PAR at each state in a given sample (73).
Disassociation constants for NDM-1 and all the L3 variants were determined at 25°C by titrations curves on 40 mM morpholinepropanesulfonic acid–0.1 M NaCl (pH 7.3) (previously treated with Chelex) supplemented with 1.5 μM ZnSO4 and PAR at 3 and 6 μM. Apo enzyme was added in increasing amounts at each point until a final concentration of 3 μM was reached, and the absorption spectra between 300 and 600 nm were recorded. Absorbances at 414 and 500 nm were corrected by subtracting the absorbance at 600 nm, which was taken as the baseline value. Metal binding to wild-type NDM-1, L3VIM, and L3IMP could be described by a two-step binding model. Data were fit with DynaFit 3 (Biokin) (74) to the equilibrium shown in scheme 1; where Kd1 and Kd2 are the disassociation constants of the enzyme:Zn(II) complex, and Kd PAR corresponds to the dissociation of the PAR:Zn(II) complex. For the L3Pro variant, a second model with one Kd (Kd1,2; see Table S1 in the supplemental material) for the enzyme-Zn(II) complex was proposed (scheme 2).
Scheme 1:
Scheme 2:
Steady-state kinetics.
β-Lactamase activity was measured in a JASCO V-670 spectrophotometer at 30°C in 10 mM HEPES (pH 7.5) and 200 mM NaCl supplemented with 20 μM ZnSO4 and 20 μg/ml bovine serum albumin. Substrates were used in the micromolar range, whereas the enzymes were used in the nanomolar range in order to ensure pseudo-first-order kinetics. We only considered the concentration of the metallated protein. The following differential extinction coefficients were used: nitrocefin, Δɛ482 = 17,400 M−1 cm−1; PenG, Δɛ235 = −775 M−1 cm−1; piperacillin, Δɛ235 = −820 M−1 cm−1; ceftazidime, Δɛ256 = −7,600 M−1 cm−1; cefepime, Δɛ260 = −750 M−1 cm−1; imipenem, Δɛ300 = −9,000 M−1 cm−1; meropenem, Δɛ300 = − 6,500 M−1 cm−1; and ertapenem, Δɛ299 = −9,970 M−1 cm−1.
Stopped-flow experiments.
The variations in the visible spectra of NDM-1 and its L3 variants during hydrolysis of nitrocefin, imipenem and meropenem were followed with an Applied Photophysics SX.18-MVR stopped-flow system associated with a photodiode-array detector (Applied Photophysics, United Kingdom). The measurements were performed in 100 mM HEPES (pH 7.5), 200 mM NaCl, and 0.3 mM ZnSO4 at 6°C. Data were corrected for the instrument dead time (2 ms). In all cases, a 1:1 ratio of metallated enzyme and substrate was used.
Thermal denaturation assays.
Protein stability to thermal denaturation was determined on both holo- and apoproteins by using a protein thermal shift assay (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s instructions. Data were fit to a two-step model as previously described (75).
Preparation of apo and Co(II) substituted enzymes.
The nonmetallated forms of apo-NDM-1, apo-L3VIM, apo-L3IMP, and apo-L3Pro were prepared by successive dialysis of the purified holoproteins against chelators as described previously (49). All buffer solutions used to prepare the apoenzymes were treated by extensive stirring with Chelex 100 (Sigma). Metal content of the apoprotein preparations was checked using PAR as described above. Co(II)-substituted enzymes were obtained after titration on apo derivatives with CoSO4 (49).
X-ray crystallography.
The proteins were purified as described previously with an additional size exclusion chromatography step in a final buffer composed of 10 mM HEPES (pH 7.5) and 200 mM NaCl concentrated to 30 mg/ml. Crystals were grown with the hanging-drop vapor diffusion method at 18°C. Drops were set by mixing equal volumes of protein and reservoir solution. L3IMP crystals were grown in 100 mM HEPES (pH 7.55), 0.1 M NaCl, and 1.35 M (NH4)2SO4, applying microseeding. L3Pro crystals were grown in 100 mM HEPES (pH 7.0), 500 mM (NH4)2SO4, 5 mM CoCl2-NiCl2-MgCl2-CdCl2, and 12 to 30% (wt/vol) PEG 3350. All crystals were flash-frozen either in mother liquor supplemented with 35% glycerol or in 50% paraffin oil and then stored in liquid nitrogen.
X-ray diffraction data were collected at 100 K at the Proxima 1 beamline (Synchrotron Soleil, Saint-Aubin, France). Data reduction was carried out using XDS (76) and Aimless from the CCP4 program suite (77). The crystal structures of both proteins were solved by molecular replacement using the programs Molrep (78) or Phaser (79) and a previously determined NDM-1 structure (PDB entry 3SPU, chain C) as a search probe. The final crystallographic models were obtained through iterative rounds of refinement with Buster (80) and manual rebuilding with COOT (81). Data collection and refinement statistics are summarized in Table S4. Both crystallographic models were validated with MolProbity (82), and root mean square deviation (RMSD) calculations were performed with PDBeFold (83). Illustrations were made with PyMOL (Schrödinger, New York, NY).
To ascertain the presence of Zn(II) metal ions in the active site of L3Pro crystals, two complete data sets were collected at the ESRF beamline id23eh1 using X-ray wavelengths immediately above (hr, λ = 1.27241 Å) and below (lr, λ = 1.28348 Å) the Zn K-edge, as determined with a fluorescence scan. Using phases from the refined protein model, double difference anomalous maps [Dano(hr) – Dano(lr)] were produced with the programs SFTOOLS and FFT from the CCP4 suite (77) (Fig. S4). The presence of Ni(II) ions (from the crystallization buffer) mediating protein-protein interactions in L3Pro was confirmed in a similar way (hr, λ = 1.48030 Å; lr, λ = 1.48840 Å). In all cases, data processing and reduction were carried out as described before (Table S4).
Accession number(s).
Structural data are available in Protein Data Bank database under accession codes 6C6I (L3IMP) and 6CAC (L3Pro).
Supplementary Material
ACKNOWLEDGMENTS
A.R.P. and E.G. received doctoral fellowships from CONICET. M.F.M. received a doctoral scholarship from COLCIENCIAS. L.H.O., S.K., L.I.L., and A.J.V. are CONICET staff members. This study was supported by a grant from ANPCyT (A.J.V.), National Institutes of Health (NIH) grant R01 AI100560 (A.J.V. and R.A.B.), NIH grant R01 AI063517, NIH grant R01 AI072219 (R.A.B.), the ECOS-MinCyT collaborative project (A.J.V. and P.M.A.), the Cleveland Department of Veterans Affairs and Development (1I01BX001974 [R.A.B.]), and the Geriatric Research Education and Clinical Center (R.A.B.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors declare that they have no conflicts of interest with the contents of this article.
We thank Ahmed Haouz and Patrick Weber (Institut Pasteur) for help with the robot-driven crystallization screenings. We acknowledge the synchrotron sources Soleil (Saint-Aubin, France) and ESRF (Grenoble, France) for granting access to their facilities, and we thank their staff members for helpful assistance.
A.R.P. and M.F.M. purified protein samples, performed the biochemical characterization of the proteins, and performed and analyzed the activity measurements and stopped-flow experiments. E.G. crystalized the proteins, collected the X-ray diffraction data and solved the structures. S.K., L.H.O., and P.M.A. performed data collection and resolution of the X-ray structures. M.A.T. and C.R.B. helped with the construction of the protein variants. E.G., S.K., L.H.O., and P.M.A. analyzed and discussed the crystallographic data. A.R.P., M.F.M., R.A.B., L.I.L., and A.J.V. analyzed and discussed the data. A.R.P., M.F.M., E.G., and A.J.V. wrote the paper, and all authors discussed the results and commented on the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01754-18.
REFERENCES
- 1.Fisher JF, Meroueh SO, Mobashery S. 2005. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev 105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 2.Bonomo RA, Tolmasky M. 2007. Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition, 1st ed, vol 83 ASM Press, Washington, DC. [Google Scholar]
- 3.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King AM, Reid-Yu SA, Wang W, King DT, De Pascale G, Strynadka NC, Walsh TR, Coombes BK, Wright GD. 2014. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature 510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez MM, Kosmopoulou M, Mojica MF, Castillo V, Hinchliffe P, Pettinati I, Brem J, Schofield CJ, Mahler G, Bonomo RA, Llarrull LI, Spencer J, Vila AJ. 2015. Bisthiazolidines: a substrate-mimicking scaffold as an inhibitor of the NDM-1 carbapenemase. ACS Infect Dis 1:544–554. doi: 10.1021/acsinfecdis.5b00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinchliffe P, González MM, Mojica MF, González JM, Castillo V, Saiz C, Kosmopoulou M, Tooke CL, Llarrull LI, Mahler G, Bonomo RA, Vila AJ, Spencer J. 2016. Cross-class metallo-β-lactamase inhibition by bisthiazolidines reveals multiple binding modes. Proc Natl Acad Sci U S A 113:E3745–E3754. doi: 10.1073/pnas.1601368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mojica MF, Mahler SG, Bethel CR, Taracila MA, Kosmopoulou M, Papp-Wallace KM, Llarrull LI, Wilson BM, Marshall SH, Wallace CJ. 2015. Exploring the role of residue 228 in substrate and inhibitor recognition by VIM metallo-β-lactamases. Biochemistry 54:3183–3196. doi: 10.1021/acs.biochem.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brem J, Cain R, Cahill S, McDonough MA, Clifton IJ, Jimenez-Castellanos JC, Avison MB, Spencer J, Fishwick CW, Schofield CJ. 2016. Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates. Nat Commun 7:12406. doi: 10.1038/ncomms12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett M, Sprynski N, Coelho A, Castandet J, Bayet M, Bougnon J, Lozano C, Davies DT, Leiris S, Zalacain M. 2018. Discovery of a novel metallo-β-lactamase inhibitor, which can potentiate meropenem activity against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 62:e00074. doi: 10.1128/AAC.00074-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinchliffe P, Tanner CA, Krismanich AP, Labbé G, Goodfellow VJ, Marrone L, Desoky AY, Calvopiña K, Whittle EE, Zeng F. 2018. Structural and kinetic studies of the potent inhibition of metallo-β-lactamases by 6-phosphonomethylpyridine-2-carboxylates. Biochemistry 57:1880–1892. doi: 10.1021/acs.biochem.7b01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sevaille L, Gavara L, Bebrone C, De Luca F, Nauton L, Achard M, Mercuri P, Tanfoni S, Borgianni L, Guyon C. 2017. 1,2,4-Triazole-3-thione compounds as inhibitors of di-zinc metallo-β-lactamases. ChemMedChem 12:972–985. doi: 10.1002/cmdc.201700186. [DOI] [PubMed] [Google Scholar]
- 12.Wang R, Lai T-P, Gao P, Zhang H, Ho P-L, Woo PC-Y, Ma G, Kao RY-T, Li H, Sun H. 2018. Bismuth antimicrobial drugs serve as broad-spectrum metallo-β-lactamase inhibitors. Nat Commun 9:439. doi: 10.1038/s41467-018-02828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bebrone C. 2007. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol 74:1686–1701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 15.Crowder MW, Spencer J, Vila AJ. 2006. Metallo-beta-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc Chem Res 39:721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 16.Mojica MF, Bonomo RA, Fast W. 2016. B1-metallo-β-lactamases: where do we stand? Curr Drug Targets 17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen I. 2015. New promising β-lactamase inhibitors for clinical use. Eur J Clin Microbiol Infect Dis 34:1303–1308. doi: 10.1007/s10096-015-2375-0. [DOI] [PubMed] [Google Scholar]
- 18.Rotondo CM, Wright GD. 2017. Inhibitors of metallo-β-lactamases. Curr Opin Microbiol 39:96–105. doi: 10.1016/j.mib.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Bonomo RA. 2017. Beta-lactamases: a focus on current challenges. Cold Spring Harb Perspect Med 7:a025239. doi: 10.1101/cshperspect.a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Z, Jacobsen FE, Giedroc DP. 2009. Coordination chemistry of bacterial metal transport and sensing. Chem Rev 109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meini MR, Llarrull LI, Vila AJ. 2015. Overcoming differences: the catalytic mechanism of metallo-beta-lactamases. FEBS Lett 589:3419–3432. doi: 10.1016/j.febslet.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palzkill T. 2013. Metallo-beta-lactamase structure and function. Ann N Y Acad Sci 1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ. 2016. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat Chem Biol 12:516–522. doi: 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonomo RA. 2011. New Delhi metallo-β-lactamase and multidrug resistance: a global SOS? Clin Infect Dis 52:485–487. doi: 10.1093/cid/ciq179. [DOI] [PubMed] [Google Scholar]
- 25.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez JM, Meini MR, Tomatis PE, Medrano Martin FJ, Cricco JA, Vila AJ. 2012. Metallo-beta-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat Chem Biol 8:698–700. doi: 10.1038/nchembio.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacquin O, Balbeur D, Damblon C, Marchot P, De Pauw E, Roberts GC, Frere JM, Matagne A. 2009. Positively cooperative binding of zinc ions to Bacillus cereus 569/H/9 beta-lactamase II suggests that the binuclear enzyme is the only relevant form for catalysis. J Mol Biol 392:1278–1291. doi: 10.1016/j.jmb.2009.07.092. [DOI] [PubMed] [Google Scholar]
- 28.Garau G, Garcia-Saez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frere JM, Dideberg O. 2004. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob Agents Chemother 48:2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orellano EG, Girardini JE, Cricco JA, Ceccarelli EA, Vila AJ. 1998. Spectroscopic characterization of a binuclear metal site in Bacillus cereus beta-lactamase II. Biochemistry 37:10173–10180. doi: 10.1021/bi980309j. [DOI] [PubMed] [Google Scholar]
- 30.Concha NO, Rasmussen BA, Bush K, Herzberg O. 1996. Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Structure 4:823–836. doi: 10.1016/S0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- 31.Carfi A, Paul-Soto R, Martin L, Petillot Y, Frère J-M, Dideberg O. 1997. Purification, crystallization, and preliminary X-ray analysis of Bacteroides fragilis Zn2+ β-lactamase. Acta Crystallogr Sect D Biol Crystallogr 53:485–487. doi: 10.1107/S0907444997000966. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, Aitha M, Hetrick AM, Richmond TK, Tierney DL, Crowder MW. 2012. Mechanistic and spectroscopic studies of metallo-beta-lactamase NDM-1. Biochemistry 51:3839–3847. doi: 10.1021/bi300056y. [DOI] [PubMed] [Google Scholar]
- 33.Lisa MN, Palacios AR, Aitha M, Gonzalez MM, Moreno DM, Crowder MW, Bonomo RA, Spencer J, Tierney DL, Llarrull LI, Vila AJ. 2017. A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-beta-lactamases. Nat Commun 8:538. doi: 10.1038/s41467-017-00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tioni MF, Llarrull LI, Poeylaut-Palena AA, Marti MA, Saggu M, Periyannan GR, Mata EG, Bennett B, Murgida DH, Vila AJ. 2008. Trapping and characterization of a reaction intermediate in carbapenem hydrolysis by B. cereus metallo-beta-lactamase. J Am Chem Soc 130:15852–15863. doi: 10.1021/ja801169j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Materon IC, Beharry Z, Huang W, Perez C, Palzkill T. 2004. Analysis of the context dependent sequence requirements of active site residues in the metallo-beta-lactamase IMP-1. J Mol Biol 344:653–663. doi: 10.1016/j.jmb.2004.09.074. [DOI] [PubMed] [Google Scholar]
- 36.Furuyama T, Nonomura H, Ishii Y, Hanson ND, Shimizu-Ibuka A. 2016. Structural and mutagenic analysis of metallo-β-lactamase IMP-18. Antimicrob Agents Chemother 60:5521–5526. doi: 10.1128/AAC.00985-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez MM, Abriata LA, Tomatis PE, Vila AJ. 2016. Optimization of conformational dynamics in an epistatic evolutionary trajectory. Mol Biol Evol 33:1768–1776. doi: 10.1093/molbev/msw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kupper MB, Herzog K, Bennink S, Schlomer P, Bogaerts P, Glupczynski Y, Fischer R, Bebrone C, Hoffmann KM. 2015. The three-dimensional structure of VIM-31–a metallo-beta-lactamase from Enterobacter cloacae in its native and oxidized form. FEBS J 282:2352–2360. doi: 10.1111/febs.13283. [DOI] [PubMed] [Google Scholar]
- 39.Moali C, Anne C, Lamotte-Brasseur J, Groslambert S, Devreese B, Van Beeumen J, Galleni M, Frere JM. 2003. Analysis of the importance of the metallo-beta-lactamase active site loop in substrate binding and catalysis. Chem Biol 10:319–329. doi: 10.1016/S1074-5521(03)00070-X. [DOI] [PubMed] [Google Scholar]
- 40.LaCuran AE, Pegg KM, Liu EM, Bethel CR, Ai N, Welsh WJ, Bonomo RA, Oelschlaeger P. 2015. Elucidating the role of residue 67 in IMP-type metallo-beta-lactamase evolution. Antimicrob Agents Chemother 59:7299–7307. doi: 10.1128/AAC.01651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green VL, Verma A, Owens RJ, Phillips SE, Carr SB. 2011. Structure of New Delhi metallo-beta-lactamase 1 (NDM-1). Acta Crystallogr Sect F Struct Biol Cryst Commun 67:1160–1164. doi: 10.1107/S1744309111029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo Y, Wang J, Niu G, Shui W, Sun Y, Zhou H, Zhang Y, Yang C, Lou Z, Rao Z. 2011. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein Cell 2:384–394. doi: 10.1007/s13238-011-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci 20:1484–1491. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King DT, Worrall LJ, Gruninger R, Strynadka NC. 2012. New Delhi metallo-beta-lactamase: structural insights into beta-lactam recognition and inhibition. J Am Chem Soc 134:11362–11365. doi: 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi Y, Matsueda S, Matsunaga K, Takashio N, Toma-Fukai S, Yamagata Y, Shibata N, Wachino J, Shibayama K, Arakawa Y, Kurosaki H. 2015. Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2. Biol Pharm Bull 38:96–101. doi: 10.1248/bpb.b14-00594. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Hao Q. 2011. Crystal structure of NDM-1 reveals a common β-lactam hydrolysis mechanism. FASEB J 25:2574–2582. doi: 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
- 47.Morán-Barrio J, Limansky AS, Viale AM. 2009. Secretion of GOB metallo-β-lactamase in Escherichia coli depends strictly on the cooperation between the cytoplasmic DnaK chaperone system and the Sec machinery: completion of folding and Zn (II) ion acquisition occur in the bacterial periplasm. Antimicrob Agents Chemother 53:2908–2917. doi: 10.1128/AAC.01637-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neumann W, Gulati A, Nolan EM. 2017. Metal homeostasis in infectious disease: recent advances in bacterial metallophores and the human metal-withholding response. Curr Opin Chem Biol 37:10–18. doi: 10.1016/j.cbpa.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Llarrull LI, Tioni MF, Kowalski J, Bennett B, Vila AJ. 2007. Evidence for a dinuclear active site in the metallo-beta-lactamase BcII with substoichiometric Co(II). A new model for metal uptake. J Biol Chem 282:30586–30595. doi: 10.1074/jbc.M704613200. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Fast W, Benkovic SJ. 1998. Direct observation of an enzyme-bound intermediate in the catalytic cycle of the metallo-β-lactamase from Bacteroides fragilis. J Am Chem Soc 120:10788–10789. doi: 10.1021/ja982621m. [DOI] [Google Scholar]
- 51.Thomas PW, Zheng M, Wu S, Guo H, Liu D, Xu D, Fast W. 2011. Characterization of purified New Delhi metallo-beta-lactamase-1. Biochemistry 50:10102–10113. doi: 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- 52.Brem J, Struwe WB, Rydzik AM, Tarhonskaya H, Pfeffer I, Flashman E, van Berkel SS, Spencer J, Claridge TD, McDonough MA, Benesch JL, Schofield CJ. 2015. Studying the active-site loop movement of the Sao Paolo metallo-beta-lactamase-1. Chem Sci 6:956–963. doi: 10.1039/c4sc01752h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aitha M, Moritz L, Sahu ID, Sanyurah O, Roche Z, McCarrick R, Lorigan GA, Bennett B, Crowder MW. 2015. Conformational dynamics of metallo-beta-lactamase CcrA during catalysis investigated by using DEER spectroscopy. J Biol Inorg Chem 20:585–594. doi: 10.1007/s00775-015-1244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aitha M, Moller AJ, Sahu ID, Horitani M, Tierney DL, Crowder MW. 2016. Investigating the position of the hairpin loop in New Delhi metallo-beta-lactamase, NDM-1, during catalysis and inhibitor binding. J Inorg Biochem 156:35–39. doi: 10.1016/j.jinorgbio.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oelschlaeger P, Mayo SL. 2005. Hydroxyl groups in the ββ sandwich of metallo-β-lactamases favor enzyme activity: a computational protein design study. J Mol Biol 350:395–401. doi: 10.1016/j.jmb.2005.04.044. [DOI] [PubMed] [Google Scholar]
- 56.Oelschlaeger P, Schmid RD, Pleiss J. 2003. Modeling domino effects in enzymes: molecular basis of the substrate specificity of the bacterial metallo-beta-lactamases IMP-1 and IMP-6. Biochemistry 42:8945–8956. doi: 10.1021/bi0300332. [DOI] [PubMed] [Google Scholar]
- 57.Abboud MI, Hinchliffe P, Brem J, Macsics R, Pfeffer I, Makena A, Umland KD, Rydzik AM, Li GB, Spencer J, Claridge TD, Schofield CJ. 2017. (19) F-NMR reveals the role of mobile loops in product and inhibitor binding by the Sao Paulo metallo-beta-lactamase. Angew Chem Int Ed Engl 56:3862–3866. doi: 10.1002/anie.201612185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rydzik AM, Brem J, van Berkel SS, Pfeffer I, Makena A, Claridge TD, Schofield CJ. 2014. Monitoring conformational changes in the NDM-1 metallo-beta-lactamase by 19F NMR spectroscopy. Angew Chem Int Ed Engl 53:3129–3133. doi: 10.1002/anie.201310866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scrofani SD, Chung J, Huntley JJ, Benkovic SJ, Wright PE, Dyson HJ. 1999. NMR characterization of the metallo-beta-lactamase from Bacteroides fragilis and its interaction with a tight-binding inhibitor: role of an active-site loop. Biochemistry 38:14507–14514. doi: 10.1021/bi990986t. [DOI] [PubMed] [Google Scholar]
- 60.Brem J, van Berkel SS, Zollman D, Lee SY, Gileadi O, McHugh PJ, Walsh TR, McDonough MA, Schofield CJ. 2015. Structural basis of metallo-beta-lactamase inhibition by captopril stereoisomers. Antimicrob Agents Chemother 60:142–150. doi: 10.1128/AAC.01335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huntley JJ, Fast W, Benkovic SJ, Wright PE, Dyson HJ. 2003. Role of a solvent-exposed tryptophan in the recognition and binding of antibiotic substrates for a metallo-beta-lactamase. Protein Sci 12:1368–1375. doi: 10.1110/ps.0305303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huntley JJ, Scrofani SD, Osborne MJ, Wright PE, Dyson HJ. 2000. Dynamics of the metallo-β-lactamase from Bacteroides fragilis in the presence and absence of a tight-binding inhibitor. Biochemistry 39:13356–13364. doi: 10.1021/bi001210r. [DOI] [PubMed] [Google Scholar]
- 63.Yang H, Aitha M, Marts AR, Hetrick A, Bennett B, Crowder MW, Tierney DL. 2014. Spectroscopic and mechanistic studies of heterodimetallic forms of metallo-beta-lactamase NDM-1. J Am Chem Soc 136:7273–7285. doi: 10.1021/ja410376s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tripathi R, Nair NN. 2015. Mechanism of meropenem hydrolysis by New Delhi metallo β-lactamase. ACS Catalysis 5:2577–2586. doi: 10.1021/acscatal.5b00242. [DOI] [Google Scholar]
- 65.Gallagher SR. 2001. One-dimensional electrophoresis using nondenaturing conditions. Curr Protoc Protein Sci 94:e73. doi: 10.1002/cpps.73. [DOI] [PubMed] [Google Scholar]
- 66.Llarrull LI, Fabiane SM, Kowalski JM, Bennett B, Sutton BJ, Vila AJ. 2007. Asp-120 locates Zn2 for optimal metallo-beta-lactamase activity. J Biol Chem 282:18276–18285. doi: 10.1074/jbc.M700742200. [DOI] [PubMed] [Google Scholar]
- 67.Tomatis PE, Fabiane SM, Simona F, Carloni P, Sutton BJ, Vila AJ. 2008. Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proc Natl Acad Sci U S A 105:20605–20610. doi: 10.1073/pnas.0807989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez JM, Medrano Martin FJ, Costello AL, Tierney DL, Vila AJ. 2007. The Zn2 position in metallo-beta-lactamases is critical for activity: a study on chimeric metal sites on a conserved protein scaffold. J Mol Biol 373:1141–1156. doi: 10.1016/j.jmb.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 69.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed CLSI Supplement M100S. CLSI, Wayne, PA. [Google Scholar]
- 71.Bahr G, Vitor-Horen L, Bethel CR, Bonomo RA, Gonzalez LJ, Vila AJ. 2017. Clinical evolution of New Delhi metallo-beta-lactamase (NDM) optimizes resistance under Zn(II) deprivation. Antimicrob Agents Chemother 62:e01849-17. doi: 10.1128/AAC.01849-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hunt JB, Neece SH, Ginsburg A. 1985. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal Biochem 146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- 73.Skagseth S, Carlsen TJ, Bjerga GEK, Spencer J, Samuelsen Ø, Leiros H-KS. 2016. Role of residues W228 and Y233 in the structure and activity of metallo-β-lactamase GIM-1. Antimicrob Agents Chemother 60:990–1002. doi: 10.1128/AAC.02017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuzmic P. 1996. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem 237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 75.Huynh K, Partch CL. 2015. Analysis of protein stability and ligand interactions by thermal shift assay. Curr Protoc Protein Sci 79:28.9.1–28.9.14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 79.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bricogne G, Blanc E, Brandl M, Flensburg C, Kelle P, Paciorek P, Roversi P, Sharff A, Smart O, Vonrhein C, Womack T. 2010. BUSTER, version 2.9. Global Phasing, Ltd., Cambridge, United Kingdom. [Google Scholar]
- 81.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 82.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.