While carbapenem resistance in Gram-negative bacteria is mainly due to the production of efficient carbapenemases, β-lactamases with a narrower spectrum may also contribute to resistance when combined with additional mechanisms. OXA-10-type class D β-lactamases, previously shown to be weak carbapenemases, could represent such a case.
KEYWORDS: CHDL, IncQ, OXA-10, β-lactamases, carbapenemases
ABSTRACT
While carbapenem resistance in Gram-negative bacteria is mainly due to the production of efficient carbapenemases, β-lactamases with a narrower spectrum may also contribute to resistance when combined with additional mechanisms. OXA-10-type class D β-lactamases, previously shown to be weak carbapenemases, could represent such a case. In this study, two novel OXA-10 variants were identified as the sole carbapenem-hydrolyzing enzymes in meropenem-resistant enterobacteria isolated from hospital wastewater and found by next-generation sequencing to express additional β-lactam resistance mechanisms. The new variants, OXA-655 and OXA-656, were carried by two related IncQ1 broad-host-range plasmids. Compared to the sequence of OXA-10, they both harbored a Thr26Met substitution, with OXA-655 also bearing a leucine instead of a valine in position 117 of the SAV catalytic motif. Susceptibility profiling of laboratory strains replicating the natural blaOXA plasmids and of recombinant clones expressing OXA-10 and the novel variants in an isogenic background indicated that OXA-655 is a more efficient carbapenemase. The carbapenemase activity of OXA-655 is due to the Val117Leu substitution, as shown by steady-state kinetic experiments, where the kcat of meropenem hydrolysis was increased 4-fold. In contrast, OXA-655 had no activity toward oxyimino-β-lactams, while its catalytic efficiency against oxacillin was significantly reduced. Moreover, the Val117Leu variant was more efficient against temocillin and cefoxitin. Molecular dynamics indicated that Val117Leu affects the position 117-Leu155 interaction, leading to structural shifts in the active site that may alter carbapenem alignment. The evolutionary potential of OXA-10 enzymes toward carbapenem hydrolysis combined with their spread by promiscuous plasmids indicates that they may pose a future clinical threat.
INTRODUCTION
Infections caused by multidrug-resistant bacteria are threatening public health systems. Currently, the most pressing problem is the virtually universal spread of resistant Gram-negative bacteria, such as several Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa strains that are commonly implicated in life-threatening hospital infections. These bacteria can be the hosts for a wide variety of β-lactamases belonging to molecular classes A, C, D (serine β-lactamases), and B (metallo-β-lactamases [MβL] possessing Zn2+ as a cofactor). Most β-lactamases are narrow-spectrum enzymes that inactivate penicillins and older cephalosporins, yet enzymes hydrolyzing last-resort β-lactams, such as carbapenems and oxyimino-substituted β-lactams (such as ceftazidime and cefotaxime), are increasingly detected in pathogens.
So far, enzymes with significant carbapenemase activity have been documented in classes A, B, and D, with the first two being the most clinically relevant in terms of hydrolytic efficiency and host range. Carbapenem-hydrolyzing class D β-lactamases (CHDLs; main lineages, OXA-23, OXA-24/40, OXA-48, and OXA-51) are characterized by a lower efficiency and are mainly confined to Acinetobacter species, while they have a lesser role in carbapenem-resistant enterobacteria, yet highly resistant clones expressing enzymes of this group (e.g., Acinetobacter baumannii and OXA-48-producing Klebsiella pneumoniae) have been implicated in hospital outbreaks (1, 2). This, in conjunction with their relative resistance to inactivation by clinically used inhibitors of serine β-lactamases (3), underscores their importance as emerging carbapenem resistance determinants.
Carbapenem hydrolysis seems not to be restricted to the main CHDL lineages, as OXA-2 and OXA-10 also possess weak carbapenemase activity at a level that is the same as or higher than that of, e.g., OXA-58 (4). The class D β-lactamases of the OXA-10 group are commonly produced by P. aeruginosa (3), with their genes being expressed by class I integrons associated with transposons (5–11). Due to horizontal gene transfer (HGT), these genes are also encountered in Enterobacteriaceae, albeit at a lower frequency than in Pseudomonas (12–17). Most enzymes of the OXA-10 group are narrow-spectrum β-lactamases (3, 5, 9), yet several natural variants, such as OXA-11, OXA-14, OXA-16, OXA-17, OXA-19, and OXA-35, able to turn over oxyimino-β-lactams due to point mutations have been described (5, 7, 18–21). This indicates that the enzymes of the OXA-10 group possess an evolutionary potential that, under selective pressure, may result in the emergence of potent and clinically important variants. Identification of novel enzymes with carbapenemase activity among human pathogens is pivotal for the management of antibiotic resistance. In this study, we therefore focused on Enterobacteriaceae isolates from hospital sewage showing nonsusceptibility to carbapenems without carrying any known carbapenemase gene with the aim to characterize novel genes contributing to carbapenem resistance. Through genetic and biochemical characterization, we identified a Val117Leu variant of OXA-10 (OXA-655) that conferred high-level carbapenem resistance to wild-type Escherichia coli in the presence of additional resistance mechanisms. To our knowledge, OXA-655 is the first natural OXA-10 derivative with increased carbapenemase activity.
RESULTS AND DISCUSSION
Isolation and characterization of Enterobacter cloacae WW13 and E. coli WW16.
During a screening for carbapenem-resistant Enterobacteriaceae in sewage effluent, collected at Sahlgrenska University Hospital in Gothenburg, Sweden, in March of 2014, several colonies were identified on MacConkey agar plates containing meropenem. The isolates were analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) for species identification, the MIC for meropenem was determined, and the presence of known carbapenemase genes was examined through PCR. Two of the lactose-fermenting isolates exhibited elevated meropenem MICs, while they did not appear to carry any of the known carbapenemase genes. The first strain (WW13) was classified as Enterobacter cloacae/Enterobacter asburiae by its MS profile, exhibited a MIC for meropenem of 6 μg/ml, and was isolated from a plate containing 0.5-μg/ml meropenem, while the second one (WW16) was identified as E. coli, was inhibited by meropenem concentrations above 16 μg/ml, and was selected on a plate containing 4 μg/ml meropenem. These findings and, not least, the high meropenem MIC for E. coli WW16 suggested that obscure carbapenem resistance mechanisms were likely expressed by the two strains, and thus, they were selected for subsequent characterization.
Enterobacter strain WW13 was resistant to penicillins (including amoxicillin-clavulanate), first- and second-generation cephalosporins, cefotaxime, and cefotaxime-clavulanate. The ceftazidime, cefepime, and aztreonam MICs were above the epidemiological cutoffs (ECOFFs) for the genus (Table 1) (https://mic.eucast.org/Eucast2/). Furthermore, the isolate was resistant to cephamycins and carbapenems, while the MIC for the 6α-methoxy-substituted penicillin temocillin was within the wild-type range (Table 1). No antagonism was observed between inducers of chromosomal AmpC production (i.e., clavulanate and imipenem) and substrates of this enzyme. Moreover, no synergy between suicide inhibitors and third-generation cephalosporins was apparent, while the phenotypic test for MβL production was negative. Hence, the dominant β-lactam resistance phenotype of this strain resembled that observed during overproduction of the chromosomal class C β-lactamase due to derepression. The non-β-lactam resistance of the isolate was observed only for sulfamethoxazole.
TABLE 1.
Antimicrobial susceptibility profile conferred by β-lactamase-carrying plasmids of E. cloacae WW13 and E. coli WW16 under various genetic backgroundsd
| Antibiotic | Etest MIC (μg/ml) for the following strain host and plasmid β-lactamasesd: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| E. cloacae WW13a ST24 producing OXA-656 | E. coli WW16b ST401 producing OXA-655, SHV-12, TEM-1, and OXA-1 | trcWW13 E. coli CAG18439(pQGU13, IncFII) producing OXA-656 | trcWW16-1 E. coli CAG18439(pQGU16, pEA3-like) producing OXA-655 | trcWW16-2 E. coli CAG18439/IncFII(K) producing SHV-12, OXA-1, and TEM-1 | E. coli CAG18439 control | trfWW13 E. coli TOP10(pQGU13) producing OXA-656 | trfWW16 E. coli TOP10(pQGU16) producing OXA-655 | E. coli TOP10 control | |
| β-Lactams | |||||||||
| Amoxicillin | >256 | >256 | >256 | >256 | >256 | 6 | >256 | >256 | 4 |
| Amoxicillin-clavulanate | 64 | 24 | 12 | 8 | 12 | 6 | 12 | 12 | 4 |
| Temocillin | 16 | 128 | 12 | 32 | 16 | 12 | 16 | 64 | 12 |
| Cephalothin | >256 | >256 | 16 | 12 | 256 | 12 | 12 | 8 | 8 |
| Cefuroxime | >256 | >256 | 6 | 3 | 8 | 3 | 6 | 3 | 2 |
| Cefotaxime | 16 | >32 | 0.125 | 0.094 | 3 | 0.094 | 0.125 | 0.047 | 0.047 |
| Cefotaxime-clavulanate | >1 | 0.19 | 0.032 | 0.032 | 0.032 | 0.032 | 0.032 | 0.023 | 0.023 |
| Ceftazidime | 3 | >256 | 0.094 | 0.094 | 6 | 0.094 | 0.19 | 0.19 | 0.125 |
| Cefepime | 2 | 16 | 0.094 | 0.032 | 0.38 | 0.032 | 0.094 | 0.023 | 0.023 |
| Cefoxitin | >256 | >256 | 4 | 6 | 4 | 3 | 8 | 8 | 8 |
| Aztreonam | 3 | >256 | 0.125 | 0.064 | 16 | 0.064 | 0.38 | 0.094 | 0.064 |
| Imipenem | 12 | 1.5 | 0.25 | 0.38 | 0.19 | 0.19 | 0.25 | 0.25 | 0.25 |
| Meropenem | 6 | 16 | 0.032 | 0.125 | 0.016 | 0.016 | 0.023 | 0.125 | 0.023 |
| Ertapenem | 8 | >32 | 0.016 | 0.25 | 0.004 | 0.003 | 0.016 | 0.19 | 0.002 |
| Doripenem | 4 | 6 | 0.047 | 0.125 | 0.023 | 0.023 | 0.032 | 0.125 | 0.032 |
| Non β-lactams | |||||||||
| Gentamicin | 0.5 | >256 | 0.25 | 0.19 | 16 | 0.25 | 0.38 | 0.38 | 0.19 |
| Streptomycin | 8 | >1,024 | 6 | 6 | 1.5 | 1.5 | >1,024 | >1,024 | >1,024c |
| Sulfamethoxazole | >1,024 | >1,024 | >1,024 | >1,024 | 8 | 8 | >1,024 | >1,024 | 8 |
| Trimethoprim | 0.25 | >32 | 0.125 | 0.125 | >32 | 0.125 | 0.19 | 0.19 | 0.19 |
| Chloramphenicol | 1.5 | >256 | 8 | 8 | >256 | 8 | 2 | 2 | 2 |
| Tetracycline | 1.5 | 2 | >256 | >256 | >256 | >256c | 1.5 | 1.5 | 1.5 |
| Ciprofloxacin | 0.75 | 0.5 | ND | ND | ND | ND | ND | ND | ND |
Chromosomal AmpC derepression and OmpC, OmpD, and OmpF loss. See the text.
OmpF loss. See the text.
Resistant host.
trc, transconjugant; trf, transformant; ND, not determined.
The β-lactam resistance profile of E. coli WW16 suggested production of an extended-spectrum β-lactamase (ESBL), as the strain was resistant to oxyimino-cephalosporins and aztreonam, while it was inhibited by cefotaxime-clavulanate and gave a positive double-disk synergy test (DDST) result. Additionally, it was resistant to cephamycins and carbapenems and exhibited a high temocillin MIC (Table 1). Regarding carbapenem resistance, the highest MIC values were observed for ertapenem and meropenem, while the strain was more susceptible to imipenem, opposite the findings for strain WW13 (Table 1). The MβL phenotypic test was again negative. The MICs of cephamycin and carbapenems for E. coli WW16 suggested that non-β-lactamase mechanisms contributed to its carbapenem resistance phenotype. Moreover, the strain was susceptible to tetracycline and ciprofloxacin, while it was resistant to aminoglycosides, sulfamethoxazole, trimethoprim, and chloramphenicol (Table 1).
Analysis of the assembled Illumina reads classified Enterobacter cloacae WW13 as sequence type 24 (ST24) and E. coli WW16 as ST401 according to the respective multilocus sequence typing (MLST) schemes.
E. cloacae WW13 carried, apart from the chromosomal class C β-lactamase, a variant of blaOXA-10 that differed by one nonsynonymous mutation from the parental gene (GenBank accession number U37105, blaOXA-10 nucleotides 1307 to 2107). This was due to a C-to-T transition in position 77, resulting in the replacement of the threonine at position 26 (standard class D β-lactamase numbering [22]) of the produced protein with methionine (assigned variant, OXA-656 [GenBank accession number MH384611]). The same mutation was found in a blaOXA-10 variant detected in the whole-genome sequencing (WGS) data for E. coli WW16; in addition, the blaOXA-10 variant carried a C-to-G transversion at position 350, leading to a leucine-for-valine substitution in residue 117 (OXA-655; GenBank accession number MH384610). The Thr26Met substitution of the novel OXA-10 variants OXA-656 and OXA-655 is located at the amino terminus of the mature protein, while the Val117Leu observed in OXA-655 is within the second catalytically important conserved motif (S115XX; SAV in OXA-10-type enzymes) of class D β-lactamases (23). Apart from a common mutation, evidence for a close relation between blaOXA-656 and blaOXA-655 was provided by their presence on similar contigs, both of which are positioned as the first gene cassettes of class I integrons linked to a novel Tn3-like transposon inserted in the streptomycin kinase gene (strB) of IncQ1 plasmids akin to RSF1010 (24). E. coli WW16 produced additional acquired β-lactamases that were identified as SHV-12, TEM-1, and OXA-1.
Non-β-lactam resistance determinants were found in the WGS data for E. cloacae WW13 and E. coli WW16, in accordance with the observed phenotypes (see Table S2 in the supplemental material). Furthermore, both strains expressed additional intrinsic β-lactam resistance mechanisms concerning (i) derepressed overexpression of the AmpC of E. cloacae WW13 due to alterations in the carboxyl end of the ampD gene (which contained 14 amino acid substitutions for the produced N-acetylmuramyl-l-alanine amidase compared to that of E. cloacae NCTC 13405 and which was 4 residues shorter [25–28]) and (ii) reduced outer membrane permeability resulting from porin loss due to premature stop codons in the respective genes (ompC, ompF, and ompD in E. cloacae WW13 and ompF in E. coli WW16).
In silico analysis of the likely plasmid content of the isolates using PlasmidFinder revealed that, apart from the IncQ1 plasmids harboring the novel blaOXA variants, they carried several additional replicons (Table S3 in the supplemental material). E. coli WW16 harbored an additional replicon not detected by PlasmidFinder that was 99% identical to and that covered 99% of the pEA3 29.5-kb plasmid found in the plant pathogen Erwinia amylovora CFBP 2585 (GenBank accession number NC_020920) and related to molecules found in E. coli strains isolated from swine (strains FSEC-01 and FSEC-02) (29). Application of the commercial PCR-based replicon typing (PBRT) method identified the FII incompatibility group in E. cloacae WW13 and IncHI1 and IncFII(K) in E. coli WW16. Electrophoresis of plasmid DNA extracted by an alkaline lysis protocol confirmed the presence of several small to medium-size plasmids in both strains that were undetectable by PBRT.
Both strains could readily serve as donors of β-lactam resistance during mating assays. Transconjugant clones obtained by a conjugation experiment with E. cloacae WW13 exhibited a phenotype typical for class D β-lactamases (i.e., resistance to penicillins, reduced susceptibility to penicillin-inhibitor combinations, and susceptibility to cephalosporins, cephamycins, and carbapenems; Table 1, strain trcWW13), and they produced the OXA-656 variant located on the IncQ1 plasmid of strain WW13(pQGU13). The mobilization of pQGU13 was facilitated by an IncFII plasmid, as indicated by PBRT. WW13 transconjugants were also resistant to sulfonamides and exhibited elevated MICs for streptomycin (Table 1), in line with the gene content of pQGU13, further indicating that the IncFII replicon did not carry any antibiotic resistance gene. The conjugation mixture of E. coli WW16 yielded two types of transconjugants differing in their β-lactam resistance phenotypes. The first corresponded to production of an OXA-type enzyme (Table 1, strain trcWW16-1), with PCR and sequencing confirming the presence of blaOXA-655 and the IncQ1 plasmid (pQGU16). The replicon that enabled its mobilization was the pEA3-like plasmid. The second phenotype was typical for expression of an ESBL (Table 1, strain trcWW16-2). trcWW16-2 produced, in addition to the SHV-12 ESBL, the narrow-spectrum enzymes OXA-1 and TEM-1 and, moreover, exhibited resistances to aminoglycosides, chloramphenicol, and trimethoprim that were expressed by the IncFII(K) plasmid (Table 1).
Characterization of the novel OXA-expressing IncQ1 plasmids.
Carriage of the novel OXA-10 variants by mobilizable IncQ1 plasmids highlighted their spreading potential in diverse species and various habitats (30). Indeed, the prototype plasmid of the group RSF1010 is able to replicate and be maintained not only in gammaproteobacterial species but also in members of the alpha-, beta-, and deltaproteobacteria of both clinical and environmental origin (24, 31). The likely promiscuous nature of the OXA-expressing replicons prompted their detailed characterization, and transformant clones were prepared using E. coli TOP10 as a host.
(i) Structure of the novel replicons. Overall, the two plasmids were highly similar. They differed in their accessory regions, with pQGU13 being slightly larger (15.5 kb versus 14.1 kb for pQGU16) due to an additional IS4321 element, and by a nonsynonymous mutation in the repC gene of pQGU13 (Ala108Val substitution) (Fig. 1). Their backbones were nearly identical to that of RSF1010. The differences concerned the repC gene, which carried the mutations observed in other recently described IncQ1 replicons (32–34). As the origins of replication of the novel plasmids were identical to the origin of replication of RSF1010, the three plasmids should be considered incompatible. Moreover, their backbones, containing all the genes required for their double-displacement replication mechanism (30), indicated that they could be maintained in a wide range of hosts, as is the case for their parental molecule.
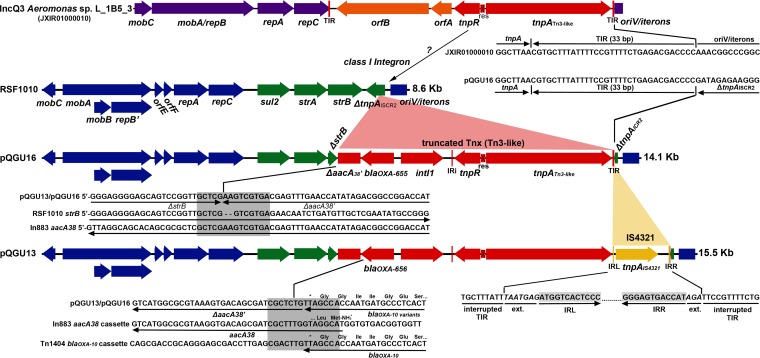
FIG 1.
Structure of the novel plasmids pQGU16 and pQGU13 and comparison with their relative molecules. Genes comprising the IncQ1 backbone are colored blue, while those of the acquired region of RSF1010 (GenBank accession number M28829) are colored green. The novel class I integron containing Tn3-like transposon is colored red, while the IS4321 element of pQGU13 is colored yellow. A map of contig 10 of Aeromonas sp. L_1B5_3 WGS (GenBank accession number JXIR01000010; reverse complement) carrying a Tn3-like transposon having a transposition module (identity > 99%) highly similar to the one found on the IncQ1 plasmids is shown in the top row. Reannotation and sequence comparison of this contig revealed that it contains a typical IncQ3 backbone (purple), with nucleotides 8676 to 11093 expressing a chimeric MobA/RepB relaxase/mobilization nuclease (92% identical to the one of pQ7; GenBank accession number FJ696404) (85) and nucleotides 11077 to 11508 corresponding to a MobC ORF (96% identical to the one of pQ7). Nucleotides 1 to 206 make up the OriV/iteron region that at its 3′ end is partially captured and at its 5′ end is interrupted by the Tn3-like transposon. Both the right and left terminal inverted repeats (IRR and IRL, respectively) of the transposon are present, with their sequences being identical to the right TIR identified in pQGU16 (shown at the junction points of the Aeromonas IncQ3 and pQGU16 plasmids). In the TIR of pQGU13, an IS4321 element has been inserted, with its left and right inverted repeat sequences extended by 7 and 3 nucleotides, respectively (35), being identified at the junctions. The sequence overlap between strB and aacA38 at the ΔstrB-ΔaacA38 region of the pQGU plasmids is highlighted by a gray box. The high similarity between the beginning of the aacA38 gene of In883 (GenBank accession number KJ668593) and the end of blaOXA-10 in Tn1404 (GenBank accession number U37105) that most likely led to the collapse of the 59-bp element in pQGU plasmids through homologous recombination was also detected.
A remnant of the accessory region of RSF1010 comprising the sul2 and strA genes and the first 223 nucleotides of the strB gene was found downstream of the repC gene on both pQGU13 and pQGU16 (Fig. 1). The strB gene was disrupted by a novel truncated Tn3-like transposon that contained a class I integron expressing the OXA-10 variants as the first cassette. Moreover, the integron contained a nonfunctional second cassette consisting of a truncated AAC(I) aminoglycoside acetyltransferase open reading frame (ORF) from which both the 5′ and 3′ ends were missing and that was similar to the aacA38 gene found in In883 (GenBank accession number KJ668593). The intergenic region between the two cassettes had collapsed, while the integron 3′ conserved sequence (CS) was absent (Fig. 1). The integron 5′ CS, containing the attI1 site and the class I integrase gene (intI1), was 100% identical to the one found on the Tn1404 element identified on plasmid R151 of P. aeruginosa POW 151 (8). Downstream of intI1, the integron inverted repeat IRi was found to adjoin the transposition module that consisted of a resolvase gene and a transposase gene in the opposite orientation. In pQGU16, a 33-bp terminal inverted repeat (TIR) sequence was detected downstream of tnpA, forming the right extremity of the transposon and interrupting the remnant of the ISCR2 transposase gene found on the variable region of RSF1010 at the vicinity of the plasmid’s replication origin (Fig. 1). In E. cloacae WW13, the TIR region of the IncQ plasmid was interrupted by the IS4321 element (Fig. 1).
Sequences analyses indicated that pQGU13 may have originated from a molecule that had the structure of pQGU16, as IS4321 elements are known to recognize and be inserted into terminal inverted repeat sequences of Tn21/Tn3-like transposons without forming any target site duplication (35). The progenitor of pQGU13 and pQGU16 most likely emerged during an initial transposition event inside the truncated ISCR2 transposase gene of RSF1010, as it is indicated by the intact right end of the Tn3-like transposon in pQGU16. Although the structures of the novel plasmids do not permit the reconstruction of the exact events that led to their emergence, some assumptions based on sequence comparisons can be made. The transposition module of the novel Tn3 element was 99% identical to the one identified in WGS data of an Aeromonas sp. isolate recovered from an artificial lake in Malaysia (Aeromonas sp. strain L_1B5_3 [BioProject accession number PRJNA270791]). The Aeromonas Tn3-like transposon appears to be intact, as both TIR regions are present bracketing the transposition module along with two accessory metabolic genes and, most likely, resides on an IncQ3 plasmid (contig 10; GenBank accession number JXIR01000010; Fig. 1). Based on the information presented above, the ancestor of pQGU13 and pQGU16 probably emerged through transposition of the IncQ3 Tn3-like transposon into the accessory region of an RSF1010-like plasmid. The subsequent replacement of the two transposon accessory ORFs by the OXA-expressing class I integron may have occurred at a single step through homologous recombination. Indeed, there are already described complex structures where class I integrons are colocalized with strA-strB-carrying Tn3-like transposons (e.g., Tn1403 carrying the Tn5393c strA-strB Tn3-like transposon located on plasmid RPL11 of P. aeruginosa [36]). Evidence of such crossovers can be observed in the sequences of the novel OXA-expressing plasmids, such as those of the ΔstrB-ΔaacA38 region, where the respective intact genes share a high homology (Fig. 1). Similarly, collapse of the 59-bp element of the blaOXA cassette along with the start codon of aacA38' may have resulted from homologous recombination (Fig. 1).
These observations strengthen the notion that recombination events have a key role in shaping the structure of IncQ plasmids (32, 37). Indeed, their unique replication mechanism, where extended parts of the molecule remain single stranded for prolonged periods of time (30), favors both intra- and intermolecular priming, leading to the excision and exchange of genetic structures. In the case of the novel OXA-expressing IncQ1 plasmids, some hypotheses can be made regarding the potential habitats and bacterial species that hosted the above-described genetic exchanges. Both the Aeromonas sp. and P. aeruginosa, which contain mobile genetic structures highly similar to those identified in pQGU13 and pQGU16, are important components of aquatic microbial communities (38–42). Thus, while RSF1010-like plasmids are widespread and are recovered from various sites, the acquired region of the novel IncQ1 plasmids suggests that these plasmids could have emerged in aquatic ecosystems.
(ii) Effects on β-lactam resistance. Under each genetic background tested (i.e., E. coli CAG18439, in the presence of a helper cryptic plasmid, and E. coli TOP10), carriage of blaOXA-655-harboring pQGU16 significantly increased the MICs of meropenem, doripenem, and ertapenem (5 to 8, 4 to 5, and 80 to 95 times, respectively), while imipenem susceptibility was not affected (Table 1). Production of OXA-655 also increased the MICs of temocillin by 2.7 to 5.3 times compared to those observed for the host strains without plasmids (Table 1). On the contrary, blaOXA-656 in an identical immediate genetic context as blaOXA-655 did not affect the carbapenem and temocillin MICs, although there was a 4 to 8 times increase for ertapenem. Oxyimino-substituted β-lactams retained their potency against the pQGU16-carrying E. coli clones, whereas a 6-fold increase in the MIC of aztreonam and minor changes for cefotaxime and ceftazidime were observed for the clones carrying pQGU13 (Table 1).
The findings presented above and, especially, the susceptibility profiles of E. coli transformants suggest that the Met26/Leu117 variant of OXA-10 (OXA-655) is more efficient against carbapenems and temocillin than OXA-656 containing only the Thr26Met substitution. Therefore, E. coli WW16, for which carbapenem MICs were unusually high (considering the resistance mechanisms that it expressed), produced an OXA-10 type β-lactamase that hydrolyzed carbapenems faster than its parental enzyme due to a substitution in the SAV catalytic motif.
Effects of Thr26Met and Val117Leu on interactions with β-lactams.
In order to compare the resistance levels conferred by the two novel OXA variants as well as by their parental OXA-10 under strict isogenic conditions and to examine in detail their hydrolytic properties, the respective genes were cloned into the pZE21 vector, with their expression being controlled by the inducible pTetOL1 promoter in a tetR-positive E. coli strain (E. coli C600Z1).
(i) β-Lactam resistance profile. In the absence of a pTetOL1 inducer, production of the three OXA enzymes by the respective recombinant clones was minimal, as the β-lactam MICs were not affected (Table 2). Induction with anhydrotetracycline resulted in the overproduction of the cloned β-lactamases, as indicated by the resistance levels observed for the various β-lactams (Table 2). OXA-10 and OXA-656 caused identical effects on β-lactam MICs that concerned resistance to amoxicillin and its combination with clavulanic acid as well as to the second-generation oxyimino-cephalosporin cefuroxime. Resistance against cephalothin was also affected, but to a lesser extent. Overproduction of both enzymes significantly increased the MICs of cefotaxime, aztreonam, and cefepime (16-, 42-, and 16-fold, respectively), while there were no effects on susceptibility toward ceftazidime (Table 2). OXA-10 and OXA-656 caused the same increases in the MICs of temocillin, meropenem, ertapenem, and doripenem, while cefoxitin and imipenem action was not altered, probably due to the reduced susceptibility of the host strain used (Table 2).
TABLE 2.
β-Lactam resistance profile conferred by OXA-10 and the new variants overproduced in E. coli C600Z1 under isogenic conditions
| β-Lactam | Etest MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| pZE21-blaOXA-10a producing OXA-10 (Thr26, Val117) | pZE21-blaOXA-656a producing OXA-656 (Met26, Val117) | pZE21-blaOXA-655a producing OXA-655 (Met26, Leu117) | pZE21-blaOXA, no induction | Control | |
| Amoxicillin | >256 | >256 | >256 | 16 | 6 |
| Amoxicillin-clavulanate | 64 | 64 | 24 | 6 | 6 |
| Temocillin | 64 | 64 | 192 | 12 | 12 |
| Cephalothin | 24 | 32 | 16 | 12 | 12 |
| Cefuroxime | 64 | 64 | 4 | 4 | 4 |
| Cefotaxime | 1.5 | 1.5 | 0.094 | 0.094 | 0.094 |
| Cefotaxime-clavulanate | 0.094 | 0.094 | 0.094 | 0.094 | 0.094 |
| Ceftazidime | 0.125 | 0.125 | 0.094 | 0.125 | 0.125 |
| Cefepime | 1.5 | 1.5 | 0.032 | 0.047 | 0.047 |
| Cefoxitin | 8 | 8 | 8 | 8 | 8 |
| Aztreonam | 8 | 8 | 0.19 | 0.094 | 0.094 |
| Imipenem | 0.25 | 0.25 | 0.5 | 0.25 | 0.25 |
| Meropenem | 0.125 | 0.125 | 0.5 | 0.016 | 0.016 |
| Ertapenem | 0.19 | 0.25 | 0.75 | 0.004 | 0.004 |
| Doripenem | 0.125 | 0.125 | 0.5 | 0.032 | 0.032 |
Determined in Mueller-Hinton agar plates containing 200 ng/ml anhydrotetracycline.
The OXA-655 recombinant strain exhibited a β-lactam susceptibility profile different from that of the OXA-10 and OXA-656 recombinant strains. Carbapenem MICs indicated elevated resistance compared to that for the parental enzymes, with the MICs of meropenem, ertapenem, and doripenem exhibiting the highest increases (4-, 3-, and 4-fold, respectively). Oxyimino-cephalosporins and aztreonam were as potent against the blaOXA-655-carrying clone as against the empty host, while the temocillin MIC was, again, higher than that for OXA-10 enzymes with a valine at position 117 (Table 2). Comparison of the resistance levels conferred by the three OXA-10-type enzymes when expressed under isogenic conditions suggested that the Thr26Met substitution (seen in both OXA-656 and OXA-655) had no detectable phenotypic effect, while the Val117Leu substitution caused significant alterations of the interactions with β-lactams. The substitution had a clear negative effect on the turnover of β-lactams carrying an oxyimino R1 side chain, while it appeared to increase the enzyme's efficiency against carbapenems and temocillin, which share a structural feature concerning an oxygen-containing 6α substituent.
(ii) Hydrolysis constants. Purification of the three β-lactamases to near homogeneity (purity, >95%) and assessment of their Michaelis-Menten catalytic constants revealed that the Leu117 variant indeed interacted with the various classes of β-lactams differently than it did with the Val117 enzymes (Table 3). While OXA-10 and OXA-656 shared nearly identical hydrolysis constants, OXA-655 exhibited statistically significant alterations of kcat and Km values for every β-lactam tested. Carbapenem hydrolysis by the three enzymes was characterized by low kcat and low Km values, pointing to deacylation as the reaction's rate-limiting step. Carbapenem Michaelis constants were lower than the lowest concentration of substrate that could be assayed, and thus, for these reactions, only kcat could be accurately measured. OXA-655 hydrolyzed both imipenem and meropenem faster at every substrate concentration tested than OXA-10 and OXA-656 did (Table 3). The effect was stronger in the case of meropenem, where a 4-fold increase in the kcat value was observed (Table 3). Benzylpenicillin and oxacillin hydrolysis was more efficient in the Val117 variants, mostly due to increased turnover rates, which, in the case of the latter substrate, were higher by approximately 7 times compared to those for OXA-655 (Table 3). The Michaelis constant was also affected during oxacillin hydrolysis, and it was 2 times higher in the case of OXA-655, thus leading to a >10-fold decrease in kcat/Km compared to that for the Val117 enzymes.
TABLE 3.
Steady-state kinetic constants of β-lactam hydrolysisd
| β-Lactam | OXA-10 (Thr26, Val117) |
OXA-656 (Met26, Val117) |
OXA-655 (Met26, Leu117) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (M−1·s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1·s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1·s−1) | |
| Imipenem | 0.047 ± 0.001 | <2 | >2.3 × 104 | 0.046 ± 0.002 | <2 | >2.3 × 104 | 0.085 ± 0.003 | <2 | >4.3 × 104 |
| Meropenem | 0.023 ± 0.001 | <2 | >1.2 × 104 | 0.024 ± 0.002 | <2 | >1.2 × 104 | 0.089 ± 0.006 | <2 | >4.5 × 104 |
| Benzylpenicillin | 91 ± 1 | 14 ± 1 | 6.7 × 106 | 92 ± 11 | 14 ± 6 | 6.7 × 106 | 50 ± 6 | 22 ± 6 | 2.3 × 106 |
| Oxacillin | 346 ± 3 | 65 ± 5 | 5.3 × 106 | 331 ± 11 | 58 ± 2 | 5.7 × 106 | 50 ± 3 | 113 ± 20 | 4.4 × 105 |
| Nitrocefin | 110 ± 15 | 13 ± 8 | 8.5 × 106 | 111 ± 3 | 16 ± 2 | 6.9 × 106 | 77 ± 14 | 60 ± 24 | 1.3 × 106 |
| Cephalothin | 1.07 ± 0.03 | 4.4 ± 1.4 | 2.5 × 105 | 0.99 ± 0.05 | 3.3 ± 1.2 | 3.0 × 105 | 2.3 ± 0.6 | 152 ± 24 | 1.5 × 104 |
| Cefotaxime | 2.06 ± 0.19 | 104 ± 17 | 1.9 × 104 | 2.09 ± 0.20 | 98 ± 17 | 2.1 × 104 | >0.03 | >120 | 2.4 × 102 ± 22 |
| Cefoxitin | >0.07 | >200 | 3.8 × 102 ± 12 | >0.07 | >200 | 3.9 × 102 ± 25 | >0.12 | >200 | 7.4 × 102 ± 65 |
The data represent the means ± standard deviations from three independent experiments.
First-generation cephalosporins were also hydrolyzed less efficiently by OXA-655. Regarding nitrocefin, Val117Leu affected both kcat and Km (a 1.4-fold higher kcat and a 4-fold lower Km in OXA-10 and OXA-656), while during cephalothin hydrolysis, only the Michaelis constant was significantly altered, as it was 46 times higher in OXA-655 (Table 3). The increase in Km suggested, furthermore, that Leu117 has a profound negative effect on the affinity of the enzyme for the latter substrate. OXA-10 and OXA-656 exhibited measurable cefotaxime hydrolysis, characterized by low kcat and high Km values, indicating that the hydrolysis of cefotaxime was 270 times less efficient than that of oxacillin (Table 3). In contrast, cefotaxime hydrolysis by OXA-655 was barely detectable, due to very low turnover rates in conjunction with high Km values. The estimate for kcat/Km during hydrolysis of cefotaxime by OXA-655 indicated that the Val117Leu substitution reduced the catalytic efficiency by 100 times. Cefoxitin was a poor substrate for all three OXA-10-type enzymes, reflected by low kcat and high Km values exceeding the highest concentration of the substrate that could be measured by UV spectrophotometry. Nonetheless, at every substrate concentration tested, OXA-655 hydrolyzed the molecule with initial velocities that were double those of its parental enzymes, a feature also reflected in the kcat/Km estimates (Table 3).
Enzyme kinetics indicated that the Thr26Met substitution had no effect on OXA-10 function, while the Val117Leu substitution induced major alterations, including an increased hydrolytic efficiency for the carbapenems imipenem and meropenem. Regarding the latter substrate, the increase in kcat was much higher. Considering the differences observed during MIC experiments under isogenic conditions, it is highly likely that the hydrolytic efficiency is significantly increased for ertapenem and doripenem as well, which, in contrast to imipenem, carry an 1β-methyl group and are substituted at C-2 by bulkier side chains. Increased hydrolysis was also observed for cefoxitin. On the other hand, the substitution caused reduction in hydrolysis of the preferred substrates for these type of enzymes (i.e., oxacillin and other penicillins) as well as of cephalosporins and, especially, oxyimino-substituted molecules.
(iii) Structural bases of the observed phenomena. As previously shown, position 117 has a key functional role in class D β-lactamases (23). It is the third amino acid in the second conserved motif, SXX (i.e., a motif equivalent to the S130DN motif of class A β-lactamases and the Y150AN motif of class C enzymes), with its side chain being positioned in the vicinity of Lys70. The latter amino acid acts as the general base during β-lactam hydrolysis, deprotonating the catalytic Ser67 and the deacylation water through its carboxylated ε-amino group (43, 44). Mutation of valine 117 to threonine in OXA-10 caused significant activity loss, and it has been proposed that the hydrophobic nature of that residue is important in lowering the pKa of Lys70, thus facilitating its carboxylation (23). Position 117 is relatively conserved in class D enzymes, with the majority of them containing valine (Fig. S1 in the supplemental material). Notable exceptions concern the OXA-51 carbapenemase group (45), where isoleucine is most frequent, and the OXA-62-like carbapenemases (46), where position 117 is occupied by leucine.
The data presented herein indicate that replacement of valine by leucine in position 117 of OXA-10 induces the carbapenemase character of the enzyme, sacrificing the activity against its preferred substrates along with that against newer cephalosporins. The substitution introduces a somewhat more hydrophobic core in the environment of Lys70 due to the extra carbon of the leucine side chain in relation to valine. The kinetic data, though, indicated that the observed functional differences are not due to an increased number of carboxylated species, as the activity for several substrates was decreased. In an initial effort to unveil the likely effects of the substitution on the enzyme’s structural properties, molecular dynamics (MD) simulations were employed. OXA-10, OXA-656, and OXA-655 were simulated in their free forms with Lys70 carboxylated. For each enzyme, three 10-ns simulations were performed in order to assert a relatively expanded sampling of the phase space.
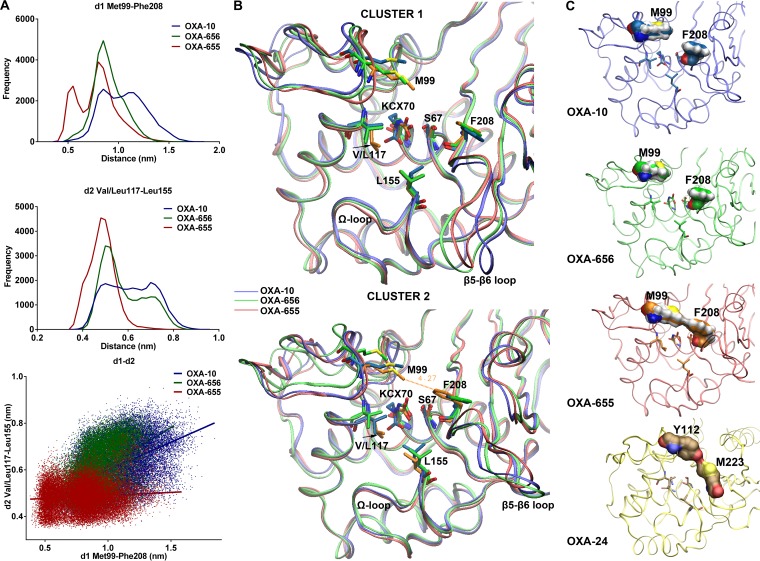
Analysis of the obtained MD trajectories identified two sites in the active center cavity where OXA-655 exhibited notable differences from OXA-10 and OXA-656 that were present in all three sets of simulations. The first one concerned the mutated position and its interactions with Leu155 (standard class D β-lactamase numbering), located in the Ω loop. While the plane of the two methyl groups of the Val117 isopropyl side chain was positioned in parallel with the active site's bottom in OXA-10 and OXA-656, the plane formed by the equivalent groups of the isobutyl side chain in Leu117 of OXA-655 was rotated by 90° (Fig. 2B). The side chain of Leu117 exhibited at this altered position more frequent contacts with the side chain of Leu155, in contrast to OXA-10 and OXA-656, where Val117 did not form a bridge with that residue for a significant part of each simulation (Fig. 2A, middle).
FIG 2.
Structural differences between the three enzymes observed during molecular dynamics simulations. (A) Distribution of distances between the C-ε atom of the Met99 side chain and the center of mass of Phe208 ring carbon atoms (d1; top) and between the methyl groups of Val/Leu117 and Leu155 (d2; middle). Frequency denotes the actual number of structures. The relation of d1 and d2 is depicted in the lower plot. The two distances in OXA-10 and OXA-656 exhibited a weak positive correlation (R2 = 0.2406 and 0.2698, respectively), in contrast to OXA-655 (R2 = 0.0079). Results correspond to combined data from the three sets of simulations (see Fig. S2 in the supplemental material). (B) Superimposition of the average minimized structures of the two main clusters for each enzyme focusing on the active sites. The different positioning of the side chain methyl groups of Leu117 in OXA-655 compared to those of Val117 in OXA-10 and OXA-656 is denoted by arrows. The proximity of the Met99 and Phe208 side chains in the second cluster of OXA-655 is also evident. (C) Surface representations of Met99 and Phe208 in the secondary clusters of OXA-10 variants. In OXA-655, the two residues form a hydrophobic bridge atop the active site. This surface is analogous to the one formed by residues Tyr112 and Met223 in the OXA-24 carbapenemase (PDB accession number 2JC7).
The second significant difference regarded the distances between the side chains of Met99 and Phe208 (B5 β-sheet), located at opposite edges of the active site. The side chains of the two residues were distantly positioned in OXA-10 and OXA-656, and contacts between them were quite rare. In OXA-655, on the other hand, an increased number of species in which the two residues interacted, forming a bridge, were sampled. In fact, OXA-655 was clustered between two main structural populations, in one of which the two residues were in contact (Fig. 2A, top, and Fig. 2B). The difference described above was mostly due to the movements of Phe208 (Fig. 2B). This effect of Val117Leu most probably resulted from the different mode of interactions between residue 117 and Leu155. The side chain of the latter residue also interacted with the C-α and C-β atoms of Ser209, located at the carboxy end of the B5 β-sheet, thus participating in the movements of that segment. In OXA-10 and OXA-656, the distances between Val117-Leu155 and Met99-Phe208 exhibited a degree of correlation, with longer distances between the former residues being associated with remote positioning of the latter and vice versa, while in OXA-655, no such correlation was observed (Fig. 2A, bottom). It therefore seems that the stronger interactions between Leu117-Leu155 in OXA-655 decoupled the effects of Leu155 on the movements of B5, permitting the side chain of Phe208 to frequently adopt conformations closer to Met99, forming a tunnel-like structure (Fig. 2C).
Interestingly, a similar tunnel-like structure has been observed in the crystals of the OXA-23 and OXA-24 carbapenemases formed by Phe110-Met221 and Tyr112-Met223 (Fig. 2C), respectively (47, 48). The above-described structural feature has been proposed to be the factor responsible for the carbapenem-hydrolyzing activity of the above-described enzymes as well as for their low efficiency against oxacillin due to steric hindrance (47). The finding that OXA-655 hydrolyzed carbapenems faster and hydrolyzed oxacillin less efficiently than its parental enzymes due to a substitution that leads to the formation of an equivalent surface points to a likely role of this feature in the observed phenomena. Carbapenems form stable acyl-enzymes with class D enzymes that undergo very slow hydrolysis (49). Deacylation of carbapenem acyl-enzymes proceeds at higher rates in OXA carbapenemases, but the underlying mechanisms have not yet been deciphered. In OXA-24, the tunnel-like structure has been proposed to achieve faster deacylation by favoring tautomerization of the pyrroline ring to its Δ2 tautomer, which could be hydrolyzed faster than Δ1 (which is more prevalent in the OXA-1 acyl-enzyme) (50). The absence of an equivalent bridge in OXA-48, though (51), which is one of the most efficient class D carbapenemases identified so far, indicated that the various enzymes of the group exhibiting high sequence variation may utilize different mechanisms in order to increase efficiency (51) or, alternately, that this surface may not be the factor that leads to increased hydrolysis rates. In a mutagenesis study of OXA-10, it has been shown that the carboxy end of B5, along with the β5-β6 loop, is an important determinant for the carbapenemase function of class D enzymes. Replacement of that segment with the respective ones from OXA-23, OXA-24, and OXA-48 (which differ from OXA-10 both in amino acid composition and in length) significantly increased the catalytic efficiency toward carbapenems (52).
In the MD simulations presented herein, the relation of B5 movements to the position 117/Leu155 bridge as well as to the presence of a hydrophobic surface atop the active site may provide a link between the hypotheses that have been proposed so far. The Val117-Leu155 surface (Val128-Leu166 in OXA-23 and Val130-Leu168 in OXA-24) in carbapenem acyl-enzymes participates in the binding of the 6α-hydroxyethyl substituent (48). The positioning of the latter seems to control the approach of the catalytic water to a hydrolytically competent position (48). Furthermore, in a recent study, the movement of the Val117-Leu155 surface, which could affect the hydration of the active site in carbapenem acyl-enzymes of CHDLs, has been shown (49). It can be hypothesized that the Val117Leu substitution leads to a catalytically competent positioning of the 6α-hydroxyethyl group, permitting the hydrolytic attack, as one of its significant effects concerned the structure and dynamics of the position 117-Leu155 surface, in line with a recently proposed hypothesis (49). This is also supported by the improved efficiency against cefoxitin (a 1.9-fold kcat/Km increase) and the likely increased hydrolytic activity against temocillin (a 3- to 4-fold MIC increase), both of which contain in the α face of the β-lactam ring an equivalent C-6(7) substitution. Further structural and functional studies of OXA-655 interactions with carbapenems and other β-lactams carrying 6α oxygen-containing substituents would validate the hypothesis presented above, and a crystallization effort is under way.
Resistance selection in the hospital wastewater interface.
The role of the environmental gene pool in the emergence of antibiotic resistance in clinical bacteria has been long discussed (53, 54). In hospital sewage, environmental and clinical strains coexist under exposure from a diversity of antimicrobials that may reach concentrations capable of stimulating HGT as well as selecting for resistant strains. Thus, hospital sewage has been pointed out to be an environment that presents a risk for antibiotic resistance development (55). In such a niche, the movement of genes could be bidirectional; i.e., clinical strains could also pass genetic elements to environmental ones, apart from receiving them.
Evidence on the likely association of E. coli WW16 with the hospital environment was provided by a screening for meropenem-resistant enterobacteria in sewage collected at another site of the hospital sewer system performed 6 months after the isolation of WW16. From this screening, an E. coli isolate (E. coli WW41) exhibiting a high meropenem MIC (16 μg/ml) was collected and found by WGS to carry the pGU16 plasmid encoding the OXA-655 carbapenemase. This strain was again classified as ST401 and was genetically nearly identical to E. coli WW16 in terms of plasmid content and resistance determinants (the sole differences being the absence of the pEA3-like plasmid and an additional small plasmid). It therefore seems that E. coli WW16 is a reasonably common strain type here, although its wider geographic spread is not yet known.
It is possible that the strain originates from the intestinal flora of patients at the local hospital, an interpretation in line with the broad resistance profile of the strain as well as its carriage of an IncFII(K) plasmid harboring blaSHV-12, blaTEM-1, and blaOXA-1, yet several characteristics rather link it to nonclinical settings. First, E. coli ST401 has not been implicated in infections, and so far it has been isolated from retail meat (56) and sea-surface water from the Antarctic (57). Furthermore, the plasmid content of E. coli WW16 indicated contact with nonclinical bacteria, as several of its plasmids could not be classified by the PBRT scheme (developed for plasmids from clinical enterobacteria) and the relatives of one of its plasmids, the pEA3-like plasmid, has been isolated from a plant pathogen and E. coli strains causing infections in animals. These features of both clinical and environmental bacteria suggest a possible role of this strain as an intermediate in antibiotic resistance transfer events. Conditions prevailing in the hospital wastewater may yield genetic structures having the potential to become future threats, and strains carrying these could facilitate the transfer of novel resistance determinants in the clinic (either in a direct manner or by passing the novel structures to already established clinical strains).
OXA-10-type enzymes appear to be widespread in wastewater (58, 59), likely due to the high abundance of the P. aeruginosa strains that usually host them. The gene has been spread in clinical enterobacterial populations as well, and recent studies indicate increasing isolation trends (14, 15). The initial steps of these HGT events were most probably facilitated by broad-host-range plasmids, and those of the IncQ group seem to be the most likely vector. Indeed, sequence analysis of pQGU13 and pQGU16 indicated that some of the acquisition events that built up their variable region took place in P. aeruginosa, while a relation with an IncQ3 plasmid found in an Aeromonas sp. was also established. A previous finding, where an OXA-10 variant was expressed as the first cassette of a class 3 integron carried by an IncQ3 plasmid, further strengthens this notion (60). Moreover, as this plasmid was also found in an enterobacterial strain isolated from hospital wastewater (E. cloacae LIM73), this habitat might be of importance for the spread of the gene. It is therefore plausible that the transfer of OXA-10-type enzymes from P. aeruginosa to clinical enterobacteria is an ongoing process in hospital wastewater mediated by broad-host-range IncQ plasmids.
Conclusions.
OXA-10-type enzymes are among the most widespread acquired β-lactamases. Their measurable rates of hydrolysis of newer β-lactams, such as third-generation cephalosporins and carbapenems, suggest a spectrum broader than that of other oxacillinases, like OXA-1. Here, a novel OXA-10 variant with enhanced carbapenemase activity has been found on a broad-host-range replicon that can be readily transferred by helper conjugative plasmids and replicated at high copy numbers (31, 61). It is known that the hydrolysis of expanded-spectrum cephalosporins by OXA-10-type enzymes can be further enhanced through point mutations. Here, we show that the carbapenemase activity can also be enhanced by a single substitution, Val117Leu, located in the SAV catalytic motif. It is likely that production of this novel enzyme (OXA-655) by the wild-type E. coli strain, combined with porin loss, led to the unusually high meropenem MICs. The wild-type E. cloacae strain producing the parental OXA-656 was also resistant to carbapenems due to both porin loss and AmpC overproduction. It therefore seems that the ability of OXA-10-type enzymes for carbapenem turnover can contribute to clinically important resistance in enterobacteria when additional mechanisms are present. The finding that these enzymes possess an evolving potential toward hydrolysis of carbapenems may prove valuable in understanding the function of this enigmatic group of enzymes. OXA-655 is the first reported OXA-10 natural mutant exhibiting enhanced carbapenemase activity, and resolving the structural bases of its function may help further decipher how class D β-lactamases work.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Enterobacter cloacae WW13 and E. coli WW16 were isolated from wastewater collected at the Sahlgrenska University Hospital, Gothenburg, Sweden, in March of 2014 in a parallel surveillance study. Briefly, 30 ml of raw hospital wastewater was sampled at 40-min intervals for 6 days, and the samples were pooled. MacConkey agar plates containing either 0.5 μg/ml or 4 μg/ml meropenem were inoculated with 100 μl of the pooled sample and incubated at 37°C for 24 h. Colonies exhibiting good growth and lactose fermentation were picked and subjected to species identification by MALDI-TOF MS using a Vitek MS system (bioMérieux, Marcy l’Étoile, France) before subsequent characterization. E. coli WW41 was isolated as described above from a wastewater sample collected in September of 2014 at another site of the hospital’s sewage system.
E. coli CAG18439 (Tetr F− lacZ118(Oc) lacI3042::Tn10 λ− rph-1 [62]) was utilized as the recipient in conjugal plasmid transfer assays, while chemically competent E. coli One Shot TOP10 cells [Strr F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL endA1 nupG] (Thermo Scientific, MA, USA) were used as hosts during transformation by natural plasmids. Expression of recombinant blaOXA-10, blaOXA-656, and blaOXA-655 was carried out in E. coli C600Z1 (Spr lacIq PN25-tetR lacY1 leuB6 mcrB+ supE44 thi-1 thr-1 tonA21) (Exxpressys, Bammental, Germany).
The open reading frames of OXA-10, OXA-656, and OXA-655 were synthesized by GeneArt gene synthesis service (Thermo Scientific, Regensburg, Germany), with KpnI (5′-GGGGTACC) and BamHI (GGATCCCG-3′) motifs (indicated in boldface) being attached to their 5′ and 3′ ends, respectively. Then, each gene was cloned into the KpnI and BamHI sites of the pZE21-MCS vector (Kanr), which has been previously described (63). In the resulting constructs (pZE21-blaOXA-10, pZE21-blaOXA-656, and pZE21-blaOXA-655), the cloned genes were under the control of the pTetOL1 promoter/operator, with their transcription being suppressed by TetR in the absence of an inducer and overexpressed in the presence of tetracycline or a tetracycline analogue (64). The OXA-expressing recombinant plasmids were used to transform TetR-producing E. coli C600Z1 cells through electroporation, with transformant clones being selected on Luria-Bertani (LB) agar containing 50 μg/ml kanamycin. The sequence integrity of the cloned region was verified by Sanger sequencing. The resulting clones were utilized for antimicrobial susceptibility testing as well as for enzyme purification.
Whole-genome sequencing and bioinformatics analyses.
DNA libraries (paired end 2 × 300 bp) were prepared using genomic DNA from Enterobacter cloacae WW13, E. coli WW16, and E. coli WW41 and a TruSeq DNA sample kit (Illumina). DNA sequencing was performed on an Illumina MiSeq system at the facilities of the Science for Life Laboratory (Royal Institute of Technology, Sweden). Demultiplexed paired-end reads were trimmed and filtered using Trim Galore software [−q 20 −length 20] (65). Assembling the paired-end reads was performed by using SPAdes (v.2.9.0) in the careful mode and with k-mers ranging from 21 to 127 bases (66). The assembled sequences were analyzed using the in silico multilocus sequence typing (MLST) tool MLST (v.1.8; https://cge.cbs.dtu.dk/services/MLST/; Center for Genomic Epidemiology [CGE]) (67). Acquired resistance genes were detected using the ResFinder (v.2.1) tool of the CGE server (https://cge.cbs.dtu.dk/services/ResFinder/) (68), while the likely plasmid content was assessed using PlasmidFinder (v.1.2; https://cge.cbs.dtu.dk/services/PlasmidFinder/) (69). Bacterial genome annotation was performed using the RAST server (http://www.nmpdr.org/FIG/wiki/view.cgi/FIG/RapidAnnotationServer) (70), and the predicted ORFs were further analyzed, using the BLASTp program, for additional putative β-lactamases and plasmid proteins. Contigs containing acquired β-lactamase genes were compared to the sequences in the NCBI reference nucleic acid database using the BLASTn program. The porin and penicillin binding protein genes of the studied strains were compared with those of E. coli ATCC 25922 (GenBank accession number NZ_CP009072) (71) and E. cloacae ATCC 13047 (GenBank accession number CP001918) (72). The ampR and ampD genes participating in the control of Enterobacter chromosomal cephalosporinase expression were compared with the respective genes of E. cloacae NCTC 13405 (GenBank accession numbers KT780419 and KT780420, respectively) (25). The promoter/attenuator region controlling the expression of the E. coli ampC gene was compared to that of E. coli ATCC 25922 exhibiting basal cephalosporinase production (73), and the detected polymorphisms did not belong to those yielding increased transcription.
Detection of plasmids and β-lactamases.
Genomic DNA was isolated from E. cloacae WW13, E. coli WW16, and E. coli WW41 using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany). The purity and concentration of the DNA preparations were assessed with a NanoDrop ND-1000 spectrophotometer and a Qubit instrument, respectively. The purified DNA was screened for the presence of known carbapenemase genes using PCR primers specific for the VIM, IMP, NDM, KPC, GES, and OXA-48 β-lactamases described earlier (74), although the reverse primer for IMP was slightly modified due to detected mismatches (see Table S1 in the supplemental material).
The transferability of β-lactam resistance was assessed through conjugation experiments, as described previously (75). Conjugation was performed for 4 h at 30°C without shaking, and transconjugants were selected on MacConkey agar plates containing 25 μg/ml tetracycline and 50 μg/ml amoxicillin.
The plasmid content (small to medium-size molecules) of wild-type strains as well as of transconjugant clones was examined electrophoretically on 0.8% agarose gels using purified preparations obtained by an alkaline lysis protocol (76). Incompatibility typing was performed using a commercial PCR-based replicon typing (PBRT) kit (Diatheva, Cartoceto, Italy) and genomic DNA from wild-type and transconjugant strains as the template. The IncQ1 plasmids were detected by PCR using the OXA-10-R, Tn3x-tnpR-F, mobC-F, and Tn3x-tnpA-R primers (Table S1), hybridizing specifically with the respective contigs, while the pEA3-like plasmid, not detected by the commercial PBRT scheme, was identified using the repB_pEA3-F and repB_pEA3-R oligonucleotides (Table S1).
Plasmid preparations from transconjugant clones carrying the IncQ1 plasmids were used to transform chemically competent E. coli One Shot TOP10 cells. Transformation was performed through heat shock using 10 ng of plasmid DNA, and selection was carried out with 50 μg/ml amoxicillin. Carriage of the IncQ1 plasmids was confirmed by PCR and electrophoresis of plasmid DNA, isolated as described above. The whole sequences of the IncQ1 plasmids were obtained by Sanger sequencing using primer walking and PCR mapping utilizing the OXA-10-F, OXA-10-R, mobC-F, tnpA-Tn3x-F, tnpR-Tn3x-F, IntI1F, IS4321-F, and IS4321-R primers targeting the assembled sequences of the respective contigs (Table S1).
The presence of WGS-detected β-lactamase genes in the wild-type and laboratory-generated strains was verified using the OXA-10-F, OXA-10-R, M-SHV-F, M-SHV-R, TEM-A, TEM-B, M-OXA-1-F, and M-OXA-1-R primers (Table S1). The sequence of the OXA-10 variants as well as that of the integron-provided promoter region in the respective transconjugant and transformant clones was verified through Sanger sequencing using the IntI1-F, OXA-10-F, and OXA-10-R primers.
Antimicrobial susceptibility testing.
The β-lactam resistance profile was routinely assessed by disc diffusion on Muller-Hinton agar plates using Bio-Rad antibiotic discs. Phenotypic tests detecting ESBL (the double-disc synergy test [DDST]) and metallo-β-lactamase production were performed as previously described (77, 78). MICs were determined using Etest strips (bioMérieux) on Mueller-Hinton (MH) agar inoculated with a 0.5 MacFarland standard of bacterial suspensions prepared from fresh cultures. The suspensions of E. coli C600Z1 clones expressing the recombinant OXA-10 variants were additionally inoculated on MH agar plates containing 200 ng/ml anhydrotetracycline in order to achieve β-lactamase overexpression. The results were documented after 18 h of incubation at 37°C.
Expression and purification of β-lactamases.
The E. coli C600Z1 clones carrying the recombinant plasmids pZE21-blaOXA-10, pZE21-blaOXA-656, and pZE21-blaOXA-655 were utilized for the large-scale production and purification of the three OXA-10 variants. Overnight bacterial cultures were used to inoculate 500 ml of LB containing 50 μg/ml kanamycin, and then the cells were grown at 37°C with shaking until late logarithmic phase (optical density at 600 nm = 0.8). Anhydrotetracycline was added to each culture to a final concentration of 200 ng/ml, and induction was performed at room temperature for 20 h with shaking. The cells were washed twice, and the protein content was released through mild sonication in 20 mM 2-(N-morpholino)ethanesulfonic acid solution (MES), pH 6. Cell lysates were clarified by centrifugation, and the enzymes were purified using two steps of ion-exchange chromatography as previously described (4) with some modifications. Crude extracts were loaded on a Q-Sepharose column (Bio-Rad, CA, USA) equilibrated with 20 mM MES, pH 6, and the unbound proteins were washed with the same buffer. The loading and washing effluents were then loaded on an S-Sepharose cation exchanger (Bio-Rad) equilibrated with 20 mM MES, pH 6, and the bound β-lactamases were eluted through linearly increased ionic strengths using 0.5 M K2SO4 in 20 mM MES, pH 5.5. Fractions containing β-lactamase activity were pooled, diluted 1:1 with 100 mM PiNa+, 50 mM NaHCO3, pH 7, in order to increase protein solubility, and subjected to buffer exchange and concentration through ultrafiltration using the buffer described above and 10-kDa-cutoff Amicon Ultracel filters (Millipore, MA, USA). Purity was assessed using SDS-polyacrylamide gel electrophoresis (PAGE), and the enzyme concentration was determined by spectrophotometry at 280 nm using a molecular absorption coefficient of 47,565 M−1·cm−1 (calculated for the mature proteins [signal peptide positions 1 to 21] using the ProtParam tool of the ExPASy server [http://web.expasy.org/protparam/]).
Enzyme kinetics.
Purified enzyme preparations were used for determining the steady-state kinetic constants during interaction with important β-lactam classes. β-Lactam hydrolysis was monitored using spectrophotometry on a Cary 60 UV/visible instrument (Agilent, CA, USA). For each enzyme, initial hydrolysis rates were determined at various substrate concentrations under identical experimental conditions, and the kcat (turnover rate) and Km (Michaelis constant) values were obtained through nonlinear regression of the data by the Michaelis-Menten equation, using the plot V/[E] versus [S] (where V is the initial velocity, and [E] and [S] are the enzyme and substrate concentrations, respectively), using Prism software (v.6; GraphPad). In cases where the Km values were higher than the highest concentration of the substrate that could be assayed, only the kcat/Km ratios were estimated through linear regression, as has been done previously (79). All reactions were performed in 100 mM PiNa+, 50 mM NaHCO3, pH 7, at a constant temperature of 25°C using a Peltier thermostatted cell holder (Agilent). Wavelengths and Δε values for the various substrates were as follows: for cephalothin, λ = 262 nm and Δε = −7,660 M−1·cm−1; for nitrocefin, λ = 482 nm and Δε = +17,400 M−1·cm−1; for cefotaxime, λ = 266 nm and Δε = −6,700 M−1·cm−1; for cefoxitin, λ = 260 nm and Δε = −7,600 M−1·cm−1; for oxacillin, λ = 260 nm and Δε = +440 M−1·cm−1; for benzylpenicillin, λ = 233 nm and Δε = −900 M−1·cm−1; for imipenem, λ = 300 nm and Δε = −9,000 M−1·cm−1; and for meropenem, λ = 298 nm and Δε = −7,200 M−1·cm−1. Each kcat and Km measurement was repeated in three independent experiments.
Molecular simulations.
Three-dimensional structures of the OXA-656 and OXA-655 enzymes were built through homology modeling using as a reference the crystal structure of OXA-10 (PDB accession number 4WZ5; resolution, 1.6 Å) (80) and the MODELLER package, as has been previously described (81, 82). The three enzymes were simulated using molecular dynamics (MD) (82) with the GROMACS (v.3.3) package and the AMBER99 force field and with Lys70 carboxylated at its ε-amino group. Partial atomic charges of the modified lysine residue (KCX) were obtained by a two-step RESP fitting procedure using AmberTools and the electrostatic potential points of the tripeptide +NH3-Phe-KCX-Ile-COO−, calculated at the Hartee-Fock/6-31G* level of molecular orbital theory through GAUSSIAN03. Van der Waals and bonded parameters for the carboxylated lysine atoms were extracted from the general amber force field (GAFF) for organic molecules and converted in order to be compatible with the units and energy function terms used by GROMACS (83, 84). For each system, 3 separate simulations were performed using different seeds to generate the Maxwellian distributions of initial velocities at 300 K. The obtained trajectories were analyzed in terms of structural clustering, using a cutoff root mean square deviation of 1 Å for backbone atoms and the GROMOS algorithm. An average structure for each cluster was calculated and minimized using 5,000 steps of steepest descents energy minimization. For the side chain of KCX70, the solvent accessible surface area, water contacts, and water residence times (τres) were calculated (82), without revealing any statistically significant difference between the three enzymes. Structural differences observed during superimposition of the minimized average structures of the main clusters for each enzyme were quantified by measuring the distances between the respective atoms throughout the trajectories.
Accession number(s).
The novel OXA-10 variants OXA-655 and OXA-656 have been submitted to NCBI’s β-lactamase database under GenBank accession numbers MH384610 and MH384611, respectively. The sequences of the plasmids pQGU13 and pQGU16 have been submitted to GenBank under accession numbers MH718731 and MH718732, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Swedish Research Council VR (grant numbers 521-2013-8633 and 2015-02492) and FORMAS (942-2015-750) to D.G.J.L., the Swedish Research Council FORMAS (219-2014-1575) to C.-F.F., as well as the Center for Antibiotic Resistance Research (CARe).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01817-18.
REFERENCES
- 1.Opazo A, Dominguez M, Bello H, Amyes SG, Gonzalez-Rocha G. 2012. OXA-type carbapenemases in Acinetobacter baumannii in South America. J Infect Dev Ctries 6:311–316. [DOI] [PubMed] [Google Scholar]
- 2.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 3.Antunes NT, Fisher JF. 2014. Acquired class D beta-lactamases. Antibiotics (Basel) 3:398–434. doi: 10.3390/antibiotics3030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. 2014. Class D beta-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 58:2119–2125. doi: 10.1128/AAC.02522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aubert D, Poirel L, Ali AB, Goldstein FW, Nordmann P. 2001. OXA-35 is an OXA-10-related beta-lactamase from Pseudomonas aeruginosa. J Antimicrob Chemother 48:717–721. doi: 10.1093/jac/48.5.717. [DOI] [PubMed] [Google Scholar]
- 6.Danel F, Hall LM, Duke B, Gur D, Livermore DM. 1999. OXA-17, a further extended-spectrum variant of OXA-10 beta-lactamase, isolated from Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:1362–1366. doi: 10.1128/AAC.43.6.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mugnier P, Podglajen I, Goldstein FW, Collatz E. 1998. Carbapenems as inhibitors of OXA-13, a novel, integron-encoded beta-lactamase in Pseudomonas aeruginosa. Microbiology 144:1021–1031. doi: 10.1099/00221287-144-4-1021. [DOI] [PubMed] [Google Scholar]
- 8.Partridge SR, Collis CM, Hall RM. 2002. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R151. Antimicrob Agents Chemother 46:2400–2408. doi: 10.1128/AAC.46.8.2400-2408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philippon AM, Paul GC, Jacoby GA. 1983. Properties of PSE-2 beta-lactamase and genetic basis for its production in Pseudomonas aeruginosa. Antimicrob Agents Chemother 24:362–369. doi: 10.1128/AAC.24.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel L, Naas T, Nordmann P. 2010. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother 54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa—a phenomenon of bacterial resistance. J Med Microbiol 58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 12.Aibinu IE, Pfeifer Y, Ogunsola F, Odugbemi T, Koenig W, Ghebremedhin B. 2011. Emergence of beta-lactamases OXA-10, VEB-1 and CMY in Providencia spp. from Nigeria. J Antimicrob Chemother 66:1931–1932. doi: 10.1093/jac/dkr197. [DOI] [PubMed] [Google Scholar]
- 13.Giakkoupi P, Tryfinopoulou K, Polemis M, Pappa O, Miriagou V, Vatopoulos A. 2015. Circulation of a multiresistant, conjugative, IncA/C plasmid within the nosocomial Providencia stuartii population in the Athens area. Diagn Microbiol Infect Dis 82:62–64. doi: 10.1016/j.diagmicrobio.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Kiratisin P, Apisarnthanarak A, Laesripa C, Saifon P. 2008. Molecular characterization and epidemiology of extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother 52:2818–2824. doi: 10.1128/AAC.00171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurya AP, Dhar D, Basumatary MK, Paul D, Ingti B, Choudhury D, Talukdar AD, Chakravarty A, Mishra S, Bhattacharjee A. 2017. Expansion of highly stable bla OXA-10 beta-lactamase family within diverse host range among nosocomial isolates of Gram-negative bacilli within a tertiary referral hospital of northeast India. BMC Res Notes 10:145. doi: 10.1186/s13104-017-2467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papagiannitsis CC, Miriagou V, Kotsakis SD, Tzelepi E, Vatopoulos AC, Petinaki E, Tzouvelekis LS. 2012. Characterization of a transmissible plasmid encoding VEB-1 and VIM-1 in Proteus mirabilis. Antimicrob Agents Chemother 56:4024–4025. doi: 10.1128/AAC.00470-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porto A, Ayala J, Gutkind G, Di Conza J. 2010. A novel OXA-10-like beta-lactamase is present in different Enterobacteriaceae. Diagn Microbiol Infect Dis 66:228–229. doi: 10.1016/j.diagmicrobio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Hall LM, Livermore DM, Gur D, Akova M, Akalin HE. 1993. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 37:1637–1644. doi: 10.1128/AAC.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danel F, Hall LM, Gur D, Livermore DM. 1995. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 39:1881–1884. doi: 10.1128/AAC.39.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danel F, Hall LM, Gur D, Livermore DM. 1998. OXA-16, a further extended-spectrum variant of OXA-10 beta-lactamase, from two Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother 42:3117–3122. doi: 10.1128/AAC.42.12.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mugnier P, Casin I, Bouthors AT, Collatz E. 1998. Novel OXA-10-derived extended-spectrum beta-lactamases selected in vivo or in vitro. Antimicrob Agents Chemother 42:3113–3116. doi: 10.1128/AAC.42.12.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mossakowska D, Ali NA, Dale JW. 1989. Oxacillin-hydrolysing beta-lactamases. A comparative analysis at nucleotide and amino acid sequence levels. Eur J Biochem 180:309–318. doi: 10.1111/j.1432-1033.1989.tb14649.x. [DOI] [PubMed] [Google Scholar]
- 23.Vercheval L, Bauvois C, di Paolo A, Borel F, Ferrer JL, Sauvage E, Matagne A, Frère JM, Charlier P, Galleni M, Kerff F. 2010. Three factors that modulate the activity of class D beta-lactamases and interfere with the post-translational carboxylation of Lys70. Biochem J 432:495–504. doi: 10.1042/BJ20101122. [DOI] [PubMed] [Google Scholar]
- 24.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 25.Babouee Flury B, Ellington MJ, Hopkins KL, Turton JF, Doumith M, Loy R, Staves P, Hinic V, Frei R, Woodford N. 2016. Association of novel nonsynonymous single nucleotide polymorphisms in ampD with cephalosporin resistance and phylogenetic variations in ampC, ampR, ompF, and ompC in Enterobacter cloacae isolates that are highly resistant to carbapenems. Antimicrob Agents Chemother 60:2383–2390. doi: 10.1128/AAC.02835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genereux C, Dehareng D, Devreese B, Van Beeumen J, Frere JM, Joris B. 2004. Mutational analysis of the catalytic centre of the Citrobacter freundii AmpD N-acetylmuramyl-l-alanine amidase. Biochem J 377:111–120. doi: 10.1042/BJ20030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson ND, Sanders CC. 1999. Regulation of inducible AmpC beta-lactamase expression among Enterobacteriaceae. Curr Pharm Des 5:881–894. [PubMed] [Google Scholar]
- 28.Kopp U, Wiedemann B, Lindquist S, Normark S. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob Agents Chemother 37:224–228. doi: 10.1128/AAC.37.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Sun B, Wang Y, Lei L, Schwarz S, Wu C. 2015. Characterization of a cfr-carrying plasmid from porcine Escherichia coli that closely resembles plasmid pEA3 from the plant pathogen Erwinia amylovora. Antimicrob Agents Chemother 60:658–661. doi: 10.1128/AAC.02114-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loftie-Eaton W, Rawlings DE. 2012. Diversity, biology and evolution of IncQ-family plasmids. Plasmid 67:15–34. doi: 10.1016/j.plasmid.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Rawlings DE, Tietze E. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev 65:481–496. doi: 10.1128/MMBR.65.4.481-496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotsakis SD, Tzouvelekis LS, Lebessi E, Doudoulakakis A, Bouli T, Tzelepi E, Miriagou V. 2015. Characterization of a mobilizable IncQ plasmid encoding cephalosporinase CMY-4 in Escherichia coli. Antimicrob Agents Chemother 59:2964–2966. doi: 10.1128/AAC.05017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hooton SP, Timms AR, Cummings NJ, Moreton J, Wilson R, Connerton IF. 2014. The complete plasmid sequences of Salmonella enterica serovar Typhimurium U288. Plasmid 76:32–39. doi: 10.1016/j.plasmid.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Kehrenberg C, Wallmann J, Schwarz S. 2008. Molecular analysis of florfenicol-resistant Pasteurella multocida isolates in Germany. J Antimicrob Chemother 62:951–955. doi: 10.1093/jac/dkn359. [DOI] [PubMed] [Google Scholar]
- 35.Partridge SR, Hall RM. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J Bacteriol 185:6371–6384. doi: 10.1128/JB.185.21.6371-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stokes HW, Elbourne LD, Hall RM. 2007. Tn1403, a multiple-antibiotic resistance transposon made up of three distinct transposons. Antimicrob Agents Chemother 51:1827–1829. doi: 10.1128/AAC.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yau S, Liu X, Djordjevic SP, Hall RM. 2010. RSF1010-like plasmids in Australian Salmonella enterica serovar Typhimurium and origin of their sul2-strA-strB antibiotic resistance gene cluster. Microb Drug Resist 16:249–252. doi: 10.1089/mdr.2010.0033. [DOI] [PubMed] [Google Scholar]
- 38.Khan NH, Ishii Y, Kimata-Kino N, Esaki H, Nishino T, Nishimura M, Kogure K. 2007. Isolation of Pseudomonas aeruginosa from open ocean and comparison with freshwater, clinical, and animal isolates. Microb Ecol 53:173–186. doi: 10.1007/s00248-006-9059-3. [DOI] [PubMed] [Google Scholar]
- 39.Nonaka L, Inubushi A, Shinomiya H, Murase M, Suzuki S. 2010. Differences of genetic diversity and antibiotics susceptibility of Pseudomonas aeruginosa isolated from hospital, river and coastal seawater. Environ Microbiol Rep 2:465–472. doi: 10.1111/j.1758-2229.2010.00178.x. [DOI] [PubMed] [Google Scholar]
- 40.Corno G, Coci M, Giardina M, Plechuk S, Campanile F, Stefani S. 2014. Antibiotics promote aggregation within aquatic bacterial communities. Front Microbiol 5:297. doi: 10.3389/fmicb.2014.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hazen TC, Fliermans CB, Hirsch RP, Esch GW. 1978. Prevalence and distribution of Aeromonas hydrophila in the United States. Appl Environ Microbiol 36:731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarria-Guzmán Y, López-Ramírez MP, Chávez-Romero Y, Ruiz-Romero E, Dendooven L, Bello-López JM. 2014. Identification of antibiotic resistance cassettes in class 1 integrons in Aeromonas spp. strains isolated from fresh fish (Cyprinus carpio L.). Curr Microbiol 68:581–586. doi: 10.1007/s00284-013-0511-6. [DOI] [PubMed] [Google Scholar]
- 43.Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. 2001. Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases. Proc Natl Acad Sci U S A 98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Cross JB, Vreven T, Meroueh SO, Mobashery S, Schlegel HB. 2005. Lysine carboxylation in proteins: OXA-10 beta-lactamase. Proteins 61:246–257. doi: 10.1002/prot.20596. [DOI] [PubMed] [Google Scholar]
- 45.Evans BA, Amyes SG. 2014. OXA beta-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider I, Queenan AM, Bauernfeind A. 2006. Novel carbapenem-hydrolyzing oxacillinase OXA-62 from Pandoraea pnomenusa. Antimicrob Agents Chemother 50:1330–1335. doi: 10.1128/AAC.50.4.1330-1335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santillana E, Beceiro A, Bou G, Romero A. 2007. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc Natl Acad Sci U S A 104:5354–5359. doi: 10.1073/pnas.0607557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith CA, Antunes NT, Stewart NK, Toth M, Kumarasiri M, Chang M, Mobashery S, Vakulenko SB. 2013. Structural basis for carbapenemase activity of the OXA-23 beta-lactamase from Acinetobacter baumannii. Chem Biol 20:1107–1115. doi: 10.1016/j.chembiol.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toth M, Smith CA, Antunes NT, Stewart NK, Maltz L, Vakulenko SB. 2017. The role of conserved surface hydrophobic residues in the carbapenemase activity of the class D β-lactamases. Acta Crystallogr D Struct Biol 73:692–701. doi: 10.1107/S2059798317008671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider KD, Ortega CJ, Renck NA, Bonomo RA, Powers RA, Leonard DA. 2011. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J Mol Biol 406:583–594. doi: 10.1016/j.jmb.2010.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L, Tafi A, Nordmann P, Botta M, Rossolini GM, Mangani S. 2009. Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol 16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 52.De Luca F, Benvenuti M, Carboni F, Pozzi C, Rossolini GM, Mangani S, Docquier JD. 2011. Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D beta-lactamase OXA-10 by rational protein design. Proc Natl Acad Sci U S A 108:18424–18429. doi: 10.1073/pnas.1110530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amos GC, Zhang L, Hawkey PM, Gaze WH, Wellington EM. 2014. Functional metagenomic analysis reveals rivers are a reservoir for diverse antibiotic resistance genes. Vet Microbiol 171:441–447. doi: 10.1016/j.vetmic.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 54.Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, Johnson-Rollings AS, Jones DL, Lee NM, Otten W, Thomas CM, Williams AP. 2013. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis 13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 55.Rowe WPM, Baker-Austin C, Verner-Jeffreys DW, Ryan JJ, Micallef C, Maskell DJ, Pearce GP. 2017. Overexpression of antibiotic resistance genes in hospital effluents over time. J Antimicrob Chemother 72:1617–1623. doi: 10.1093/jac/dkx017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, Galanakis C, Reid-Smith RJ, Tellier PP, Tellis PA, Ziebell K, Manges AR. 2010. Food reservoir for Escherichia coli causing urinary tract infections. Emerg Infect Dis 16:88–95. doi: 10.3201/eid1601.091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernandez J, Stedt J, Bonnedahl J, Molin Y, Drobni M, Calisto-Ulloa N, Gomez-Fuentes C, Astorga-España MS, González-Acuña D, Waldenström J, Blomqvist M, Olsen B. 2012. Human-associated extended-spectrum beta-lactamase in the Antarctic. Appl Environ Microbiol 78:2056–2058. doi: 10.1128/AEM.07320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fróes AM, da Mota FF, Cuadrat RR, Dávila AM. 2016. Distribution and classification of serine beta-lactamases in Brazilian hospital sewage and other environmental metagenomes deposited in public databases. Front Microbiol 7:1790. doi: 10.3389/fmicb.2016.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Y, Zhang T, Zhang XX, Liang DW, Zhang M, Gao DW, Zhu HG, Huang QG, Fang HH. 2012. Quantification and characterization of beta-lactam resistance genes in 15 sewage treatment plants from East Asia and North America. Appl Microbiol Biotechnol 95:1351–1358. doi: 10.1007/s00253-011-3810-5. [DOI] [PubMed] [Google Scholar]
- 60.Barraud O, Casellas M, Dagot C, Ploy MC. 2013. An antibiotic-resistant class 3 integron in an Enterobacter cloacae isolate from hospital effluent. Clin Microbiol Infect 19:E306–E308. doi: 10.1111/1469-0691.12186. [DOI] [PubMed] [Google Scholar]
- 61.Becker EC, Meyer RJ. 1997. Acquisition of resistance genes by the IncQ plasmid R1162 is limited by its high copy number and lack of a partitioning mechanism. J Bacteriol 179:5947–5950. doi: 10.1128/jb.179.18.5947-5950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertram R, Hillen W. 2008. The application of Tet repressor in prokaryotic gene regulation and expression. Microb Biotechnol 1:2–16. doi: 10.1111/j.1751-7915.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krueger F. 2015. Trim Galore!: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Babraham Bioinformatics, Cambridge, United Kingdom: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. [Google Scholar]
- 66.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minogue TD, Daligault HA, Davenport KW, Bishop-Lilly KA, Broomall SM, Bruce DC, Chain PS, Chertkov O, Coyne SR, Freitas T, Frey KG, Gibbons HS, Jaissle J, Redden CL, Rosenzweig CN, Xu Y, Johnson SL. 2014. Complete genome assembly of Escherichia coli ATCC 25922, a serotype O6 reference strain. Genome Announc 2:e00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ren Y, Ren Y, Zhou Z, Guo X, Li Y, Feng L, Wang L. 2010. Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047. J Bacteriol 192:2463–2464. doi: 10.1128/JB.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. 2012. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother 67:906–909. doi: 10.1093/jac/dkr563. [DOI] [PubMed] [Google Scholar]
- 74.Tracz DM, Boyd DA, Hizon R, Bryce E, McGeer A, Ofner-Agostini M, Simor AE, Paton S, Mulvey MR, Canadian Nosocomial Infection Surveillance Program. 2007. ampC gene expression in promoter mutants of cefoxitin-resistant Escherichia coli clinical isolates. FEMS Microbiol Lett 270:265–271. doi: 10.1111/j.1574-6968.2007.00672.x. [DOI] [PubMed] [Google Scholar]
- 75.Datta N, Hedges RW, Shaw EJ, Sykes RB, Richmond MH. 1971. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol 108:1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maniatis T, Fritsch EF, Sambrook J. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 77.Garrec H, Drieux-Rouzet L, Golmard JL, Jarlier V, Robert J. 2011. Comparison of nine phenotypic methods for detection of extended-spectrum beta-lactamase production by Enterobacteriaceae. J Clin Microbiol 49:1048–1057. doi: 10.1128/JCM.02130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsakris A, Poulou A, Pournaras S, Voulgari E, Vrioni G, Themeli-Digalaki K, Petropoulou D, Sofianou D. 2010. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J Antimicrob Chemother 65:1664–1671. doi: 10.1093/jac/dkq210. [DOI] [PubMed] [Google Scholar]
- 79.Mehta SC, Rice K, Palzkill T. 2015. Natural variants of the KPC-2 carbapenemase have evolved increased catalytic efficiency for ceftazidime hydrolysis at the cost of enzyme stability. PLoS Pathog 11:e1004949. doi: 10.1371/journal.ppat.1004949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McKinney DC, Zhou F, Eyermann CJ, Ferguson AD, Prince DB, Breen J, Giacobbe RA, Lahiri S, Verheijen JC. 2015. 4,5-Disubstituted 6-aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles are broad-spectrum serine beta-lactamase inhibitors. ACS Infect Dis 1:310–316. doi: 10.1021/acsinfecdis.5b00031. [DOI] [PubMed] [Google Scholar]
- 81.Kotsakis SD, Miriagou V, Vetouli EE, Bozavoutoglou E, Lebessi E, Tzelepi E, Tzouvelekis LS. 2015. Increased hydrolysis of oximino-beta-lactams by CMY-107, a Tyr199Cys mutant form of CMY-2 produced by Escherichia coli. Antimicrob Agents Chemother 59:7894–7898. doi: 10.1128/AAC.01793-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kotsakis SD, Tzouvelekis LS, Petinaki E, Tzelepi E, Miriagou V. 2011. Effects of the Val211Gly substitution on molecular dynamics of the CMY-2 cephalosporinase: implications on hydrolysis of expanded-spectrum cephalosporins. Proteins 79:3180–3192. doi: 10.1002/prot.23150. [DOI] [PubMed] [Google Scholar]
- 83.Sorin EJ, Pande VS. 2005. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys J 88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. 2005. GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 85.Poirel L, Carattoli A, Bernabeu S, Bruderer T, Frei R, Nordmann P. 2010. A novel IncQ plasmid type harbouring a class 3 integron from Escherichia coli. J Antimicrob Chemother 65:1594–1598. doi: 10.1093/jac/dkq166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.