Combination therapy is a successful approach to treat tuberculosis in patients with susceptible strains of Mycobacterium tuberculosis. However, the emergence of resistant strains requires identification of new, effective therapies.
KEYWORDS: Mycobacterium tuberculosis, acid-phase-growth bacteria, combination therapy, log-phase-growth bacteria, moxifloxacin, nonreplicating-persister-phase-growth bacteria, pretomanid
ABSTRACT
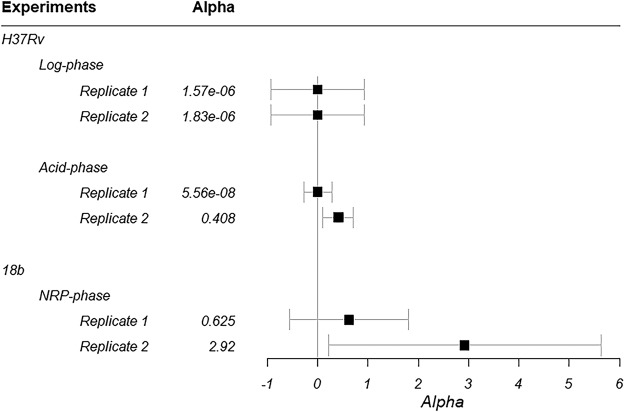
Combination therapy is a successful approach to treat tuberculosis in patients with susceptible strains of Mycobacterium tuberculosis. However, the emergence of resistant strains requires identification of new, effective therapies. Pretomanid (PA824) and moxifloxacin (MXF) are promising options currently under evaluation in clinical trials for the treatment of susceptible and resistant mycobacteria. We applied our recently described screening strategy to characterize the interaction between PA824 and MXF toward the killing of M. tuberculosis in logarithmic growth phase (log phase), acid phase, and nonreplicating-persister (NRP) phase. Respective in vitro data generated for the H37Rv and 18b strains were evaluated in a microdilution plate system containing both drugs in combination. The Universal Response Surface Approach model from Greco et al. (W. R. Greco, G. Bravo, and J. C. Parsons, Pharmacol Rev 47:331–385, 1995) was used to characterize the nature of the interaction between both drugs; synergistic or additive combinations would prompt additional evaluation in the hollow-fiber infection model (HFIM) and in animal studies. The interaction between MXF and PA824 was additive against M. tuberculosis organisms in acid phase (interaction parameter [α] = 5.56e−8 [95% confidence interval {CI} = −0.278 to 0.278] and α = 0.408 [95% CI = 0.105 to 0.711], respectively), NRP phase (α = 0.625 [95% CI = −0.556 to 1.81] and α = 2.92 [95% CI = 0.215 to 5.63], respectively), and log phase (α = 1.57e−6 [95% CI = −0.930 to 0.930] and α = 1.83e−6 [95% CI = −0.929 and 0.929], respectively), prompting further testing of this promising combination for the treatment of tuberculosis in the HFIM and in animal studies.
INTRODUCTION
The traditional treatment of drug-susceptible Mycobacterium tuberculosis takes advantage of combination therapy during both the intensive phase (rifampin [RIF], isoniazid, pyrazinamide, and ethambutol) and the continuation phase (rifampin and isoniazid) (1). The most common in vitro states of M. tuberculosis used to represent this in vivo scenario are the logarithmic growth phase (log phase), the slowly replicating acid-phase growth (acid phase), and the nonreplicating-persister (NRP) phase. The rate of treatment success with drug combination therapy ranges from 90% to as low as 30%, according to the presence of drug-sensitive or drug-resistant strains (1, 2). The increase in drug-resistant strains has added urgency to the search for new drug combinations that promote bacterial kill and suppress the emergence of resistance (2).
One of the recent efforts to improve the treatment of tuberculosis (TB) was based on a clinical trial for evaluation of the combination of rifampin (RIF) and moxifloxacin (MXF) (the REMox trial). The goal of the REMox trial was to shorten the duration of therapy to 4 months in patients with fully susceptible M. tuberculosis by substituting MXF for isoniazid in both the intensive and continuation phases in one of the trial arms. The trial results showed that the combination of RIF and MXF cleared the sputum significantly faster than the time to clearance for the control in the intensive phase but failed to decrease the therapy duration in the continuation phase (3). Those clinical outcomes can be related to in vitro scenarios: the clearance of M. tuberculosis from the sputum in the intensive phase may be related to the killing of bacteria in log phase, while the results in the continuation phase may be related to the killing of slower-growing or nongrowing bacteria. The results from a previous in vitro hollow-fiber infection model (HFIM) study corroborate the clinical trial outcomes, since an antagonistic interaction between those two drugs for the killing of NRP-phase organisms was observed (4). Therefore, identification of new combination regimens should be focused on combinations with clear synergistic or additive effects on bacterial kill and resistance suppression.
MFX and PA824 were chosen for this evaluation because both drugs have promising activity toward rapidly growing (log-phase) organisms (5–7). However, little has been done to evaluate their effect against mycobacteria at acid phase or NRP phase. MXF is a fluoroquinolone licensed by the U.S. Food and Drug Administration for the treatment of community-acquired pneumonia, skin infections, intra-abdominal infections, sinusitis, and chronic bronchitis caused by susceptible microbes (8). Early clinical trials on tuberculosis patients also revealed the utility of MXF as a single drug with an early-phase killing of mycobacteria similar to that of isoniazid (6, 7). Ongoing phase II/III trials are evaluating the safety and efficacy of MXF-containing regimens for the treatment of multidrug-resistant TB (MDR-TB) and extensively resistant TB (XDR-TB) (9). Although its use for early-phase killing is promising, a recent study in mice reported that MXF failed to kill persistent mycobacteria when used as a substitute for isoniazid and ethambutol in standard anti-TB regimens (10). PA824 is a bicyclic nitroimidazole currently under evaluation in a phase II study as part of combination therapy for the treatment of both drug-susceptible and drug-resistant TB (9). In vitro studies have demonstrated the promising activity of PA824 toward both log-phase and NRP-phase bacteria (5).
Therefore, given the promising activity of MXF and PA824 during early-phase trials for the treatment of both susceptible and resistant TB and the activity of PA824 toward NRP-phase bacteria, we decided to evaluate whether use of the combination of these drugs would be an even more promising strategy. We used our recently developed in vitro strategy (11) coupled to the Universal Response Surface Approach (URSA) model of Greco et al. (12) to characterize the nature of the pharmacodynamic interaction between the two drugs. Combinations characterized as additive and synergistic will prompt further evaluation in the HFIM and animal studies.
RESULTS
MIC determination.
The MXF MIC was 0.25 mg/liter for the M. tuberculosis H37Rv and 18b strains in log phase and 0.5 mg/liter for M. tuberculosis H37Rv in acid phase. The PA824 MIC was 0.125 mg/liter for the H37Rv strain in log phase and acid phase and the 18b strain in log phase. Since M. tuberculosis bacteria in the NRP phase do not replicate, the MICs of MXF and PA824 could not be determined for bacteria in this metabolic state.
In vitro drug interaction studies in the plate system.
Estimated values and associated confidence intervals (CI) for the interaction parameter (α) are shown in Fig. 1 for H37Rv in log and acid phases as well as for 18b in NRP phase. All estimates for α were positive. Associated CIs overlapped 0 in all metabolic states in at least one of the replicates. Overall, these results suggest that the interaction between MXF and PA824 against M. tuberculosis is beneficial (additive) in all three metabolic states.
FIG 1.
Estimated values for the interaction parameter (α) for each M. tuberculosis phase. Black boxes, mean estimates; error bars, 95% CI.
Parameter estimates and the associated 95% CIs for the remaining model parameters for each studied strain and phenotype are listed in Table 1. The estimated 50% inhibitory concentration (IC50) values showed that there are significant differences between strains and metabolic states for each drug; however, there were no differences in potency between the two drugs. Both PA824 and MXF were less potent against NRP-phase mycobacteria than against log- and acid-phase organisms.
TABLE 1.
Estimated parameter values from the URSA model of Greco et al. in ADAPT 5 for each strain/phenotypea
| Parameter (drug) | Replicate | Unit | Value (95% CI) for the following strain, phenotype: |
||
|---|---|---|---|---|---|
| H37RV, log phase | H37RV acid phase | 18b NRP | |||
| ECON | 1 | log10 (CFU/ml) | 7.63 (6.70–8.55)b | 6.20 (5.96–6.44) | 4.35 (4.05–4.66) |
| 2 | log10 (CFU/ml) | 7.03 (6.16–7.89)b | 5.84 (5.66–6.02) | 4.44 (4.13–4.74) | |
| IC50,1 (PA824) | 1 | mg/liter | 0.0714 (0.046–0.0968)c | 0.108 (0.0987–0.118)d | 0.445 (0.355–0.534) |
| 2 | mg/liter | 0.0649 (0.0493–0.0805)c | 0.129 (0.122–0.136)d | 0.409 (0.307–0.51) | |
| IC50,2 (MXF) | 1 | mg/liter | 0.0640 (0.0571–0.0708)e | 0.0881 (0.0818–0.0945)d ,f | 0.514 (0.389–0.639) |
| 2 | mg/liter | 0.0662 (0.0559–0.0766)e | 0.121 (0.111–0.130)d | 0.602 (0.449–0.755) | |
| m1 (PA824) | 1 | 1.57 (0.966–2.17)d | 5.55 (3.00–8.1) | 2.37 (1.49–3.25) | |
| 2 | 4.19 (0.649–7.74) | 5.56 (2.46–8.66) | 1.70 (1.15–2.26) | ||
| m2 (MXF) | 1 | 25.08 (−16.8–67.0) | 4.35 (3.62–5.08) | 1.41 (1.05–1.77) | |
| 2 | 14.2 (−6.61–35) | 37.2 (−50–124.4) | 1.46 (1.07–1.85) | ||
CI, 95% confidence interval; IC50,1 and IC50,2, concentration for half-maximal effect for PA824 and MXF, respectively; m1 and m2, Hill constants for PA824 and MXF, respectively; ECON, effect for the control.
P < 0.05 between the H37Rv strain at log phase, the H37Rv strain at acid phase, and the 18b strain at NRP phase.
P < 0.05 between the H37Rv strain at log phase, the H37Rv strain at acid phase, and the 18b strain at NRP phase for PA824.
P < 0.05 between the H37Rv strain at acid phase and the 18b strain at NRP phase for PA824.
P < 0.05 between the H37Rv strain at log phase, the H37Rv strain at acid phase, and the 18b strain at NRP phase for MXF.
P < 0.05 between IC50,1 (PA824) and IC50,2 (MXF).
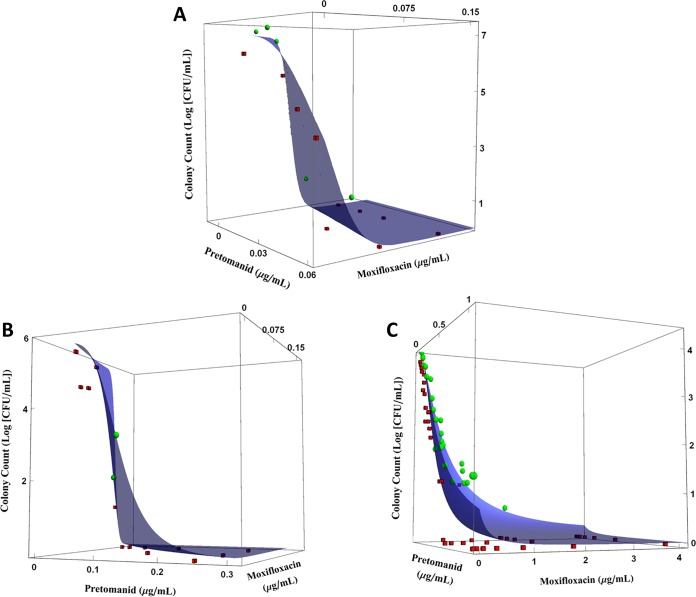
Figure 2 shows the predicted response surface effect of the PA824 and MXF combination on quantitative culture counts for M. tuberculosis strain H37Rv in log and acid phases and strain 18b in NRP phase. With the exception of those in Fig. 2C (strain 18b at NRP phase), the data observations are evenly distributed around the prediction surfaces. With strain 18b in NRP phase, colony counts associated with PA824 concentrations above 0.5 mg/liter are located below the fitted response surface, indicating that bacterial killing is underpredicted for those PA824 concentrations.
FIG 2.
Effect of MXF and PA824 on the total colony counts of M. tuberculosis metabolic states and pH environments according to the URSA model of Greco et al. (12). (A) The H37Rv strain in log-phase growth; (B) the H37Rv strain in acid phase; (C) the 18b strain in the NRP-phase phenotype. Blue surfaces, model predictions; green spheres, observations above the fitted surface; red cubes, observations below the fitted surface. Experiments were performed in duplicate. The plots show the results of a representative replicate.
Mathematical model.
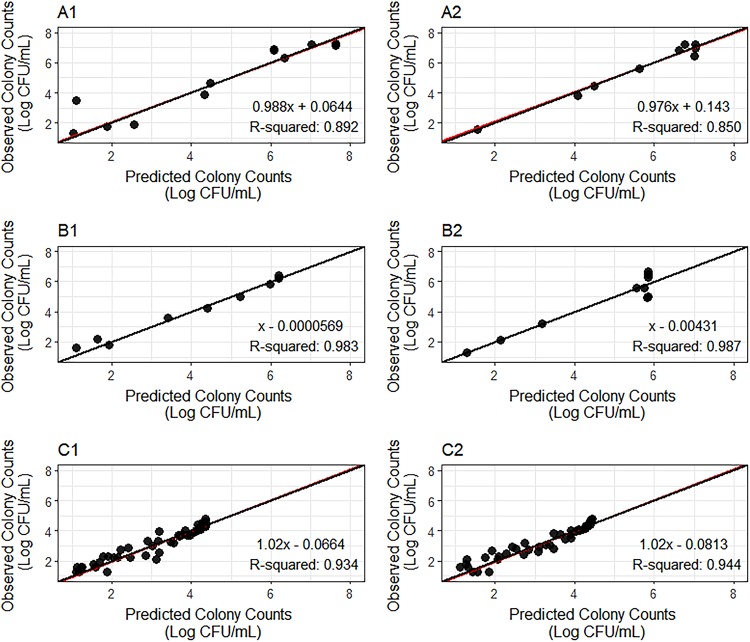
Observed versus predicted colony counts are reported for each replicate of strain H37Rv at log and acid phases and strain 18b at NRP phase (Fig. 3). Observation/prediction pairs are evenly distributed around the unit line. In addition, the regression lines are practically superimposed on the unit lines. All coefficients of determination (R2) are above 0.85, and the slopes of the regression lines range from 0.976 to 1.02. These results indicate that the model adequately describes the data.
FIG 3.
Observed versus predicted colony counts for the H37Rv strain at log phase (A1 and A2) and acid phase (B1 and B2) and for the 18b strain at NRP phase (C1 and C2). Panels 1 and 2 are replicates for the same experiment. Red lines, the regression line over the observed versus model-predicted colony counts; black lines, unity lines; black full circles, observation/prediction pairs.
DISCUSSION
This study aimed to evaluate the combined effect of PA824 and MXF on bacterial killing of the main metabolic states of M. tuberculosis. The drug combination was evaluated using our recently developed screening strategy (11), which consists of evaluating a two-drug combination in a static in vitro 9- by 8-well checkerboard plate system where constant fractions and multiples of the MICs for both compounds were incubated with H37Rv in log phase and acid phase and streptomycin (STR)-starved M. tuberculosis 18b in NRP phase. Characterization of the interaction was performed by estimating the interaction parameter (α) by use of the URSA model of Greco et al. and culture data (12). The surface plots obtained for the effect of combination therapy on strain H37Rv in log and acid phases and strain 18b in NRP phase (Fig. 2) showed no surface mosaicism, with no areas with either underprediction or overprediction surrounded by unaltered areas (i.e., a trough or a peak), which could indicate resistance suppression or amplification of resistance in some wells of the 9- by 8-well plate assay.
Our study findings for α estimates suggest that the effect of the interaction between PA824 and MXF against M. tuberculosis in acid, NRP, and log phases is additive. One replicate for acid phase and one replicate for NRP phase had estimates of α significantly different from 0 (the 95% CIs did not overlap 0). Each of these estimates differed from those for the second replicate. The variability in the model estimates of α and its 95% CI in NRP- and acid-phase mycobacteria reflects the variability in the experimental data at the lowest colony counts. This behavior is expected for data that follow a Poisson distribution, where variability is increased for events with a low incidence rate (i.e., low colony counts). However, the key point of this work was to apply a low-complexity in vitro system, such as the in vitro 9- by 8-well plate assay, where mycobacteria are exposed to fixed concentrations of drugs, in order to preselect promising combinations to be tested in a more complex in vitro system where exposure to drugs mimics the pharmacokinetics in vivo. Our goal was to select a drug combination that could lead to either an additive or a synergistic effect. Therefore, despite the variability in the α estimates for the NRP and acid phases, our results indicate that this drug combination should be selected for further testing in more complex systems. Those results indicate that PA824 and MXF are a promising combination for the killing of M. tuberculosis in NRP phase, while this combination also displays promising activity toward bacteria in the log and acid phases.
This combination will be further evaluated in HFIM, where human pharmacokinetic profiles and exposures can be simulated for PA824 and MXF and the combined effect against both sensitive and resistant strains can be evaluated. Updated estimates of α would be obtained through the Drusano-Greco model. This approach enables the testing of different combinations of doses and, due to the parametric behavior of the data, the simulation of several therapeutic regimens (2). Should additivity and synergy prevail, the combination will be evaluated in preclinical animal models of tuberculosis (13, 14). The parameter estimates shown in Table 1 (IC50,1 and IC50,2) indicate the relative potencies of PA824 and MXF; both compounds were more potent in killing the H37Rv strain in log phase, followed by H37Rv in acid phase. The relative potency of both drugs was the least for strain 18b in NRP phase.
MXF is a fluoroquinolone and acts on bacterial cells by blocking DNA replication and repair through inhibition of the DNA gyrase (topoisomerase II) and DNA topoisomerase IV (only topoisomerase II in M. tuberculosis) enzymes (15, 16). On the other hand, PA824 presents a novel mechanism of action: after activation by the deazaflavin-dependent reductase of M. tuberculosis, PA824 promotes mycobacterial killing through the inhibition of cytochrome/cytochrome oxidase, mediated by the generation of nitric oxide. Furthermore, PA824 also inhibits cell wall synthesis similarly to isoniazid (5, 17, 18). Since PA824 acts as a respiratory poison and inhibits cell wall synthesis and MXF targets DNA replication, it is reasonable to assume that they would have a lower potency against the metabolic phase with the lowest growth/turnover rates, such as the NRP phase in M. tuberculosis (19, 20). This hierarchy of effects was previously observed in the same assay for bedaquiline (11), a drug that affects energy consumption and is therefore expected to present a greater impact on cells at a higher consumption state, such as log phase.
A limitation of this study pertains to the absence of resistance suppression data due to the small bacterial load in each 9- by 8-well plate (4). This aspect will be further evaluated in future HFIM experiments. However, previous studies indicated that the combination of MXF and PA824 might suppress resistance amplification because of their nonoverlapping mechanisms of action (2, 4). The results of combination studies with MXF-RIF as well as linezolid (LZD)-RIF support this hypothesis. While MXF plus RIF led to resistance suppression, RIF plus LZD did not. This observation can be explained by the overlap of the mechanisms of action of RIF and LZD. Both drugs inhibit protein synthesis (RIF via the interaction with RNA polymerase, LZD via inhibition of the ribosomal 30S subunit), whereas this is not an issue for RIF plus MXF (2, 4, 15, 21–24).
In conclusion, we identified the combination of PA824 and MXF to be a promising option for bacterial killing of M. tuberculosis in log, acid, and NRP phases. This combination will be evaluated for resistance suppression in the HFIM.
MATERIALS AND METHODS
Bacteria and generation of metabolic phases.
M. tuberculosis bacteria in log and acid phases were created using strain H37Rv, and M. tuberculosis bacteria in NRP phase were generated with M. tuberculosis 18b. M. tuberculosis 18b is a streptomycin (STR)-resistant, STR auxotroph obtained from a clinical isolate that requires the addition of at least 10 mg/liter of STR to the culture medium to convert it from the NRP state into log-phase growth. Removal of STR promotes its back conversion into the NRP phase (25). Bacterial stocks were stored at −80°C. Bacterial stocks of the H37Rv strain were thawed, and log-phase growth was induced by incubation of the stocks at 37°C in 5% CO2 with shaking in Middlebrook 7H9 broth supplemented with 10% albumin, dextrose, and catalase (ADC) and 0.05% Tween 80 (referred to as TB broth) for 7 to 10 days. The generation of log-phase M. tuberculosis 18b bacteria was performed through incubation of a thawed aliquot in TB broth with STR at 50 mg/liter. The transition of M. tuberculosis 18b into the NRP state was induced by removal of STR through washing of the culture medium thrice by centrifugation with phosphate-buffered saline (PBS) containing 0.05% Tween 80, followed by addition of STR-free TB broth. STR-free 18b cultures were submitted to further incubation at 37°C in 5% CO2 with shaking for 10 days to allow the microbe to transition to the NRP phase. M. tuberculosis bacteria in acid phase were generated by transferring 100 µl of log-phase H37Rv into 40 ml of TB broth at acidic pH (pH 6), followed by additional incubation at 37°C in 5% CO2 for 7 to 10 days to allow the bacteria to convert into acid phase (2, 20, 25, 26).
Drugs.
MXF was purchased from BOC Sciences (Shirley, NY), pharmaceutical PA824 was obtained from TB Alliance (New York, NY), and STR was from Sigma-Aldrich (St. Louis, MO). All compounds were prepared at a concentration of 1 mg/liter and stored according to the manufacturers' instructions. MXF and STR were dissolved in sterile water, and PA824 was dissolved in dimethyl sulfoxide (DMSO). Subsequent dilutions were performed in TB medium. The final concentration of DMSO in the checkerboard plate was 2%.
MIC determination.
H37Rv bacteria in log and acid phases, obtained as described above, were used to determine the microdilution MIC values of PA824 and MXF in broth. Determination of the MIC for MXF and PA824 for each M. tuberculosis strain was performed to establish the range of MICs for both drugs used in our 9- by 8-well microdilution plate assay. The bacterial inoculum was 1 × 102 CFU/well for susceptibility testing with PA824 and 1 × 104 CFU/well for susceptibility testing with MXF. Each drug was incubated separately in round-bottom wells with drugs at concentrations within geometric 2-fold dilutions. The TB broth was adjusted to pH 6 for studies with acid-phase M. tuberculosis. For PA824 and MXF, the MICs were read after 14 days and 21 days of incubation, respectively, at 37°C in 5% CO2. The MIC was defined as the lowest concentration that resulted in no visible growth.
In vitro drug interaction studies in the plate system.
M. tuberculosis bacteria in the log and acid phases were inoculated at 5 × 102 CFU/well, while M. tuberculosis bacteria in the NRP phase (prepared as described above) were inoculated at 5 × 103 CFU/well into 96-well round-bottom microdilution plates (Falcon; Corning, Corning, NY). A combination of no drug or serial 2-fold increments of MXF (as 0, 0.25, 0.5, 1, 2, 4, 8, 16, and 32× MIC) and PA824 (as 0, 0.125, 0.25, 0.50, 1, 2, 4, and 8× MIC) as a single drug and drugs in combination were added to the microdilution plates in a 9- by 8-well matrix. Due to the impossibility of determining the MICs for M. tuberculosis strain 18b in the NRP state, we used the MICs generated from 18b in log-phase growth to establish a range of MXF and PA824 concentrations to be tested. Following 21 days of incubation with the antibiotic(s) for the log- and acid-phase phenotypes and 14 days for the NRP metabolic state, the microdilution plates containing the M. tuberculosis suspensions were washed twice with PBS plus 0.05% Tween 80 to remove drug carryover, and the contents of each well were quantitatively cultured onto 7H10 agar supplemented with 10% oleic acid, albumin, dextrose, and catalase (OADC) and 0.05% Tween 80 for log-phase and acid-phase M. tuberculosis and 7H10 agar supplemented with 10% OADC, 0.05% Tween 80, and 75 mg/liter STR for NRP-phase strain 18b. The colonies were counted after 4 weeks of incubation at 37°C in 5% CO2 (2, 20, 25, 26).
Mathematical model.
Data from the quantitative culture counts at log, acid, and NRP phases were analyzed using the Universal Response Surface Approach (URSA) equation of Greco and colleagues (12) in ADAPT 5 (27), as described previously, with the weighted least-squares estimation method (11):
where Drug1 and Drug2 represent the PA824 and MXF concentrations, respectively; IC50,1 and IC50,2 are the concentrations of PA824 and MXF that lead to half-maximal effects, respectively; m1 and m2 are the respective Hill coefficients; ECON represents the effect for the control; α is the interaction parameter; and E is the fractional effect. Characterization of the drug combination as additive, synergistic, and antagonistic was performed through estimation of the α interaction value and its associated confidence interval. The combined effect is characterized as additive if α and its 95% confidence interval include 0, as synergistic if α and its 95% confidence interval are positive and do not include 0, and as antagonistic if α and its 95% confidence interval are negative and do not include 0. Diagnostic plots were generated using the Mathematica (v.11.1; Wolfram Research Inc., Champaign, IL) and R (The R Foundation, R Core Team) programs.
ACKNOWLEDGMENTS
This study was funded by P01 grant AI123036 from NIAID.
We declare no conflict of interest.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.World Health Organization. 2017. Diagnosis and treatment: TB, HIV-associated TB and drug-resistant TB, p 63–96. In Global tuberculosis report 2017. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/en/. Accessed 23 March 2018. [Google Scholar]
- 2.Drusano GL, Neely M, Van Guilder M, Schumitzky A, Brown D, Fikes S, Peloquin C, Louie A. 2014. Analysis of combination drug therapy to develop regimens with shortened duration of treatment for tuberculosis. PLoS One 9:e101311. doi: 10.1371/journal.pone.0101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillespie SH, Crook AM, McHugh TD, Mendel CM, Meredith SK, Murray SR, Pappas F, Phillips PPJ, Nunn AJ. 2014. Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N Engl J Med 371:1577–1587. doi: 10.1056/NEJMoa1407426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drusano GL, Sgambati N, Eichas A, Brown DL, Kulawy R, Louie A. 2010. The combination of rifampin plus moxifloxacin is synergistic for suppression of resistance but antagonistic for cell kill of Mycobacterium tuberculosis as determined in a hollow-fiber infection model. mBio 1:e00139-10. doi: 10.1128/mBio.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stover CK, Warrener P, VanDevanter DR, Sherman DR, Arain TM, Langhorne MH, Anderson SW, Towell JA, Yuan Y, McMurray DN, Kreiswirth BN, Barry CE, Baker WR. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962–9660. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 6.Gosling RD, Uiso LO, Sam NE, Bongard E, Kanduma EG, Nyindo M, Morris RW, Gillespie SH. 2003. The bactericidal activity of moxifloxacin in patients with pulmonary tuberculosis. Am J Respir Crit Care Med 168:1342–1345. doi: 10.1164/rccm.200305-682OC. [DOI] [PubMed] [Google Scholar]
- 7.Pletz MW, De Roux A, Roth A, Neumann KH, Mauch H, Lode H. 2004. Early bactericidal activity of moxifloxacin in treatment of pulmonary tuberculosis: a prospective, randomized study. Antimicrob Agents Chemother 48:780–782. doi: 10.1128/AAC.48.3.780-782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Food and Drug Administration, Center for Drug Evaluation and Research. 2016. NDA 21-085. Food and Drug Administration, Silver Spring, MD: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/021085s061s062,021277s057s058lbl.pdf Accessed 3 July 2018. [Google Scholar]
- 9.World Health Organization. 2017. TB research and development, p 137–147. In Global tuberculosis report 2017. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/en/ Accessed 3 July 2018. [Google Scholar]
- 10.Liu Y, Pertinez H, Davies GR, Gillespie SH, Coates AR, Hu Y. 2018. Moxifloxacin replacement in contemporary tuberculosis drug regimens is ineffective against persistent Mycobacterium tuberculosis in the Cornell mouse model. Antimicrob Agents Chemother 62:e00190-18. doi: 10.1128/AAC.00190-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Miranda Silva C, Hajihosseini A, Myrick J, Nole J, Louie A, Schmidt S, Drusano GL. 2018. Effect of linezolid plus bedaquiline against Mycobacterium tuberculosis in log phase, acid phase and nonreplicating-persister (NRP) phase in an in vitro assay. Antimicrob Agents Chemother 62:e00856-18. doi: 10.1128/AAC.00856-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greco WR, Bravo G, Parsons JC. 1995. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev 47:331–385. [PubMed] [Google Scholar]
- 13.Balasubramanian V, Solapure S, Gaonkar S, Mahesh Kumar KN, Shandil RK, Deshpande A, Kumar N, Vishwas KG, Panduga V, Reddy J, Ganguly S, Louie A, Drusano GL. 2012. Effect of coadministration of moxifloxacin and rifampin on Mycobacterium tuberculosis in a murine aerosol infection model. Antimicrob Agents Chemother 56:3054–3057. doi: 10.1128/AAC.06383-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White AG, Maiello P, Coleman MT, Tomko JA, Frye LJ, Scanga CA, Lin PL, Flynn JL. 2017. Analysis of 18FDG PET/CT imaging as a tool for studying Mycobacterium tuberculosis infection and treatment in non-human primates. J Vis Exp 2017:56375. doi: 10.3791/56375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willmott CJ, Critchlow SE, Eperon IC, Maxwell A. 1994. The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J Mol Biol 242:351–363. doi: 10.1006/jmbi.1994.1586. [DOI] [PubMed] [Google Scholar]
- 16.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 17.Manjunatha U, Boshoff HIM, Barry IIICE. 2009. The mechanism of action of PA-824: novel insights from transcriptional profiling. Commun Integr Biol 2:215–218. doi: 10.4161/cib.2.3.7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, Kang S, Keller TH, Jiricek J, Barry CE III.. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barry CE III, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol 7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sala C, Dhar N, Hartkoorn RC, Zhang M, Ha YH, Schneider P, Cole ST. 2010. Simple model for testing drugs against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 54:4150–4158. doi: 10.1128/AAC.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiasa H, Yousef D, Marians K. 1996. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J Biol Chem 271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- 22.Shea ME, Hiasa H. 1999. Interactions between DNA helicases and frozen topoisomerase IV-quinolone-DNA ternary complexes. J Biol Chem 274:22747–22754. doi: 10.1074/jbc.274.32.22747. [DOI] [PubMed] [Google Scholar]
- 23.Wehrli W. 1983. Rifampin: mechanisms of action and resistance. Rev Infect Dis 5:S407–S411. doi: 10.1093/clinids/5.Supplement_3.S407. [DOI] [PubMed] [Google Scholar]
- 24.Swaney SM, Aoki H, Ganoza MC, Shinabarger D. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother 42:3251–3255. doi: 10.1128/AAC.42.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M, Sala C, Hartkoorn RC, Dhar N, Mendoza-Losana A, Cole ST. 2012. Streptomycin-starved Mycobacterium tuberculosis 18b, a drug discovery tool for latent tuberculosis. Antimicrob Agents Chemother 56:5782–5789. doi: 10.1128/AAC.01125-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gumbo T, Dona CS, Meek C, Leff R. 2009. Pharmacokinetics-pharmacodynamics of pyrazinamide in a novel in vitro model of tuberculosis for sterilizing effect: a paradigm for faster assessment of new antituberculosis drugs. Antimicrob Agents Chemother 53:3197–3204. doi: 10.1128/AAC.01681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Argenio DZ, Schumitzky A, Wang X. 2009. ADAPT 5 user’s guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles, CA. [Google Scholar]