Although the Sensititre Yeast-One (SYO) and Etest methods are widely utilized, interpretive criteria are not available for triazole susceptibility testing of Candida or Aspergillus species. We collected fluconazole, itraconazole, posaconazole, and voriconazole SYO and Etest MICs from 39 laboratories representing all continents for (method/agent-dependent) 11,171 Candida albicans, 215 C. dubliniensis, 4,418 C. glabrata species complex, 157 C. guilliermondii (Meyerozyma guilliermondii), 676 C. krusei (Pichia kudriavzevii), 298 C. lusitaniae (Clavispora lusitaniae), 911 C. parapsilosis sensu stricto, 3,691 C. parapsilosis species complex, 36 C. metapsilosis, 110 C. orthopsilosis, 1,854 C. tropicalis, 244 Saccharomyces cerevisiae, 1,409 Aspergillus fumigatus, 389 A. flavus, 130 A. nidulans, 233 A. niger, and 302 A. terreus complex isolates.
KEYWORDS: Aspergillus spp., Candida glabrata, Candida albicans, Etest MICs for fungal mutants, Etest method ECVs, SYO MICs for fungal mutants, SYO method ECVs, antifungal resistance, triazole ECVs
ABSTRACT
Although the Sensititre Yeast-One (SYO) and Etest methods are widely utilized, interpretive criteria are not available for triazole susceptibility testing of Candida or Aspergillus species. We collected fluconazole, itraconazole, posaconazole, and voriconazole SYO and Etest MICs from 39 laboratories representing all continents for (method/agent-dependent) 11,171 Candida albicans, 215 C. dubliniensis, 4,418 C. glabrata species complex, 157 C. guilliermondii (Meyerozyma guilliermondii), 676 C. krusei (Pichia kudriavzevii), 298 C. lusitaniae (Clavispora lusitaniae), 911 C. parapsilosis sensu stricto, 3,691 C. parapsilosis species complex, 36 C. metapsilosis, 110 C. orthopsilosis, 1,854 C. tropicalis, 244 Saccharomyces cerevisiae, 1,409 Aspergillus fumigatus, 389 A. flavus, 130 A. nidulans, 233 A. niger, and 302 A. terreus complex isolates. SYO/Etest MICs for 282 confirmed non-wild-type (non-WT) isolates were included: ERG11 (C. albicans), ERG11 and MRR1 (C. parapsilosis), cyp51A (A. fumigatus), and CDR2 and CDR1 overexpression (C. albicans and C. glabrata, respectively). Interlaboratory modal agreement was superior by SYO for yeast species and by the Etest for Aspergillus spp. Distributions fulfilling CLSI criteria for epidemiological cutoff value (ECV) definition were pooled, and we proposed SYO ECVs for S. cerevisiae and 9 yeast and 3 Aspergillus species and Etest ECVs for 5 yeast and 4 Aspergillus species. The posaconazole SYO ECV of 0.06 µg/ml for C. albicans and the Etest itraconazole ECV of 2 µg/ml for A. fumigatus were the best predictors of non-WT isolates. These findings support the need for method-dependent ECVs, as, overall, the SYO appears to perform better for susceptibility testing of yeast species and the Etest appears to perform better for susceptibility testing of Aspergillus spp. Further evaluations should be conducted with more Candida mutants.
INTRODUCTION
The triazoles (fluconazole, isavuconazole, itraconazole, posaconazole, and voriconazole) are the current treatments for severe candidiasis and aspergillosis (e.g., as first-line or prophylactic, adjunctive, empirical, transition from another agent, or salvage therapies) (1–3). These fungal infections may cause elevated levels of morbidity and mortality among immunocompromised patients (3–5). The impact of azole resistance and its prevalence has been widely recognized, and various mechanisms of mutational resistance have been elucidated in the four most common species of Candida, especially in Candida albicans, and in Aspergillus fumigatus (6–10). In most Candida isolates, azole resistance (or unusually high or increased MICs) are mostly associated with two main molecular mechanisms, among others: an increase (overexpression) of the azole target azole sterol demethylase or alterations (amino acid substitutions) in either the gene ERG11, as the enzyme is encoded during the fungal ergosterol biosynthesis pathway, or the MRR1 transcriptional regulator (6, 8, 9). However, in the case of C. glabrata, azole resistance has frequently been related to the overexpression or alteration of the PDR1 gene, which regulates efflux pumps (7). On the other hand, the main azole resistance mechanism in A. fumigatus is due to alterations of the cyp51A gene (10).
Azole susceptibility testing (yielding MICs) is recommended for all bloodstream and other clinically relevant Candida isolates (1). Although routine MIC determination for Aspergillus species isolates is not usually recommended during initial aspergillosis therapy, MICs have an important role in identifying potentially resistant isolates, e.g., isolates from patients failing therapy (2). There are several antifungal susceptibility methods for the determination of MICs for isolates of both Candida and Aspergillus, including the broth microdilution M27 and M38 reference methods by the Clinical and Laboratory Standards Institute (CLSI) (11, 12) and the Antifungal Subcommittee of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (13) (http://www.eucast.org/ast_of_fungi/). In addition, the colorimetric broth microdilution Sensititre Yeast-One (SYO; Trek Diagnostic System, Cleveland, OH) method as well as the agar diffusion Etest (bioMérieux, Marcy l’Etoile, France) method, among other commercial assays, are widely utilized for antifungal susceptibility testing in the clinical laboratory; these methods are more practical and less time-consuming for routine use (14–16).
The objective of earlier studies evaluating the performance of the SYO and Etest methods involved the comparison of azole MICs obtained by these methods with those obtained by the reference assays for prevalent species of Candida and Aspergillus (17–19). Some of those early studies also evaluated the agreement on the ranking of isolates within existent categorical endpoints with little attention to the critical issue of interlaboratory reproducibility. Recently, triazole MIC data for A. fumigatus and C. glabrata mutant strains obtained by these commercial methods have been reported (20–24). However, a lack of suitable clinical data has precluded the establishment of breakpoints (BPs) for the categorical interpretation of triazole MICs for either Candida or Aspergillus spp. by these two methods. Therefore, both assays rely on available CLSI BPs for Candida spp. as interpretive categories as well as for quality control (QC) (14, 16). The proposal of SYO/Etest epidemiological cutoff values (ECVs) for susceptibility testing of either Candida or Aspergillus isolates with amphotericin B or the echinocandins has revealed substantial method-dependent differences between some of those values, despite the regulatory requirement to show equivalence to the reference method before marketing (25, 26). Those results emphasize the need to establish method-dependent triazole ECVs for these two widely used commercial methods for testing the susceptibility of Candida and Aspergillus isolates to the triazoles in the clinical laboratory.
For the last 2 years, we have gathered available triazole MICs obtained by both the SYO and Etest assays for isolates of prevalent and nonprevalent yeast species (C. albicans, C. dubliniensis, C. glabrata species complex, C. guilliermondii [Meyerozyma guilliermondii], C. krusei [Pichia kudriavzevii], C. lusitaniae [Clavispora lusitaniae], and the C. parapsilosis species complex [including C. parapsilosis sensu stricto, C. orthopsilosis, and C. tropicalis]), Saccharomyces cerevisiae, and five Aspergillus species complexes (A. fumigatus [including A. fumigatus sensu stricto], A. flavus, A. nidulans, A. niger, and A. terreus). Additional SYO MIC distributions for the less prevalent or common yeast species C. famata (Debaryomyces hansenii), C. kefyr (Kluyveromyces marxianus), and C. metapsilosis were also reported when they originated from at least three laboratories and had comparable modes. From here on we use the most common clinical names. These triazole MICs were submitted from 39 independent worldwide laboratories (method/agent/species dependent) in order (i) to define the MIC distributions obtained by each commercial susceptibility testing method/agent and species; (ii) to examine the suitability of these distributions for the ECV setting, including the evaluation of interlaboratory modal agreement; and (iii) to define ECVs for each species/agent/method that fulfilled the CLSI criteria for ECV definition (modal compatibility among the laboratories, at least 100 MICs for each species/method/agent that originated in ≥3 independent laboratories) using the iterative statistical method at the 97.5% cutoff value (27–29) or the second numerical derivative method when the putative wild-type (WT) mode was at the lowest concentration in the distribution (30).
Although the majority of the isolates evaluated were not assessed for mechanisms of resistance, we also collected MIC data for 282 known or confirmed mutants (non-wild type [non-WT]) obtained by both methods, as follows: SYO and Etest MICs for C. albicans (ERG11), SYO MICs for C. parapsilosis (ERG11, MRR1) mutants and/or strains with overexpression of the CDR2 gene, SYO MICs for C. glabrata strains with overexpression of the CDR1 gene, and SYO and Etest MICs for A. fumigatus sensu stricto strains harboring cyp51A mutations. These data were submitted mostly from European laboratories as well as from Argentina, Thailand, and South Africa and one published Etest study (20). SYO data for 58 C. glabrata PDR gene mutants also were submitted, but those data were not included due to the large modal variability for the nonmutants compared with the global modes.
RESULTS AND DISCUSSION
Antimicrobial susceptibility testing for clinical isolates is most useful when either method- and species-dependent BPs or ECVs are available for the isolate and agent evaluated. The BP categorizes the isolate as either susceptible or resistant, and the ECV categorizes the isolate as either wild type (WT; no detectable phenotypic resistance) or non-WT (more likely harboring resistance mechanisms) (27). Since ECVs are based solely on in vitro data (either MIC or minimal effective concentration [MEC] results), classification of an isolate as a presumptive WT cannot directly predict a successful therapeutic outcome. Classification of an isolate as a non-WT indicates that it could harbor acquired mechanisms of resistance to the agent being evaluated and would less likely respond to contemporary therapy (27). However, the putative mechanism of resistance would not necessarily be known in order to categorize a strain as non-WT. CLSI BPs are based on in vitro and clinical data, genetic mechanisms of resistance, as well as pharmacokinetic/pharmacodynamics (PK/PD) parameters (27, 28). EUCAST ECVs and BPs are based on MIC distributions and PK/PD parameters (http://www.eucast.org/ast_of_fungi/). Therefore, when the BP is available for the isolate and agent being evaluated, that is the value that should be used. To our knowledge, method-dependent SYO or Etest ECVs or BPs for the four triazoles evaluated have not been proposed for the categorization of Candida or Aspergillus isolates. Our ECVs were defined following the criteria recently published by the CLSI (27). They were based on either SYO or Etest triazole MIC distributions that originated from 3 to 30 (SYO) or 3 to 11 (Etest) laboratories (species and agent dependent) (Tables 1 to 4) (27). As mentioned before, SYO MICs were submitted from multiple laboratories for the following mutants: 59 C. albicans ERG11 (4 laboratories) and 39 A. fumigatus sensu stricto cyp51A (5 laboratories) mutants. Etest MICs were submitted from multiple laboratories for 81 A. fumigatus cyp51A (7 laboratories and one published study [20]) mutants (Tables 1, 2, and 5). SYO MICs were received from single laboratories for the following mutants: 13 C. glabrata and 2 C. albicans mutants with overexpression of the CDR1 and CDR2 genes, respectively, and 78 C. parapsilosis mutants (49 ERG11 mutants and 29 MRR1 mutants). Etest MICs were received from single laboratories for 10 C. albicans (ERG11) mutants (not listed in Table 1, 2, or 5). The MICs for these confirmed mutants provided a preliminary assessment of the utility of our proposed ECVs in recognizing the non-WT strains. Therefore, since BPs are not available for these commercial methods, the ECVs proposed in the present study could help the clinician and laboratory personnel in identifying isolates with possible acquired resistance mechanisms or could be useful for surveillance or epidemiological studies.
TABLE 1.
SYO pooled triazole MIC distributions for species of Candida, Saccharomyces, and Aspergillusa
| Agent and speciesb | No. of isolates | No. of labs used/total no.c | No. of isolates with MIC (µg/ml) ofd: | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fluconazole | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ≥128 | ||
| C. albicans | 11,171 | 28/30 | 12 | 1,016 | 4,252 | 4,152 | 978 | 238 | 122 | 82 | 78 | 33 | 49 | 159 |
| Confirmed ERG11 mutants | 59 | 4/4 | 3 | 1 | 4 | 3 | 1 | 2 | 2 | 43 | ||||
| C. dubliniensis | 195 | 7/10 | 48 | 64 | 57 | 13 | 6 | 3 | 2 | 1 | 1 | |||
| C. famata | 23 | 3/6 | 1 | 2 | 11 | 5 | 3 | 1 | ||||||
| C. glabrata | 4,418 | 30/30 | 13 | 9 | 23 | 64 | 152 | 375 | 1,049 | 1,330 | 691 | 216 | 496 | |
| C. guilliermondii | 153 | 8/13 | 2 | 1 | 6 | 20 | 36 | 46 | 19 | 10 | 6 | 4 | 3 | |
| C. kefyr | 55 | 3/4 | 13 | 25 | 15 | 2 | ||||||||
| C. krusei | 537 | 15/16 | 1 | 1 | 1 | 3 | 8 | 43 | 193 | 220 | 67 | |||
| C. lusitaniae | 298 | 12/12 | 16 | 41 | 75 | 99 | 43 | 12 | 5 | 1 | 4 | 1 | 1 | |
| C. parapsilosis | 3,691 | 28/30 | 89 | 502 | 1,210 | 958 | 421 | 221 | 151 | 82 | 27 | 19 | 11 | |
| C. parapsilosis sensu stricto | 911 | 5/5 | 18 | 118 | 282 | 216 | 121 | 74 | 53 | 19 | 6 | 4 | ||
| C. metapsilosis | 36 | 4/4 | 1 | 2 | 17 | 10 | 5 | 1 | ||||||
| C. orthopsilosis | 110 | 5/5 | 3 | 4 | 29 | 43 | 15 | 8 | 4 | 2 | 1 | 1 | ||
| C. tropicalis | 1,854 | 24/28 | 20 | 82 | 270 | 701 | 482 | 129 | 53 | 19 | 24 | 14 | 60 | |
| S. cerevisiae | 244 | 3/3 | 4 | 3 | 9 | 40 | 70 | 76 | 26 | 10 | 4 | 2 | ||
| Itraconazole | 0.008 | 0.01 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | ||
| C. albicans | 7,843 | 27/30 | 69 | 995 | 2,696 | 2,754 | 905 | 164 | 77 | 27 | 14 | 7 | 9 | 126 |
| Confirmed ERG11 mutants | 59 | 4/4 | 2 | 2 | 2 | 5 | 11 | 3 | 7 | 4 | 23 | |||
| C. dubliniensis | 125 | 6/8 | 13 | 21 | 47 | 27 | 7 | 2 | 5 | 1 | 2 | |||
| C. famata | 18 | 3/5 | 1 | 1 | 2 | 3 | 7 | 3 | 1 | |||||
| C. glabrata | 3,594 | 29/30 | 12 | 19 | 42 | 112 | 428 | 1,335 | 910 | 195 | 71 | 26 | 444 | |
| C. guilliermondii | 149 | 9/13 | 3 | 8 | 31 | 55 | 37 | 10 | 2 | 3 | ||||
| C. kefyr | 45 | 3/3 | 5 | 10 | 17 | 12 | 1 | |||||||
| C. krusei | 574 | 13/16 | 4 | 3 | 14 | 69 | 283 | 156 | 33 | 2 | 1 | 9 | ||
| C. lusitaniae | 171 | 8/11 | 11 | 12 | 52 | 60 | 28 | 7 | 1 | |||||
| C. parapsilosis | 3,353 | 23/30 | 209 | 570 | 1,098 | 1,150 | 252 | 59 | 13 | 1 | 1 | |||
| C. parapsilosis sensu stricto | 730 | 4/5 | 68 | 79 | 237 | 254 | 83 | 6 | 3 | |||||
| C. metapsilosis | 32 | 4/4 | 3 | 3 | 12 | 11 | 1 | 2 | ||||||
| C. orthopsilosis | 88 | 3/4 | 2 | 13 | 35 | 26 | 12 | |||||||
| C. tropicalis | 1,399 | 23/29 | 14 | 51 | 138 | 513 | 508 | 126 | 16 | 4 | 1 | 5 | 23 | |
| S. cerevisiae | 41 | 3/3 | 1 | 1 | 3 | 21 | 11 | 2 | 2 | |||||
| A. niger | 233 | 6/7 | 18 | 23 | 48 | 69 | 44 | 17 | 6 | 1 | 7 | |||
| Posaconazole | 0.008 | 0.01 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | ||
| C. albicans | 6,729 | 27/30 | 596 | 2,768 | 2,318 | 587 | 175 | 96 | 56 | 32 | 10 | 3 | 60 | 28 |
| Confirmed ERG11 mutants | 59 | 1 | 3 | 1 | 4 | 9 | 8 | 7 | 1 | 1 | 24 | |||
| C. dubliniensis | 185 | 7/8 | 35 | 56 | 63 | 25 | 4 | 2 | ||||||
| C. glabrata | 2,999 | 25/29 | 4 | 5 | 28 | 39 | 50 | 153 | 590 | 1,145 | 579 | 62 | 251 | 93 |
| C. guilliermondii | 111 | 9/12 | 3 | 1 | 9 | 15 | 27 | 35 | 18 | 3 | ||||
| C. kefyr | 40 | 3/3 | 7 | 13 | 13 | 5 | 2 | |||||||
| C. krusei | 562 | 13/15 | 1 | 1 | 3 | 20 | 90 | 264 | 151 | 25 | 5 | 2 | ||
| C. lusitaniae | 172 | 11/11 | 17 | 49 | 58 | 36 | 10 | 1 | 1 | |||||
| C. parapsilosis | 3,085 | 26/30 | 136 | 538 | 1,091 | 915 | 297 | 86 | 11 | 7 | 1 | 1 | 2 | |
| C. parapsilosis sensu stricto | 670 | 5/5 | 40 | 127 | 206 | 193 | 69 | 31 | 2 | 2 | ||||
| C. metapsilosis | 17 | 3/4 | 3 | 7 | 4 | 3 | ||||||||
| C. orthopsilosis | 30 | 3/4 | 5 | 14 | 7 | 4 | ||||||||
| C. tropicalis | 1,366 | 23/29 | 16 | 50 | 107 | 250 | 408 | 336 | 147 | 22 | 6 | 17 | 7 | |
| S. cerevisiae | 41 | 3/3 | 3 | 6 | 20 | 9 | 2 | 1 | ||||||
| Voriconazole | 0.008 | 0.01 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | ||
| C. albicans | 8,747 | 29/30 | 5,947 | 1,691 | 481 | 222 | 111 | 82 | 47 | 22 | 10 | 15 | 76 | 43 |
| Confirmed ERG11 mutants | 59 | 4/4 | 3 | 4 | 4 | 1 | 4 | 1 | 7 | 6 | 3 | 5 | 2 | 19 |
| C. dubliniensis | 215 | 7/9 | 182 | 21 | 5 | 3 | 1 | 2 | 1 | |||||
| C. famata | 25 | 3/5 | 5 | 10 | 4 | 2 | 2 | 2 | ||||||
| C. glabrata | 3,255 | 24/30 | 23 | 29 | 65 | 189 | 486 | 911 | 824 | 340 | 136 | 156 | 82 | 14 |
| C. guilliermondii | 157 | 11/12 | 8 | 10 | 32 | 46 | 34 | 10 | 11 | 4 | 1 | 1 | ||
| C. kefyr | 55 | 3/3 | 46 | 8 | 1 | |||||||||
| C. krusei | 676 | 14/16 | 2 | 1 | 1 | 16 | 108 | 291 | 199 | 42 | 11 | 3 | 1 | 1 |
| C. lusitaniae | 248 | 11/12 | 120 | 70 | 32 | 15 | 4 | 1 | 2 | 4 | ||||
| C. parapsilosis | 2,670 | 26/30 | 1,213 | 695 | 364 | 210 | 103 | 50 | 16 | 10 | 9 | |||
| Voriconazole | 0.008 | 0.01 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | ||
| C. parapsilosis sensu stricto | 718 | 5/5 | 261 | 185 | 122 | 80 | 47 | 12 | 5 | 4 | 2 | |||
| C. metapsilosis | 30 | 3/4 | 2 | 10 | 11 | 4 | 2 | 1 | ||||||
| C. orthopsilosis | 20 | 3/4 | 1 | 8 | 3 | 6 | 2 | |||||||
| C. tropicalis | 1,637 | 19/28 | 45 | 92 | 227 | 466 | 443 | 200 | 70 | 25 | 20 | 9 | 23 | 17 |
| S. cerevisiae | 41 | 3/3 | 1 | 3 | 17 | 15 | 2 | 2 | 1 | |||||
| A. fumigatus | 903 | 8/8 | 2 | 7 | 35 | 64 | 157 | 396 | 179 | 33 | 8 | 7 | 7 | 8 |
| Confirmed cyp51A mutants | 39 | 5/5 | 3 | 4 | 8 | 3 | 4 | 5 | 2 | 10 | ||||
| A. flavus | 389 | 6/7 | 5 | 1 | 14 | 32 | 89 | 139 | 59 | 29 | 16 | 1 | 0 | 4 |
| A. niger | 74 | 3/6 | 1 | 9 | 19 | 33 | 12 | |||||||
| A. terreus | 302 | 5/6 | 6 | 5 | 16 | 19 | 48 | 122 | 69 | 15 | 2 | |||
Including the complexes of C. glabrata, C. parapsilosis, and Aspergillus spp. (the cyp51A mutants are A. fumigatus sensu stricto).
Newly accepted taxonomic names or reclassifications are as follows: C. famata (D. hansenii), C. guilliermondii (M. guilliermondii), C. kefyr (K. marxianus), C. krusei (P. kudriavzevii), and Candida lusitaniae (Clavispora lusitaniae).
Total number of laboratories included in the ECV definition pool/total number of laboratories that submitted data.
Data are from between 3 and 30 laboratories and were determined by the colorimetric broth microdilution SYO method (14); the highest number in each row (showing the most frequent, or mode, MIC) is in bold.
TABLE 4.
Method-dependent Etest ECOFFinder ECVs of four triazoles for species of Candida and Aspergillusa
| Agent and speciesb | No. of isolates | No. of labs used/total no.c | MIC (µg/ml) |
ECV (µg/ml)d | |
|---|---|---|---|---|---|
| Range | Mode | ||||
| Fluconazole | |||||
| C. glabrata | 356 | 7/10 | 0.12 to ≥128 | 8 | 64 |
| C. parapsilosis | 639 | 9/9 | 0.03 to ≥128 | 0.5 | 4 |
| C. tropicalis | 368 | 9/10 | 0.03 to ≥128 | 1 | 4 |
| Itraconazole | |||||
| C. albicans | 975 | 8/9 | ≤0.004 to ≥16 | 0.06 | 0.25 |
| C. krusei | 101 | 3/3 | 0.03 to ≥16 | 0.5 | 2 |
| C. tropicalis | 165 | 5/8 | ≤0.004 to 8 | 0.03 | 0.5 |
| A. fumigatus | 1,112 | 10/10 | 0.008 to ≥16 | 0.5 | 2 |
| A. flavus | 250 | 7/8 | 0.01 to 2 | 0.25 | 1 |
| A. nidulans | 130 | 4/4 | 0.01 to 8 | 0.12 | 1 |
| A. niger | 176 | 4/5 | 0.03 to ≥16 | 1 | 4 |
| Posaconazolec | |||||
| C. albicans | 305 | 4/6 | ≤0.004 to 1 | 0.03 | 0.12 |
| C. parapsilosis | 162 | 4/5 | ≤0.004 to ≥16 | 0.01 | 0.12 |
| C. tropicalis | 101 | 4/5 | 0.008 to 8 | 0.03 | 0.12 |
| A. flavus | 204 | 7/7 | 0.01 to 1 | 0.25 | 0.5 |
| A. niger | 168 | 4/5 | 0.03 to 1 | 0.25 | 0.5 |
| A. terreus | 194 | 5/5 | 0.03 to 4 | 0.12 | 0.25 |
| Voriconazole | |||||
| C. albicans | 2,159 | 8/11 | ≤0.004 to ≥16 | 0.008 | 0.03 |
| C. glabrata | 551 | 8/9 | 0.008 to ≥16 | 0.25 | 2 |
| C. krusei | 130 | 6/6 | 0.01 to 8 | 0.5 | 2 |
| C. parapsilosis | 506 | 7/9 | ≤0.004 to ≥16 | 0.06 | 0.25 |
| C. tropicalis | 260 | 6/10 | ≤0.004 to 8 | 0.12 | 0.5 |
| A. fumigatus | 1,409 | 11/11 | ≤0.004 to ≥16 | 0.12 | 0.5 |
| A. flavus | 257 | 7/7 | 0.01 to ≥16 | 0.25 | 0.5 |
| A. niger | 173 | 4/5 | 0.01 to ≥16 | 0.25 | 1 |
Including the complexes of C. parapsilosis, C. glabrata, and Aspergillus spp. Variability or insufficient data precluded the proposal of ECVs for some species of both Candida and Aspergillus.
Newly accepted taxonomic name or reclassification: C. krusei (P. kudriavzevii).
Total number of laboratories included in the ECV definition pool/total number of laboratories that submitted data (including data from one published study [20]).
TABLE 2.
Etest triazole pooled MIC distributions for species of Candida and Aspergillusa
| Agent and speciesb | No. of isolates | No. of labs used/total no.c | No. of isolates with MIC (µg/ml) of:d | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fluconazole | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ≥128 | ||
| C. glabrata | 356 | 7/10 | 1 | 4 | 13 | 34 | 50 | 79 | 88 | 36 | 13 | 28 | 10 | ||
| C. parapsilosis | 639 | 9/9 | 3 | 19 | 68 | 131 | 153 | 138 | 66 | 21 | 9 | 7 | 20 | 4 | |
| C. tropicalis | 368 | 9/10 | 3 | 5 | 11 | 61 | 96 | 120 | 47 | 11 | 4 | 1 | 4 | 4 | 1 |
| Itraconazole | ≤0.004 | 0.008 | 0.01 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | ||
| C. albicans | 975 | 8/9 | 7 | 55 | 145 | 237 | 295 | 150 | 27 | 19 | 16 | 6 | 7 | 5 | 6 |
| C. krusei | 101 | 3/3 | 1 | 2 | 9 | 36 | 35 | 7 | 7 | 1 | 3 | ||||
| C. tropicalis | 165 | 5/8 | 2 | 12 | 23 | 39 | 30 | 22 | 15 | 11 | 7 | 2 | 1 | 1 | |
| A. fumigatus | 1,112 | 10/10 | 4 | 1 | 3 | 30 | 56 | 157 | 483 | 268 | 73 | 12 | 5 | 20 | |
| Confirmed cyp51A mutants | 81 | 8/8 | 1 | 1 | 1 | 7 | 4 | 67 | |||||||
| A. flavus | 250 | 7/8 | 1 | 4 | 19 | 37 | 103 | 69 | 16 | 1 | |||||
| A. nidulans | 130 | 4/4 | 1 | 1 | 13 | 39 | 34 | 23 | 11 | 7 | 1 | ||||
| A. niger | 176 | 4/5 | 1 | 1 | 2 | 5 | 25 | 71 | 45 | 17 | 5 | 4 | |||
| Posaconazole | ≤0.004 | 0.008 | 0.01 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | ||
| C. albicans | 305 | 4/6 | 6 | 29 | 94 | 102 | 44 | 17 | 9 | 3 | 1 | ||||
| C. krusei | 48 | 3/3 | 1 | 5 | 17 | 16 | 7 | 2 | |||||||
| C. parapsilosis | 162 | 4/5 | 8 | 26 | 51 | 37 | 23 | 9 | 3 | 2 | 1 | 2 | |||
| C. tropicalis | 101 | 4/5 | 9 | 21 | 32 | 21 | 4 | 8 | 3 | 1 | 1 | 1 | |||
| A. flavus | 204 | 7/7 | 1 | 4 | 14 | 70 | 96 | 17 | 2 | ||||||
| A. niger | 168 | 4/5 | 5 | 16 | 58 | 73 | 15 | 1 | |||||||
| A. terreus | 194 | 5/5 | 8 | 47 | 105 | 27 | 4 | 2 | 1 | ||||||
| Voriconazole | ≤0.004 | 0.008 | 0.01 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | ||
| C. albicans | 2,159 | 8/11 | 485 | 803 | 491 | 158 | 104 | 42 | 22 | 20 | 11 | 5 | 7 | 1 | 10 |
| C. glabrata | 551 | 8/9 | 7 | 11 | 20 | 37 | 105 | 143 | 96 | 63 | 30 | 15 | 14 | 10 | |
| C. krusei | 130 | 6/6 | 1 | 2 | 3 | 20 | 30 | 45 | 24 | 5 | 2 | ||||
| C. parapsilosis | 506 | 7/9 | 4 | 20 | 46 | 97 | 167 | 100 | 39 | 15 | 4 | 4 | 8 | 1 | 1 |
| C. tropicalis | 260 | 6/10 | 4 | 4 | 12 | 28 | 82 | 88 | 22 | 14 | 5 | 1 | |||
| A. fumigatus | 1,409 | 11/11 | 1 | 6 | 2 | 30 | 132 | 633 | 473 | 100 | 19 | 7 | 3 | 2 | 1 |
| Confirmed cyp51A mutants | 75 | 8/8 | 5 | 6 | 7 | 8 | 13 | 15 | 3 | 1 | 17 | ||||
| A. flavus | 257 | 7/7 | 1 | 18 | 84 | 103 | 39 | 10 | 1 | 1 | |||||
| A. niger | 173 | 4/5 | 2 | 3 | 22 | 37 | 81 | 25 | 1 | 1 | 1 | ||||
Including the complexes of C. parapsilosis, C. glabrata, and Aspergillus species (the cyp51A mutants are A. fumigatus sensu stricto).
Newly accepted taxonomic name or reclassification: C. krusei (P. kudriavzevii).
Total number of laboratories included in the ECV definition pool/total number of study laboratories and one published study (20) that submitted data.
Data are from between 3 and 11 laboratories and were determined by the agar diffusion Etest method (15); the highest number in each row (showing the most frequent, or mode, MIC) is in bold.
TABLE 5.
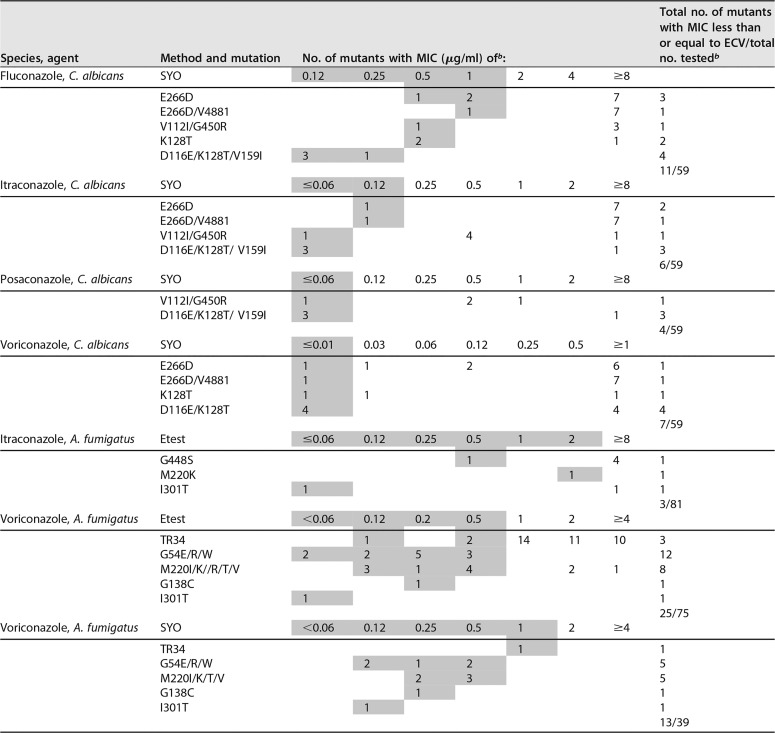
Triazole SYO and Etest MICs for selected confirmed C. albicans ERG11 and A. fumigatus sensu stricto cyp51 mutantsa
Listed are SYO and Etest MICs for C. albicans and A. fumigatus mutants that were either below or above (shaded and nonshaded, respectively) each correspondent ECV among the total data points for the 59 C. albicans mutants and 75, 81, or 39 A. fumigatus mutants. Data were submitted from multiple participant laboratories (4 to 8) and a single published study (20).
b The proposed SYO ECVs were as follows: for C. albicans versus fluconazole, 1 µg/ml; for C. albicans versus itraconazole, 0.12 µg/ml; for C. albicans versus posaconazole, 0.06 µg/ml; for C. albicans versus voriconazole, 0.01 µg/ml; and for A. fumigatus versus voriconazole, 1 µg/ml. Etest ECVs were as follows: for A. fumigatus versus itraconazole, 2 µg/ml; and for A. fumigatus versus voriconazole, 0.5 µg/ml.
Although SYO MICs for the species evaluated originated from 30 of the 39 participant centers, exclusions were made according to the CLSI criteria for ECV definition (Table 1) (27). During data consolidation, individual SYO MIC distributions for Candida and Aspergillus were not included in the ECV analysis when they were bimodal, when the particular mode for a distribution was more than 1 to 2 dilutions from the global mode, when there were less than five isolates in the distribution or when the modes were aberrant or not defined. MIC distributions were also excluded when the MIC data for the QC isolates were outside the recommended range (14, 16). The total number of isolates for which SYO MICs for the 12 Candida species and the four triazoles from 3 to 30 independent laboratories were pooled for ECV definition ranged from 11,171 to 17, with data points for C. parapsilosis sensu stricto, C. parapsilosis species complex, C. metapsilosis, C. orthopsilosis, C. famata, C. kefyr, and S. cerevisiae being included. The SYO MIC distributions for the 59 C. albicans and 39 A. fumigatus sensu stricto mutants from multiple laboratories are also listed in Table 1. In the case of SYO data for Aspergillus spp., interlaboratory modal consensus was an overall issue given that, of the submitted data for five species, ECVs were proposed only for voriconazole (3 of 4 species) and itraconazole (A. niger) (Tables 1 and 3). Of the 903 A. fumigatus listed in Table 1, 71% (640 data points) were identified as A. fumigatus sensu stricto and 29% (263 data points) were identified as A. fumigatus species complex (identification by morphological methods, matrix-assisted laser desorption ionization–time of flight [MALDI-TOF] mass spectrometry, or molecular methods [e.g., β-tubulin and calmodulin sequencing]) (31). Candida isolates also were identified to the species level by biochemical tests, MALDI-TOF mass spectrometry, and/or molecular methods in the laboratories submitting the data (31, 32); C. parapsilosis and C. glabrata were submitted mainly as species complex isolates (Table 1).
TABLE 3.
Method-dependent SYO ECOFFinder ECVs of four triazoles for species of Candida, Saccharomyces, and Aspergillusa
| Agent and speciesb | No. of isolates | No. of labs used/total no. | MIC (µg/ml) |
ECV (µg/ml)c | |
|---|---|---|---|---|---|
| Range | Mode | ||||
| Fluconazole | |||||
| C. albicans | 11,171 | 28/30 | 0.06 to ≥128 | 0.25 | 1 |
| C. dubliniensis | 195 | 7/10 | 0.12 to 64 | 0.25 | 1 |
| C. glabrata | 4,418 | 30/30 | 0.12 to ≥128 | 16 | 64 |
| C. guilliermondii | 153 | 8/13 | 0.12 to ≥128 | 4 | 16 |
| C. krusei | 537 | 15/16 | 0.12 to ≥128 | 64 | 128 |
| C. lusitaniae | 298 | 12/12 | 0.12 to ≥128 | 1 | 4 |
| C. parapsilosis | 3,691 | 28/30 | 0.12 to ≥128 | 0.5 | 2 |
| C. parapsilosis sensu stricto | 911 | 5/5 | 0.12 to 64 | 0.5 | 2 |
| C. orthopsilosis | 110 | 5/5 | 0.12 to ≥128 | 1 | 4 |
| C. tropicalis | 1,854 | 24/28 | 0.12 to ≥128 | 1 | 4 |
| S. cerevisiae | 244 | 3/3 | 0.12 to 64 | 4 | 16 |
| Itraconazole | |||||
| C. albicans | 7,843 | 27/30 | 0.008 to ≥16 | 0.06 | 0.12 |
| C. dubliniensis | 125 | 6/8 | 0.01 to ≥16 | 0.06 | 0.25 |
| C. glabrata | 3,594 | 29/30 | 0.01 to ≥16 | 0.5 | 2 |
| C. guilliermondii | 149 | 9/13 | 0.01 to ≥16 | 0.25 | 1 |
| C. krusei | 574 | 13/16 | 0.01 to ≥16 | 0.25 | 1 |
| C. lusitaniae | 171 | 8/11 | 0.01 to 1 | 0.12 | 0.5 |
| C. parapsilosis | 3,353 | 23/30 | 0.01 to 4 | 0.12 | 0.25 |
| C. parapsilosis sensu stricto | 730 | 4/5 | 0.01 to 1 | 0.12 | 0.5 |
| C. tropicalis | 1,399 | 23/29 | 0.01 to ≥16 | 0.12 | 0.5 |
| A. niger | 233 | 6/7 | 0.01 to ≥16 | 0.12 | 1 |
| Posaconazole | |||||
| C. albicans | 6,729 | 27/30 | 0.008 to ≥16 | 0.01 | 0.06 |
| C. dubliniensis | 185 | 7/8 | 0.008 to 1 | 0.03 | 0.12 |
| C. glabrata | 2,999 | 25/29 | 0.008 to ≥16 | 1 | 4 |
| C. guilliermondii | 111 | 9/12 | 0.008 to 1 | 0.25 | 1 |
| C. krusei | 562 | 13/15 | 0.008 to 8 | 0.25 | 1 |
| C. lusitaniae | 172 | 11/11 | 0.008 to 1 | 0.03 | 0.12 |
| C. parapsilosis | 3,085 | 26/30 | 0.008 to ≥16 | 0.03 | 0.12 |
| C. parapsilosis sensu stricto | 670 | 5/5 | 0.008 to 8 | 0.03 | 0.25 |
| C. tropicalis | 1,366 | 23/29 | 0.008 to ≥16 | 0.12 | 1 |
| Voriconazole | |||||
| C. albicans | 8,747 | 29/30 | 0.008 to ≥16 | 0.008 | 0.01d |
| C. dubliniensis | 215 | 7/9 | 0.008 to 2 | 0.008 | 0.01d |
| C. glabrata | 3,255 | 24/30 | 0.008 to ≥16 | 0.25 | 2 |
| C. guilliermondii | 157 | 11/12 | 0.008 to ≥16 | 0.06 | 0.5 |
| C. krusei | 676 | 14/16 | 0.008 to ≥16 | 0.25 | 1 |
| C. lusitaniae | 248 | 11/12 | 0.008 to 1 | 0.008 | 0.03d |
| C. parapsilosis | 2670 | 26/30 | 0.008 to 2 | 0.008 | 0.01d |
| C. parapsilosis sensu stricto | 718 | 5/5 | 0.008 to 2 | 0.008 | 0.03d |
| C. tropicalis | 1,637 | 19/28 | 0.008 to ≥16 | 0.06 | 0.5 |
| A. fumigatus | 903 | 8/8 | 0.008 to ≥16 | 0.25 | 1 |
| A. flavus | 389 | 6/7 | 0.008 to ≥16 | 0.25 | 1 |
| A. terreus | 302 | 5/6 | 0.008 to 2 | 0.25 | 1 |
Including the complexes of C. parapsilosis, C. glabrata, and Aspergillus species. Modal variability or insufficient data precluded the proposal of ECVs for some species of both Candida and Aspergillus.
Newly accepted taxonomic names or reclassifications are as follows: C. guilliermondii (M. guilliermondii), C. krusei (P. kudriavzevii), and Candida lusitaniae (Clavispora lusitaniae).
ECOFFinder ECVs for 97.5% of the statistically modeled population based on MICs obtained by the colorimetric broth microdilution SYO method (14, 29), except where indicated by a superscript c, referring to footnote c. The proposed method-dependent SYO ECV for A. fumigatus and posaconazole is 0.06 µg/ml, as reported elsewhere (23). C. krusei is intrinsically resistant to fluconazole regardless of the MIC.
ECV as estimated using the second derivative method (30).
Table 1 also depicts the SYO modes for Candida and Aspergillus species. The lowest SYO fluconazole modes (0.25 µg/ml) were for C. albicans, C. dubliniensis, and C. kefyr, and the highest mode was for C. krusei (64 µg/ml). A similar modal diversity was noted among posaconazole MICs (modes, 0.01 µg/ml for C. albicans and 1 µg/ml for C. glabrata). However, itraconazole and voriconazole modes were mostly 0.06 µg/ml to 0.12 µg/ml and 0.008 to 0.03 µg/ml, respectively. The exceptions were itraconazole modes for C. glabrata (0.5 µg/ml) and C. guilliermondii and C. krusei (0.25 µg/ml) and voriconazole modes for C. guilliermondii and C. tropicalis (0.06 µg/ml) and C. glabrata and C. krusei (0.25 µg/ml). Most SYO modes for the C. parapsilosis complex were ±1 doubling dilution, but all posaconazole modes for the four species in the complex were 0.03 µg/ml. SYO voriconazole modes for Aspergillus spp. and the itraconazole mode for A. niger ranged from 0.12 to 0.5 µg/ml. As expected, SYO modes for the C. albicans and A. fumigatus mutants were much higher than those for the nonmutant isolates, and we observed an overlap between both groups of MICs among the lower drug concentrations (Table 1). Therefore, the SYO data for Candida spp. showed excellent modal agreement, while most SYO data points for Aspergillus spp. were unsuitable for the ECV definition pool, as previously reported among SYO posaconazole data for A. fumigatus (23).
Eleven of the 39 laboratories contributed Etest MICs for the four more prevalent Candida spp., C. krusei, and Aspergillus spp. Eight laboratories, as well as a published study (20), contributed Etest voriconazole and itraconazole data for the 75 and 81 A. fumigatus sensu stricto mutants, respectively (Tables 2 and 5). A total of 64% (712 of the 1,112 itraconazole MICs) of the A. fumigatus isolates and most Candida isolates were identified at the species level (31, 32), but C. glabrata and C. parapsilosis were mainly identified as the species complex. Therefore, we were unable to provide the potential antifungal susceptibility differences among the species in the C. parapsilosis species complex by the Etest like we did by the SYO method (Table 1). The modal variability among the Etest MIC distributions entering the ECV definition data pool also precluded our ECV definition for C. albicans and fluconazole, C. glabrata and both itraconazole and posaconazole, C. parapsilosis and itraconazole, and C. krusei and fluconazole. However, most Etest data points for the Aspergillus/agent combinations were suitable for the ECV definition pool, although we observed modal discrepancies for itraconazole and voriconazole versus A. terreus. Consequently, we collected more suitable Etest data for Aspergillus spp., while the overall SYO data for Candida spp. were superior. The lowest Etest modes were for C. parapsilosis versus fluconazole and posaconazole (0.5 and 0.01 µg/ml, respectively), C. tropicalis versus itraconazole (0.03 µg/ml), and C. albicans versus voriconazole (0.008 µg/ml). All Etest modal values for Aspergillus spp. ranged from 0.12 to 0.25 µg/ml, except for the itraconazole modes for A. fumigatus (0.5 µg/ml) and for A. niger (1 µg/ml). Etest modes for the A. fumigatus mutants were also much higher than those for the nonmutant isolates.
Tables 3 and 4 depict the proposed ECOFFinder program SYO and Etest triazole ECVs, respectively, for 97.5% of the modeled MIC population for the species and triazole combinations that fulfilled the CLSI criteria for ECV calculation (27). There was no need to weigh the data since none of the individual distributions contributed ≥50% of the total. In addition to SYO ECVs for the prevalent Candida spp., fluconazole ECVs were proposed for C. orthopsilosis (4 µg/ml) and S. cerevisiae (16 µg/ml) (Table 3). Although fluconazole ECVs for C. parapsilosis sensu stricto and species complex were the same (2 µg/ml), the other ECVs for C. parapsilosis sensu stricto were 1 dilution higher. To our knowledge, ECVs for C. parapsilosis sensu stricto or any other member of this complex and for S. cerevisiae are not yet available for the reference methods (26, 28; http://www.eucast.org/ast_of_fungi/). Due to aberrant modes by the Etest, we only defined voriconazole Etest ECVs of 0.03 to 2 µg/ml for five Candida spp. and ECVs of 0.12 to 64 µg/ml for the other three agents and three to four species (Table 4). However, we proposed ECVs for three to four relevant Aspergillus spp. (2, 4, 5). Inconsistent itraconazole and voriconazole modes for A. terreus from four laboratories as well as insufficient posaconazole and voriconazole MICs for A. nidulans (data were submitted from only two laboratories) precluded definition of an ECV for these two species/agents (27) (data not shown in Table 2). In Table 6, we compared our SYO and Etest ECVs with the approved CLSI ECVs listed in the new edition of the M59 document (26). In general, SYO ECVs were 1 to 2 dilutions higher than those for the CLSI or Etest methods. In some instances, such as for fluconazole and voriconazole versus C. glabrata, among others, SYO and CLSI ECVs of 64 and 8 µg/ml and 2 and 0.25 µg/ml, respectively, have been defined (26). All these observations underscore the need for method-dependent ECVs in order to properly categorize the MIC for the infecting isolate being evaluated as either WT or non-WT. It also demonstrates that while commercial systems can successfully establish equivalence according to FDA criteria, the pooling of data from multiple laboratories can more easily detect differences between these assays and the reference method, at least in what is measured as the wild type.
TABLE 6.
Method-dependent ECVs of four triazoles for species of Candida, Saccharomyces, and Aspergillus by three susceptibility testing methodsa
| Speciesb | Method-dependent ECV (µg/ml) for the following agent: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fluconazole |
Itraconazole |
Posaconazole |
Voriconazole |
|||||||||
| SYO | Etest | CLSI | SYO | Etest | CLSI | SYO | Etest | CLSI | SYO | Etest | CLSI | |
| C. albicans | 1 | AM | 0.5 | 0.12 | 0.25 | NA | 0.06 | 0.12 | 0.06 | 0.01 | 0.03 | 0.03 |
| C. dubliniensis | 1 | ID | 0.5 | 0.25 | ID | NA | 0.12 | ID | 0.25 | 0.01 | ID | 0.03 |
| C. glabrata | 64 | 64 | 8 | 2 | 8 | 4 | 4 | ID | 1 | 2 | 2 | 0.25 |
| C. guilliermondii | 16 | ID | 8 | 1 | ID | NA | 1 | ID | 0.5 | 0.5 | ID | 0.12 |
| C. krusei | 128 | ID | 32 | 1 | 2 | 1 | 1 | ID | 0.5 | 1 | 2 | 0.5 |
| C. lusitaniae | 4 | ID | 1 | 0.5 | ID | 0.5 | 0.12 | ID | 0.06 | 0.03 | ID | 0.06 |
| C. parapsilosis species complex | 2 | 4 | 1 | 0.25 | AM | NA | 0.12 | 0.12 | 0.25 | 0.01 | 0.25 | 0.03 |
| C. parapsilosis sensu stricto | 2 | NA | NA | 0.5 | NA | NA | 0.25 | NA | NA | 0.03 | NA | NA |
| C. tropicalis | 4 | 4 | 1 | 0.5 | 0.5 | 0.5 | 1 | 0.12 | 0.12 | 0.5 | 0.5 | 0.12 |
| S. cerevisiae | 16 | ID | NA | ID | ID | NA | ID | ID | NA | ID | ID | NA |
| A. fumigatus | NA | NA | NA | AM | 2 | 1 | 0.06c | 0.25c | 0.25c | 1 | 0.5 | 1 |
| A. flavus | NA | NA | NA | AM | 1 | 1 | NA | 0.5 | 0.5 | 1 | 0.5 | 2 |
| A. niger | NA | NA | NA | 1 | 4 | 4 | NA | 0.5 | 2 | ID | 1 | 2 |
| A. terreus | NA | NA | NA | AM | AM | 2 | NA | 0.25 | 1 | 1 | AM | 2 |
SYO and Etest ECVs proposed in the present study and CLSI M27 and M38 methods (11, 12). AM, aberrant modes, modal variability; ID, insufficient number of laboratories/isolates entering the ECV definition pool; NA, not available or not applicable.
Newly accepted taxonomic names or reclassifications are as follows: C. guilliermondii (M. guilliermondii), C. krusei (P. kudriavzevii), and Candida lusitaniae (Clavispora lusitaniae).
As mentioned above, the main role of the ECV is to identify the strains that could harbor intrinsic or acquired resistance mechanisms (non-WT or mutant isolates) (27, 28). CLSI MICs for Candida and Aspergillus mutants are readily available in the literature (6–10, 33–36), but they are scarce by the commercial methods (20–24). A total of 162 SYO and Etest MICs for C. albicans, C. glabrata, and C. parapsilosis mutants were received. The number of SYO MICs above the ECVs of the four triazoles for the 59 ERG11 C. albicans mutants was agent dependent. The posaconazole ECV of 0.06 µg/ml recognized the highest percentage of mutants (55/59; 93%), followed by the itraconazole ECV of 0.12 µg/ml (53/59; 90%), the voriconazole ECV of 0.01 µg/ml (52/59; 88%), and the fluconazole SYO ECV of 1 µg/ml (48/59; 81%). These C. albicans mutants had the following ERG11 substitutions: F145L, Y132H, S442F, S405F, G464S, A114S, G464S, F145T, T22OL, and P98A (alone or in combination). Although high CLSI triazole MICs have been documented for most of those substitutions (6, 8, 33–35), T22OL and P98A (alone or in different combinations with E266D, G448R, V437I, V488I, K143R, and Y132H/X) have not been previously reported. Considering their high MICs of >8 µg/ml (Table 1), it seems that these strains could also harbor combined resistance mechanisms (e.g., the most common efflux pump overexpression plus erg11 overexpression and/or an erg11 mutation). These molecular combinations are due to aneuploidy (duplication of chromosome 5 or multiplication of its long arm). However, we did not receive efflux pump overexpression data for the 59 C. albicans mutants. On the other hand, in Table 5 we list the C. albicans and A. fumigatus mutants that, according to our method-dependent ECVs, could be categorized as either WT (for which MICs are less than or equal to each ECV) and/or non-WT (for which MICs are greater than the ECV). Isolates with those substitutions have been reported as both susceptible and resistant using CLSI methodologies and BPs (8, 33–35). Regarding data from the single laboratories, the SYO MICs of the four agents for the 2 C. albicans strains and 11 of the 13 C. glabrata strains from single laboratories with overexpression of the CDR2 and CDR1 gene efflux pumps, respectively, were above the four ECVs (data not shown in Table 5). However, only the fluconazole and voriconazole ECVs (2 and 0.03 µg/ml, respectively) recognized >96% of the 78 C. parapsilosis mutants. Therefore, the potential ability of our SYO ECVs to recognize ≥90% of the isolates with mechanisms of resistance among the most prevalent Candida spp. (C. albicans, C. glabrata) provided a preliminary indication of their clinical value. More data points for other Candida mutants would better assess the utility of the SYO method for yeast testing in the clinical laboratory.
In the present study, a total of 75 and 81 Etest MICs (for voriconazole and itraconazole, respectively) and 39 SYO MICs (for voriconazole) for A. fumigatus sensu stricto cyp51A mutants were evaluated. Our proposed Etest itraconazole ECV of 2 µg/ml for A. fumigatus had performance in recognizing the cyp51A mutants (78/81; 96%) superior to that of the voriconazole Etest ECV of 0.5 µg/ml (50/75; 67%) and the SYO ECV of 1 µg/ml (26/39; 67%) (Table 5). Etest itraconazole MICs were above the ECV for isolates with the following mutations: 48 isolates with TR34/L98 (59%), 12 with G54 (15%), 9 with M220 (11%), 5 with G448S (6%), and 7 with miscellaneous mutations (9%), including 2 with TR46/Y121F mutations (data not listed in Table 5). However, cyp51A G54 changes have been linked in the literature with cross-resistance to both itraconazole and posaconazole, and the M220 change has been linked with either high or low triazole MICs (36). An overlap between posaconazole MICs for nonmutants and those for a much larger number of mutants of A. fumigatus by three antifungal susceptibility methods (CLSI, EUCAST and Etest) has also been reported (23). These preliminary results for Aspergillus spp. indicated that the Etest appears to be a superior method for detecting mutations in A. fumigatus as well as for testing other Aspergillus spp. Once again, these results underscore the need for method-dependent ECVs. As far as the SYO data for Aspergillus spp. are concerned, further collaborative studies should evaluate the endpoint determination; both color change and growth inhibition have been reported in the literature.
In conclusion, we proposed method-dependent SYO and Etest ECVs for various species/triazole combinations for which suitable data were available from multiple laboratories (3 to 30). Substantial data with excellent interlaboratory modal agreement were evaluated by the SYO method for Candida and other yeast species, including MIC distributions for the C. parapsilosis complex (C. parapsilosis sensu stricto, C. metapsilosis, and C. orthopsilosis) and S. cerevisiae. Because of that, we proposed SYO ECVs for 8 to 10 yeast species and the four triazoles evaluated, including C. orthopsilosis and S. cerevisiae versus fluconazole. We also provided MIC ranges and, more importantly, modes for other less prevalent yeast species. On the other hand, interlaboratory modal agreement by the Etest was better for Aspergillus than for yeast species. As a result, we proposed Etest ECVs of itraconazole, posaconazole, and voriconazole for three to four Aspergillus spp. and voriconazole ECVs for the four most prevalent Candida spp. and C. krusei. Finally, the SYO posaconazole ECV of 0.06 µg/ml for C. albicans and the Etest itraconazole ECV of 2 µg/ml for A. fumigatus were the best predictors in recognizing the non-WT or mutants (the highest percentage of MICs for mutants that were above the ECV). Although ECVs of fluconazole and voriconazole for C. parapsilosis recognized >96% of the non-WT isolates, the results were unsatisfactory with posaconazole and itraconazole ECVs. Data for mutants of other Candida spp. would better assess the method-dependent proposed ECVs. The SYO method appears to yield more suitable MIC data for testing most Candida spp. and the Etest for Aspergillus spp.
MATERIALS AND METHODS
Isolates.
The Candida and other yeast isolates evaluated were mostly recovered from blood and other normally sterile sites from patients with candidemia or other deep infections (>90%) as well as superficial, oral, and vaginal infections and thrush. The Aspergillus isolates were also recovered from deep infections and sterile and other sites (mostly [>90%] bronchoalveolar lavage fluid and sputum) at the following medical centers: VCU Medical Center, Richmond, VA, USA; Mycology Reference Laboratory, National Centre for Microbiology, Instituto de Salud Carlos III, Majadahonda, Madrid, Spain; Unité de Parasitologie, Mycologie, Département de Bactériologie Virologie Hygiène Mycologie Parasitologie, Créteil, France; Grupo de Infección Grave, Instituto de Investigación Sanitaria La Fe, Valencia, Spain; Unidad de Gestión Clínica de Enfermedades Infecciosas y Microbiología, Hospital de Valme, Seville, Spain; Department of Internal Medicine, National Taiwan University Hospital and College of Medicine, Taipei, Taiwan; Public Health Ontario, Toronto, ON, Canada; Klinisk Mikrobiologi, Karolinska, Universitetlaboratoriet, Karolinska, Universitetssjukhuset, Stockholm, Sweden; Université Paris-Descartes, Faculté de Médecine, APHP, Hôpital Européen Georges Pompidou, Unité de Parasitologie-Mycologie, Service de Microbiologie, Paris, France; Laboratorio de Micología y Diagnóstico Molecular, Cátedra de Parasitología y Micología, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Santa Fe, Argentina; Universidad Autónoma de Nuevo León, Mexico; National Institute for Communicable Diseases (Centre for Healthcare-Associated Infections, Antimicrobial Resistance and Mycoses), a Division of the National Health Laboratory Service and Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa; Hospital General Universitario Gregorio Marañón, Madrid, Spain; SA Pathology, National Mycology Reference Centre, Adelaide, South Australia, Australia; Division of Hygiene and Medical Microbiology, Medical University of Innsbruck, Innsbruck, Austria; Departamento de Microbiología, Facultad de Medicina y Enfermería, Universidad de Córdoba, Córdoba, Spain; Servicio de Microbilogía, Hospital Universitario Cruces, Barakaldo, Spain; Servicio de Microbiología, Hospital Universitario Central de Asturias, Asturias, Spain; Departamento de Inmunología, Microbiología y Parasitología, Facultad de Medicina y Enfermería, Universidad del País Vasco/Euskal Herriko Unibertsitatea, UPV/EHU, Bilbao, Spain; Departamento de Biomedicina, Biotecnología y Salud Pública, Universidad de Cádiz, Cadiz, Spain; Hospital de Alcañiz, Alcañiz (Teruel), Spain; Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Institute of Microbiology, Università Cattolica del Sacro Cuore, Rome, Italy; University of Pittsburgh, Pittsburgh, PA, USA; Department of Pharmaceutical Technology and Biochemistry, Faculty of Chemistry, Gdańsk University of Technology, Gdańsk, Poland; Department of Pharmaceutical Technology and Biochemistry, Faculty of Chemistry, University of Technology, Gdansk, Poland; Department of Biomedical Sciences for Health, Università degli Studi di Milano, Milan, Italy; Mayo Clinic, Rochester, MN, USA; Microbiology Laboratory, Ospedale San Gerardo, Monza, Italy; Microbiology and Virology Unit IRCCS Policlinico San Matteo, Pavia, Italy; Microbiology Section, Humanitas Research Hospital, Milan, Italy; Microbiology Laboratory, A.O. Spedali Civili, Brescia, Italy; Microbiology Laboratory, Fondazione IRCCS Cà Granda O. Maggiore Policlinico, Milan, Italy; Microbiology Laboratory, Niguarda Hospital, Milan, Italy; Clinical Microbiology Laboratory, Attikon Hospital, Medical School, National and Kapodistrian, University of Athens, Athens, Greece; Laboratório Especial de Micologia, Disciplina de Infectologia, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, SP, Brazil; Microbiology Institute, ASST Papa Giovanni XXIII, Bergamo, Italy; Unidad de Micología, Servicio de Microbiología, Hospital Universitario La Fe, Valencia, Spain; Microbiology-ASST Lariana, Como, Italy; and Medicina di Laboratorio, IRCCS Policlinico San Donato, Milan, Italy.
The submitted triazole MICs of the four triazoles for yeast species from 3 to 30 laboratories obtained by the SYO and/or Etest methods and evaluated for ECV definition were for (method/agent dependent) (Tables 1 and 2) 11,171 C. albicans, 215 C. dubliniensis, 4,418 C. glabrata species complex (including 349 C. glabrata sensu stricto), 157 C. guilliermondii, 676 C. krusei, 298 C. lusitaniae, 3,691 C. parapsilosis species complex, 922 C. parapsilosis sensu stricto, and 1,854 C. tropicalis isolates (Tables 1 and 2). SYO MICs for other, less common Candida and yeast species from at least three laboratories were collected for 25 C. famata, 55 C. kefyr, 36 C. metapsilosis, and 110 C. orthopsilosis isolates, and SYO data were also collected for 244 isolates of S. cerevisiae. In addition, we pooled SYO and mostly Etest data for the five most prevalent Aspergillus complexes originating from 3 to 11 independent laboratories, as follows (method agent dependent): 1,409 A. fumigatus, 389 A. flavus, 103 A. nidulans, 233 A. niger, and 302 A. terreus isolates.
We also received a total of 282 MICs for mutants: 59 SYO and 10 Etest MICs for C. albicans mutants (Erg11 gene mutations), 2 C. albicans and 13 C. glabrata mutants (overexpression of CDR2 or CDR1 efflux pumps, respectively), and 78 C. parapsilosis mutants (Erg11 and MRR1). SYO and Etest MICs were gathered for 39 and 81 A. fumigatus sensu stricto mutant isolates, respectively, with cyp51A gene mechanisms of resistance (TR34/L98H, G54, M220, and others) from five to seven participant laboratories and one previous Etest study (20) (Tables 1, 2, and 5). The isolates were identified at each medical center by conventional and molecular methodologies that included macro- and microscopic morphology, thermotolerance (incubation at 50°C), MALDI-TOF mass spectrometry, and β-tubulin and calmodulin sequencing (31, 32). Since molecular identification was not performed for all the isolates evaluated in the present study, we listed the nonmutant isolates in the respective tables as the complexes of C. glabrata or C. parapsilosis or Aspergillus spp. Strains of A. fumigatus, C. albicans, and C. glabrata that were submitted as having mutations were screened in the participant laboratories using published protocols (31, 36–38).
At least one of following quality control (QC) isolates and/or reference isolates was evaluated by the two methods in each of the participant laboratories: QC isolates C. parapsilosis ATCC 22019, C. krusei ATCC 6258, and Paecilomyces variotii ATCC MYA-3630 and reference isolates A. fumigatus ATCC MYA-3626 and A. flavus ATCC MYA-204304 (14, 16). MIC data were not included in the study unless the participant laboratories reported that their MICs for the individual QC isolates used in each center were within the expected MIC ranges.
Antifungal susceptibility testing.
Triazole SYO and Etest MICs were obtained by the two commercial antifungal susceptibility methods by following the manufacturers’ guidelines (14–16). The SYO MIC was the first blue or purple well after 24 h (Candida) or, mostly, 48 h (Aspergillus) of incubation. The Etest MIC was the lowest drug concentration at which the border of the growth-free elliptical inhibition intercepted the scale on the antifungal strip after 24 to 48 h, as needed; trailing growth was allowed solely for the definition of Etest MICs for Candida isolates.
Definitions.
The definition of the ECV as a categorical endpoint has been widely described previously as well as above (27, 28). Briefly, the ECV is the highest MIC/MEC distribution for the WT population and is established by using reliable MIC/MEC distributions from at least three laboratories. A non-WT organism usually shows reduced susceptibility to the agent being evaluated compared to that of the WT (no phenotypic resistance) population. In addition to MIC distributions, the ECV calculation takes into account each laboratory distribution mode, the inherent variability of the test (usually within 1 doubling dilution), and the fact that the ECV should encompass 95% to 97% of isolates. We used those same criteria and requirements for establishing our proposed Etest and SYO method-dependent ECVs. Most published ECVs are based on reference MIC distributions, and ECVs based on other methods could be different, as was shown in our study (Table 6).
Data collation and analyses.
Triazole MICs were submitted from 39 independent laboratories worldwide (method/agent/species dependent) in order (i) to define the MIC distributions by each commercial susceptibility testing method/agent and species; (ii) to examine the suitability of these distributions for pooling prior to ECV setting, including the evaluation of interlaboratory modal agreement; and (iii) to estimate ECVs for each species/agent/method that fulfilled the CLSI criteria for ECV definition after pooling (at least 100 MICs for each species/method/agent that originated in ≥3 independent laboratories) (27, 28). ECVs were estimated by the iterative statistical method at the 97.5% cutoff value (29) or the second numerical derivative method when the putative wild-type mode was at the lowest concentration in the distribution (30) (Tables 2 and 4). SYO MIC distributions for less common yeast species (C. famata and C. kefyr) and C. metapsilosis were also reported when they originated from at least three laboratories and had comparable modes.
ACKNOWLEDGMENTS
We thank S. Gamarra, F. Leonardelli, C. Dudiuk, and D. Macedo of the Laboratorio de Micología y Diagnóstico Molecular, Universidad Nacional del Litoral, Santa Fe, Argentina, and H. Alexiou, S. R. Davis, and D. H. Ellis of the National Mycology Reference Centre, SA Pathology, Adelaide, Australia.
REFERENCES
- 1.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guidelines for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:1–50. doi: 10.1093/cid/civ747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patterson TF, Thompson GR III, Denning DW, Fishman JA, Hadley S, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Nguyen MH, Segal BH, Steinbach WJ, Stevens DA, Walsh TJ, Wingard JR, Young JA, Bennett JE. 2016. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 63:e1–e60. doi: 10.1093/cid/ciw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornely OA, Bassetti M, Calandra T, Garbino J, Kullberg BJ, Lortholary O, Meersseman W, Akova M, Arendrup MC, Arikan-Akdagli S, Bille J, Castagnola E, Cuenca-Estrella M, Donnelly JP, Groll AH, Herbrecht R, Hope WW, Jensen HE, Lass-Flörl C, Petrikkos G, Richardson MD, Roilides E, Verweij PE, Viscoli C, Ullmann AJ, ESCMID Fungal Infection Study Group. 2012. ESCMID guidelines for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin Microbiol Infect 18:19–37. doi: 10.1111/1469-0691.12039. [DOI] [PubMed] [Google Scholar]
- 4.Klingspor L, Saaedi B, Ljungman P, Szakos A. 2015. Epidemiology and outcomes of patients with invasive mould infections: a retrospective observational study from a single centre (2005-2009). Mycoses 58:470–477. doi: 10.1111/myc.12344. [DOI] [PubMed] [Google Scholar]
- 5.Kosmidis C, Denning DW. 2015. The clinical spectrum of pulmonary aspergillosis. Thorax 70:270–277. doi: 10.1136/thoraxjnl-2014-206291. [DOI] [PubMed] [Google Scholar]
- 6.Berkow EL, Lockhart SR. 2017. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist 10:237–245. doi: 10.2147/IDR.S118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vale-Silva LA, Moeckli B, Torelli R, Posteraro B, Sanguinetti M, Sanglard D. 2016. Upregulation of the adhesin gene EPA1 mediated by PDR1 in Candida glabrata leads to enhanced host colonization. mSphere 1:e00065-15. doi: 10.1128/mSphere.00065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanglard D, Coste AT. 2016. Activity of isavuconazole and other azoles against Candida clinical isolates and yeast model systems with known azole resistance mechanisms. Antimicrob Agents Chemother 60:229–238. doi: 10.1128/AAC.02157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forastiero A, Mesa-Arango AC, Alastruey-Izquierdo A, Alcazar-Fuoli L, Bernal-Martinez L, Pelaez T, Lopez JF, Grimalt JO, Gomez-Lopez A, Cuesta I, Zaragoza O, Mellado E. 2013. Candida tropicalis antifungal cross-resistance is related to different azole target (Erg11p) modifications. Antimicrob Agents Chemother 57:4769–4781. doi: 10.1128/AAC.00477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernal-Martínez L, Gil H, Rivero-Menéndez O, Gago S, Cuenca-Estrella M, Mellado E, Alastruey-Izquierdo A. 2017. Development and validation of a high resolution melting assay to detect azole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 61:e01083-17. doi: 10.1128/AAC.01083-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of yeasts, 4th ed CLSI standard M27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, 3rd ed CLSI standard M38. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Arendrup MC, Meletiadis J, Mouton JW, Guinea J, Cuenca-Estrella M, Lagrou K, Howard SJ, Arendrup MC, Meletiadis J, Howard SJ, Mouton J, Guinea J, Lagrou K, Arikan-Akdagli S, Barchiesi F, Hamal P, Järv H, Lass-Flörl C, Mares M, Matos T, Muehlethaler K, Rogers TR, Torp Andersen C, Verweij P, Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing. 2016. EUCAST technical note on isavuconazole breakpoints for Aspergillus, itraconazole breakpoints for Candida and updates for the antifungal susceptibility testing method documents. Clin Infect Dis 22:571.e1–571.e4. doi: 10.1016/j.cmi.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Trek Diagnostic Systems. 2012. Sensititre Yeast One: Yeast One susceptibility, v1.8. Trek Diagnostic Systems, Cleveland, OH. [Google Scholar]
- 15.bioMérieux SA. 2013. Etest antifungal susceptibility testing package insert. bioMérieux SA, Marcy-l’Etoile, France. [Google Scholar]
- 16.bioMérieux SA. 2013. Etest performance, interpretative criteria and quality control ranges table. bioMérieux SA, Marcy-l’Etoile, France. [Google Scholar]
- 17.Pfaller MA, Espinel-Ingroff A, Jones RN. 2004. Clinical evaluation of Sensititre YeastOne colorimetric antifungal plate for antifungal susceptibility testing of the new triazoles voriconazole, posaconazole, and ravuconazole. J Clin Microbiol 42:4577–4580. doi: 10.1128/JCM.42.10.4577-4580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinel-Ingroff A, Pfaller M, Erwin ME, Jones RN. 1996. Interlaboratory evaluation of Etest method for testing antifungal susceptibilities of pathogenic yeasts to five antifungal agents using a Casitone agar and solidified RPMI 1640 medium with 2% glucose. J Clin Microbiol 34:848–885. doi: 10.1128/JCM.40.6.2101-2107.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espinel-Ingroff A, Rezuzta A. 2002. E-test method for testing susceptibilities of Aspergillus spp. to the new triazoles voriconazole and posaconazole and to established antifungal agents: comparison with NCCLS broth microdilution method. J Clin Microbiol 40:2001–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgel P-R, Baixench M-T, Amsellem M, Audureau E, Chapron J, Kanaan R, Honoré I, Dupouy-Camet J, Dusser D, Klaassen CH, Meis JF, Hubert D, Paugam A. 2012. High prevalence of azole-resistant Aspergillus fumigatus in adults with cystic fibrosis exposed to itraconazole. Antimicrob Agents Chemother 56:869–874. doi: 10.1128/AAC.05077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C-J, Wang H-C, Lee J-C, Lo H-J, Dai C-T, Chou P-H, Ko W-C, Chen Y-C. 2015. Azole-resistant Aspergillus fumigatus isolates carrying TR34/L98H mutations in Taiwan. Mycoses 58:544–549. doi: 10.1111/myc.12354. [DOI] [PubMed] [Google Scholar]
- 22.Mello E, Posteraro B, Vella A, De Carolis E, Torelli R, D’Inzeo T, Verweij PE, Sanguinetti M. 2017. Susceptibility testing of common and uncommon Aspergillus species against posaconazole and other mold-active antifungal azoles using the Sensititre method. Antimicrob Agents Chemother 61:e00168-17. doi: 10.1128/AAC.00168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espinel-Ingroff A, Turnidge J, Alastruey-Izquierdo A, Dannaoui E, Garcia-Effron G, Guinea J, Kidd S, Pelaez T, Sanguinetti M, Meletiadis J, Botterel F, Bustamante B, Chen Y-C, Chakrabarti A, Chowdhary A, Chryssanthou E, Córdoba S, Gonzalez GM, Guarro J, Johnson EM, Kus JV, Lass-Flörl C, Linares-Sicilia MJ, Martín-Mazuelos E, Negri CE, Pfaller MA, Tortorano AM. 2018. Posaconazole MIC distributions for Aspergillus fumigatus species complex by four methods: impact of cyp51A mutations on estimation of epidemiological cutoff values. Antimicrob Agents Chemother 62:e01916-17. doi: 10.1128/AAC.01916-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szweda P, Gucwa K, Romanowska E, Dzierz˙anowska-Fangrat K, Naumiuk Ł, Brillowska-Da˛browska A, Wojciechowska-Koszko I, Milewski S. 2015. Mechanisms of azole resistance among clinical isolates of Candida glabrata in Poland. J Med Microbiol 64:610–619. doi: 10.1099/jmm.0.000062. [DOI] [PubMed] [Google Scholar]
- 25.Espinel-Ingroff A, Arendrup M, Cantón E, Cordoba S, Dannaoui E, García-Rodríguez J, Gonzalez GM, Govender NP, Martin-Mazuelos E, Lackner M, Lass-Flörl C, Linares Sicilia MJ, Rodriguez-Iglesias MA, Pelaez T, Shields RK, Garcia-Effron G, Guinea J, Sanguinetti M, Turnidge J. 2017. Multicenter study of method-dependent epidemiological cutoff values for detection of resistance in Candida spp. and Aspergillus spp. to amphotericin B and echinocandins for the Etest agar diffusion method. Antimicrob Agents Chemother 61:e01792-16. doi: 10.1128/AAC.01792-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute. 2018. Epidemiological cutoff values for antifungal susceptibility testing. CLSI supplement M59, 2nd ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2016. Principles and procedures for the development of epidemiological cutoff values for antifungal susceptibility testing. CLSI M57 document, 1st ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 28.Espinel-Ingroff A, Turnidge J. 2016. The role of epidemiological cutoff values (ECVs/ECOFFs) in antifungal susceptibility testing and interpretation for uncommon yeasts and moulds. Rev Iberoam Micol 33:63–75. doi: 10.1016/j.riam.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterization of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 30.Meletiadis J, Curfs-Breuker I, Meis JF, Mouton JW. 2017. In vitro antifungal susceptibility testing of Candida isolates with the EUCAST methodology, a new method for ECOFF determination. Antimicrob Agents Chemother 61:e02372-16. doi: 10.1128/AAC.02372-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alastruey-Izquierdo A, Alcazar-Fuoli L, Cuenca-Estrella M. 2014. Antifungal susceptibility profile of cryptic species of Aspergillus. Mycopathologia 178:427–433. doi: 10.1007/s11046-014-9775-z. [DOI] [PubMed] [Google Scholar]
- 32.Howell S, Hazen KC. 2011. Candida, Cryptococcus and other yeasts of medical importance In Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW (ed), Manual of clinical microbiology, 10th ed ASM Press, Washington, DC. [Google Scholar]
- 33.Chau AS, Mendrick CA, Sabatelli FL, Loebenberg D, McNicholas PM. 2004. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother 48:2124–2131. doi: 10.1128/AAC.48.6.2124-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morio F, Loge C, Besse B, Hennequin C, Le Pape P. 2010. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn Microbiol Infect Dis 66:373–384. doi: 10.1016/j.diagmicrobio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Flowers SA, Brendan CB, Whaley SG, Schuler MA, Rogers D. 2015. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 59:450–460. doi: 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivero-Menendez O, Alastruey-Izquierdo A, Mellado E, Cuenca-Estrella M. 2016. Triazole resistance in Aspergillus spp.: a worldwide problem? J Fungi 2:21. doi: 10.3390/jof2030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Effron G, Dilger A, Alcazar-Fuoli L, Park S, Mellado E, Perlin DS. 2008. Rapid detection of triazole antifungal resistance in Aspergillus fumigatus. J Clin Microbiol 46:1200–1206. doi: 10.1128/JCM.02330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. 2005. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother 49:668–679. doi: 10.1128/AAC.49.2.668-679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]