Candida auris is an emerging multidrug-resistant yeast that can cause serious invasive infections. The accurate and rapid assessment of antifungal resistance is important for effective patient management.
KEYWORDS: Candida auris, ERG11, FKS, azole, echinocandin, molecular beacons, resistance
ABSTRACT
Candida auris is an emerging multidrug-resistant yeast that can cause serious invasive infections. The accurate and rapid assessment of antifungal resistance is important for effective patient management. A novel and highly accurate diagnostic platform was established for the rapid identification of ERG11 mutations conferring azole resistance and FKS1 mutations associated with echinocandin resistance in C. auris. Using allele-specific molecular beacons and DNA melting curve analysis following asymmetric PCR, a duplex ERG11 assay and a simplex FKS1 HS1 assay were developed to identify the most prominent resistance-associated mutations (Y132F and K143R in ERG11; S639F in FKS1 HS1) within 2 h. Assays were validated by testing a panel of 94 C. auris clinical isolates in a blind manner. The molecular diagnostic results from the assays were 100% concordant with DNA sequencing results. This platform has the potential to overcome the deficiencies of existing in vitro susceptibility-based assays to identify azole- and/or echinocandin-resistant C. auris, and thus, it holds promise as a surrogate diagnostic method to direct antifungal therapy more effectively.
INTRODUCTION
Candida auris is an emerging yeast pathogen that has posed a serious challenge to global health. C. auris was first identified in 2009 from the external ear canal discharge of a patient in Japan (1) and quickly spread around the globe, with reports from five continents (2, 3). C. auris can cause invasive infections and has been associated with outbreaks in health care settings (4). The U.S. Centers for Disease Control and Prevention (CDC) issued a clinical alert to health care facilities about C. auris to local or public health authorities in June 2016 (5). Moreover, many clinical isolates of C. auris have been reported to be multidrug resistant (MDR) with reduced susceptibility to two or even three classes of antifungal drugs, exhausting available therapeutic options (6–8).
Despite multiple mechanisms of azole resistance being described in other Candida species, mutations in ERG11 (the gene encoding lanosterol 14-α-demethylase, the target enzyme in the ergosterol synthesis pathway) have been the only reported mechanism responsible for azole resistance in C. auris thus far. A number of ERG11 mutations have been observed in C. auris isolates originating from different geographic areas (7, 9); however, only the Y132F and K143R substitutions are definitely linked with the reduced azole susceptibility in C. auris (7). Of note, the direct contribution of Y132F and K143R to conferring azole resistance in C. auris has been confirmed in a recent study that expressed different C. auris ERG11 alleles in Saccharomyces cerevisiae (10). Therefore, these two substitutions have been suggested to be valuable as initial markers for C. auris azole resistance within South Asian and South American clade isolates (10).
Echinocandin resistance, although at a lower frequency than azole resistance, has been reported in isolates from both India and South Africa (9, 11–13). As in other Candida species, echinocandin resistance in C. auris is associated with hot spot (HS) mutations of FKS, the gene encoding the catalytic subunits of the drug target enzyme β-1,3-d-glucan synthase (14). In an Indian study, all four isolates containing a serine to phenylalanine amino acid substitution at codon 639 (S639F) had elevated pan-echinocandin MICs (7) and did not respond to echinocandin therapy in the mouse model of invasive candidiasis (6). A different amino acid substitution in the same position (S639P) was recently reported in echinocandin-resistant C. auris isolates (13).
Given the prevalence of MDR C. auris, resistance surveillance is recommended in patients who are infected or colonized with C. auris and are on antifungals (15). Antifungal susceptibility profiles can be assessed phenotypically, using either microdilution or disk diffusion MIC assays in accordance with the Clinical and Laboratory Standards Institute (CLSI) M27-A3 standard (16) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) definitive document EDef 7.2 (17). However, standardized susceptibility assays have limitations, such as being time-consuming, subjective for reading visual end points, and prone to interlaboratory variability, especially for caspofungin. They also show known Eagle effects with echinocandins, which interfere with accurate MIC readings (6).
Now that azole and echinocandin resistance in C. auris are closely associated with specific ERG11 and FKS1 mutations, genotyping both genes is likely to offer the most valuable information to facilitate effective resistance detection and antifungal therapy. Recently, we developed a rapid diagnostic platform to identify FKS-associated echinocandin resistance in Candida glabrata, using asymmetric PCR in conjunction with molecular beacon (MB) probe-based melting curve analysis (18). Herein, we have adopted the same strategy to develop a molecular diagnostic assay platform for the rapid genotyping of ERG11 and FKS1 HS1 of C. auris. Assay performance was evaluated in a blind manner on a panel of clinical C. auris isolates acquired from different sources.
RESULTS
Melting curve analysis-based ERG11 genotyping.
To distinguish azole-resistant ERG11 mutants from the wild type (WT), we designed two molecular beacon probes labeled with different fluorophores (6-carboxyfluorescein [FAM] and CAL Fluor Red [CR610]) to facilitate one-step genotyping at both target amino acid positions Y132 and K143. The probes were designed to perfectly match the Y132WT- and K143R-encoding DNA sequences in the target region but possess lower binding energies to non-Y132WT and K143WT sequences, respectively. To enable the efficient dissociation of probe-target hybrids for rapid genotyping, asymmetric PCR amplifications were optimized to generate an excess of the antisense strand of the target. Owing to different thermal-dynamic stabilities of the probe-target hybrids, characteristic profiles were produced for different ERG11 genotypes in the subsequent melting curve analysis. Figure 1A shows representative melting curves of the Y132 genotypes. The Y132F profile had a signature melting temperature (Tm) of 55.84°C ± 0.07°C, remarkably different from the Tm of 60.53°C ± 0.07°C of the WT. The K143 profile was captured simultaneously in the same assay through the CR610 channel. A melting curve with the Tm at 52.48°C ± 0.04°C was featured for the WT, and the Tm was obtained at 58.90°C ± 0.06°C for K143R (Fig. 1B).
FIG 1.
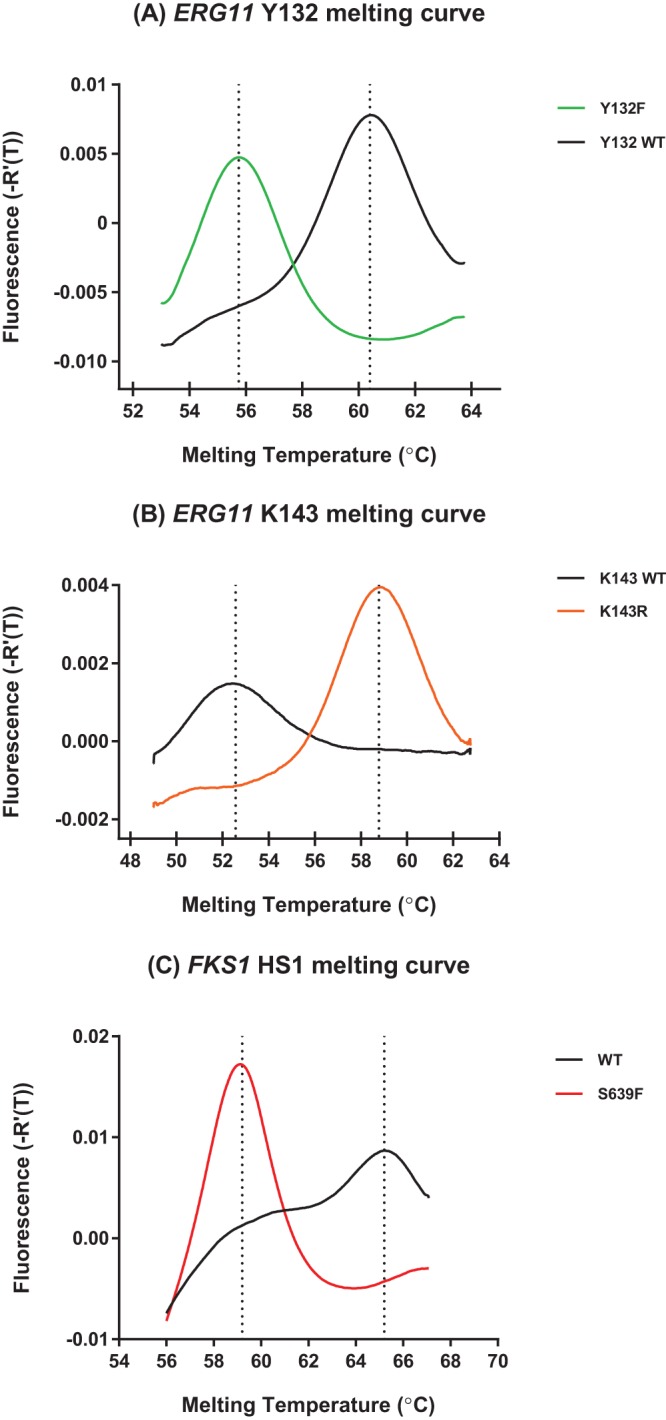
Representative melting profiles for C. auris ERG11 genotypes, including Y132WT and Y132F (A) and K143WT and K143R (B) and FKS1 HS1 genotypes, including WT and S639F (C).
Melting curve analysis-based FKS1 HS1 genotyping.
In the FKS1 HS1 assay, an MB complementing the WT sequence in the target region was used to distinguish the mutated genotype from the WT on the postamplification melting curve. Figure 1C shows melting curves of FKS1 HS1 WT and mutant fks1 encoding S639F from representative strains. The signature melting temperature for each FKS1 HS1 genotype was widely separated from each other, with an approximate 6°C difference.
Probe validation experiments.
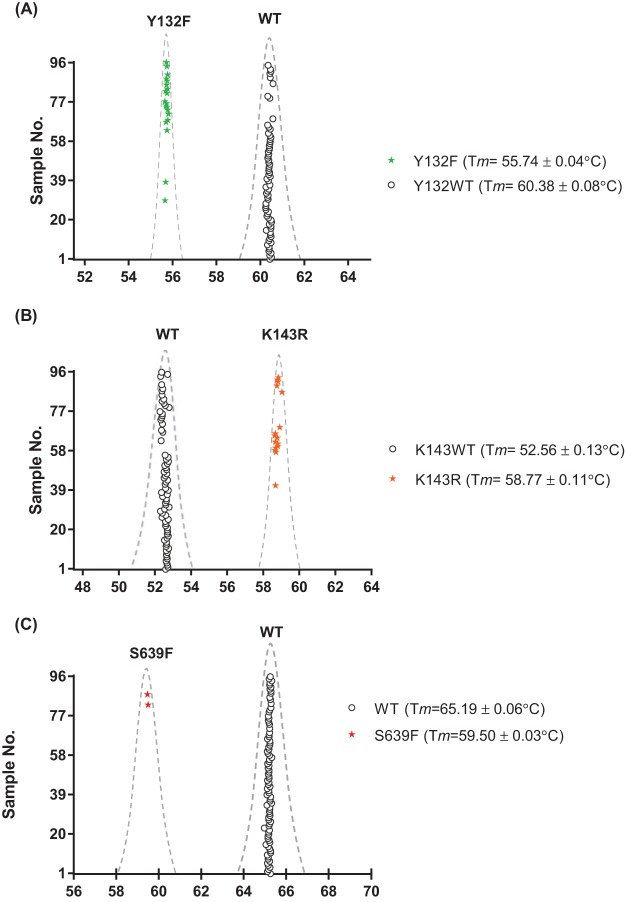
As a proof of concept, we evaluated the diagnostic performance of this novel genotyping platform using a panel of 94 C. auris clinical isolates. Assay performers were blinded to information on the ERG11 and FKS1 sequences and antifungal susceptibility at the time of molecular diagnosis. All isolates were tested in duplicate for these assays, and reference strains representing all genotypes used for platform establishment were tested in parallel in every run. The inclusion of reference strains in each test run was important, as they served as systematic quality controls to ensure the assay’s accuracy and reproducibility. Robust and reproducible melting profiles were observed for all tested isolates. The ERG11 duplex assay identified 73 Y132WT sequences (Tm = 60.38°C ± 0.08°C), 21 Y132F mutations (55.74°C ± 0.04°C) (Fig. 2A), 78 K143WT sequences (Tm = 52.56°C ± 0.13°C), and 16 K143R mutations (58.77°C ± 0.11°C) (Fig. 2B). The FKS1 HS1 assay picked up 92 WT sequences (Tm = 65.19°C ± 0.06°C) and captured two S639F alleles (Tm = 59.50°C ± 0.03°C) (Fig. 2C). DNA sequencing results confirmed all predicted genotypes. In summary, both ERG11 and FKS1 HS1 assays are highly accurate in differentiating prominent azole- and echinocandin-resistant mutants from the WT population, with 100% specificity and 100% sensitivity. The assays in this study are designed to be specific to ERG11 or FKS1 HS1 of C. auris; we tested closely related species and no cross-reactivity to any other species was detected.
FIG 2.
ERG11 Y132 (A), ERG11 K143 (B), and FKS1 HS1 (C) Tm distributions of 94 C. auris clinical isolates tested in a blind manner.
DISCUSSION
C. auris has become a global concern, with existing multidrug resistance to fluconazole and amphotericin B, and is characterized by a strong potential for nosocomial transmission (4, 9). Studies examining the susceptibility of this organism to antifungals have used a variety of methods, including CLSI and EUCAST broth microdilution (16, 17) or their commercial counterparts, such as Sensititre YeastOne, Etest and the Vitek 2 yeast susceptibility system. At present, there are no antifungal clinical breakpoints reported for C. auris by CLSI or EUCAST. The tentative epidemiological cut off values (ECVs) proposed in a recent study were determined based on the analysis of MIC distributions of 123 clinical isolates of C. auris from India (19). These ECVs are valuable in the analysis of MICs of isolates from the South Asian clade, yet application of the ECVs to isolates from other clades may lead to incorrect estimation of resistance, since there are strong indications that MIC distributions can vary substantially for C. auris isolates from different clades. Therefore, an analysis of susceptibility results obtained by these phenotypic tests is not straightforward and may have difficulties defining susceptible or resistant isolates.
There is an emerging need for alternative diagnostic tools, which allow rapid and accurate identification of resistance-associated mutations in C. auris. These tests have the potential to become part of the routine clinical laboratory tests for resistance detection to help direct therapy and enhance epidemiological surveillance. Encoding the primary drug targets of which specific mutations account for the most prominent azole or echinocandin resistance in C. auris, ERG11 and FKS1 are perfect targets for the molecular diagnosis of drug-resistant C. auris. Along this line, we developed the first molecular diagnostic platform for rapid ERG11 and FKS1 HS1 genotyping of C. auris isolates. The assays are designed to detect known mechanisms of resistance and are not intended at this stage to be a surrogate for the standard antifungal susceptibility test. This platform, taking advantage of the single nucleotide polymorphism sensitive nature of molecular beacons and incorporating asymmetric PCR, accurately discriminated WT from drug-resistant ERG11 and FKS1 HS1 mutants within 2 h, without the need of DNA sequencing. Both ERG11 and FKS1 HS1 assays are highly robust, as each genotype presented a signature melting curve with a Tm highly distinguishable from others. Bearing a robust discrimination power, this novel platform demonstrated 100% accuracy in identifying C. auris clinical isolates in a blind manner as either WT or drug resistant genotype in the subsequent proof-of-concept diagnostic performance evaluation.
There are some limitations in this study. First, only Y132 and K143 were included in the ERG11 genotyping assay. We recognize the possibility of other mechanisms being involved in azole resistance in C. auris. However, given that Y132F and K143R are currently the most prevalent and the only mutations confirmed to confer azole resistance in C. auris, the present ERG11 assay is of proper clinical value. Second, there was only one mutated genotype (S639F) included in the FKS1 HS1 assay development and evaluation. To date, only two FKS1 HS1 mutations have been found in echinocandin-resistant C. auris isolates around the world, S639F and the recently reported S639P (13). It is unfortunate that we did not have the S639P mutant strain encompassed in our study due to limited strain access. There is a chance that the Tm value of the S639P allele may not be distinguishable from S639F, since these mutations occur on the same codon and their melting profiles are presumably similar. However, it is anticipated that the S639P melting signature will be remarkably different from that of the WT, given what has been generated with the S639F allele and our previous experience with C. glabrata FKS alleles (18), ensuring that the assay is a useful tool for discriminating echinocandin-resistant mutants from the susceptible WT population. Nevertheless, further work to include the S639P allele is warranted upon availability of the strain. It should be noted that the FKS1 HS1 assay in this study is an open platform that allows the easy incorporation of any unknown HS1 alleles without the need of assay redesign. The assay becomes applicable to any novel FKS1 HS1 mutations as soon as the corresponding genotype is captured and added into the reference genotype library.
In summary, we have developed a rapid and accurate diagnostic platform that has the potential to overcome the deficiencies of existing MIC-based assays to identify azole- and echinocandin-resistant C. auris. It holds the promise to be a robust diagnostic method to better detect resistance-associated mutations and to direct more effective antifungal therapy against infections caused by C. auris, and it can be used for epidemiologic surveillance.
MATERIALS AND METHODS
Candida auris strains and culture conditions.
A total of 98 C. auris isolates were included in the present study. Forty C. auris isolates were obtained from Vallabhbhai Patel Chest Institute, University of Delhi (Delhi, India); 48 isolates were obtained from Clinica General del Norte (Barranquilla, Colombia); 8 isolates were obtained from a clinic of high complexity (Santa Marta, Colombia); and 2 isolates were from Westerdijk Fungal Biodiversity Institute (formerly CBS-KNAW, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands). Four strains with different ERG11 (Y132 and K143) and FKS1 HS1 (S639) genotypes were used for assay development and reference library establishment, and the remaining 94 C. auris isolates were used for the validation study in a blind fashion. Strains were grown on yeast extract-peptone-dextrose (YPD) agar plates, and species identification of all isolates was performed by sequencing of the ribosomal DNA (rDNA) region (6).
Antifungal susceptibility testing.
Echinocandin (caspofungin, micafungin, and anidulafungin) and azole (fluconazole and voriconazole) MICs were determined according to CLSI protocol M27-A3 (Table 1) (16).
TABLE 1.
Distribution of different Candida auris EGR11 and FKS1 HS1 variant MICs
| Gene variant | MIC range (mode) (mg/liter) for: |
||||
|---|---|---|---|---|---|
| Fluconazole | Voriconazole | Micafungin | Anidulafungin | Caspofungin | |
| ERG11 substitutions | |||||
| WT | 2 to 64 (2) | 0.03 to 1 (0.06) | |||
| Y132F | 64 to >128 (>128) | 2 to >16 (4) | |||
| K143R | 32 to >128 (>128) | 0.12 to 1 (0.5) | |||
| FKS1 HS1 substitutions | |||||
| WT | 0.03 to 0.5 (0.125) | 0.03 to 1 (0.5) | 0.125 to 1 (0.25) | ||
| S639F | 16 | 4 to >16 (>16) | 8 to 16 (8) | ||
DNA extraction.
C. auris DNA was prepared by incubating a single colony of a strain in 100 µl of extraction buffer (60 mM NaHCO3, 250 mM KCl, 50 mM Tris; pH 9.5) at 95°C for 10 min, followed by an addition of 100 µl of 2% bovine serum albumin (20).
Primers and probe design.
Two sets of asymmetric PCR primers were designed to amplify an F132- and K143-containing region of ERG11 and the FKS1 HS1 region of C. auris. The ERG11 region was amplified by using excess primer CauERG11-X (5′-ACTTCCTCTTGGATTCTGGG-3′) and limiting primer CauERG11-L (5′- TGCTTATTCCCACTTGACCACTCCAG-3′). Two molecular beacons were designed targeting the Y132WT and K143R allele sequences of ERG11, and they were CauERG11-Y132WT (5′-FAM-CGCGATTGTCATTTACGACTGTCCCATCGCG-DABCYL-3′, where DABCYL is N-[4-(4-dimethylamino)phenylazo]benzoic acid) and CauERG11-K143R (5′-CR610-CGCGATATGGAGCAGAGGAAATTTGATCGCG-BHQ1-3′ [BHQ1, black hole quencher 1]). The excess primer for FKS1 HS1 amplification was CauF1H1-X (5′-CGTCATGGTGGACAAGTTTCTA-3′) and the limiting primer was CauF1H1-L (5′-GGGTCACTGTGTTTGCTGCTAAGTTGG-3′). The FKS1 HS1 MB was CauF1H1-WT (5′-FAM-CGCGACTTCTTGACTTTGTCCTTGAGAGATCCTGTCGCG-DABCYL-3′) (underlining signifies the stem portion of the molecular beacon).
Asymmetric PCR and molecular beacon-based melting curve analysis.
Asymmetric PCR was carried out on the Mic real-time PCR cycler (Bio Molecular Systems) in a 10-µl reaction volume using SensiFAST probe no-ROX mix (Bioline, London, UK). The ERG11 duplex assay contained 800 nM CauERG11-X, 40 nM CauERG11-L, 500 nM CauERG11-Y132WT MB, 500 nM CauERG11-K143R MB, and 10 to 25 ng of DNA template. ERG11 PCR conditions were 95°C for 3 min; 45 cycles of 10 s at 95°C, 20 s at 60°C, and 30 s at 72°C; and 2 min at 72°C. Immediately after amplification, melting curve analysis was initiated as a minute incubation at 95°C, after which it was melted from 53°C to 64°C with a ramp rate of 0.1°C/s for the Y132WT probe and melted from 49°C to 63°C with a ramp rate of 0.1°C/s for the K143R probe. The FKS1 HS1 assay contained 1 µM CauF1H1-X, 40 nM CauF1H1-L, and 500 nM CauF1H1-WT MB. FKS1 HS1 PCR thermal cycling consisted of a 3-min incubation at 95°C; 45 cycles of 10 s at 95°C, 20 s at 60°C, and 30 s at 72°C; and then incubation at 72°C for 2 min. Immediately after amplification, melting curve analysis was performed at 95°C for 2 min, after which it was melted from 54°C to 67°C with a ramp rate of 0.025°C/s.
DNA sequencing.
ERG11 and FKS1 HS1 regions were amplified and sequenced in both directions as previously described (7).
Statistical analysis.
Tm values for ERG11 and FKS genotypes were determined by melting curve analysis using the Mic real-time PCR system software (Bio Molecular Systems). Genotyping results by rapid molecular diagnostic assays were compared with the DNA sequencing results. The Tm distribution of clinical isolates was analyzed by GraphPad Prism 7.01 software. The accuracy of the novel assays discriminating WT from mutated ERG11 and FKS genotypes was evaluated by calculating sensitivity and specificity for each assay.
Accession number(s).
The ERG11 and FKS1 sequences of representative strains of each genotype in this study were deposited in GenBank with accession numbers MK059959 to MK059966 and MK059967 to MK059974, respectively (Table S1).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by CDC’s investments to combat antibiotic resistance under contract number 200-2017-96195 (BAA FY2017-OADS-01) and Astellas Pharma (Reference Center for Molecular Evaluation of Drug Resistance to Echinocandin and Triazole Antifungal Drugs) to D.S.P. X.H. is supported by the China Scholarship Council (CSC; number 201706210414).
We thank Anuradha Chowdhary and Indira Berrio for providing the strains.
D.S.P. receives funding from the U.S. National Institutes of Health and contracts from Astellas, Scynexis, Cidara, and Amplyx. He serves on advisory boards for Astellas, Cidara, Amplyx, Scynexis, and Matinas. In addition, D.S.P. has an issued U.S. patent concerning echinocandin resistance. The remaining authors have no potential conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01811-18.
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Candida auris Incident Management T, Manuel R, Brown CS. 2018. Candida auris: a review of the literature. Clin Microbiol Rev 31:e00029-17. doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2016. Clinical alert to U.S. healthcare facilities—June 2016. Emergence of invasive infections caused by the multidrug-resistant yeast Candida auris. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/fungal/diseases/candidiasis/candida-auris-alert.html. [Google Scholar]
- 6.Kordalewska M, Lee A, Park S, Berrio I, Chowdhary A, Zhao Y, Perlin DS. 2018. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother 62:e00238-18. doi: 10.1128/AAC.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhary A, Voss A, Meis JF. 2016. Multidrug-resistant Candida auris: “new kid on the block” in hospital-associated infections? J Hosp Infect 94:209–212. doi: 10.1016/j.jhin.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Healey KR, Kordalewska M, Jimenez Ortigosa C, Singh A, Berrio I, Chowdhary A, Perlin DS. 2018. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 62:e01427-18. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 12.Sharma C, Kumar N, Pandey R, Meis JF, Chowdhary A. 2016. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berkow EL, Lockhart SR. 2018. Activity of CD101, a long-acting echinocandin, against clinical isolates of Candida auris. Diagn Microbiol Infect Dis 90:196–197. doi: 10.1016/j.diagmicrobio.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Perlin DS. 2015. Echinocandin resistance in Candida. Clin Infect Dis 61:S612–S617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, Vallabhaneni S. 2018. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol doi: 10.1093/mmy/myy054. [DOI] [PubMed] [Google Scholar]
- 16.CLSI. 2012. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard—3rd ed. CLSI document M27-A3 CLSI, Wayne, PA. [Google Scholar]
- 17.Arendrup MC, Cuenca-Estrella M, Lass-Florl C, Hope W, Eucast A. 2012. EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin Microbiol Infect 18:E246–E247. doi: 10.1111/j.1469-0691.2012.03880.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Nagasaki Y, Kordalewska M, Press EG, Shields RK, Nguyen MH, Clancy CJ, Perlin DS. 2016. Rapid detection of FKS-associated echinocandin resistance in Candida glabrata. Antimicrob Agents Chemother 60:6573–6577. doi: 10.1128/AAC.01574-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. 2017. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother 61:e00485-17. doi: 10.1128/AAC.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brillowska-Dabrowska A, Saunte DM, Arendrup MC. 2007. Five-hour diagnosis of dermatophyte nail infections with specific detection of Trichophyton rubrum. J Clin Microbiol 45:1200–1204. doi: 10.1128/JCM.02072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.