Increased expression of drug efflux pumps and changes in the target enzyme Erg11p are known to contribute to azole resistance in Candida albicans, one of the most prevalent fungal pathogens. Mutations that inactivate ERG3, which encodes sterol Δ5,6-desaturase, also confer in vitro azole resistance.
KEYWORDS: Candida albicans, ERG3, azoles, resistance, trailing growth
ABSTRACT
Increased expression of drug efflux pumps and changes in the target enzyme Erg11p are known to contribute to azole resistance in Candida albicans, one of the most prevalent fungal pathogens. Mutations that inactivate ERG3, which encodes sterol Δ5,6-desaturase, also confer in vitro azole resistance. However, it is unclear whether the loss of Erg3p activity is sufficient to confer resistance within the mammalian host, and relatively few erg3 mutants have been reported among azole-resistant clinical isolates. Trailing growth (residual growth in the presence of the azoles) is a phenotype observed with many C. albicans isolates and, in its extreme form, can be mistaken for resistance. The purpose of this study was to determine whether the growth of Erg3p-deficient C. albicans mutants in the presence of the azoles possesses the characteristics of azole resistance or of an exaggerated form of trailing growth. Our results demonstrate that, similar to trailing isolates, the capacity of an erg3Δ/Δ mutant to endure the consequences of azole exposure is at least partly dependent on both temperature and pH. This contrasts with true azole resistance that results from enhanced drug efflux and/or changes in the target enzyme. The erg3Δ/Δ mutant and trailing isolates also appear to sustain significant membrane damage upon azole treatment, further distinguishing them from resistant isolates. However, the insensitivity of the erg3Δ/Δ mutant to azoles is unaffected by the calcineurin inhibitor cyclosporin A, distinguishing it from trailing isolates. In conclusion, the erg3 mutant phenotype is qualitatively and quantitatively distinct from both azole resistance and trailing growth.
INTRODUCTION
Mortality rates associated with invasive fungal infections (IFIs) remain alarmingly high, despite the availability of three major classes of antifungal drugs (1). The azoles remain the most widely used antifungal treatments, acting through inhibition of lanosterol demethylase (Erg11p), an enzyme required for ergosterol biosynthesis. Depletion of cellular ergosterol and the accumulation of abnormal sterol species, such as 14α-methylergosta-8,24(28)-dien-3β,6α-diol, are both thought to cause plasma membrane dysfunction and ultimately to arrest fungal growth following azole treatment (2). In Candida albicans, one of the most prevalent fungal pathogens, several well-described mechanisms are known to contribute to azole resistance, including increased expression of drug efflux pumps belonging to the major facilitator and ABC transporter superfamilies, as well as the target enzyme (Erg11p) itself (3, 4). Mutations that reduce the azole binding affinity of the target enzyme are also important determinants of susceptibility (5–7). However, a combination of the aforementioned mechanisms is usually necessary to confer clinically relevant levels of azole resistance (3, 4, 8, 9).
A less common mechanism of azole resistance arises through inactivation of sterol Δ5,6-desaturase (Erg3p), the enzyme responsible for converting the lanosterol/14α-methylfecosterol that accumulates upon azole-mediated inhibition of Erg11p into the toxic diol species (2). As a result, ERG3-deficient mutants of C. albicans accumulate 14α-methylfecosterol instead of 14α-methylergosta-8,24(28)-dien-3β,6α-diol following azole treatment, which is apparently compatible with continued growth (2). In contrast to the aforementioned resistance mechanisms, inactivation of the ERG3 gene results in complete azole insensitivity in a single step. Curiously, while azole-resistant erg3 mutants can be readily selected under in vitro conditions (10), they are detected only rarely among azole-resistant clinical isolates (2, 11–13). Furthermore, Miyazaki et al. reported that, while an erg3Δ/Δ mutant was apparently azole resistant in vitro, it was susceptible in a mouse model of disseminated candidiasis (14). Thus, while loss of Erg3p activity enables C. albicans to grow in the presence of the azoles in vitro, the contribution of this mechanism to azole resistance in a clinical sense is less certain. Trailing growth is a phenotype observed in a significant proportion of C. albicans isolates, and it can be easily mistaken for azole resistance (15). Using the standard CLSI broth microdilution susceptibility testing method, trailing isolates typically appear azole susceptible after 24 h of incubation, but continued growth makes them appear resistant by the 48-h time point. Nonetheless, patients and experimental animals infected with trailing isolates generally respond well to treatment with the azoles (16, 17). The purpose of this study was to determine whether the continued growth of Erg3p-deficient C. albicans mutants in the presence of the azoles represents true azole resistance or resembles an exaggerated form of the trailing growth phenotype.
RESULTS
A C. albicans erg3Δ/Δ mutant exhibits characteristics of both azole resistance and trailing growth.
In this study, we used a previously reported C. albicans erg3Δ/Δ mutant and its isogenic control strain, in which a wild-type (WT) copy of ERG3 has been reintroduced (18). These strains were compared with the well-characterized azole-resistant clinical isolate TW17, which overexpresses the Mdr1p and Cdr1p efflux pumps, as well as a variant of the target enzyme Erg11p with reduced affinity for azoles (4). Additional reference strains included the azole-susceptible isolate SC5314 and several other C. albicans clinical isolates that exhibit either trailing growth or azole resistance. We also constructed a panel of isogenic strains, each possessing a single mechanism known to confer azole resistance, specifically (i) overexpression of the Cdr1p azole efflux pump (strain OCDR), (ii) overexpression of the Mdr1p azole efflux pump (strain OMDR), and (iii) overexpression of the azole target protein, Erg11p (strain OERG), as well as a strain overexpressing all three proteins (strain OTR). Overexpression of MDR1, CDR1, and ERG11 was confirmed by quantitative real-time PCR (qRT-PCR) (see Fig. S1A in the supplemental material).
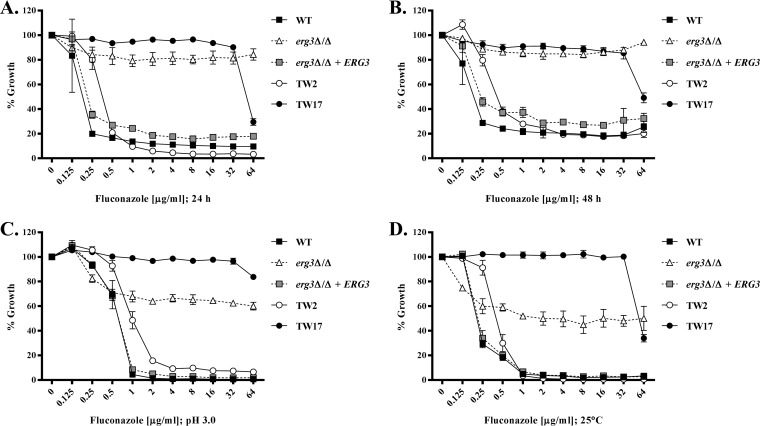
We initially compared the fluconazole susceptibility of each isolate using the standard CLSI broth microdilution protocol (19), and we quantified fungal growth at each drug concentration as the optical density at 600 nm (OD600). As expected, the erg3Δ/Δ mutant was apparently insensitive to fluconazole at either the 24-h or 48-h time point (Fig. 1A and B), with a growth profile similar to that of the azole-resistant isolate TW17. One notable difference was that TW17 growth was partly suppressed at very high fluconazole concentrations (64 µg/ml), whereas growth of the erg3Δ/Δ mutant was unaffected. A different response was seen for SC5314 and the JS7, JS17, JS22, and JS23 clinical isolates, each of which exhibited a low fluconazole MIC (≤0.5 µg/ml) but exhibited significant and reproducible levels of residual growth at suprainhibitory concentrations, at both 24 and 48 h (Fig. 2A and B). Therefore, we classified these as trailing isolates. A third group of isolates (JS28, JS29, and JS40) had intermediate MICs (8 to 32 µg/ml) and were classified as resistant, according to the CLSI guidelines (19). The OCDR, OMDR, and OERG strains had 4- to 16-fold increases in fluconazole MICs, compared with the isogenic control, while the OTR strain had a MIC approximately 128-fold greater than that of the control strain (Fig. S1B and C). Interestingly, both the OCDR and OTR strains had noticeably more trailing growth at supra-MIC concentrations of fluconazole than did the control strain (Fig. S1B and C), possibly indicating a relationship between elevated CDR1 expression and trailing growth.
FIG 1.
A Candida albicans erg3Δ/Δ mutant displays features of both azole resistance and trailing growth. (A and B) The fluconazole susceptibilities of a WT strain (GP1), the erg3Δ/Δ mutant, erg3Δ/Δ + ERG3, and a matched pair of azole-susceptible and azole-resistant clinical isolates (TW2 and TW17) were evaluated using the CLSI broth microdilution protocol; growth was measured as the OD600 after 24 h (A) and 48 h (B) of incubation, and results are expressed as a percentage of the growth in the no-drug (DMSO alone) control wells for each strain. (C and D) The same experiment was also conducted with RPMI 1640 medium adjusted to pH 3.0 (C) or with incubation at 25°C instead of 35°C (D), and growth was assessed after 48 h of incubation. Data in all panels are the means ± standard deviations of three biological replicates.
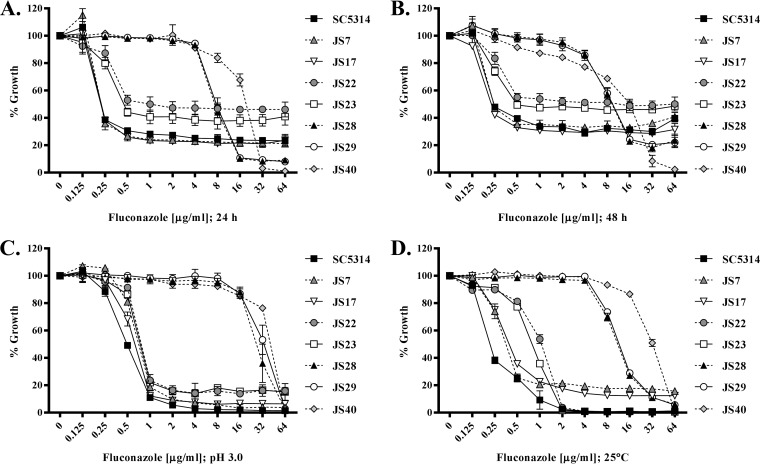
FIG 2.
Trailing growth but not azole resistance is eliminated by adjusting the incubation temperature or the pH of the medium. (A and B) The fluconazole susceptibilities of the WT isolate SC5314 and a set of clinical isolates (JS strains) were evaluated using the CLSI broth microdilution protocol; growth was measured as the OD600 after 24 h (A) and 48 h (B) of incubation, and results are expressed as a percentage of the growth in the no-drug (DMSO alone) control wells for each strain. (C and D) The same experiment was also conducted with RPMI 1640 medium adjusted to pH 3.0 (C) or with incubation at 25°C (D), and growth was assessed after 48 h of incubation. The means ± standard deviations of three biological replicates are indicated for all panels.
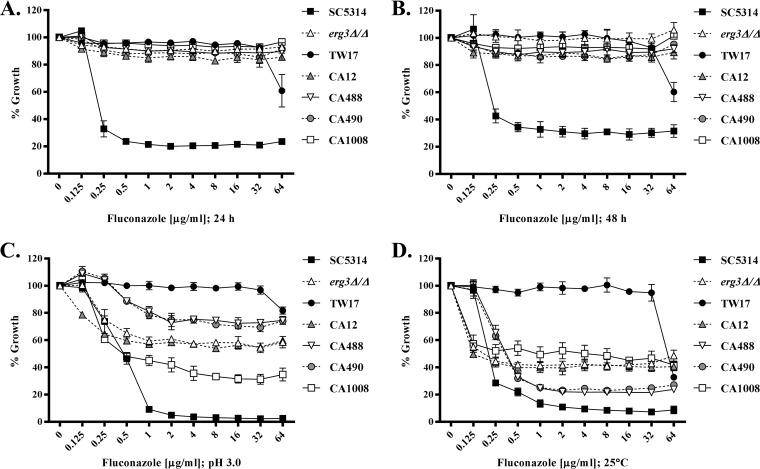
It was shown previously that acidifying the growth medium or reducing the incubation temperature was sufficient to eliminate trailing growth but not true azole resistance (20). Reducing the pH of the RPMI 1640 medium to 3.0 did not dramatically affect the MIC for any of the clinical C. albicans isolates but significantly reduced the trailing growth observed with isolates SC5314, JS7, JS17, JS22, and JS23 (Fig. 2C). Similarly, although there was a slight increase (2- to 4-fold) in the MIC for each strain, the relative susceptibilities of the OCDR, OMDR, OERG, and OTR strains were unaltered in the acidified medium (Fig. S1D) but all trailing growth was eliminated. Almost identical results were observed with reduction of the incubation temperature from 35°C to 25°C, in that MICs were not substantially affected but trailing growth was reduced or eliminated (Fig. 1D and 2D; also see Fig. S1E). Interestingly, in acidified medium, fluconazole was able to inhibit the growth of the erg3Δ/Δ mutant to a measurable degree at all concentrations ≥0.5 µg/ml, which dramatically reduced the MIC (Fig. 1C). However, the mutant maintained extremely high levels of residual growth at suprainhibitory concentrations of fluconazole, a phenotype resembling the trailing growth phenotype normally observed at pH 7. Similarly, the MIC for the erg3Δ/Δ mutant also decreased substantively at 25°C but, in contrast to the trailing isolates, the mutant sustained very high levels of residual growth at suprainhibitory concentrations of fluconazole. Thus, while the erg3Δ/Δ mutant appears fully resistant to fluconazole under the conditions of the standard CLSI susceptibility testing protocol, its capacity for continued growth is partly dependent on the specific culture conditions used. Similar results were found for several clinically derived Erg3p-deficient mutants (11), for which growth in the presence of fluconazole is also partly temperature and pH dependent (Fig. 3). In this respect, the continued growth of the erg3 mutants in the presence of fluconazole shares at least some characteristics with the trailing growth phenomenon. However, the phenotype of the erg3Δ/Δ mutant is distinct from that of the trailing isolates, as the growth observed in the presence of fluconazole is substantially more robust under all conditions tested. The erg3 phenotype also contrasts with that of the truly resistant isolates TW17, OTR, JS28, JS29, and JS40, for which growth in the presence of fluconazole is largely independent of the culture conditions used.
FIG 3.
The growth of Candida albicans Erg3p-deficient clinical isolates in the presence of fluconazole is partly temperature and pH dependent. (A and B) The fluconazole susceptibilities of C. albicans SC5314, the erg3Δ/Δ mutant, the azole-resistant isolate TW17, and the previously reported Erg3p-deficient clinical isolates CA12, CA1008, CA488, and CA490 were compared using the CLSI broth microdilution protocol; growth was quantified as the OD600 after 24 h (A) and 48 h (B) of growth, and results are expressed as a percentage of the no-drug (DMSO alone) control wells. (C and D) The same experiment was conducted with RPMI 1640 medium adjusted to pH 3.0 (C) or with incubation at 25°C (D), and growth was assessed after 48 h. The means ± standard deviations of three biological replicates are shown for each panel.
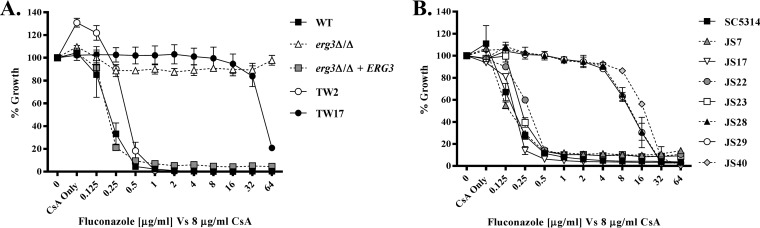
Finally, the azole antifungals are fungistatic rather than fungicidal, with C. albicans survival in the presence of growth inhibitory concentrations of fluconazole depending on signaling through the calcineurin phosphatase (21–23). Inhibition of the calcineurin pathway with cyclosporine A thus renders the azoles fungicidal and eliminates trailing growth (21–23). Therefore, we determined whether the capacity of the erg3Δ/Δ mutant to grow in the presence of fluconazole was affected by cyclosporine A. While cyclosporine A did not significantly alter the MIC for any C. albicans strain, it eradicated the trailing growth observed previously for SC5314, JS7, JS17, JS22, and JS23 (Fig. 4B; also see Fig. S2B). These data confirm that, while trailing growth is dependent on calcineurin signaling, true azole resistance is not. Cyclosporin A treatment was not sufficient to restore fluconazole sensitivity or to reduce the growth of the erg3Δ/Δ mutant in the presence of fluconazole (Fig. 4A; also see Fig. S2A). This capacity to survive and to proliferate in the absence of calcineurin signaling is consistent with true azole resistance, rather than an exaggerated form of the trailing growth phenotype. Thus, while the erg3Δ/Δ mutant shares at least some characteristics with the trailing growth phenomenon, other characteristics more closely resemble true azole resistance.
FIG 4.
Growth of the Candida albicans erg3Δ/Δ mutant in the presence of fluconazole is not dependent on calcineurin signaling. (A) The growth of the WT strain (GP1), the erg3Δ/Δ mutant, and a pair of matched azole-susceptible and azole-resistant clinical isolates (TW2 and TW17) was compared in fluconazole dose-response experiments in the absence or presence of 8 µg/ml cyclosporine A (CsA), according to the CLSI broth microdilution protocol. The growth of each strain was measured as the OD600 after 48 h, and results are expressed as a percentage of growth in the no-drug (DMSO) control wells. (B) The same experiment was performed with strain SC5314 and a set of clinical isolates (JS strains). The means ± standard deviations of three biological replicates are shown in each panel.
The C. albicans erg3Δ/Δ mutant sustains significant membrane damage upon fluconazole treatment.
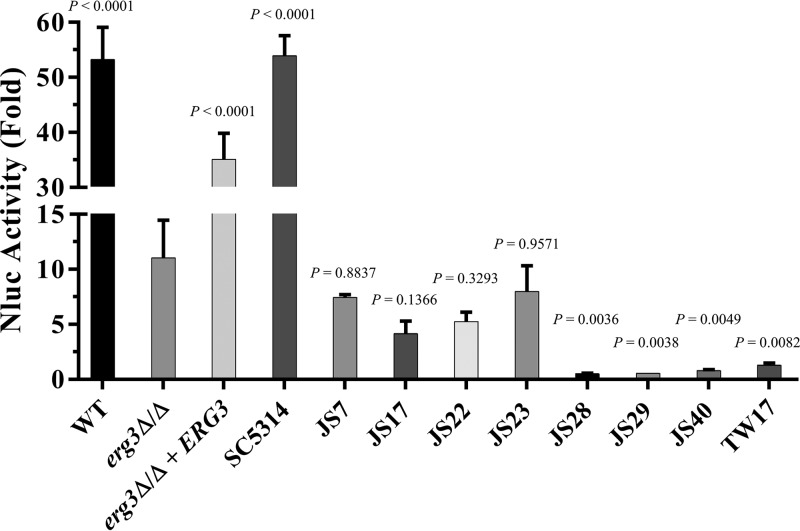
We next compared the amount of membrane damage sustained upon azole treatment. A cytoplasmic version of NanoLuc luciferase (Nluc) (Promega Corp.) (24) was expressed in the erg3Δ/Δ mutant and reference strains, so that Nluc release into the culture supernatant could be used to detect loss of plasma membrane integrity (Fig. 5). Substantial amounts of Nluc activity were detected in the culture supernatant of the WT reference strain SC5314 after treatment with 5 µg/ml fluconazole for 24 h at 35°C (∼50- to 60-fold increases over untreated cells) (Fig. 5), indicating loss of plasma membrane integrity. More modest amounts of Nluc were released from the trailing isolates JS7, JS17, JS22, and JS23 (∼5- to 10-fold increases over untreated control cells), while no increase in Nluc release was detected for the resistant isolates JS28, JS29, JS40, and TW17. Interestingly, upon fluconazole exposure, the erg3Δ/Δ mutant released significantly more Nluc than the resistant isolates and in fact released levels similar to those of the trailing isolates. Nonetheless, the amount of Nluc released was substantially less than those for the isogenic ERG3+ revertant strain, the WT reference strain (GP1), and the corresponding clinical isolate from which all of these laboratory-derived strains were ultimately derived (SC5314). Thus, while the erg3Δ/Δ mutant tolerates fluconazole treatment better than the WT strain, it is not unaffected. In conclusion, these data confirm that the ability of the erg3Δ/Δ mutant to endure the consequences of azole exposure is qualitatively and quantitatively distinct from true azole resistance conferred by enhanced drug efflux or changes in the target enzyme Erg11p.
FIG 5.
The Candida albicans erg3Δ/Δ mutant sustains membrane damage following fluconazole treatment. The Candida albicans WT strain (GP1), the erg3Δ/Δ mutant, erg3Δ/Δ + ERG3, a group of trailing isolates (SC5314, JS7, JS17, JS22, and JS23), and a set of azole-resistant isolates (JS28, JS29, JS40, and TW17) were transformed with plasmid pDUPNluc. The resulting strains were then grown in 96-well plates in YNB medium with 5 µg/ml fluconazole or 0.5% DMSO (no-drug control). After 24 h of incubation at 35°C, culture supernatant was removed from each well and Nluc activity was measured using a luminescent substrate. Nluc activity was normalized with respect to growth (OD600), and results are expressed relative to the no-drug controls for each strain. The means ± standard deviations for three independent transformants of each strain are shown. The means for the experimental groups were compared using one-way analysis of variance and Tukey's multiple-comparison test. The P values for each strain, compared to the erg3Δ/Δ mutant, are indicated.
DISCUSSION
As various research groups and different fields of microbiology use the terms drug “resistance” and “tolerance” inconsistently, or even interchangeably, we will use the following definitions based on the underlying molecular mechanisms. On a molecular level, resistance implies that a drug is unable to engage or to modulate sufficiently the activity of its target protein and thus is ineffective in inhibiting microbial growth. In contrast, drug tolerance implies that a microbe is able to endure the consequences of a drug acting upon its target, and thus it can survive and/or proliferate in the presence of that drug. According to these definitions, drug efflux mechanisms (which prevent the azoles from accessing their intracellular target), elevated target protein expression, and mutations that reduce azole binding or inhibition of Erg11p all represent mechanisms of drug resistance. However, the residual growth observed with trailing isolates should be considered a form of azole tolerance, as the isolates are able to survive and to proliferate in the presence of the fungistatic azoles despite Erg11p inhibition (15, 25, 26). In this context, loss of Erg3p activity should also be considered a form of azole tolerance, as the azoles engage and inhibit Erg11p in both the WT strain and erg3 null mutants, altering the sterol composition of both. The implications of these distinctions become apparent when the physiological consequences are considered. As reported by others, we find that changing the temperature or pH under which the CLSI assay is conducted is sufficient to eliminate the trailing growth observed with the azoles. Thus, while trailing isolates are to some degree able to proliferate in the presence of the azoles under standard conditions, the physiological consequences of ergosterol depletion and the accumulation of abnormal sterols can be more severe when the culture conditions are changed. Trailing growth also depends on the induction of stress responses that are activated by calcineurin (23). In contrast, azole resistance conferred by increased efflux pump or target protein expression is independent of the culture conditions and calcineurin signaling. The results of this study underscore the fact that, while Erg3p-deficient mutants appear completely insensitive to the azoles in standard MIC assays, there are significant physiological consequences that compromise membrane integrity and function. Specifically, following azole-mediated inhibition of Erg11p activity, erg3 null mutants accumulate 14α-methylfecosterol instead of 14α-methylergosta-8,24(28)-dien-3β,6α-diol, which accumulates in the WT strain (2, 11, 27). While 14α-methylfecosterol can apparently provide adequate function to support full growth under the conditions of the standard CLSI assay (19), the level of dysfunction is sufficient to impinge upon growth rates when the conditions (e.g., pH or temperature) are altered. Unlike many of the trailing isolates, however, erg3 null mutants are able to persist and to proliferate to a significant cell density in the presence of fluconazole under all conditions tested. Also, in contrast to trailing isolates, the capacity of erg3 mutants to proliferate in the presence of the azoles does not depend upon calcineurin signaling. Finally, the Nluc release assay revealed that C. albicans erg3 null mutants are not invulnerable to azole treatment but sustain substantively less membrane damage than the WT strain. We reported previously that azole exposure leads to fragmentation of the fungal vacuole, a large acidified intracellular organelle (25). Azole treatment also induces vacuolar fragmentation in the C. albicans erg3Δ/Δ mutant, but to a much lesser extent than for the ERG3+ control strains (Fig. S3). These observations confirm that the level of membrane dysfunction resulting from azole-induced shifts in the cellular sterol content is less severe in erg3 null mutants than ERG3+ strains. In summary, C. albicans erg3 null mutants do not completely conform to the characteristics of either trailing growth or true azole resistance, possessing some qualities of both phenotypes.
Of course, from the perspective of clinicians, the only pertinent point is whether a patient will respond to treatment with an azole antifungal. When interpreted in this light, the results of this study may be revealing. It has been well established that elevated expression of drug efflux pumps and Erg11p contributes to azole resistance and treatment failure in clinical settings (4). In contrast, most C. albicans isolates that exhibit trailing growth have been associated with favorable outcomes following treatment with azoles (28). In a mouse model of disseminated infection, C. albicans trailing isolates were reported to be susceptible to fluconazole treatment (17, 26). In addition, we recently reported that a C. albicans endosomal trafficking mutant that exhibited significant levels of trailing growth in vitro was susceptible to fluconazole treatment in a mouse model of vaginal candidiasis (16).
However, a recent study suggested that the trailing phenotype results from the slow growth of subpopulations of cells that are able to endure the deleterious effects of drug treatment, which may have implications in clinical settings (29). Clinical isolates from patients with persistent candidemia were also found to have higher levels of trailing than those readily cleared by azole treatment. The repeated isolation of trailing isolates from patients with oropharyngeal candidiasis following treatment with azoles could also indicate that such isolates are more difficult to eradicate from infected patients than are nontrailing isolates (28).
Similarly, the relative importance of ERG3-inactivating mutations in azole resistance in clinical settings has been difficult to determine. It is clear that, while erg3 mutants can be readily selected in vitro following exposure to azoles (10, 30, 31), relatively few have been reported among azole-resistant clinical isolates. Loss of Erg3p activity itself blocks ergosterol biosynthesis, resulting in membrane abnormalities, hypersensitivity to some physiological stresses, defective hyphal growth, and reduced virulence in a mouse model of disseminated infection (13, 14, 18, 32). The resulting reduction in pathogenic fitness may result in the rapid elimination of erg3 mutants from infected patients, effectively imposing a counter-selective pressure against ERG3 inactivation. In a recent study, we reported that, despite severe defects in virulence (i.e. an inability to cause lethality), an erg3Δ/Δ deletion mutant was relatively resistant to fluconazole treatment in a mouse model of disseminated infection, as treatment did not significantly reduce levels of erg3Δ/Δ colonization of kidney tissue. Strikingly, in a mouse model of vaginal candidiasis, the erg3Δ/Δ mutant not only was able to readily colonize and to induce pathology but also was resistant to fluconazole treatment (18). These findings clearly confirm that loss of Erg3p activity does result in azole resistance within the mammalian host, although it also results in niche or context-specific defects in pathogenicity. Importantly, our previous study used immunocompetent mice for the model of disseminated infection, and it is possible that the virulence of the erg3Δ/Δ mutant would be greater in immunosuppressed mice, i.e., in the absence of stress imposed by the immune defenses of the mammalian host. If this proved to be the case, then it would imply that erg3 mutants not only are azole resistant but also are able to cause productive infections in patients at greatest risk of IFIs, namely, those with profound immune dysfunction. To date, just one study has used immunosuppressed mice to test the virulence of erg3 mutants (13). The study revealed that such mutants appeared to have attenuated virulence. However, other genetic factors unrelated to Erg3p dysfunction could not be ruled out as the cause of the virulence defects, since isogenic strains complemented with a WT ERG3 allele were not available for comparison. Moreover, the azole susceptibility of those strains was not tested in that model of infection (13). Either way, our previous in vivo data support the idea that loss of Erg3p activity is sufficient to confer resistance to the azole antifungals within the mammalian host and, by inference, patients (18).
The low frequency with which erg3 azole-resistant mutants have been reported to date likely reflects several factors. First, diminished virulence of erg3 null mutants has been observed (see above). Second, while loss of Erg3p function seems to afford some protection against the antifungal activity of azoles, it is less than that conferred by enhanced drug efflux and/or changes in the target protein. Specifically, erg3 null mutants are not invulnerable to the physiological consequences of azole exposure and in fact sustain significant membrane damage upon treatment. Third, the frequency of Erg3p-deficient clinical isolates may be underestimated, as a detailed biochemical analysis of cellular sterol contents is needed to confirm deficiencies in C-5 desaturase activity, and this is not routinely performed. Furthermore, most reports to date have focused exclusively on the analysis of clinical isolates with mutations that completely inactivate the ERG3 gene. However, relatively small adjustments in Erg3p activity may be sufficient to confer azole resistance (18), and such subtle changes are difficult to detect, even through analysis of cellular sterol contents.
In summary, our results establish that, while loss of C-5 sterol desaturase activity enables C. albicans to grow in the presence of the azole antifungals, this phenotype is both qualitatively and quantitatively distinct from azole resistance conferred by enhanced drug efflux or changes in the target protein, Erg11p. Our results also clearly distinguish the erg3 mutant phenotype from the trailing growth phenomenon.
MATERIALS AND METHODS
Growth conditions.
C. albicans was routinely grown at 30°C on YPD medium (1% yeast extract, 2% peptone, 2% dextrose), supplemented with uridine (50 μg/ml) when necessary. Transformant selection was carried out on minimal YNB medium (6.75 g/liter yeast nitrogen base without amino acids, 2% dextrose, 2% Bacto agar), supplemented with the appropriate auxotrophic requirements described for S. cerevisiae (33) or 50 μg/ml uridine.
Plasmid construction.
All oligonucleotides used in this study are listed in Table S1 in the supplemental material. Plasmids pKE1 (34), pKE1Nluc (35), pKE4 (16), pKE4CDR1 (16), pAR8 (36), pAR8-ERG11 (36), pKE1:GFP-YPT72 (25), and pDUP3 (37) were described previously.
pGHT3 was produced by amplifying the TEF1-MCS-ADH1 3'UTR (where UTR stands for “untranslated region” and MCS stands for “multiple cloning site”) cassette from pKE4 using the primer pair TEF1prF-BamHI and ADH1-3′UTRR2-ApaI and was cloned into the BamHI and ApaI restriction sites of pGEMHIS1. For construction of the plasmid pGHT-MDR1, the MDR1 open reading frame (ORF) was amplified from SC5314 genomic DNA with HiFi Platinum Taq (Invitrogen) and the primer pair MDR1ORFF-EagI and MDR1ORFR-MluI and then was cloned between the EagI and MluI restriction sites of pGHT3. Plasmid pDUPNluc was produced by amplifying the ACT1 promoter, the NLUC ORF, and the ADH1 3′ UTR from pKE1Nluc using the primer pair ACT1prom-ClaI-F and ADH1term-SpeI-R and was cloned into pDUP3 using the ClaI and SpeI restriction sites.
C. albicans strains.
All strains used in this study are listed in Table S2. The erg3Δ/Δ mutant was constructed in a previous study (18). Transformation of C. albicans with DNA constructs was performed using the lithium acetate method (38). Gene deletion strains were constructed with the PCR-based approach described by Wilson et al. (39), using the ura3Δ/Δ his1Δ/Δ arg4Δ/Δ strain BWP17 (kindly provided by Aaron Mitchell, Carnegie Mellon University). Strains TW2 and TW17 (4) were kindly provided by Theodore C. White (University of Missouri-Kansas City). Strains CA12, CA488, CA490, and CA1008 were kindly provided by Steven Kelly (Swansea University, UK). Strains JS7, JS17, JS22, JS23, JS28, JS29, and JS40 were kindly provided by Jack Sobel (Wayne State University).
The WT strain (GP1), the erg3Δ/Δ mutant, erg3Δ/Δ + ERG3, and the TW17, JS7, JS17, JS22, JS23, JS28, JS29, and JS40 strains were transformed with NgoMIV-linearized pDUP3 or pDUPNluc and selected on YPD plates containing 200 µg/ml nourseothricin. Strains overexpressing Cdr1p (OCDR strain), Mdr1p (OMDR strain), Erg11p (OERG strain), or all three proteins (OTR strain) were constructed by sequentially transforming BWP17 with plasmids carrying the URA3, HIS1, or ARG4 markers, as follows. The OCDR strain was made by sequential transformation with pKE4CDR1 (linearized with NheI), pGHT3 (linearized with NruI), and pAR8 (linearized with ClaI). The OMDR strain was made by serial transformation with pKE4 (linearized with NheI), pGHT-MDR1 (linearized with NruI), and pAR8 (linearized with ClaI). The OERG strain was made by sequential transformation with pKE4 (linearized with NheI), pGHT3 (linearized with NruI), and pAR8-ERG11 (linearized with ClaI). The OTR strain was made by sequential transformation with pKE4CDR1 (linearized with NheI), pGHT-MDR1 (linearized with NruI), and pAR8-ERG11 (linearized with ClaI). A WT control strain (XCTRL) carrying the vectors only was made by successive transformation with pKE4 (linearized with NheI), pGHT3 (linearized with NruI), and pAR8 (linearized with ClaI). Correct integration of pKE4/pKE4CDR1, pAR8/pAR8-ERG11, and pGHT3/pGHT-MDR1 was confirmed by PCR analysis using the primer pairs LUXINTDETF/LUXINTDETR, ARG4DETF7/ARG4DETR7, and HIS1DETF9/HIS1DETR5, respectively.
RNA extraction.
Each C. albicans strain was grown overnight at 30°C in YPD medium, subcultured to an OD600 of 0.2, and incubated for 6 h at 30°C, with shaking. Cells were collected by centrifugation at 3,500 rpm for 5 min. The supernatant was poured off, and the cell pellets were stored at −80°C. RNA was extracted from the cell pellets using the hot phenol method described by Schmitt et al. (40). RNA pellets were eluted in nuclease-free water, and the quantity and purity were determined spectrophotometrically by measuring the absorbance at 260 nm and 280 nm.
Quantitative RT-PCR.
cDNA was synthesized using the Verso cDNA synthesis kit (Thermo Scientific) with random hexamers, according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed with primer pairs that bind within the ACT1 (ACT1-FWD-qRT-PCR and ACT1-RVS-qRT-PCR), CDR1 (CaCDR1-F-qPCR and CaCDR1-R-qPCR), MDR1 (BMR1-F and BMR1-R), or ERG11 (CaERG11-RVS-qRT-PCR and CaERG11-FWD-qRT-PCR) (Table S1) coding sequences, using the Maxima SYBR Green/ROX qPCR master mix (2×; Thermo Scientific), as indicated by the manufacturer. Reactions were completed in a CFX96 real-time system (Bio-Rad). Expression levels of CDR1, MDR1, and ERG11 among the strains were compared to those of ACT1 (normalizing gene) using the 2−ΔΔCT method (41).
Antifungal susceptibility testing.
Antifungal susceptibility testing of all the strains included in this study was performed using the broth microdilution method described in CLSI document M27-A3 (19), in a 96-well plate format. All drugs for susceptibility testing used in this study were diluted 2-fold in dimethyl sulfoxide (DMSO) at 200 times the final concentration. RPMI 1640 medium (Sigma-Aldrich) was prepared according to the CLSI protocol; the medium was buffered with morpholinepropanesulfonic acid (MOPS), and the pH was adjusted using NaOH and HCl. Plates were incubated without shaking for 24 or 48 h at 25°C or 35°C. The content of each well was carefully resuspended with pipetting up and down before the OD600 was measured using a BioTek Cytation 5 plate reader.
Luciferase-based membrane integrity assay.
C. albicans strains expressing Nluc were cultured overnight at 30°C in YPD medium. Cells were then washed and resuspended in distilled water, and cell density was adjusted to 1 × 107 cells/ml in YNB broth. One hundred microliters of each cell suspension was added to wells of a round-bottom, 96-well plate containing 100 µl of YNB medium plus 5 µg/ml fluconazole or 0.5% DMSO. After incubation for 24 h in a 35°C standing incubator, the cells in each well were resuspended by mixing using a multichannel pipette. Plates were then centrifuged at 1,800 rpm for 5 min to pellet the cells. Twenty-five microliters of supernatant was transferred to a white flat-bottomed 96-well plate, and Nluc activity was determined using the Nano-Glo luciferase assay reagent (Promega Corp.), according to the manufacturer’s instructions. Plates were further centrifuged at 1,500 rpm for 2 min, and luminescence was read using a Cytation 5 plate reader (BioTek Instruments). Cell growth for each strain was determined as the OD600 of 1:10 dilutions of the cell cultures. Luminescence readings were normalized to the corresponding OD600 for each sample.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (grant R01AI099080). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Promega Corporation for permission to produce and to utilize the C. albicans adapted NanoLuc coding sequence. We also thank Jack Sobel (Wayne State University) for providing strains used in this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01337-18.
REFERENCES
- 1.Roemer T, Krysan DJ. 2014. Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harbor Perspect Med 4:a019703. doi: 10.1101/cshperspect.a019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly SL, Lamb DC, Kelly DE, Manning NJ, Loeffler J, Hebart H, Schumacher U, Einsele H. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett 400:80–82. doi: 10.1016/S0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 3.Perea S, Lopez-Ribot JL, Kirkpatrick WR, McAtee RK, Santillan RA, Martinez M, Calabrese D, Sanglard D, Patterson TF. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob Agents Chemother 45:2676–2684. doi: 10.1128/AAC.45.10.2676-2684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 41:1482–1487. doi: 10.1128/AAC.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly SL, Lamb DC, Loeffler J, Einsele H, Kelly DE. 1999. The G464S amino acid substitution in Candida albicans sterol 14α-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem Biophys Res Commun 262:174–179. doi: 10.1006/bbrc.1999.1136. [DOI] [PubMed] [Google Scholar]
- 6.Kelly SL, Lamb DC, Kelly DE. 1999. Y132H substitution in Candida albicans sterol 14α-demethylase confers fluconazole resistance by preventing binding to haem. FEMS Microbiol Lett 180:171–175. [DOI] [PubMed] [Google Scholar]
- 7.Lamb DC, Kelly DE, White TC, Kelly SL. 2000. The R467K amino acid substitution in Candida albicans sterol 14α-demethylase causes drug resistance through reduced affinity. Antimicrob Agents Chemother 44:63–67. doi: 10.1128/AAC.44.1.63-67.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Ribot JL, McAtee RK, Lee LN, Kirkpatrick WR, White TC, Sanglard D, Patterson TF. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother 42:2932–2937. doi: 10.1128/AAC.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morschhauser J, Barker KS, Liu TT, Bla BWJ, Homayouni R, Rogers PD. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog 3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson JB, Sirjusingh C, Parsons AB, Boone C, Wickens C, Cowen LE, Kohn LM. 2003. Mode of selection and experimental evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics 163:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Rolley N, Kelly DE, Kelly SL. 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob Agents Chemother 54:4527–4533. doi: 10.1128/AAC.00348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vale-Silva LA, Coste AT, Ischer F, Parker JE, Kelly SL, Pinto E, Sanglard D. 2012. Azole resistance by loss of function of the sterol Δ5,6-desaturase gene (ERG3) in Candida albicans does not necessarily decrease virulence. Antimicrob Agents Chemother 56:1960–1968. doi: 10.1128/AAC.05720-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morio F, Pagniez F, Lacroix C, Miegeville M, Le Pape P. 2012. Amino acid substitutions in the Candida albicans sterol Δ5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J Antimicrob Chemother 67:2131–2138. doi: 10.1093/jac/dks186. [DOI] [PubMed] [Google Scholar]
- 14.Miyazaki T, Miyazaki Y, Izumikawa K, Kakeya H, Miyakoshi S, Bennett JE, Kohno S. 2006. Fluconazole treatment is effective against a Candida albicans erg3/erg3 mutant in vivo despite in vitro resistance. Antimicrob Agents Chemother 50:580–586. doi: 10.1128/AAC.50.2.580-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arthington-Skaggs BA, Lee-Yang W, Ciblak MA, Frade JP, Brandt ME, Hajjeh RA, Harrison LH, Sofair AN, Warnock DW. 2002. Comparison of visual and spectrophotometric methods of broth microdilution MIC end point determination and evaluation of a sterol quantitation method for in vitro susceptibility testing of fluconazole and itraconazole against trailing and nontrailing Candida isolates. Antimicrob Agents Chemother 46:2477–2481. doi: 10.1128/AAC.46.8.2477-2481.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters BM, Luna-Tapia A, Tournu H, Rybak JM, Rogers PD, Palmer GE. 2017. An azole-tolerant endosomal trafficking mutant of Candida albicans is susceptible to azole treatment in a mouse model of vaginal candidiasis. Antimicrob Agents Chemother 61:e00084-17. doi: 10.1128/AAC.00084-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rex JH, Nelson PW, Paetznick VL, Lozano-Chiu M, Espinel-Ingroff A, Anaissie EJ. 1998. Optimizing the correlation between results of testing in vitro and therapeutic outcome in vivo for fluconazole by testing critical isolates in a murine model of invasive candidiasis. Antimicrob Agents Chemother 42:129–134. doi: 10.1128/AAC.42.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luna-Tapia A, Willems HME, Parker JE, Tournu H, Barker KS, Nishimoto AT, Rogers PD, Kelly SL, Peters BM, Palmer GE. 2018. Loss of Upc2p-inducible ERG3 transcription is sufficient to confer niche-specific azole resistance without compromising Candida albicans pathogenicity. mBio 9:e00225-18. doi: 10.1128/mBio.00225-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard—3rd ed CLSI document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Marr KA, Rustad TR, Rex JH, White TC. 1999. The trailing end point phenotype in antifungal susceptibility testing is pH dependent. Antimicrob Agents Chemother 43:1383–1386. doi: 10.1128/AAC.43.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J 21:546–559. doi: 10.1093/emboj/21.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol 48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- 23.Luna-Tapia A, Tournu H, Peters TL, Palmer GE. 2016. Endosomal trafficking defects can induce calcium dependent azole tolerance in Candida albicans. Antimicrob Agents Chemother 60:7170–7177. doi: 10.1128/AAC.01034-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luna-Tapia A, Kerns ME, Eberle KE, Jursic BS, Palmer GE. 2015. Trafficking through the late endosome significantly impacts Candida albicans tolerance of the azole antifungals. Antimicrob Agents Chemother 59:2410–2420. doi: 10.1128/AAC.04239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arthington-Skaggs BA, Warnock DW, Morrison CJ. 2000. Quantitation of Candida albicans ergosterol content improves the correlation between in vitro antifungal susceptibility test results and in vivo outcome. Antimicrob Agents Chemother 44:2081–2085. doi: 10.1128/AAC.44.8.2081-2085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Kelly DE. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem Biophys Res Commun 207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 28.Revankar SG, Kirkpatrick WR, McAtee RK, Fothergill AW, Redding SW, Rinaldi MG, Patterson TF. 1998. Interpretation of trailing endpoints in antifungal susceptibility testing by the National Committee for Clinical Laboratory Standards Method. J Clin Microbiol 36:153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg A, Ene IV, Bibi M, Zakin S, Segal ES, Ziv N, Dahan AM, Colombo AL, Bennett RJ, Berman J. 2018. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat Commun 9:2470. doi: 10.1038/s41467-018-04926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinjon E, Moran GP, Jackson CJ, Kelly SL, Sanglard D, Coleman DC, Sullivan DJ. 2003. Molecular mechanisms of itraconazole resistance in Candida dubliniensis. Antimicrob Agents Chemother 47:2424–2437. doi: 10.1128/AAC.47.8.2424-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan L, Zhang J, Li M, Cao Y, Xu Z, Cao Y, Gao P, Wang Y, Jiang Y. 2008. DNA microarray analysis of fluconazole resistance in a laboratory Candida albicans strain. Acta Biochim Biophys Sin (Shanghai) 40:1048–1060. doi: 10.1111/j.1745-7270.2008.00483.x. [DOI] [PubMed] [Google Scholar]
- 32.Chau AS, Gurnani M, Hawkinson R, Laverdiere M, Cacciapuoti A, McNicholas PM. 2005. Inactivation of sterol Δ5,6-desaturase attenuates virulence in Candida albicans. Antimicrob Agents Chemother 49:3646–3651. doi: 10.1128/AAC.49.9.3646-3651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke DA, Dawson DS, Stearns T. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 34.Johnston DA, Luna-Tapia A, Eberle KE, Palmer GE. 2013. Three prevacuolar compartment Rab GTPases impact Candida albicans hyphal growth. Eukaryot Cell 12:1039–1050. doi: 10.1128/EC.00359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luna-Tapia A, Peters BM, Eberle KE, Kerns ME, Foster TP, Marrero L, Noverr MC, Fidel PL Jr, Palmer GE. 2015. ERG2 and ERG24 are required for normal vacuolar physiology as well as Candida albicans pathogenicity in a murine model of disseminated but not vaginal candidiasis. Eukaryot Cell 14:1006–1016. doi: 10.1128/EC.00116-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butts A, DeJarnette C, Peters TL, Parker JE, Kerns ME, Eberle KE, Kelly SL, Palmer GE. 2017. Target abundance-based fitness screening (TAFiS) facilitates rapid identification of target-specific and physiologically active chemical probes. mSphere 2:e00379-17. doi: 10.1128/mSphere.00379-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerami-Nejad M, Zacchi LF, McClellan M, Matter K, Berman J. 2013. Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans. Microbiology 159:565–579. doi: 10.1099/mic.0.064097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gietz D, St Jean A, Woods RA, Schiestl RH. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson RB, Davis D, Mitchell AP. 1996. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt ME, Brown TA, Trumpower BL. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.