The high rates of antifungal resistance in Candida glabrata may be facilitated by the presence of alterations in the MSH2 gene. We aimed to study the sequence of the MSH2 gene in 124 invasive C. glabrata isolates causing incident episodes of candidemia (n = 81), subsequent candidemia episodes (n = 9), endocarditis (n = 2), and in vitro-generated echinocandin-resistant isolates (n = 32) and assessed its relationship with genotypes, acquisition of antifungal resistance in vivo and in vitro, and patient prognosis.
KEYWORDS: Candida glabrata, MSH2 gene, antifungal resistance
ABSTRACT
The high rates of antifungal resistance in Candida glabrata may be facilitated by the presence of alterations in the MSH2 gene. We aimed to study the sequence of the MSH2 gene in 124 invasive C. glabrata isolates causing incident episodes of candidemia (n = 81), subsequent candidemia episodes (n = 9), endocarditis (n = 2), and in vitro-generated echinocandin-resistant isolates (n = 32) and assessed its relationship with genotypes, acquisition of antifungal resistance in vivo and in vitro, and patient prognosis. The MSH2 gene was sequenced, and isolates were genotyped using six microsatellite markers and multilocus sequence typing (MLST) based on six housekeeping genes. According to EUCAST, isolates causing candidemia (n = 90) were echinocandin susceptible, and four of them were fluconazole resistant (MIC ≥64 mg/liter). One isolate obtained from a heart valve was resistant to micafungin and anidulafungin (MICs, 2 mg/liter and 1 mg/liter, respectively). MSH2 gene mutations were present in 44.4% of the incident isolates, the most common being V239L. The presence of MSH2 mutations was not correlated with in vitro or in vivo antifungal resistance. Microsatellite and MLST revealed 27 genotypes and 17 sequence types, respectively. Fluconazole-resistant isolates were unrelated. Most MSH2 mutations were found in cluster isolates; conversely, some mutations were found in more than one genotype. No clinical differences, including previous antifungal use, were found between patients infected by wild-type MSH2 gene isolates and isolates with any point mutation. The presence of MSH2 gene mutations in C. glabrata isolates causing candidemia is not correlated with specific genotypes, the promotion of antifungal resistance, or the clinical outcome.
INTRODUCTION
Candida bloodstream infections have been increasing in recent years (1), posing a substantial economic burden for hospitals and high mortality among patients (2, 3). Since fluconazole resistance is becoming more common, echinocandins are currently preferred as first-line treatment (4). However, strains with resistance to echinocandins and/or to multiple antifungals have been sporadically reported (5).
The incidence of infections due to Candida glabrata has notoriously grown in Spain (1). In comparison to other Candida species, C. glabrata is characterized by the ability to develop fluconazole resistance and a certain degree of resistance to echinocandins in some hospitals (1, 6–9). In a previous work, we showed that C. glabrata can acquire in vitro resistance to echinocandins after exposure to low and/or increasing echinocandin concentrations (10–12). There is little information on why C. glabrata acquires resistance more rapidly to multiple antifungal classes than other Candida species. The potential for acquisition of resistance is unclear; however, organisms with a haploid genome, e.g., C. glabrata, might accumulate a number of mutations in key genes, such as FKS, during genome replication (13). DNA polymerase mismatches coupled with failures in DNA repair can lead to multiple antifungal resistance in Candida spp. (14, 15). An MSH2 gene mismatch repair in C. glabrata and Candida albicans may lead to a hypermutable phenotype prone to developing antifungal resistance (14, 15). Furthermore, certain C. glabrata genotypes seem to harbor specific polymorphisms in the MSH2 gene (16).
There are other aspects that require attention. First, only a small number of invasive isolates have been studied (16, 17). Second, studies have been reported without genotyping procedures (15, 17). Third, very few cases of MSH2 gene sequences from antifungal-susceptible isolates that have shifted to resistant have been studied (15). Finally, the studies include scarce clinical data from infected patients. Here, we aim to study the sequence of the MSH2 gene of a collection of C. glabrata isolates obtained in Spain causing fungemia and to assess the potential relationship with genotypes, possible acquisition of antifungal resistance in vivo and in vitro, and patient prognosis.
RESULTS
Antifungal susceptibility and MSH2 gene sequencing.
As per EUCAST breakpoints, the 90 blood culture isolates were echinocandin susceptible (geometric mean MICs of micafungin and anidulafungin, 0.015 and 0.023 mg/liter, respectively), and four of them (4.9%) were fluconazole resistant (patients 11, 20, 44, and 63; Fig. 1). Only one of the isolates, which was collected from the heart valve, was resistant to micafungin and anidulafungin (2 mg/liter and 1 mg/liter, respectively) and harbored an S663P mutation in hot spot 1 of the FKS2 gene. The 21 in vitro-generated echinocandin-resistant colonies carried the following FKS2 mutations: ΔF658, S663P, W715L, and E655A (12).
FIG 1.
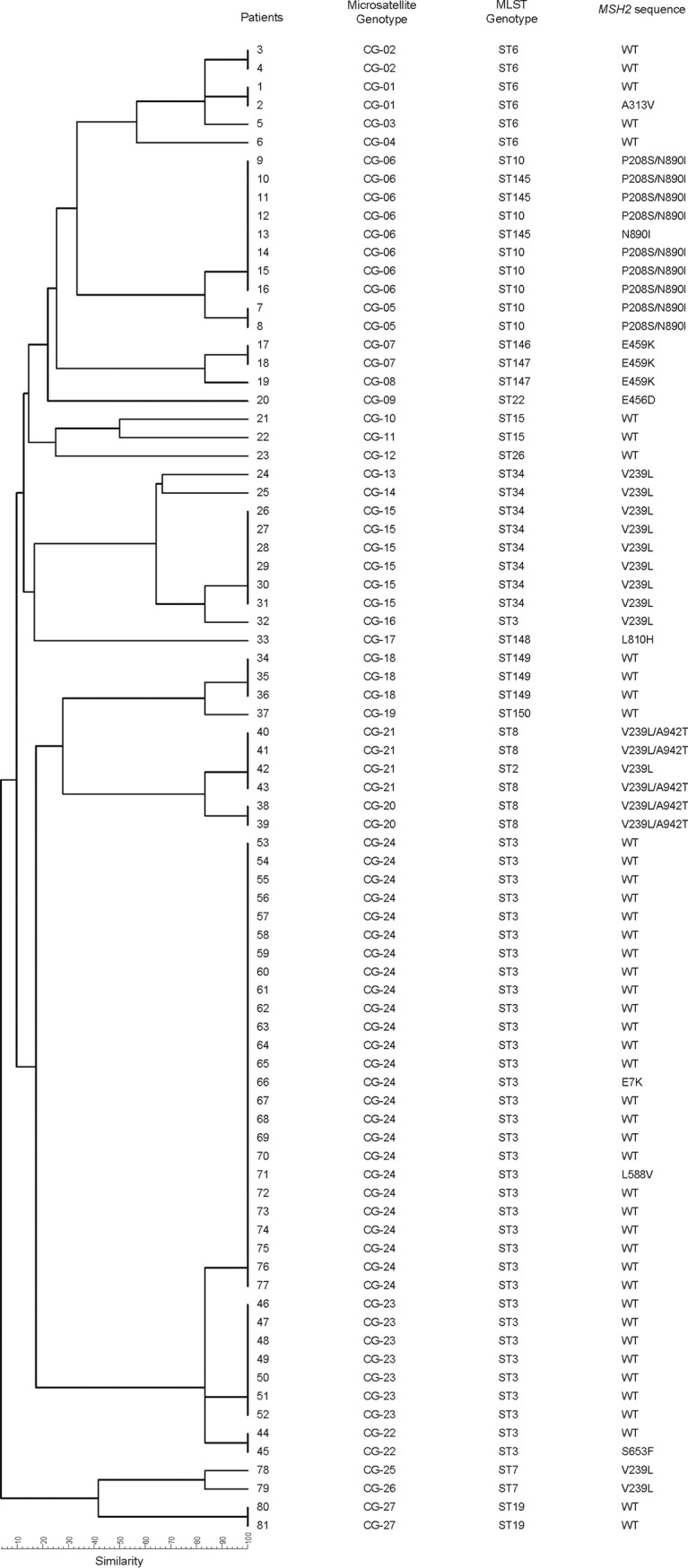
Genetic relationship with microsatellite markers between the incident 81 Candida glabrata isolates with the MSH2 sequence and the sequence type (ST). WT, wild type.
Thirty-six incident isolates from 81 patients with candidemia (44.4%) had one or more of the following MSH2 gene point mutations: V239L, P208S, A942T, S653F, A313V, E456D, E459K, E7K, L588V, N890I, and/or L810H. Here, we report the S653F mutation for the first time. Mutation frequencies were 33.3% for V239L, 25.6% for P208S/N890I, 13.9% for V239L/A942T, 8.3% for E459K, and 2.7% for each of the following: S653F, A313V, E456D, E7K, L588V, N890I, and L810H. We observed a match between MSH2 sequences of incident isolates and those causing second episodes. Two point mutations in the gene (P208S/N890I) were found in the four isolates from a patient who suffered two episodes of candidemia and endocarditis.
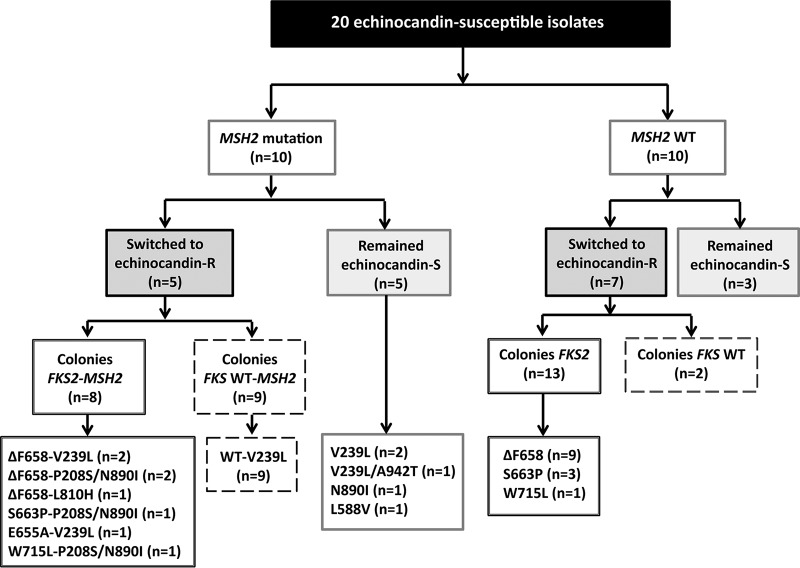
In our study, only two out of the four fluconazole-resistant isolates (from patients 11 and 20) harbored MSH2 gene mutations (P208S/N890I and E456D, respectively) (Fig. 1). The sequence of the in vitro-generated echinocandin-resistant MSH2 gene and the respective parent isolates were identical. Eight (38.1%) out of the 21 colonies that acquired FKS2 mutations harbored MSH2 gene mutations (P208S/N890I [n = 4], V239L [n = 3], and L810H [n = 1]). Nine (81.82%) out of 11 echinocandin-resistant colonies (i.e., those with an FKS mutation) had the same MSH2 gene mutation (V239L) and were sourced from a single isolate, whereas the remaining two isolates were from different isolates and the MSH2 wild type (Fig. 2).
FIG 2.
Description of the 20 echinocandin-susceptible isolates previously exposed in vitro to echinocandins and the generated colonies phenotypically resistant to echinocandins. R, resistant; S, susceptible; WT, wild type.
Genotyping analysis and MSH2 gene sequencing.
Figure 1 shows the genotyping of the 81 incident isolates. Isolates responsible for second episodes, the case of endocarditis, and in vitro-generated isolates were isogenic to parent strains, regardless of the used method. Microsatellite markers revealed 27 genotypes (CG-1 to CG-27), 13 of which were clusters involving 82.7% of the isolates. Multilocus sequence typing (MLST) revealed 17 sequence types (STs), including six newly described ones (Table 1), and 11 STs being clusters that involved 92.6% of the isolates. The clusters with the highest number of isolates were microsatellites (CG-24 [30.86%], CG-6 [9.88%], and CG-23 [8.64%]) and MLST (ST3 [43.21%], ST34 [9.88%], ST10 [8.64%], and ST6 [7.41%]). In general, clusters detected by any method mostly matched, although there was a tendency for the microsatellites to split up ST clusters into clonally related genotypes and, exceptionally, few microsatellite clusters were broken into clonally related sub-STs (Fig. 1). Fluconazole-resistant isolates were unrelated.
TABLE 1.
Newly characterized STs and description of the alleles in the six housekeeping genesa
| Patient(s) | New ST | Allele by gene |
|||||
|---|---|---|---|---|---|---|---|
| FKS | LEU2 | NMT1 | TRP1 | UGP1 | URA3 | ||
| 10, 11, 13 | 145 | 8 | 5 | 3 | 5 | 1 | 2 |
| 17 | 146 | 51 | 24 | 49 | 7 | 51 | 6 |
| 18,19 | 147 | 51 | 24 | 49 | 30 | 51 | 6 |
| 33 | 148 | 3 | 9 | 26 | 4 | 17 | 4 |
| 34, 35, 36 | 149 | 7 | 25 | 51 | 7 | 51 | 4 |
| 37 | 150 | 7 | 25 | 51 | 30 | 51 | 4 |
ST 145 is a new combination of previously identified alleles. STs 146 to 150 are new combinations of previously identified alleles or the following new alleles (reported in this work for the first time).
Most MSH2 mutations were found in cluster isolates. In general, the bulk of isolates within each cluster showed the same MSH2 polymorphism, although a few genotypes (CG-1, CG-22, and CG-24) involved wild-type and MSH2 mutant isolates. Conversely, some mutations were found in more than one genotype (Fig. 1).
Association of MSH2 mutations and genotypes with clinical characteristics of the patients.
Table 2 summarizes the characteristics of the study patients. There were no differences in demographics, underlying conditions, risk factors for candidemia, or sources of candidemia between patients infected by wild-type MSH2 isolates and those with any point mutation. Most patients infected with MSH2 mutants were adults (94.4%), with a mean age of 71 years, and 63.9% were male. Gastrointestinal disease was the most frequent comorbidity in 50% of patients, and the most frequent department of admission was a medical ward (47.2%) (Table 2). Most cases had an intra-abdominal source (38.9%) or were central venous catheter-related infections (25%).
TABLE 2.
Demographics and clinical status of patients based on MSH2 gene sequencea
| Patient characteristicb | Total (n = 81) | Wild-type MSH2 gene isolates (n = 45) | Isolates with MSH2 gene mutations (n = 36) | P value |
|---|---|---|---|---|
| Demographics | ||||
| Age (median [IQR]) (yr) | 71 (0–91) | 72 (0–90) | 71 (0–91) | 0.556 |
| Male | 50 (61.7) | 27 (60.0) | 23 (63.9) | 0.819 |
| Neonate (≤28 days) | 4 (4.9) | 2 (4.4) | 2 (5.6) | 1 |
| Department | ||||
| Medical | 36 (44.4) | 19 (42.2) | 17 (47.2) | 0.822 |
| Surgical | 21 (25.9) | 12 (26.7) | 9 (25.0) | 1 |
| Onco-hematology | 3 (3.7) | 1 (2.2) | 2 (5.6) | 0.582 |
| ICU | 21 (25.9) | 13 (28.9) | 8 (22.2) | 0.612 |
| Underlying disease | ||||
| Cardiovascular disease | 32 (39.5) | 18 (40.0) | 14 (38.9) | 1 |
| Neurological disorder | 15 (18.5) | 10 (22.2) | 5 (13.9) | 0.398 |
| Chronic renal failure | 22 (27.2) | 11 (24.4) | 11 (30.6) | 0.619 |
| Diabetes mellitus | 18 (22.2) | 13 (28.9) | 5 (13.9) | 0.178 |
| Pulmonary disease | 19 (23.5) | 9 (20.0) | 10 (27.8) | 0.440 |
| Liver disease | 13 (16.0) | 8 (17.8) | 5 (13.9) | 0.765 |
| Gastrointestinal disease | 38 (46.9) | 20 (44.4) | 18 (50.0) | 0.659 |
| Solid-organ malignancy | 28 (34.6) | 14 (31.1) | 14 (38.9) | 0.490 |
| Hematologic malignancy | 2 (2.5) | 1 (2.2) | 1 (2.8) | 0.694 |
| Solid-organ transplant | 1 (1.2) | 0 | 1 (2.8) | 0.444 |
| HIV | 5 (6.2) | 2 (4.4) | 3 (8.3) | 0.651 |
| Charlson index (median [IQR]) | 6 (0–12) | 6 (0–10) | 6 (0–12) | 0.388 |
| Risk factor | ||||
| Central venous catheter | 64 (79.0) | 35 (77.8) | 29 (80.6) | 0.791 |
| Total parenteral nutrition | 44 (54.3) | 24 (53.3) | 20 (55.6) | 1 |
| Chemotherapy | 9 (11.1) | 5 (11.1) | 4 (11.1) | 1 |
| Neutropenia (<500 cells/mm3) | 2 (2.5) | 1 (2.2) | 1 (2.8) | 1 |
| Corticosteroids (prior mo) | 15 (18.5) | 8 (17.8) | 7 (19.4) | 1 |
| Immunosuppressive therapy | 4 (4.9) | 3 (6.7) | 1 (2.8) | 0.625 |
| Surgery | 33 (40.7) | 16 (35.6) | 17 (47.2) | 0.364 |
| Abdominal surgery | 26 (32.1) | 13 (28.9) | 13 (36.1) | 0.633 |
| Previous antimicrobial use | 76 (93.8) | 42 (93.3) | 34 (94.4) | 1 |
| Antifungal use | ||||
| Previous AF used | 17 (22.4) | 9 (22.0) | 8 (22.9) | 1 |
| Voriconazole | 2 (2.5) | 2 (4.4) | 0 | 0.5 |
| Fluconazole | 15 (18.5) | 7 (15.6) | 8 (22.2) | 0.567 |
| Echinocandins | 4 (4.9) | 1 (2.2) | 3 (8.3) | 0.318 |
| Amphotericin B | 1 (1.2) | 1 (2.2) | 0 | 1 |
| Duration of AF (days) | 15 (3–63) | 21 (9–44) | 14.5 (3–63) | 0.933 |
| Time from previous AF to episode (days) | 1 (−11–73) | 0 (−6–73) | 2.5 (−11–73) | 0.755 |
| Breakthrough fungemia | 7 (8.6) | 3 (6.7) | 4 (11.1) | 0.694 |
| Fluconazole resistance | 4 (4.9) | 2 (4.4) | 2 (5.6) | 1 |
| Clinical manifestation | ||||
| Septic shock | 24 (29.6) | 18 (40.0) | 6 (16.7) | 0.028 |
| Pitt score (median [IQR]) | 1 (0–12) | 2 (0–10) | 0 (0–12) | 0.093 |
| Infection source | ||||
| Primary | 14 (17.3) | 9 (20.0) | 5 (13.9) | 0.562 |
| Central venous catheter | 27 (33.3) | 18 (40.0) | 9 (25.0) | 0.235 |
| Urinary tract | 4 (4.9) | 3 (6.7) | 1 (2.8) | 0.625 |
| Abdomen | 28 (34.6) | 14 (31.1) | 14 (38.9) | 0.490 |
| Other | 3 (3.7) | 0 | 3 (8.3) | 0.084 |
| Unknown | 5 (6.2) | 1 (2.2) | 4 (11.1) | 0.166 |
| Therapeutic management | ||||
| No antifungals | 14 (17.3) | 9 (20.0) | 5 (13.9) | 0.562 |
| Echinocandin as initial AF | 29 (43.3) | 15 (41.7) | 14 (45.2) | 0.809 |
| Adequate targeted AF | 62 (76.5) | 34 (75.6) | 28 (77.8) | 1 |
| Infection source control | 56 (81.2) | 31 (81.6) | 25 (80.6) | 1 |
| Outcomes | ||||
| Metastatic complications | 13 (18.1) | 6 (15.0) | 7 (21.9) | 0.543 |
| ICU due to candidemia | 10 (12.3) | 6 (13.3) | 4 (11.1) | 1 |
| 7-day mortality | 13 (16.0) | 10 (22.2) | 3 (8.3) | 0.129 |
| 30-day mortality | 24 (29.6) | 18 (40.0) | 6 (16.7) | 0.028 |
| Mortality >30 days | 11 (13.6) | 5 (11.1) | 6 (16.7) | 0.526 |
| Overall mortality | 35 (43.2) | 23 (51.1) | 12 (33.3) | 0.121 |
| Hospital length of stay (days) | 26 (0–165) | 25 (0–165) | 26.5 (0–112) | 0.711 |
All data reported as number (%), unless otherwise specified.
IQR, interquartile range; ICU, intensive care unit; HIV, human immunodeficiency virus; AF, antifungal.
Only one out of the four patients infected by fluconazole-resistant isolates was previously treated with fluconazole and caspofungin. The patient with endocarditis caused by the echinocandin-resistant isolate had previously received caspofungin, micafungin, and fluconazole. In vitro echinocandin-resistant-generated isolates came from 12 patients, of whom only four had received previous antifungal treatment. Eight patients with alterations in the MSH2 gene isolates received systemic antifungals before the diagnosis, including fluconazole (n = 8) and echinocandins (n = 3). No statistical differences were determined in the median time of exposure to previous antifungals between patients infected by MSH2 mutants and patients infected by wild-type isolates (14.5 and 21 days, respectively). Adequate targeted treatment and appropriate source control were equally ensured in both groups. Concerning clinical outcomes, there were no differences between groups with respect to metastatic complications, intensive care unit admission due to the severity of the candidemia, hospital length of stay, and early 7-day mortality. However, there was a higher proportion of patients with septic shock and 30-day mortality in those infected by wild-type MSH2 gene isolates (P = 0.028) (Table 2). The same trend was observed when evaluating the mortality rates among patients with less severe clinical presentations (Pitt score, ≤1), although no statistical significance was found (data not shown). Furthermore, the ST3 genotype, which included mostly wild-type MSH2 gene isolates (31/35), was detected more frequently in patients with septic shock (40% versus 21.7%; P = 0.09) and poor outcome (40% versus 21.7%; P = 0.09).
DISCUSSION
The analysis of a large number of C. glabrata isolates causing candidemia showed that the presence of MSH2 gene mutations is not correlated with the previous use of antifungal agents, with specific genotypes (either defined using MLST or microsatellite markers), or with the promotion of antifungal resistance in vitro and in the patients.
Defects in DNA mismatch repair in multiple fungal species, including C. glabrata, have been reported to be associated with the acquisition of antifungal resistance (hypermutable phenotype) (14, 15, 18). A recent study identified a prevalent mutator C. glabrata phenotype caused by a mismatch repair defect in the MSH2 gene (15). This phenotype may acquire resistance to multiple antifungals with the consequent added complication in patient care; the detection of MSH2 point mutations was thus proposed as a marker of potential development of resistance. We did not find any correlation between the presence of MSH2 gene mutations and the acquisition of antifungal resistance either in vitro or in clinical settings. This is in line with previous studies in which this lack of correlation was also highlighted (16, 17, 19).
A limitation of previous similar studies is the low number of resistant isolates. To solve this, in this study, we analyzed echinocandin-susceptible isolates that switched to a resistant phenotype after being exposed to candins in vitro (n = 20) (12) and naturally occurring in a patient with candidemia and endocarditis who had previously undergone treatment with echinocandins. We observed the following (Fig. 2): first, parent echinocandin-susceptible isolates and their respective resistant isolates had the same MSH2 gene sequence. Second, echinocandin-susceptible isolates with either MSH2 gene mutations or a wild-type MSH2 gene were able to switch to a resistant phenotype following drug exposure. Third, isolates with MSH2 gene mutations were able to yield both echinocandin-resistant and echinocandin-susceptible isolates, as shown in the patient with endocarditis. Fourth, the MSH2 gene sequence did not affect the acquired mechanism of resistance to echinocandins, as shown by the three isolates that were exposed to candins and yielded echinocandin-resistant isolates (FKS2 mutants and wild-type FKS). Finally, mutations in the MSH2 gene did not influence fluconazole resistance. The MSH2 gene sequences of incident isolates and isolates causing second episodes were identical. Moreover, mutations were commonly found in incident susceptible isolates that did not turn resistant in further episodes. Here, the rate of isolates harboring mutations in the MSH2 gene (44.4%) is similar to that reported in some other studies (15, 17) but lower than the 77.2% reported by Hou et al. (19). Certain mutations, such as V239L and P208S/N890I, have been associated with resistance (15, 17), whereas others that are less common, E7K, E456D, or E459K, have not (15–17).
The results of the two genotyping techniques are comparable, although a higher number of genotypes was obtained with microsatellite markers (19). Previous studies have tried to correlate specific genotypes with MSH2 gene mutations (16, 19). Dellière and colleagues found a correlation between some microsatellite genotypes and MSH2 gene mutations; however, the mutations were not genotype specific, as some of them were found in more than one genotype (16). Hou et al. reported a correlation between ST7 and ST10 and the presence of V239L and/or K583N and P208S/N890I mutations in the MSH2 gene, respectively; however, the authors did not disclose if the same mutations were also found in other STs (19). Finally, the results described by Deshpande and Castanheira are in line with those of Hou et al., except that the V239L mutation was also found in ST2 and ST7, whereas ST3 isolates carried a wild-type MSH2 gene (20). In considering the results of all these studies, including ours, there seems to be a relationship between STs and the MSH2 gene sequence; e.g., ST7 and ST10 are found in different geographic places, including our isolates, and harbor specific mutations. However, there is no genotype specificity regarding the mutations; for example, we found that V239L was conspicuously present in ST2, ST3, ST7, ST8, and ST34 (Fig. 1). This suggests that a number of mutations may be potential MSH2 gene constitutive polymorphisms rather than resistance-causing mutations. A recent study using whole-genome sequencing data suggests that these MSH2 mutations are natural genetic variations rather than a mutator phenotype (21). Furthermore, MSH2 gene sequences were mostly identical in the isolates belonging to a given ST or microsatellite genotype, with some exceptions (Fig. 1) (19, 22).
We report not only on the microbiological characterization of C. glabrata isolates but also the clinical description of the infected patients, which is a strength of our study. Patients infected by C. glabrata carrying MSH2 gene mutations or with the wild-type MSH2 gene presented similar demographic and clinical characteristics. Moreover, no differences in terms of previous antifungal use between groups were found. Our observations are in line with those from a previous study (16) but diverge with results from another work (15). Despite the comparable host characteristics and therapeutic management, a higher number of severe cases (higher median Pitt scores) was detected in the group of patients infected by strains with the wild-type MSH2 gene, resulting in a higher incidence of septic shock and 30-day mortality. In fact, such differences could be related to the remarkable prevalence of ST3 genotype isolates among these patients. Other authors have reported that the most frequently found ST3 and ST7 may be correlated with a poor outcome (23), supporting our findings. In fact, our observations show that patients infected by ST3 show higher 30-day mortality than do patients infected by any of the other STs (40% versus 21.7%, respectively; P = 0.09). Further studies, including a higher number of episodes, should be carried out to evaluate the clinical impact of infections by ST3 isolates.
The low numbers of FKS mutant and fluconazole-resistant isolates found in the patients are in line with previous studies and the main limitation of our study (1, 24, 25). However, our conclusions are supported by a number of C. glabrata resistant isolates in vitro generated from a set of incident isolates.
We conclude that the presence of MSH2 gene mutations in C. glabrata isolates causing candidemia does not correlate with the previous use of antifungal agents by the patients, with specific genotypes, or with the promotion of antifungal resistance. Furthermore, not differences in clinical outcomes were observed.
MATERIALS AND METHODS
Yeast isolates, antifungal susceptibility, and patients.
One hundred twenty-four C. glabrata isolates were studied. The distribution of isolates was as follows: isolates causing candidemia in 81 patients (n = 81 incident isolates and n = 9 second episodes; n = 90), isolates from the heart valve of one patient who developed endocarditis (patient no. 15, Fig. 1; n = 2), and a group of phenotypically echinocandin-resistant isolates that had been exposed in vitro (12) to echinocandins and were either FKS2 mutants (n = 21) or wild-type FKS (n = 11) (Fig. 2). Second episode was defined as the isolation of C. glabrata from blood cultures taken ≥7 days after the diagnosis. Patients were admitted to Gregorio Marañón Hospital (Madrid, Spain) between January 2007 and December 2016, and isolates were molecularly identified and tested for susceptibility to micafungin, anidulafungin, and fluconazole following the EUCAST 7.3.1 microdilution procedure (26, 27). General characteristics (demographic, department of admission, underlying diseases, risk factors, and antifungal use) and clinical condition (clinical manifestation, source of infection, therapeutic management, and outcome) were collected, as well as the use of antifungals in the six months before the diagnosis.
MSH2 sequencing and genotyping.
The MSH2 gene of the 124 isolates was amplified and sequenced as described by Dellière et al. (16), with some modifications: 1.25 U of AmpliTaq gold (Applied Biosystems), 0.2 mM deoxynucleoside triphosphates, 2 mM MgCl2, and 100 ng of the extracted DNA were used (12). MSH2 sequences were compared with the reference wild-type MSH2 gene sequence of C. glabrata (GenBank accession number XM_447585.1) (16).
The isolates were further genotyped by two previously described methods, a panel of six microsatellite markers (MT1, ERG3, GLM4, CG10, CG7, and CG4) (28–30) and MLST based on six housekeeping genes, including the loci FKS, LEU2, NMT1, TRP1, UGP1, and URA3 (31). PCR products of the six housekeeping gene loci were sequenced in both directions. The combination of the alleles of the six loci defined an MLST genotype or sequence type (ST) according to the MLST database (http://pubmlst.org/cglabrata). Singleton genotypes were defined as those found only once, whereas a cluster was defined as the presence of ≥2 strains showing identical genotypes.
Data analysis.
Normally distributed clinical variables are expressed as means ± standard deviation. Variables with nonnormal distribution are expressed as medians and interquartile ranges. Categorical variables are expressed as percentages and compared using Fisher’s exact test or the χ2 test, as appropriate. Continuous variables were compared with the Mann-Whitney U test. The comparisons were considered statistically significant with P values of <0.05 (IBM SPSS Statistics for Windows, version 21.0; Armonk, NY, USA).
Ethical considerations.
This study was approved by the ethics committee of Hospital Gregorio Marañón (CEIC-A1; study no. 203/18).
ACKNOWLEDGMENTS
We are grateful to Dainora Jaloveckas for editing and proofreading assistance.
This work was supported by grants PI14/00740, PI16/01012, and MSI15/00115 from the Fondo de Investigación Sanitaria (FIS; Instituto de Salud Carlos III, Plan Nacional de I+D+I 2013-2016) and cofunded by the European Regional Development Fund (FEDER) “A way of making Europe.” P.E. (grant CPI15/00115) and J.G. (grant CPII15/00006) are recipients of a Miguel Servet contract supported by the FIS; MB received a predoctoral grant from the Instituto de Investigación Sanitaria Gregorio Marañón (grant II-Predoc-2016-IISGM).
The funders had no role in the study design, data collection and analysis decision to publish, or preparation of the manuscript.
We declare no conflicts of interest.
REFERENCES
- 1.Guinea J, Zaragoza Ó, Escribano P, Martín-Mazuelos E, Pemán J, Sánchez-Reus F, Cuenca-Estrella M, CANDIPOP Project, GEIH-GEMICOMED (SEIMC), REIPI. 2014. Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob Agents Chemother 58:1529–1537. doi: 10.1128/AAC.02155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrell M, Fraser VJ, Kollef MH. 2005. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother 49:3640–3645. doi: 10.1128/AAC.49.9.3640-3645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garey KW, Rege M, Pai MP, Mingo DE, Suda KJ, Turpin RS, Bearden DT. 2006. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin Infect Dis 43:25–31. doi: 10.1086/504810. [DOI] [PubMed] [Google Scholar]
- 4.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, Jones RN. 2012. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J Clin Microbiol 50:1199–1203. doi: 10.1128/JCM.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vallabhaneni S, Cleveland AA, Farley MM, Harrison LH, Schaffner W, Beldavs ZG, Derado G, Pham CD, Lockhart SR, Smith RM. 2015. Epidemiology and risk factors for echinocandin nonsusceptible Candida glabrata bloodstream infections: data from a large multisite population-based candidemia surveillance program, 2008–2014. Open Forum Infect Dis 2:ofv163. doi: 10.1093/ofid/ofv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassetti M, Merelli M, Righi E, Diaz-Martin A, Rosello EM, Luzzati R, Parra A, Trecarichi EM, Sanguinetti M, Posteraro B, Garnacho-Montero J, Sartor A, Rello J, Tumbarello M. 2013. Epidemiology, species distribution, antifungal susceptibility, and outcome of candidemia across five sites in Italy and Spain. J Clin Microbiol 51:4167–4172. doi: 10.1128/JCM.01998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klotz U, Schmidt D, Willinger B, Steinmann E, Buer J, Rath PM, Steinmann J. 2016. Echinocandin resistance and population structure of invasive Candida glabrata isolates from two university hospitals in Germany and Austria. Mycoses 59:312–318. doi: 10.1111/myc.12472. [DOI] [PubMed] [Google Scholar]
- 10.Bordallo-Cardona MA, Escribano P, de la Pedrosa EG, Marcos-Zambrano LJ, Canton R, Bouza E, Guinea J. 2017. In vitro exposure to increasing micafungin concentrations easily promotes echinocandin resistance in Candida glabrata isolates. Antimicrob Agents Chemother 61:e01542-16. doi: 10.1128/AAC.01542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bordallo-Cardona MÁ, Escribano P, Marcos-Zambrano LJ, Díaz-García J, de la Pedrosa EG, Cantón R, Bouza E, Guinea J. 2018. Low and constant micafungin concentrations may be sufficient to lead to resistance mutations in FKS2 gene of Candida glabrata. Med Mycol 56:903–906. doi: 10.1093/mmy/myx124. [DOI] [PubMed] [Google Scholar]
- 12.Bordallo-Cardona MA, Marcos-Zambrano LJ, Sanchez-Carrillo C, de la Pedrosa EGG, Canton R, Bouza E, Escribano P, Guinea J. 2018. Mutant prevention concentration and mutant selection window of micafungin and anidulafungin in clinical Candida glabrata isolates. Antimicrob Agents Chemother 62:e01982-17. doi: 10.1128/AAC.01982-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlin DS. 2014. Echinocandin resistance, susceptibility testing and prophylaxis: implications for patient management. Drugs 74:1573–1585. doi: 10.1007/s40265-014-0286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. 2007. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot Cell 6:2194–2205. doi: 10.1128/EC.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Healey KR, Zhao Y, Perez WB, Lockhart SR, Sobel JD, Farmakiotis D, Kontoyiannis DP, Sanglard D, Taj-Aldeen SJ, Alexander BD, Jimenez-Ortigosa C, Shor E, Perlin DS. 2016. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun 7:11128. doi: 10.1038/ncomms11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dellière S, Healey K, Gits-Muselli M, Carrara B, Barbaro A, Guigue N, Lecefel C, Touratier S, Desnos-Ollivier M, Perlin DS, Bretagne S, Alanio A. 2016. Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a French cohort of patients harboring low rates of resistance. Front Microbiol 7:2038. doi: 10.3389/fmicb.2016.02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Healey KR, Yadav P, Upadhyaya G, Sachdeva N, Sarma S, Kumar A, Tarai B, Perlin DS, Chowdhary A. 2018. Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob Agents Chemother 62:e00195-18. doi: 10.1128/AAC.00195-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyce KJ, Wang Y, Verma S, Shakya VPS, Xue C, Idnurm A. 2017. Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. mBio 8:e00595-17. doi: 10.1128/mBio.00595-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou X, Xiao M, Wang H, Yu SY, Zhang G, Zhao Y, Xu YC. 2018. Profiling of PDR1 and MSH2 in Candida glabrata bloodstream isolates from a multicenter study in China. Antimicrob Agents Chemother 62:e00153-18. doi: 10.1128/AAC.00153-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshpande LMPM, Castanheira M. 2018. Prevalence of MSH2 mutator genotype in echinocandin-resistant Candida glabrata from a 3-year global surveillance program. 28th European Congress of Clinical Microbiology and Infectious Diseases, 21 to 24 April 2018, Madrid, Spain. [Google Scholar]
- 21.Carreté L, Ksiezopolska E, Pegueroles C, Gómez-Molero E, Saus E, Iraola-Guzmán S, Loska D, Bader O, Fairhead C, Gabaldón T. 2018. Patterns of genomic variation in the opportunistic pathogen Candida glabrata suggest the existence of mating and a secondary association with humans. Curr Biol 28:15–27.e17. doi: 10.1016/j.cub.2017.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amanloo S, Shams-Ghahfarokhi M, Ghahri M, Razzaghi-Abyaneh M. 2018. Genotyping of clinical isolates of Candida glabrata from Iran by multilocus sequence typing and determination of population structure and drug resistance profile. Med Mycol 56:207–215. doi: 10.1093/mmy/myx030. [DOI] [PubMed] [Google Scholar]
- 23.Byun SA, Won EJ, Choi MJ, Kwon YJ, Shin JH, Kim SH, Lee WG, Kim MN, Lee K, Lee HS, Lee J. 2018. Comparison of the clinical characteristics of Candida glabrata bloodstream isolates with different genotypes as determined by multilocus sequence typing. 28th European Congress of Clinical Microbiology and Infectious Diseases, 21 to 24 April 2018, Madrid, Spain. [Google Scholar]
- 24.Espinel-Ingroff A, Alvarez-Fernandez M, Cantón E, Carver PL, Chen SC-A, Eschenauer G, Getsinger DL, Gonzalez GM, Govender NP, Grancini A, Hanson KE, Kidd SE, Klinker K, Kubin CJ, Kus JV, Lockhart SR, Meletiadis J, Morris AJ, Pelaez T, Quindós G, Rodriguez-Iglesias M, Sánchez-Reus F, Shoham S, Wengenack NL, Borrell Solé N, Echeverria J, Esperalba J, Gómez-G de la Pedrosa E, García García I, Linares MJ, Marco F, Merino P, Pemán J, Pérez del Molino L, Roselló Mayans E, Rubio Calvo C, Ruiz Pérez de Pipaon M, Yagüe G, Garcia-Effron G, Guinea J, Perlin DS, Sanguinetti M, Shields R, Turnidge J. 2015. Multicenter study of epidemiological cutoff values and detection of resistance in Candida spp. to anidulafungin, caspofungin, and micafungin using the Sensititre YeastOne colorimetric method. Antimicrob Agents Chemother 59:6725–6732. doi: 10.1128/AAC.01250-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.da Matta DA, Souza ACR, Colombo AL. 2017. Revisiting species distribution and antifungal susceptibility of Candida bloodstream isolates from Latin American medical centers. J Fungi (Basel) 3:E24. doi: 10.3390/jof3020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White T, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA. [Google Scholar]
- 27.Arendrup MC, Meletiadis J, Mouton JW, Lagrou K, Hamal P, Guinea J. 2017. EUCAST Definitive document E.DEF 7.3.1. Method for the determination of broth dilution minimum Inhibitory concentrations of antifungal agents for yeasts. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_7_3_1_Yeast_testing__definitive.pdf.
- 28.Foulet F, Nicolas N, Eloy O, Botterel F, Gantier JC, Costa JM, Bretagne S. 2005. Microsatellite marker analysis as a typing system for Candida glabrata. J Clin Microbiol 43:4574–4579. doi: 10.1128/JCM.43.9.4574-4579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbes S, Sellami H, Sellami A, Hadrich I, Amouri I, Mahfoudh N, Neji S, Makni F, Makni H, Ayadi A. 2012. Candida glabrata strain relatedness by new microsatellite markers. Eur J Clin Microbiol Infect Dis 31:83–91. doi: 10.1007/s10096-011-1280-4. [DOI] [PubMed] [Google Scholar]
- 30.Grenouillet F, Millon L, Bart JM, Roussel S, Biot I, Didier E, Ong AS, Piarroux R. 2007. Multiple-locus variable-number tandem-repeat analysis for rapid typing of Candida glabrata. J Clin Microbiol 45:3781–3784. doi: 10.1128/JCM.01603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol 41:5709–5717. doi: 10.1128/JCM.41.12.5709-5717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]