LETTER
Acinetobacter baumannii is one of the most important and threatening pathogens for health care-associated infections (HAI), and the treatments for multidrug-resistant A. baumannii (MDRAB) are limited (1, 2). Sulbactam serves as an alternative and effective option to combat these infections because of its intrinsic antimicrobial activity against the Acinetobacter genus (2). Unfortunately, increasing resistance to sulbactam compound in A. baumannii has been reported during the last decade (3, 4). Previous studies have demonstrated that β-lactamases such as TEM-1 and ADC confer sulbactam resistance in A. baumannii; however, they did not fully explain the sulbactam resistance phenomenon in each clinical strain (5–7). Therefore, we screened all β-lactamase genes in clinical isolates by whole-genome sequencing and aimed to assess the role of other β-lactamases apart from TEM-1 and ADC in sulbactam resistance of A. baumannii.
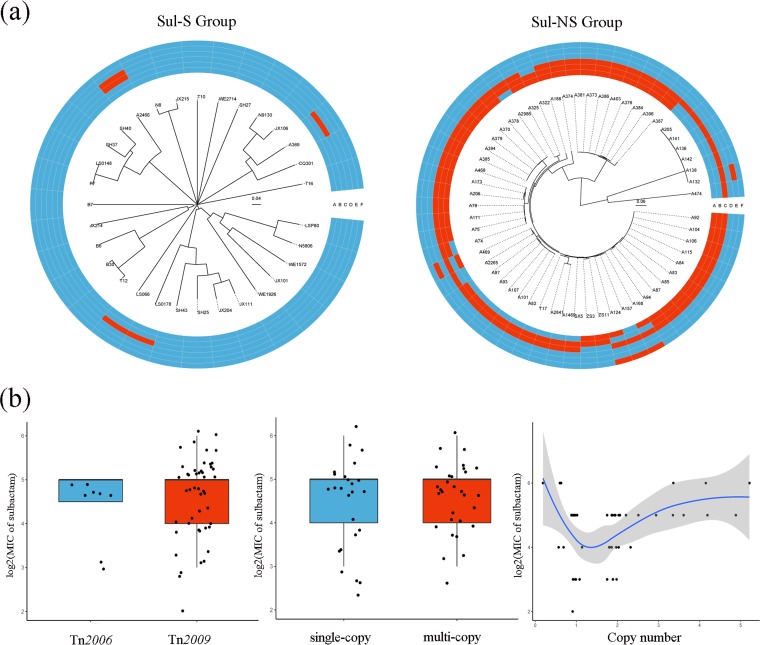
Eighty-eight clinical isolates, including 57 sulbactam-nonsusceptible isolates and 31 sulbactam-susceptible isolates, were selected from a broad geographic distribution across China (7). The presence of the β-lactamase genes blaTEM-1, ISAba1-blaADC, blaOXA-23, blaOXA-58, blaPER-1, and blaCARB-2 were screened by whole-genome sequencing (Fig. 1a). Of note, in the sulbactam-nonsusceptible group, 54 (94.74%) strains possessed the blaOXA-23 gene, while in the sulbactam-susceptible group, only 4 (12.90%) isolates were positive for the blaOXA-23 gene (P < 0.001). These results indicated that blaOXA-23 might facilitate sulbactam resistance in A. baumannii.
FIG 1.
(a) Heatmap of the presence of the β-lactamase genes of all selected clinical A. baumannii strains. Due to the lack of a breakpoint for sulbactam alone, we chose ≤4 mg/liter as the temporary susceptibility breakpoint on the basis of the CLSI susceptibility breakpoint for ampicillin-sulbactam (≤8/4 mg/liter) in A. baumannii. On the basis of the breakpoint, all clinical isolates were divided into sulbactam-nonsusceptible or sulbactam-susceptible groups. Sul-S group, sulbactam-susceptible group; Sul-NS group, sulbactam-nonsusceptible group; A, blaTEM-1; B, ISAba1-blaADC; C, blaOXA-23; D, blaOXA-58; E, blaPER-1; F, blaCARB-2. Red color represents positive, and the blue color represents negative. (b) Correlation of the characteristics of the blaOXA-23 gene associated with sulbactam MIC. All the blaOXA-23-positive strains were divided into two groups according to the transposase types carrying blaOXA-23 or copy number of blaOXA-23: Tn2009 and Tn2006, as well as single-copy group (copy number <1.5) and multicopy group (copy number >1.5).
To further elucidate the correlation between blaOXA-23 and sulbactam resistance, deletion and complementation of this gene were performed. See Table S1 in the supplemental material for a list of the strains and plasmids used in this study. The blaOXA-23 gene was deleted in a clinical A. baumannii isolate, A2265, which carried one copy of blaOXA-23 without blaTEM-1 or ISAba1-blaADC. The strain A2265 ΔblaOXA-23 was complemented with the recombinant plasmid pYMAb2-Hygr::ISAba1-blaOXA-23. The changes in the MICs were determined (Table 1). Notably, A2265 ΔblaOXA-23 exhibited increased susceptibility to sulbactam (8-fold) compared to that of A2265, and a similar trend was observed in the MICs of cefoperazone-sulbactam, imipenem, and meropenem, but no obvious change in MIC values was found in other agents. The MICs of A2265 ΔblaOXA-23(pYMAb2-Hygr::ISAba1-blaOXA-23) for sulbactam, cefoperazone-sulbactam, imipenem, and meropenem returned to levels at or above those of strain A2265.
TABLE 1.
MICs of the knockouts and the transformants
| Isolatea | MIC (mg/liter)b |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SUL | CPS | IPM | MEM | CTX | CAZ | CIP | MIN | GEN | CST | TGC | ATM | |
| A2265 | 8 | 16 | 32 | 32 | 16 | 2 | >32 | 4 | >256 | 0.25 | 1 | 12 |
| A2265 ΔblaOXA-23 | 1 | 1.5 | 1 | 1 | 8 | 4 | >32 | 2 | >256 | 0.25 | 1 | 12 |
| A2265 ΔblaOXA-23(pYMAb2-Hygr) | 1 | 0.75 | 1 | 1 | 4 | 6 | >32 | 8 | >256 | 0.25 | 1 | 6 |
| A2265 ΔblaOXA-23(pYMAb2-Hygr::ISAba1-blaOXA-23) | 64 | 64 | 128 | 128 | >32 | 3 | >32 | 4 | >256 | 0.25 | 1 | 24 |
A2265 ΔblaOXA-23, the clinical strain A2265 without the blaOXA-23 gene; A2265 ΔblaOXA-23(pYMAb2-Hygr), the strain A2265 was transformed with a plasmid pYMAb2-Hygr; A2265 ΔblaOXA-23(pYMAb2-Hygr::ISAba1-blaOXA-23), the strain A2265 ΔblaOXA-23 was complemented with the recombinant plasmid pYMAb2-Hygr::ISAba1-blaOXA-23.
SUL, sulbactam; CPS, cefoperazone-sulbactam; IPM, imipenem; MEM, meropenem; CTX, cefotaxime; CAZ, ceftazidime; CIP, ciprofloxacin; MIN, minocycline; GEN, gentamicin; CST, colistin; TGC, tigecycline; ATM, aztreonam.
Among the 58 blaOXA-23-positive strains, 47 isolates were correlated with Tn2009, 8 isolates were associated with Tn2006, and the remaining 3 isolates in the sulbactam-susceptible group were found to carry only a partial sequence of blaOXA-23. Tn2007 and Tn2008 were not detected. No significant difference in the sulbactam MICs (log2) of the Tn2009-group and Tn2006-group was found (P > 0.05) (Fig. 1b). The copy numbers of all blaOXA-23-positive strains were measured by quantitative real-time PCR. A total of 25 isolates were found to carry a single copy of blaOXA-23, while 30 isolates had at least two copies of blaOXA-23. The sulbactam MICs of the multicopy group were similar to that of the single-copy group (P > 0.05) (Fig. 1b). Furthermore, the MIC of sulbactam did not correlate with the copy number of blaOXA-23 (r = 0.263) (Fig. 1b).
OXA-23 is notoriously not inhibited by sulbactam, but sulbactam is also a well-known agent used directly against the Acinetobacter genus as an inhibitor. Although OXA-23 manifests considerable hydrolytic activity on a variety of β-lactam antibiotics, its effect on sulbactam has not been fully established (8, 9). Our research described the effect of OXA-23 on the sulbactam MIC in a clinical A. baumannii strain by gene deletion and complementation experiments. Nevertheless, we found that the sulbactam MIC did not relate to the transposon type or copy number of blaOXA-23. Similar results were found in studies concerning blaOXA-23 associated with carbapenem resistance (10, 11). The unstable copy number due to the plasmid location of blaOXA-23 mentioned in prior research may be one of the factors (11). Another possible explanation may be the coexistence of other mechanisms, such as blaTEM-1, ISAba1-blaADC, mutations of penicillin-binding proteins, and overexpression of the efflux pump, which can affect the sulbactam MIC simultaneously (12).
Altogether, the above-described findings suggest that sulbactam resistance in A. baumannii is multifactorial and can indeed be affected via OXA-23.
Supplementary Material
ACKNOWLEDGMENTS
We thank Te-Li Chen (National Yang-Ming University), Kim Lee Chua (National University of Singapore), and Pep Charusanti (University of California) for the kind gifts of the plasmids pYMAb2, pMo130-Telr, and pACBSR-Hygr, respectively. We also thank Carina Dexter (Monash University) for helpful comments on the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (grant no. 81501777 and 81601799) and the Natural Scientific Foundation of Zhejiang Province, China (LY16H190003). A.Y.P was supported by an Australian National Health and Medical Research Practitioner Fellowship.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01676-18.
REFERENCES
- 1.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neonakis IK, Spandidos DA, Petinaki E. 2011. Confronting multidrug-resistant Acinetobacter baumannii: a review. Int J Antimicrob Agents 37:102–109. doi: 10.1016/j.ijantimicag.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Zilberberg MD, Kollef MH, Shorr AF. 2016. Secular trends in Acinetobacter baumannii resistance in respiratory and blood stream specimens in the United States, 2003 to 2012: a survey study. J Hosp Med 11:21–26. doi: 10.1002/jhm.2477. [DOI] [PubMed] [Google Scholar]
- 4.Hu FP, Guo Y, Zhu DM, Wang F, Jiang XF, Xu YC, Zhang XJ, Zhang CX, Ji P, Xie Y, Kang M, Wang CQ, Wang AM, Xu YH, Shen JL, Sun ZY, Chen ZJ, Ni YX, Sun JY, Chu YZ, Tian SF, Hu ZD, Li J, Yu YS, Lin J, Shan B, Du Y, Han Y, Guo S, Wei LH, Wu L, Zhang H, Kong J, Hu YJ, Ai XM, Zhuo C, Su DH, Yang Q, Jia B, Huang W. 2016. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect 22:S9–S14. doi: 10.1016/j.cmi.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Krizova L, Poirel L, Nordmann P, Nemec A. 2013. TEM-1 beta-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J Antimicrob Chemother 68:2786–2791. doi: 10.1093/jac/dkt275. [DOI] [PubMed] [Google Scholar]
- 6.Kuo SC, Lee YT, Yang Lauderdale TL, Huang WC, Chuang MF, Chen CP, Su SC, Lee KR, Chen TL. 2015. Contribution of Acinetobacter-derived cephalosporinase-30 to sulbactam resistance in Acinetobacter baumannii. Front Microbiol 6:231. doi: 10.3389/fmicb.2015.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Fu Y, Lan P, Xu Q, Jiang Y, Chen Y, Ruan Z, Ji S, Hua X, Yu Y. 2018. Molecular epidemiology and mechanism of sulbactam resistance in Acinetobacter baumannii isolates with diverse genetic background in China. Antimicrob Agents Chemother 62:e01947-17. doi: 10.1128/AAC.01947-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaitany KC, Klinger NV, June CM, Ramey ME, Bonomo RA, Powers RA, Leonard DA. 2013. Structures of the class D Carbapenemases OXA–23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam. Antimicrob Agents Chemother 57:4848–4855. doi: 10.1128/AAC.00762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torol S, Kasap M. 2013. Purification and characterization of OXA-23 from Acinetobacter baumannii. J Enzyme Inhib Med Chem 28:836–842. doi: 10.3109/14756366.2012.689296. [DOI] [PubMed] [Google Scholar]
- 10.Hua X, Shu J, Ruan Z, Yu Y, Feng Y. 2016. Multiplication of blaOXA-23 is common in clinical Acinetobacter baumannii, but does not enhance carbapenem resistance. J Antimicrob Chemother 71:3381–3385. doi: 10.1093/jac/dkw310. [DOI] [PubMed] [Google Scholar]
- 11.Yoon EJ, Kim JO, Yang JW, Kim HS, Lee KJ, Jeong SH, Lee H, Lee K. 2017. The blaOXA-23-associated transposons in the genome of Acinetobacter spp. represent an epidemiological situation of the species encountering carbapenems. J Antimicrob Chemother 72:2708–2714. doi: 10.1093/jac/dkx205. [DOI] [PubMed] [Google Scholar]
- 12.Penwell WF, Shapiro AB, Giacobbe RA, Gu RF, Gao N, Thresher J, McLaughlin RE, Huband MD, DeJonge BL, Ehmann DE, Miller AA. 2015. Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob Agents Chemother 59:1680–1689. doi: 10.1128/AAC.04808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.