LETTER
Most of the variants of Salmonella genomic island 1 (SGI1) are multidrug resistance (MDR) DNA elements that can be transferred by horizontal gene transfer (HGT) (1). SGI1 was initially described in Salmonella enterica serovar Typhimurium DT104 (1). It consists of a 27.4-kb backbone (28 open reading frames [ORFs]) with an MDR region consisting of a 15-kb complex class 1 integron of the family In104 (1). The SGI1 variant SGI1-L was detected for the first time in Proteus mirabilis and Morganella morganii strains from Palestine in 2006 and from France in 2017, respectively (2, 3). The 3′ end of the chromosomal trmE gene is the SGI1-specific integration site in the SGI1-carrying strains (3, 4).
Providencia stuartii is Gram-negative bacterium of the family Morganellaceae commonly associated with urinary tract infections or indwelling urinary catheters in hospitalized and nursing home patients (5). It has intrinsic resistance to polymyxin, tigecycline, aminopenicillin, and first-generation cephalosporins (5).
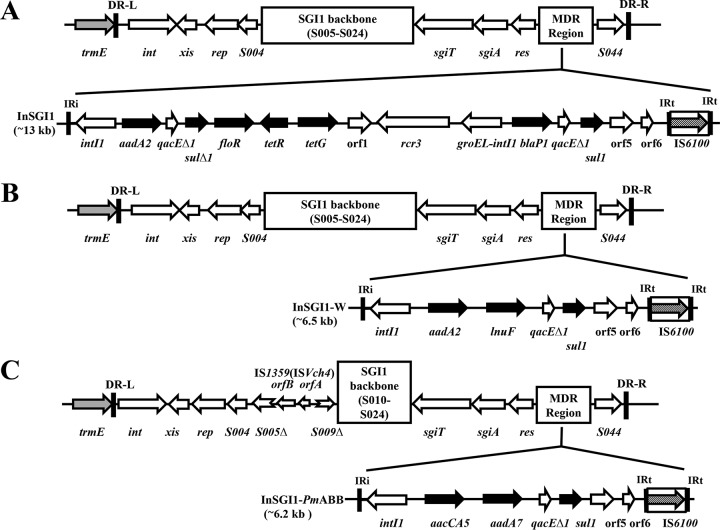
Here, we report the first identification (to our knowledge) of SGI1 in a multidrug-resistant clinical isolate of P. stuartii. The strain, designed PsTAN3, was isolated from a wound pus swab from a male patient in a teaching hospital in Tanta City, Egypt, in 2014. PsTAN3 was resistant to aztreonam (MIC, ≥512 μg/ml), ciprofloxacin (MIC, ≥16 μg/ml), chloramphenicol (MIC, ≥512 μg/ml), colistin (MIC, ≥128 μg/ml), gentamicin (MIC, ≥128 μg/ml), tetracycline (MIC, ≥128 μg/ml), streptomycin (MIC, ≥512 μg/ml), cefoxitin (MIC, ≥256 μg/ml), cefoperazone (MIC, ≥512 μg/ml), trimethoprim (MIC, ≥512 μg/ml), cefotaxime (MIC, ≥512 μg/ml), meropenem (MIC, ≥16 μg/ml), and nalidixic acid (MIC, ≥512 μg/ml) but sensitive to doripenem (MIC, ≤1 μg/ml). PsTAN3 was negative by the modified carbapenem inactivation method (mCIM), indicating the absence of carbapenemase production. PCR and DNA sequencing were used to map the entire SGI1 (for a list of PCR primers, see Table S1 in the supplemental material) and identified variant SGI1-W (6, 7). The multidrug resistance region was 6.54 kb with the genetic arrangement intI1-aadA2-lnuF-qacEΔ1-sul1-orf5-orf6-IS6100 (PsTAN3 class 1 integron GenBank accession no. LC370342) (Fig. 1). SGI1-W was integrated at the 3′ end of the trmE gene (Fig. 1). The strain also carried blaTEM-1, blaDHA-1, qnrD, and floR genes conferring resistance to third-generation cephalosporins, ciprofloxacin, florfenicol, and chloramphenicol, respectively, and the class 1 integrons aadA2 and lnuF. The MDR region of SGI1 in PsTAN3 was 100% identical to that of SGI1-PmC105 reported in a P. mirabilis isolate from a poultry farm in China (7). Furthermore, the circular form was detected in PsTAN3 after two steps of DNA amplification using primers SGI1circ1 and SGI1circ2 with the expected target size (364 bp) (GenBank accession no. LC370343) (see Fig. S1A in the supplemental material). The sequence of the SGI1-W attP site in PsTAN3 (5′-TTCTGTATCGGGAAGTAA-3′) differs in 1 bp (C→T) from that of S. enterica DT104 and P. mirabilis (5′-TTCTGTATTGGGAAGTAA-3′) (2, 6). (The bold underlined letters in the sequences indicate the differences in sequence between the SGI1-W attP site in PsTAN3 and that of S. enterica DT104 and P. mirabilis.) The same sequence of the attB site as that of the SGI1-W attP site in PsTAN3 was detected in the IncA/C2-blaTEM-24-carrying P. stuartii strain 719, which was used as an experimental recipient for SGI1 mobilization (8). The left and right junctions of SGI1-W have been amplified using primers PstLJ1(+) (5′-GCACTTAGCTCAAGGTCATGA-3′)/LJ-R1 (5′-AGTTCTAAAGGTTCGTAGTCG-3′) and 104RJ (+) (5′-CTGACGAGCTGAAGCGAATTG-3′)/Transf-Pstuartii-outR (5′-TTGTGCATTCGGCTCTTC-3′), respectively (SGI1-W left junction GenBank accession no. LC411942) (2, 6, 8).
FIG 1.
(A) Genetic mapping of SGI1 from S. Typhimurium DT104 (from GenBank accession no. AF261825 and references 1 and 2). (B) Genetic mapping of SGI-W in PsTAN3 showing the arrangement of the different gene cassettes (from PCR mapping, DNA sequencing, and GenBank accession no. KJ186151). (C) Genetic mapping of SGI1-PmABB (from GenBank accession no. JX121638) showing the arrangement of the different gene cassettes. Gene sizes are not to scale. Genes and ORFs are shown as arrows, with their transcriptional orientations indicated by arrowheads. Antibiotic resistance genes are shown as black-filled arrows. Dotted arrows within boxes indicate insertion sequence (IS) elements. Conserved SGI1 genes are shown as white-filled arrows. Thick vertical bars represent inverted repeats (IRi and IRt). Complex class 1 integrons were drawn with PCR mapping and DNA sequencing, and GenBank accession no. AF261825 (SGI1), JX121638 (SGI1-PmABB), and KJ186151 (SGI1-W) are shown.
To detect the presence of any deletion or insertion in the SGI1-W backbone, primer sets demonstrating almost all SGI1 backbone genes were used in the PCR amplification, as previously described (see Table S1) (2, 6, 9). The results indicated that PsTAN3 carried the entire SGI1 sequence. Additionally, all the SGI1-W-positive strains reported globally were characterized by the absence of any deletion or insertion in the SGI1-W backbone genes (6, 7, 9). The absence of any deletion in ORF S005 (traN) in SGI1-W of PsTAN3 and the presence of two intact ORFs (S006 and S007) coding for homologs of FlhDCSGI1 will result in increasing SGI1 maintenance, transfer, and/or spreading. Consequently, we tried to mobilize SGI1-W to the azide-resistant (AZr) strain Escherichia coli J53 (transconjugant containing the IncA/C plasmid). Transconjugants were selected on MacConkey agar plates containing 16 µg/ml trimethoprim or 4 µg/ml gentamicin and 150 µg/ml sodium azide. Unfortunately, conjugation was unsuccessful. In addition, the detection of blaVEB-6 and blaNDM-1 in PGI1-PmPEL and blaVEB-6-carrying SGI1-V in P. mirabilis strains isolated in France are a serious public health threat (10). Recently, we reported the detection of SGI1-PmABB in the P. mirabilis strain PmTAN59 in the same hospital (6). In addition, we reported the detection of clonally related SGI1-W-carrying P. mirabilis strains isolated from chickens and patients in Egypt (9). These findings indicated that SGI1-W might be distributed in the Egyptian sector. Therefore, we suggest that SGI1 has a broad bacterial host range and has the potential to spread to several enterobacterial strains or even more broadly among Gram-negative species.
In conclusion, we report the first identification of an SGI1-positive P. stuartii strain; this strain carried SGI1 variant SGI1-W. Active SGI1-screening policies are essential for preventing the HGT of this mobile island to other human pathogens.
The GenBank accession numbers of PsTAN3 class 1 integron, SGI1-W attP site, and the left junction of SGI1-W are LC370342, LC370343, and LC411942, respectively.
Supplementary Material
ACKNOWLEDGMENTS
A.M.S. was supported by a doctoral fellowship from the Ministry of Education, Culture, Sports, Science and Technology of Japan (fellowship no. 153532).
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01793-18.
REFERENCES
- 1.Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, Imberechts H, Mulvey MR. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 183:5725–5732. doi: 10.1128/JB.183.19.5725-5732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed AM, Hussein AI, Shimamoto T. 2006. Proteus mirabilis clinical isolate harboring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J Antimicrob Chemother 59:184–190. doi: 10.1093/jac/dkl471. [DOI] [PubMed] [Google Scholar]
- 3.Schultz E, Barraud O, Madec JY, Haenni M, Cloeckaert A, Ploy MC, Doublet B. 2017. Multidrug resistance Salmonella genomic island 1 in a Morganella morganii subsp. morganii human clinical isolate from France. mSphere 2:e00118-17. doi: 10.1128/mSphere.00118-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doublet B, Poirel L, Praud K, Nordmann P, Cloeckaert A. 2010. European clinical isolate of Proteus mirabilis harbouring the Salmonella genomic island 1 variant SGI1-O. J Antimicrob Chemother 65:2260–2262. doi: 10.1093/jac/dkq283. [DOI] [PubMed] [Google Scholar]
- 5.Mao YC, Chang CL, Huang YC, Su LH, Lee CT. 2018. Laboratory investigation of a suspected outbreak caused by Providencia stuartii with intermediate resistance to imipenem at a long-term care facility. J Microbiol Immunol Infect 51:214–219. doi: 10.1016/j.jmii.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Soliman AM, Ahmed AM, Shimamoto T, El-Domany RA, Nariya H, Shimamoto T. 2017. First report in Africa of two clinical isolates of Proteus mirabilis carrying Salmonella genomic island (SGI1) variants, SGI1-PmABB and SGI1-W. Infect Genet Evol 51:132–137. doi: 10.1016/j.meegid.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 7.Lei CW, Zhang AY, Liu BH, Wang HN, Guan ZB, Xu CW, Xia QQ, Cheng H, Zhang DD. 2014. Molecular characteristics of Salmonella genomic island 1 in Proteus mirabilis isolates from poultry farms in China. Antimicrob Agents Chemother 58:7570–7572. doi: 10.1128/AAC.03992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siebor E, de Curraize C, Amoureux L, Neuwirth C. 2016. Mobilization of the Salmonella genomic island SGI1 and the Proteus genomic island PGI1 by the A/C2 plasmid carrying blaTEM-24 harboured by various clinical species of Enterobacteriaceae. J Antimicrob Chemother 71:2167–2170. doi: 10.1093/jac/dkw151. [DOI] [PubMed] [Google Scholar]
- 9.Soliman AM, Saad AM, Ahmed AM, Al-Baqir A, Hussein A, Shimamoto T, Nariya H, Shimamoto T. 2018. Occurrence of Salmonella genomic island 1 (SGI1) in two African Proteus mirabilis strains isolated from diseased chicken flocks. Infect Genet Evol 62:8–10. doi: 10.1016/j.meegid.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Schultz E, Cloeckaert A, Doublet B, Madec JY, Haenni M. 2017. Detection of SGI1/PGI1 elements and resistance to extended-spectrum cephalosporins in Proteae of animal origin in France. Front Microbiol 8:32. doi: 10.3389/fmicb.2017.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.