Abstract
Control of neuronal precursor cell proliferation is essential for normal brain development, and deregulation of this fundamental developmental event contributes to brain diseases. Typically, neuronal precursor cell proliferation extends over long periods of time during brain development. However, how neuronal precursor proliferation is regulated in a temporally specific manner remains to be elucidated. Here, we report that conditional KO of the transcriptional regulator SnoN in cerebellar granule neuron precursors robustly inhibits the proliferation of these cells and promotes their cell cycle exit at later stages of cerebellar development in the postnatal male and female mouse brain. In laser capture microdissection followed by RNA-Seq, designed to profile gene expression specifically in the external granule layer of the cerebellum, we find that SnoN promotes the expression of cell proliferation genes and concomitantly represses differentiation genes in granule neuron precursors in vivo. Remarkably, bioinformatics analyses reveal that SnoN-regulated genes contain binding sites for the transcription factors N-myc and Pax6, which promote the proliferation and differentiation of granule neuron precursors, respectively. Accordingly, we uncover novel physical interactions of SnoN with N-myc and Pax6 in cells. In behavior analyses, conditional KO of SnoN impairs cerebellar-dependent learning in a delayed eye-blink conditioning paradigm, suggesting that SnoN-regulation of granule neuron precursor proliferation bears functional consequences at the organismal level. Our findings define a novel function and mechanism for the major transcriptional regulator SnoN in the control of granule neuron precursor proliferation in the mammalian brain.
SIGNIFICANCE STATEMENT This study reports the discovery that the transcriptional regulator SnoN plays a crucial role in the proliferation of cerebellar granule neuron precursors in the postnatal mouse brain. Conditional KO of SnoN in granule neuron precursors robustly inhibits the proliferation of these cells and promotes their cycle exit specifically at later stages of cerebellar development, with biological consequences of impaired cerebellar-dependent learning. Genomics and bioinformatics analyses reveal that SnoN promotes the expression of cell proliferation genes and concomitantly represses cell differentiation genes in vivo. Although SnoN has been implicated in distinct aspects of the development of postmitotic neurons, this study identifies a novel function for SnoN in neuronal precursors in the mammalian brain.
Keywords: cell proliferation, cerebellar development, granule neuron precursors, SnoN, transcriptional regulator
Introduction
Control of neuronal precursor cell proliferation is essential for normal brain development, and abnormalities of this process cause neurological diseases (Dehay and Kennedy, 2007). The proliferation of cerebellar granule neuron precursors has been the subject of intense interest. Granule neurons originate from precursor cells in the rhombic lip, which migrate during embryogenesis over the surface of the cerebellar anlage to form the external granule layer (EGL) (Hatten and Heintz, 1995; Sillitoe and Joyner, 2007). Granule neuron precursors remain mitotically active in the outer zone of the EGL in the first 2 weeks postnatally in the mouse cerebellum and give rise to postmitotic granule neurons that complete their maturation in the internal granule layer (IGL) (Ramon y Cajal, 1911; Altman, 1997). Although mechanisms that govern granule neuron precursor proliferation have been characterized (Leto et al., 2016), these mechanisms remain incompletely understood.
Extrinsic cues, including BMPs, sonic hedgehog (SHH), and Notch activation as well as transcriptional regulators play critical roles in the proliferation of granule neuron precursors during embryogenesis and in the postnatal brain (Ben-Arie et al., 1997; Wechsler-Reya and Scott, 1999; Solecki et al., 2001; Rios et al., 2004). Rhombic lip-derived granule neuron precursors express the transcriptional factor Atoh1/Math1 (referred from here as Math1) (Machold and Fishell, 2005). KO of Math1 leads to loss of granule neuron precursors (Ben-Arie et al., 1997; Flora et al., 2009). KO studies have implicated a number of other transcription factors, including Zic1, N-myc, Meis1, and Zfp423, in the proliferation of granule neuron precursors (Kenney et al., 2003; Alcaraz et al., 2006; Frank et al., 2015; Owa et al., 2018).
The transcriptional regulator SnoN plays critical roles in the development of postmitotic granule neurons in the developing cerebellum (de la Torre-Ubieta and Bonni, 2011; Bonni and Bonni, 2012). SnoN promotes the growth and stability of granule neuron parallel fiber axons in the rodent cerebellar cortex (Stegmüller et al., 2006, 2008; Ikeuchi et al., 2009). In addition, SnoN regulates in an isoform-specific manner granule neuron migration and dendrite branching (Huynh et al., 2011). Whereas the roles and mechanisms of SnoN have been characterized in postmitotic granule neurons, SnoN functions in granule neuron precursors have remained unknown. Notably, outside the nervous system, SnoN has been implicated in the control of cell proliferation, epithelial-mesenchymal transition, and oncogenesis (Stroschein et al., 1999; Sun et al., 1999; Sarker et al., 2005; Netherton and Bonni, 2010; Ikeuchi et al., 2014), but the roles of SnoN in neuronal precursor cells have remained unexplored.
In this study, we have discovered a novel function for the transcriptional regulator SnoN in the control of granule neuron precursor proliferation in the postnatal mammalian brain. Conditional KO of SnoN in granule neuron precursors using the Math1-Cre driver inhibits the proliferation of these cells and promotes their cell cycle exit, leading to structural defects in the EGL at later stages of cerebellar development. To facilitate mechanistic studies, we establish laser capture microdissection followed by RNA-seq (LCMRNA-seq) to profile gene expression specifically in the EGL of the developing cerebellum, where granule neuron precursors reside. In LCMRNA-seq analyses of the EGL in SnoN KO and control littermate mice, we find that SnoN regulates the expression of cell proliferation and differentiation genes in the EGL. Strikingly, bioinformatics analyses reveal that SnoN-regulated genes are enriched for binding sites for the transcription factors N-myc and Pax6, which have been implicated in granule neuron precursor proliferation and differentiation (Kenney et al., 2003; Owa et al., 2018). Accordingly, SnoN forms a complex with N-myc and Pax6 in cells, suggesting that SnoN may regulate N-myc and Pax6 function in granule neuron precursors. In behavior analyses, conditional KO of SnoN triggers impairment of cerebellar-dependent learning, suggesting that SnoN activity in granule neuron precursors bears consequences for cerebellar function. Our findings define a novel role for SnoN in the control of cerebellar granule neuron precursor proliferation in the postnatal mammalian brain.
Materials and Methods
Animals.
Rodents were purchased or maintained under pathogen-free conditions. All animal experiments were done according to protocols approved by the Animal Studies Committee of Washington University School of Medicine and in accordance with the National Institutes of Health guidelines. ES cell line containing a “KO-first” SnoN allele (Skiltm1a(EUCOMM)Hmgu) was obtained from the European Conditional Mouse Mutagenesis Program (EUCOMM). After obtaining germ line transmission, SnoNLacZ-neo-loxp mice were bred to mice with the C57BL/6J background. The LacZ-neo cassette was removed by crossing with Flp deleter strain, followed by mating to C57BL/6J mice at least three generations. We induced KO of SnoN specifically in granule neuron precursors of the mouse cerebellum by crossing SnoNloxp/loxp mice with a transgenic mouse line in which the expression of the recombinase Cre is driven by the Math1 promoter. Conditional SnoN KO was confirmed by PCR analysis of genomic DNA, qRT-PCR, and immunoblotting. Genotyping for the SnoN loxp allele was performed with the following primers: Loxp forward (G-F), 5′-ACCAGTTATTATTCCCCTGTTCCT-3′; and Loxp reverse (G-R), 5′-GGCATGGCTTACCAGAAACC-3′.
Gender-matched male and female littermate mice were used for all experiments.
Antibodies.
Antibodies to SnoN (Santa Cruz Biotechnology, sc-9141), calbindin (Millipore, AB1778), Ki67 (Abcam, ab15580), P27 (BD Biosciences, 610241), phospho-histone H3 (Cell Signaling Technology, 53348), GFP (Abcam, 13970), cleaved caspase-3 (Asp175) (Cell Signaling Technology, 9661), Math1 (Developmental Studies Hybridoma Bank), Flag (Millipore, F1804), HA (BioLegend, 901515), SnoN (Proteintech, 19218-1-AP), BrdU (Abcam, ab6326), ERK1/2 (Cell Signaling Technology, 9102), and Cre (Millipore, 69050) were purchased.
Immunoblotting analyses.
Whole-cell lysates were separated on 8% SDS-polyacrylamide gel, transferred to 0.2 μm nitrocellulose blotting membrane (GE Healthcare), and probed with primary antibodies (SnoN, ERK, and Cre) and HRP-coupled secondary antibodies (Millipore).
RNA in situ hybridization.
In situ RNA hybridization was performed as described previously (Hua et al., 2018). A 509 bp mouse SnoN cDNA was amplified by PCR with the forward primer GGAACTGAGAACAACATGCCAG and reverse primer ATAGACTCCCCTTCCAAAAGAG. The cDNA was then ligated to the T Easy Vector (A362A, Promega) and verified by sequencing. A secondary PCR was performed with T7 (TAATACGACTCACTATAGGG) and SP6 (ATTTAGGTGACACTATAGAA), and the amplified DNA fragments served as the template to synthesize DIG-labeled RNA probe through in vitro transcription. Antisense and sense probes were individually synthesized via T7 RNA polymerase (P207E, Promega) and SP6 RNA polymerase (P108E, Promega), respectively. Sagittal sections of the brain (40 μm) were washed 3 times with 1 × PBST (0.01 m PBS containing 0.1% Tween 20), 10 min each time. Prehybridization was performed by incubating sections at 50°C for 2 h in the prehybridization solution (50% deionized formamide, 5 × SSC, 0.1% Tween 20). For hybridization performed at 50°C for 16–20 h, a final concentration of 5 μg/ml RNA probe was dissolved in the hybridization solution (50 μg/ml heparin and 0.5 mg/ml yeast tRNA in prehybridization solution). The probe was denatured at 80°C for 10 min and then immediately chilled on ice for 5 min. After hybridization, sections were washed with 5 × SSC-50% formamide (v/v) with 0.1% Tween 20 (5 × SSCT-50% formamide) at room temperature for 5 min, and then 2 × SSCT-50% formamide at 50°C for 1 h. Sections were washed twice with 2 × SSCT followed by 20 μg/ml RNase A (R1253, Thermo Fisher Scientific) treatment for 30 min at room temperature. After washing with 2 × SSCT, sections were incubated with 2 × SSCT-50% formamide at 50°C for 1 h and then washed with 2 × SSCT, 0.2 × SSCT, and 1 × PBST. AP-conjugated anti-DIG antibody (1093274, Roche Diagnostics, 1:500) was then applied overnight at 4°C in the blocking buffer (1 × PBST containing 10% normal donkey serum and 0.2% BSA). After adequate washing with 1 × PBST, sections were washed with fresh AP buffer (0.1 m Tris-HCl, pH 9.5, 0.05 m MgCl2, 0.1 m NaCl, and 20 μm levamisole hydrochloride), and then detected with NBT/BCIP (11681451001, Roche Diagnostics, 1:200 in AP buffer) for 30 min.
Coimmunoprecipitation.
Coimmunoprecipitation analyses were performed as described previously (Pot et al., 2013). HEK293T cells were lysed in TNTE (50 mm Tris, 150 mm NaCl, and 1 mm EDTA) buffer containing 0.5% Triton X-100 along with protease and phosphatase inhibitors and subjected to immunoprecipitation using mouse anti-HA or mouse anti-Flag followed by immunoblotting with specific antibodies as indicated, including anti-HA, anti-Flag or rabbit anti-SnoN, and HRP-conjugated anti-mouse and anti-rabbit IgG antibodies. Immunoblotting-generated enhanced chemiluminescence signals were visualized using a Versadoc 5000 Imager (Bio-Rad). Cell lysates were subjected to immunoblotting, including using anti-actin antibody as loading control.
For Renilla luciferase interaction studies, equivalent amounts of protein from cell lysates were subjected to immunoprecipitations using appropriate antibodies. Immunoprecipitates and aliquots of lysates were subjected to immunoblotting or Renilla luciferase assays using the Renilla luciferase kit (E1910, Promega) and the Orion II luminometer (Titertek-Berthold) detection system. Immunoprecipitates containing Rluc fusion or Rluc protein alone were resuspended in TNE buffer containing 0.1% Triton X-100 before measuring luciferase activity. For each experiment, Rluc values in immunoprecipitates were normalized to the respective input Rluc values, which were then normalized to the global average of each experiment and finally plotted relative to the empty vector sample. The pCDNA3-HA-Nmyc was obtained from Addgene (Vo et al., 2016). The pCMV5B-HA-Pax6 and pCMV5B-Flag-Pax6 plasmids were generated by subcloning a PCR-amplified Pax6 open reading frame DNA, using as template the Teto-FUW-Pax6 plasmid obtained from Addgene (Cassady et al., 2014), into pCMV5B-HA- and pCMV5-Flag-expressing vectors, respectively, and verified by sequencing. The pCMV5B-Flag-SnoN, pCMV5B-HA-SnoN, pCMV5B-Rluc, and pCMV5B-Rluc-SnoN plasmids have been described previously (Stroschein et al., 2001).
In vivo electroporation.
In vivo electroporation of postnatal mouse pups was performed as described previously (Konishi et al., 2004; Shalizi et al., 2006; Yang et al., 2009; Yamada et al., 2013). P11 mouse pups were injected with the pCAG-GFP plasmid and subjected to four electrical pulses of 135 mV with 950 ms intervals. Electroporated pups were returned to moms and examined 3 d later with immunohistochemical analyses. Only mitotic granule precursor cells were labeled with GFP, which was subsequently retained after differentiation into postmitotic granule neurons.
Immunohistochemical analyses.
The cerebellum from animals perfused with PFA was fixed overnight in 4% PFA, cryoprotected in 15% sucrose followed by 30% sucrose, frozen in tissue optimal cutting temperature embedding medium (Thermo Fisher Scientific) and cut into 20 μm sagittal sections. Sections were incubated for 1 h with PBS containing 0.2%–0.4% Triton X-100, 10% goat serum, 3% BSA, and subsequently with primary antibodies overnight at 4°C, followed with secondary antibodies (Invitrogen) for 2 h at room temperature. Sections were counterstained with the DNA dye bisbenzimide (Hoechst), mounted with Fluoromount-G, and visualized using an Olympus FV1200 scanning confocal microscope or Carl Zeiss Axio Imager Z2 Fluorescence Microscope with ApoTome-2.
5-Ethynyl-2′-deoxyuridine (EdU) analysis.
EdU incorporation analyses were used to detect DNA synthesis during the S phase of the cell cycle. EdU (10 mg/ml) in PBS was injected intraperitoneally (20 mg/kg body weight) in P11 and P14 mice, and 30 min later the cerebellum from these mice was subjected to an assay for EdU detection using the Click-iT EdU AlexaFluor-647 Kit (Invitrogen C10340).
For fate mapping assay, mice were injected with EdU (100 mg/kg body weight). At 24 h, the cerebellum was subjected to histochemical analyses using the EdU detection reagent and Ki67 antibody. EdU+/Ki67+ cells represent granule neuron precursors, which remain in the cell cycle, whereas EdU+/Ki67− cells represent granule neuron precursors that have exited the cell cycle and differentiated into postmitotic granule neurons within 24 h.
Apoptosis assay.
Analyses of apoptosis were performed as described previously (Whittaker et al., 2017). Sections of the cerebellum were treated with 3% H2O2 and 10% methanol in PBS for 15 min to block endogenous peroxidase, antigen retrieved with 0.1 m citrate buffer, pH 6.0, at 80°C, incubated for 1 h with PBS containing 0.1% Triton X-100, 10% goat serum, 3% BSA, and subsequently with the cleaved caspase-3 antibody overnight at 4°C, followed with biotinylated secondary antibodies (1/200; Dako; E0432) for 1 h at room temperature. Signal was amplified by Vectastain ABC kit (Vector Laboratories) and incubated with the DAB peroxidase substrate (Vector Laboratories, SK-4105). Sections were dehydrated and mounted with Fluoromount-G and visualized using Carl Zeiss Axio Imager Z2 Fluorescence Microscope with ApoTome-2.
qRT-PCR.
Reverse transcription reactions were performed with Superscript III (Invitrogen) according to the manufacturer's protocol. PCRs were performed using lightcycle480 (Roche Diagnostics). The following primers were used: Gapdh forward, 5′-ATGACATCAAGAAGGTGGTG-3′; Gapdh reverse, 5′-CATACCAGGAAATGAGCTTG-3′; SnoN forward (KE-F), 5′-ACCAAAAATACCTAGGGACACCC-3′; SnoN reverse (KE-R), 5′-GGGAACATGGTCACCTTCCTGTT-3′; Math1 forward, 5′-GAGTGGGCTGAGGTAAAAGAGT-3′; and Math1 reverse, 5′- GGTCGGTGCTATCCAGGAG-3′.
Laser capture microdissection (LCM).
The cerebellum from P6 mice was rapidly dissected, snap frozen in OCT on liquid nitrogen, and stored at −80°C. The frozen cerebellum was transferred to a cryostat (Leica Microsystems), and 60 μm sagittal sections were collected on PEN slides (2.0 μm, Thermo Fischer Scientific). To enhance visualization of the EGL while minimizing RNA degradation, we used the following rapid cresyl violet staining protocol under RNase-free conditions: slides were fixed for 2 min in ice-cold 75% EtOH, incubated for 30 s twice in 75% EtOH, stained for 1 min in 0.5% cresyl violet in 75% EtOH, serially dehydrated for 30 s each in 75%, 95%, and 100% EtOH, and finally immersed for 10 s in ice-cold 75% EtOH. Slides were immediately transferred to a desiccant chamber and mounted for LCM by a LMD7000 (Leica Microsystems). The EGL was easily identified by its dense population of heavily stained cresyl violet cells. Cells isolated by LCM were collected directly in RNA-stabilizing buffer, RLT buffer (QIAGEN) with 1% β-mercaptoethanol, and stored at −80°C.
RNA-seq analyses.
Total RNA was extracted from cerebellum of P6 mice using Trizol (Invitrogen) according to the manufacturer's instructions. The EGL from the central lobules of P6 mice cerebellum was microdissected and RNA was extracted using RNeasy Plus Micro Kit with gDNA eliminator column. After BioAnalyzer quality control (Agilent Technologies), intact RNA was used for library using NEBNext Ultra RNA library preparation kit for E7530 (Illumina). The library was sequenced on a Hiseq3000 (Illumina, Genome Technology Access Center at Washington University). Briefly, sequences are adapter-trimmed using Cutadapt and subjected to quality control using PRINSEQ and aligned to mouse genome GRCm38/mm10 using STAR (Schmieder and Edwards, 2011; Dobin et al., 2013). Sequencing performance was assessed for total number of aligned reads, total number of uniquely aligned reads, genes and transcripts detected, ribosomal fraction, known junction saturation, and reads distribution over known gene models with RSeQC (Wang et al., 2012). Reads in features were counted using HTseq (Anders et al., 2015). Genes with <10 reads in all samples were excluded from further analysis. Outlier samples identified using robust principal component analysis according to the ROBPCA, and GRID algorithms implemented in rrcov R package were removed from further analysis. Genes differentially expressed in the EGL in SnoN conditional KO mice and littermate control were identified using DESeq2 with a false discovery rate (FDR) adjusted p values ≤0.1. rlog transformation normalized counts were calculated using DESeq2, and normalized gene counts were converted to z scores for plotting. Heatmaps were generated using heatmap.2 function of the gplots R package (Chen et al., 2016). DAVID was used for gene ontology enrichment analysis.
To identify candidate transcription factors that regulate differentially expressed genes, the oPossum-3 and Motif-ORI algorithms (oPossum-3: 5 kb upstream of TSS; and Motif-ORI: 5 kb upstream of ATG) were used to identify transcription factor binding sites enriched in the promoters of differentially expressed genes compared with promoter sequences of background genes (Zhao et al., 2007; Kwon et al., 2012). Raw sequencing data and samples information related to this study have been archived in the Gene Expression Omnibus database under accession number GSE120279.
Granule neuron precursor cultures.
Granule neuron precursor cultures were established as described previously (Kenney et al., 2003). The cerebellum of P6 mice was dissected in calcium and magnesium-free Hanks buffered saline solutions with glucose, and lobules IX and X were removed. Cells were dissociated with Trypsin-DNase solution followed by trituration. Granule neurons and granule neuron precursors were isolated by Percoll fractionation with a 35% and 65%, pH 7.4, acid Percoll gradient (Sigma-Aldrich, P1644). To remove glia cells, isolated cells were cultured on poly-d-lysine (Sigma-Aldrich, P72280) coated plate for 30 min. After incubation, enriched granule neuron and precursors were transferred to poly-ornithine (Sigma-Aldrich, P3655) treated glass coverslips. Three hours later, 10% FBS medium was exchanged for granule neuron precursor culture medium (DMEM-F12/N2 supplement/25 mm KCl) with or without SHH (R&D Systems).
Delay eye-blink conditioning assay.
The delay eye-blink conditioning assay was performed as described previously (Yang et al., 2016; Valnegri et al., 2017). Conditional SnoN KO and littermate control mice at 6–8 weeks of age were used. Mice anesthetized with ketamine/xylazine (100 mg/kg; 10 mg/kg) were implanted with a head plate placed over the skull bregma and secured with screws using Metabond cement (Parkell). Five days following recovery from surgery, head-fixed mice were habituated on a cylindrical treadmill over 2 h. After training, mice were tested with the head-fixed eye-blink conditioning apparatus. Mice learned to blink in response to an initially neutral conditioned stimulus (CS; blue LED) that was paired with an eyeblink-eliciting unconditioned stimulus (US, 20 psi periocular air puff through a flattened 25-gauge needle; CS-US interstimulus interval, 150 ms); 100 trials of CS and US pairing were performed each day, and learned eyelid conditioned response (CR) was tracked over 6 consecutive days using a high-speed monochrome camera (Allied Vision). The eyelid position was calculated on each frame by the “pixel area” method as described, and quantified in units of fraction eyelid closure, ranging from 0 (fully open) to 1 (fully closed). Eyelid closure >0.2 during the interstimulus interval period was defined as a CR. The percentage of trials during each session containing CRs was used as the index of motor learning.
Open field assay.
The open field assay was performed using 6- to 8-week-old conditional SnoN KO or control littermate mice. Activity was quantified over a 30 or 60 min period in an open field (41 × 41 × 38.5 cm high) constructed of Plexiglas and containing computerized photobeam instrumentation (Kinder Scientific). The apparatus contains a 16 × 16 matrix of photocell pairs as well as a rack containing photocell pairs to measure vertical rearing. Variables related to general activity and exploration (total ambulation, rearing frequency, and rearing time), center-of-the field behaviors (entries made, time spent, distance traveled in the center zone), as well as distance traveled in the peripheral zone during successive 10 or 5 min periods were analyzed. The dimensions of the center and peripheral zones may be varied in a post hoc manner. The smaller peripheral zone was a 5.13-cm-wide zone around the perimeter of the field while the smaller center was a 10.2 × 10.2 cm square in the center of the field.
DigiGait.
The DigiGait assay was performed using 6 week conditional SnoN KO or control littermate mice. Mice were placed on a DigiGait apparatus (Mouse Specific) and run on a transparent moving belt (30 cm/s, 12.5° incline) for 1–3 min. A movie of the paws was recorded during running using a high-speed video camera and analyzed using DigiGait software.
Statistics.
Statistical analyses for qRT-PCR or immunohistochemistry analyses were done using Prism 7.0 software (GraphPad). Bar graphs are presented as the mean ± SEM. For experiments in which only two groups were analyzed, the t test was used. Pairwise comparisons within multiple groups were done by ANOVA followed by the Fisher's PLSD post hoc test. Gait dynamics were analyzed by repeated-measures ANOVA, planned comparisons.
Results
Generation of conditional SnoN KO mice in cerebellar granule neuron precursors
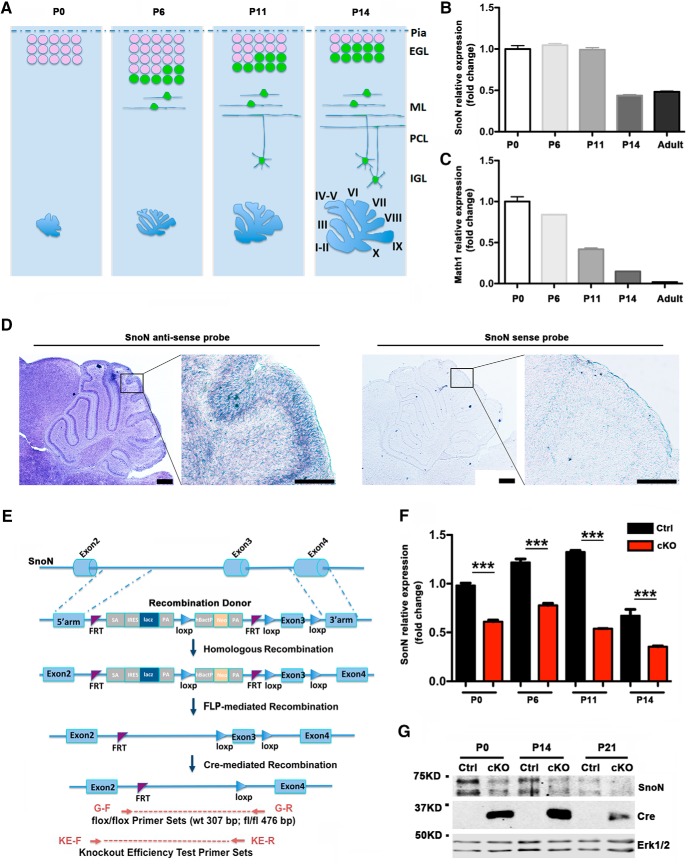
To determine whether SnoN might have a role in granule neuron precursor development, we first characterized the expression of SnoN mRNA in the developing mouse cerebellum. The EGL, where granule neuron precursors proliferate and differentiate into postmitotic neurons, is composed mostly of mitotic precursors at postnatal day 0 (P0) in rodents. Over the course of a week, the thickness of the EGL increases, with peak proliferation occurring around P6-P7 (Altman, 1997). With further development after P12, the EGL diminishes in thickness as mitotic precursors in the outer zone of the EGL (oEGL) are depleted and postmitotic granule neurons in the inner zone of the EGL (iEGL) migrate to the IGL (Fig. 1A) (Ramon y Cajal, 1911; Altman, 1997). In qRT-PCR analyses, SnoN was highly expressed in the mouse cerebellum at P0 through P11 and then declined to lower levels of expression at P14 and in the adult brain (Fig. 1B). In control analyses, Math1 mRNA levels were high at P0 in the cerebellum, and then gradually declined in a stepwise manner at P6, P11, and P14, with little or no expression in the adult (Fig. 1C). In situ RNA hybridization studies revealed that SnoN mRNA was expressed throughout the developing cerebellar cortex, with robust expression in the EGL in P6 mice (Fig. 1D). Together, these data reveal that SnoN is highly expressed in granule neuron precursors in the developing mouse cerebellum at a time when these cells undergo proliferation and differentiation.
Figure 1.
Generation of conditional SnoN KO mice in cerebellar granule neuron precursors. A, A schematic representing timeline of cerebellar granule neuron development. Granule neuron precursors are depicted in pink and postmitotic granule neurons in green in the EGL. After migrating from the EGL to the IGL via the molecular layer (ML) and Purkinje cell layer (PCL), granule neurons complete their maturation. Four representative time points (P0, P6, P11, and P14) and their lobule formation are shown. B, Lysates of mouse cerebellum at P0, P6, P11, P14, and adult were subjected to mRNA extraction followed by qRT-PCR analyses. The expression of SnoN was highly expressed in the mouse cerebellum at P0 through P11 and then declined to lower levels of expression at P14 and adult brain (n = 3). C, In control analyses, the mRNA expression of Math1 was high at P0 in the cerebellum and then declined in a stepwise manner at P6, P11, and P14, with little or no expression in the adult (n = 3). D, The cerebellum from P6 mice was subjected to in situ hybridization analyses using DIG-labeled RNA probe generated by in vitro transcription. SnoN mRNA was robustly expressed in the EGL of P6 mice (left), but no signal was detected using the sense control (right). Scale bars: low magnification, 250 μm; high magnification, 100 μm. E, A schematic representation of the strategy for constructing conditional SnoN KO (cKO) mice. Two loxp sites flanking exon 3 were designed in the recombination donor vector. Two FRT sites encompassing a Lacz reporter and neomycin cassette were inserted within intron 2. After Flp-mediated recombination, the Lacz and drug selection expression cassettes were removed. Cre-mediated recombination removes exon 3 and therefore disrupts SnoN expression specifically in cells in which Cre is expressed. Primer sets for genotyping (G-F and G-R) and KO efficacy test (KE-F and KE-R) were marked with arrows at representative positions. F, Lysates of the cerebellum from conditional SnoN KO (cKO) or littermate control (Ctrl) mice at indicated ages were subjected to qRT-PCR analyses using primers for SnoN and GAPDH, the latter serving as internal control. ***p < 0.001 (ANOVA followed by Fisher's PLSD post hoc test). n = 3 independent experiments. G, Lysates of the cerebellum from conditional SnoN KO or littermate control mice at indicated ages were subjected to immunoblotting using antibodies to SnoN, Cre and Erk1/2, the latter serving as loading control.
To study the function of SnoN in the development of granule neuron precursors, we generated a mouse in which exon 3 of the SnoN gene is flanked with loxp sites. To induce the KO of SnoN in granule neuron precursors, we crossed the SnoNloxp/loxp animals with the Math1-Cre driver mice that express the recombinase Cre in granule neuron precursors within the cerebellar cortex (Fig. 1E). We confirmed that induction of SnoN KO in granule neuron precursors led to downregulation of SnoN mRNA in the mouse cerebellum at multiple postnatal ages from P0 to P14 and the downregulation of SnoN protein from P0 to P21 (Fig. 1F,G; Table 1). These results indicate that targeting the SnoN gene in granule neuron precursors leads to SnoN KO in these cells in the developing mouse brain.
Table 1.
SnoN mRNA expression in the cerebellum of control and conditional SnoN knockout mice
| Age | Ctrl (mean ± SEM) | cKO (mean ± SEM) | No. of mice | p |
|---|---|---|---|---|
| P0 | 0.98 ± 0.02517 | 0.609 ± 0.01762 | 3 | p < 0.0001 |
| P6 | 1.217 ± 0.03667 | 0.7767 ± 0.02186 | 3 | p < 0.0001 |
| P11 | 1.323 ± 0.01667 | 0.5380 ± 0.001 | 3 | p < 0.0001 |
| P14 | 0.7033 ± 0.0318 | 0.3533 ± 0.008819 | 3 | p < 0.0001 |
Conditional KO of SnoN in granule neuron precursors triggers defects in EGL development
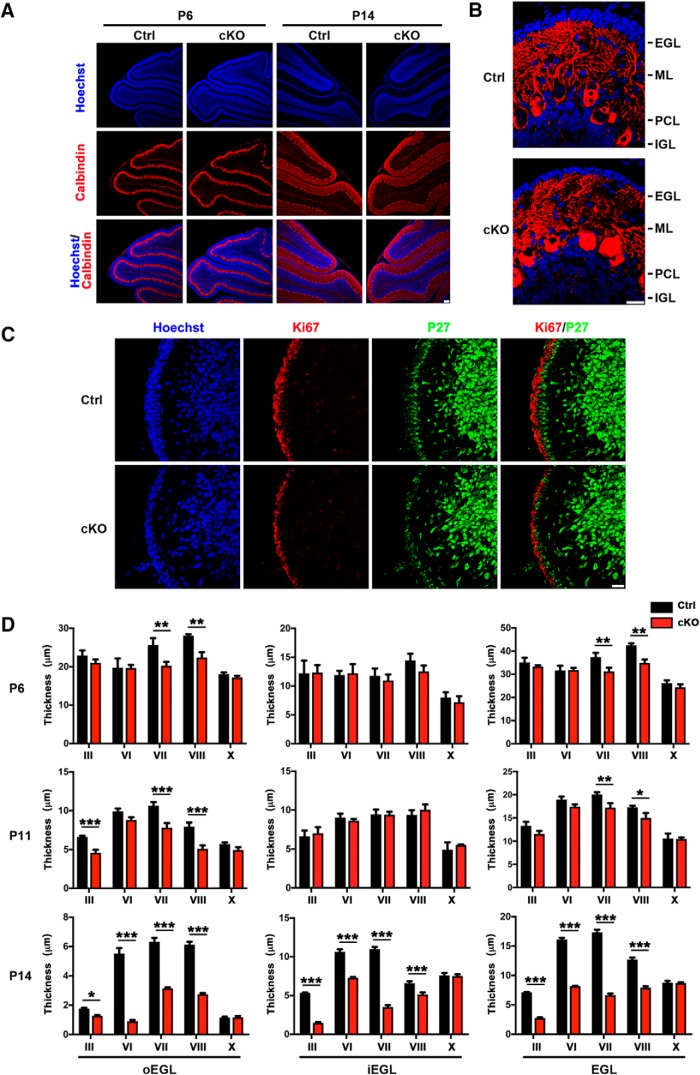
We characterized the effect of SnoN KO in granule neuron precursors on the development of the cerebellar cortex. We performed immunohistochemical analyses of the cerebellar cortex from control and conditional SnoN KO mice at P6 and P14 d of age using an antibody to the Purkinje cell marker calbindin and the DNA dye bisbenzimide (Hoechst). In these analyses, although conditional KO of SnoN in granule neuron precursors had little or no effect on the overall architecture of the cerebellar cortex in P6 mice (Fig. 2A), the EGL was substantially reduced in thickness in the cerebellar cortex of P14 mice (Fig. 2B). These results suggest that SnoN plays a critical role in the development of granule neuron precursors in the EGL at late stages of its development.
Figure 2.
Conditional SnoN KO in granule neuron precursors triggers defects in EGL development. A, The cerebellum from P6 and P14 conditional SnoN KO and control mice were subjected to immunohistochemical analyses using an antibody to calbindin and the DNA dye bisbenzimide (Hoechst). Conditional SnoN KO had little or no effect on the overall layer positioning and lobule foliation in the cerebellum. Scale bar, 100 μm. B, Higher-magnification images of the cerebellar cortex revealed that the thickness of EGL was remarkably reduced in conditional SnoN KO mice at P14 compared with control littermate animals. Scale bar, 20 μm. C, The cerebellum from P14 conditional SnoN KO and control mice was subjected to immunohistochemical analyses using antibodies to the proteins Ki67 and P27, which label proliferative and postmitotic granule cells in the oEGL and iEGL, respectively. Scale bar, 20 μm. D, Quantification of the thickness of oEGL, iEGL, and total EGL in the cerebellum of conditional SnoN KO and control littermate mice subjected to immunohistochemical analyses as in C. At P14, the thickness of oEGL, iEGL, and total EGL was significantly reduced in lobules III, VI, VII, and VIII upon conditional SnoN KO. At P11 and P6, conditional SnoN KO had little or no effect on iEGL thickness, but significantly reduced oEGL thickness, with concomitant reduction of total EGL thickness. In control analyses, EGL thickness was not reduced in lobule X upon conditional KO. All analyses were performed on midsagittal sections. Graphical data are presented as mean ± SEM. ***p < 0.001 (ANOVA followed by Fisher's PLSD post hoc test). **p < 0.01 (ANOVA followed by Fisher's PLSD post hoc test). *p < 0.05 (ANOVA followed by Fisher's PLSD post hoc test). n = 4 mice.
To determine the specific aspects of EGL development controlled by SnoN, we performed immunohistochemical analyses of the mouse cerebellar cortex using antibodies to Ki67 and p27, which label proliferating granule neuron precursors in the oEGL and postmitotic granule neurons in the iEGL, respectively (Fig. 2C) (Wojcinski et al., 2017). Using this approach, we measured the thickness of the EGL, oEGL, and iEGL in control and conditional SnoN KO mice at P14 of age. We found that the EGL and its sublayers, the oEGL and iEGL, were significantly reduced in thickness in lobules III, VI, VII, and VIII (i.e., within both the anterior and central cerebellar lobules) in conditional SnoN KO mice at P14 (Fig. 2D; Table 2). Notably, EGL thickness was not reduced in lobule X in SnoN conditional KO mice, presumably because the Math1-Cre driver is not expressed in the flocculonodular lobe (Orvis et al., 2012).
Table 2.
Quantification of the thickness of oEGL, iEGL, and total EGL at P6, P11 and P14 in control and conditional SnoN knockout mice
| Age | Lobule | Genotype | oEGL thickness (μm) | iEGL thickness (μm) | EGL thickness (μm) | No. of mice |
|---|---|---|---|---|---|---|
| P6 | III | Ctrl | 22.93 ± 1.338 | 12.2 ± 2.229 | 35.15 ± 2.028 | 4 |
| cKO (mean ± SEM) | 21.03 ± 0.8518 | 12.33 ± 1.314 | 33.35 ± 0.5439 | 4 | ||
| p | p = 0.2929 | p = 0.9462 | p = 0.3973 | |||
| VI | Ctrl | 19.75 ± 2.395 | 11.9 ± 0.745 | 31.65 ± 2.042 | 4 | |
| cKO (mean ± SEM) | 19.65 ± 0.8431 | 12.2 ± 1.602 | 31.85 ± 0.9314 | 4 | ||
| p | p = 0.9554 | p = 0.8713 | p = 0.9246 | |||
| VII | Ctrl | 25.63 ± 1.821 | 11.78 ± 1.287 | 37.4 ± 1.867 | 4 | |
| cKO (mean ± SEM) | 20.25 ± 1.002 | 10.95 ± 1.056 | 31.23 ± 1.684 | 4 | ||
| p | p = 0.005 | p = 0.6565 | p = 0.0062 | |||
| VIII | Ctrl | 28.08 ± 0.3966 | 14.45 ± 1.156 | 42.53 ± 0.8835 | 4 | |
| cKO (mean ± SEM) | 22.4 ± 1.391 | 12.53 ± 1.026 | 34.95 ± 1.427 | 4 | ||
| p | p = 0.0033 | p = 0.3029 | p = 0.0011 | |||
| X | Ctrl | 18.08 ± 0.4498 | 8.025 ± 0.8929 | 26.13 ± 1.26 | 4 | |
| cKO (mean ± SEM) | 17.2 ± 0.4143 | 7.2 ± 1.042 | 24.35 ± 1.341 | 4 | ||
| p | p = 0.6256 | p = 0.6565 | p = 0.4038 | |||
| P11 | III | Ctrl | 6.650 ± 0.1443 | 6.625 ± 0.762 | 13.3 ± 0.8396 | 4 |
| cKO (mean ± SEM) | 4.575 ± 0.3945 | 7.025 ± 0.792 | 11.55 ± 0.6702 | 4 | ||
| p | p = 0.0009 | p = 0.6517 | p = 0.1012 | |||
| VI | Ctrl | 9.925 ± 0.3376 | 9.025 ± 0.5218 | 18.98 ± 0.6223 | 4 | |
| cKO (mean ± SEM) | 8.8 ± 0.3559 | 8.625 ± 0.2175 | 17.43 ± 0.4871 | 4 | ||
| p | p = 0.0555 | p = 0.6517 | p = 0.1446 | |||
| VII | Ctrl | 10.65 ± 0.4573 | 9.425 ± 0.625 | 20.08 ± 0.4871 | 4 | |
| cKO (mean ± SEM) | 7.825 ± 0.5851 | 9.4 ± 0.3851 | 17.23 ± 0.9464 | 4 | ||
| p | p < 0.0001 | p = 0.9775 | p = 0.0099 | |||
| VIII | Ctrl | 7.975 ± 0.5006 | 9.375 ± 0.606 | 17.28 ± 0.3351 | 4 | |
| cKO (mean ± SEM) | 5.1 ± 0.4564 | 10.03 ± 0.7087 | 14.98 ± 1.057 | 4 | ||
| p | p < 0.0001 | p = 0.4645 | p = 0.0339 | |||
| X | Ctrl | 5.725 ± 0.1931 | 4.9 ± 0.9531 | 10.6 ± 1.053 | 4 | |
| cKO (mean ± SEM) | 4.95 ± 0.3594 | 5.525 ± 0.06292 | 10.48 ± 0.3224 | 4 | ||
| p | p = 0.1800 | p = 0.4817 | p = 0.9047 | |||
| P14 | III | Ctrl | 1.775 ± 0.0479 | 5.35 ± 0.02887 | 7.1 ± 0.09129 | 4 |
| cKO (mean ± SEM) | 1.275 ± 0.04787 | 1.475 ± 0.1031 | 2.75 ± 0.119 | 4 | ||
| p | p = 0.0446 | p < 0.0001 | p < 0.0001 | |||
| VI | Ctrl | 5.525 ± 0.366 | 10.63 ± 0.3301 | 16.13 ± 0.275 | 4 | |
| cKO (mean ± SEM) | 0.9 ± 0.09129 | 7.275 ± 0.1109 | 8.175 ± 0.06292 | 4 | ||
| p | p < 0.0001 | p < 0.0001 | p < 0.0001 | |||
| VII | Ctrl | 6.325 ± 0.2689 | 10.98 ± 0.275 | 17.33 ± 0.4626 | 4 | |
| cKO (mean ± SEM) | 3.15 ± 0.06455 | 3.5 ± 0.2677 | 6.65 ± 0.2533 | 4 | ||
| p | p < 0.0001 | p < 0.0001 | p < 0.0001 | |||
| VIII | Ctrl | 6.125 ± 0.2056 | 6.6 ± 0.2449 | 12.73 ± 0.3038 | 4 | |
| cKO (mean ± SEM) | 2.75 ± 0.0866 | 5.15 ± 0.2466 | 7.9 ± 0.2345 | 4 | ||
| p | p < 0.0001 | p = 0.0001 | p < 0.0001 | |||
| X | Ctrl | 1.15 ± 0.06455 | 7.575 ± 0.3351 | 8.75 ± 0.3227 | 4 | |
| cKO (mean ± SEM) | 1.175 ± 0.08539 | 7.525 ± 0.2016 | 8.7 ± 0.1414 | 4 | ||
| p | p = 0.9172 | p = 0.8816 | p = 0.8908 |
We next characterized the development of the oEGL and iEGL in conditional SnoN KO mice at P11 and P6. Interestingly, there was little or no effect on the thickness of the iEGL at P11 or P6. By contrast, oEGL and EGL thickness was significantly reduced in lobules III, VII, and VIII at P11 and in lobules VII and VIII at P6 upon conditional SnoN KO (Fig. 2D; Table 2). These results reveal that impairment of oEGL development in conditional SnoN KO mice temporally precedes the effects of the conditional SnoN KO on iEGL development. Thus, SnoN appears to regulate the development of the oEGL, and the effects on iEGL development in conditional KO of SnoN might occur as a consequence of the impairment of granule neuron precursor development in the oEGL.
SnoN promotes proliferation and suppresses cell cycle exit of granule neuron precursors in the postnatal mouse brain
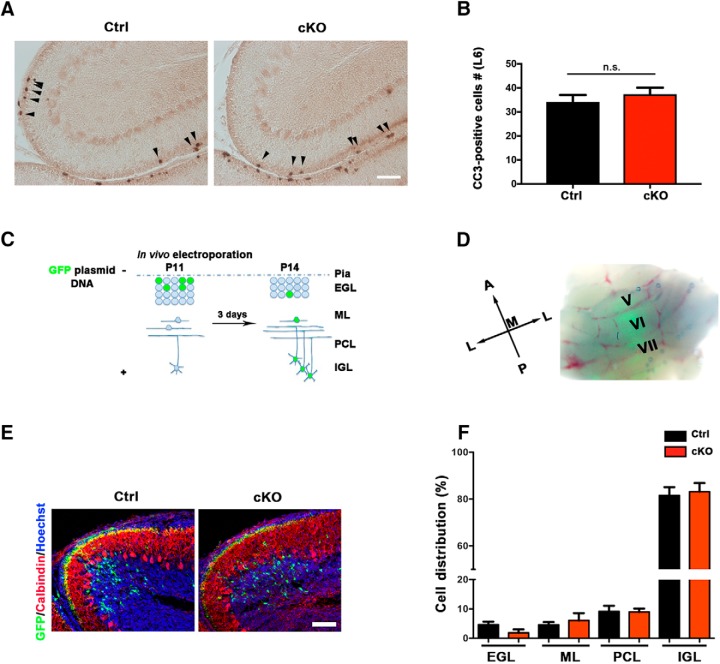
Having identified a novel role for SnoN in EGL development, we next assessed the cellular functions of SnoN in granule neuron precursors. We reasoned that the reduced thickness of the EGL in conditional SnoN KO mice might occur as a result of impaired proliferation of granule neuron precursors, increased cell death of granule neurons or their precursors, or augmented migration of granule neurons out of the EGL. In cell death analyses of the cerebellar cortex using cleaved caspase-3 immunohistochemistry (Whittaker et al., 2017), conditional KO of SnoN had little or no effect on cell death in the EGL (Fig. 3A,B), arguing against a role for cell death in SnoN regulation of granule neuron development. In an in vivo electroporation approach in postnatal mouse pups in which we expressed the GFP in granule neuron precursors at P11 and analyzed the position of granule neurons 3 d later in these pups at P14, the majority of granule neurons were in the IGL (Fig. 3C–E). In these assays, conditional KO of SnoN had little or no effect on the positioning of granule neurons (Fig. 3F; Table 3). Together, these data suggest that SnoN regulates the development of the EGL independently of changes in cell survival or migration of granule cells or their precursors.
Figure 3.
Conditional SnoN KO in granule neuron precursors has little or no effect on apoptosis or migration of granule cells. A, The cerebellum from P14 conditional SnoN KO and control littermate mice was subjected to immunohistochemical analyses using the cleaved caspase-3 analyses. Scale bar, 100 μm. B, Quantification of the number of cleaved caspase-3-positive cells within the EGL in lobule VI revealed that conditional KO of SnoN had little or no effect on cell death in the EGL. Graphical data are presented as mean ± SEM, and significance determined by t test (n = 5 mice). C, A schematic representation of in vivo electroporation. D, The cerebellum of P11 mice electroporated with a GFP expression plasmid, which targets granule cell precursors, was analyzed at P14 by fluorescence microscopy. Robust GFP fluorescence was evident. E, The cerebellum of conditional SnoN KO and control littermate mice was subjected to immunohistochemical analyses using an antibody to GFP. GFP-labeled granule neurons were similarly positioned in conditional SnoN KO and control littermate mice. Scale bar, 100 μm. F, Quantification of the percentage of GFP-labeled granule cells within the EGL, molecular layer (ML), Purkinje cell layer (PCL), and IGL revealed that conditional SnoN KO had little or no effect on the positioning of granule neurons. Graphical data are presented as mean ± SEM, and significance determined by ANOVA followed by Fisher's PLSD post hoc test (n = 7 mice).
Table 3.
Quantification of GFP-electroporated granule neuron precursors in the cerebellum of control and conditional SnoN knockout mice
| Distribution (%) | Ctrl (mean ± SEM) | cKO (mean ± SEM) | No. of mice | p |
|---|---|---|---|---|
| EGL | 4.585 ± 1.025 | 1.852 ± 1.164 | 7 | 0.1034 |
| ML | 4.578 ± 0.9592 | 6.044 ± 2.528 | 7 | 0.5976 |
| PCL | 9.156 ± 1.918 | 8.970 ± 1.169 | 7 | 0.9352 |
| IGL | 81.56 ± 3.535 | 83.12 ± 3.739 | 7 | 0.7661 |
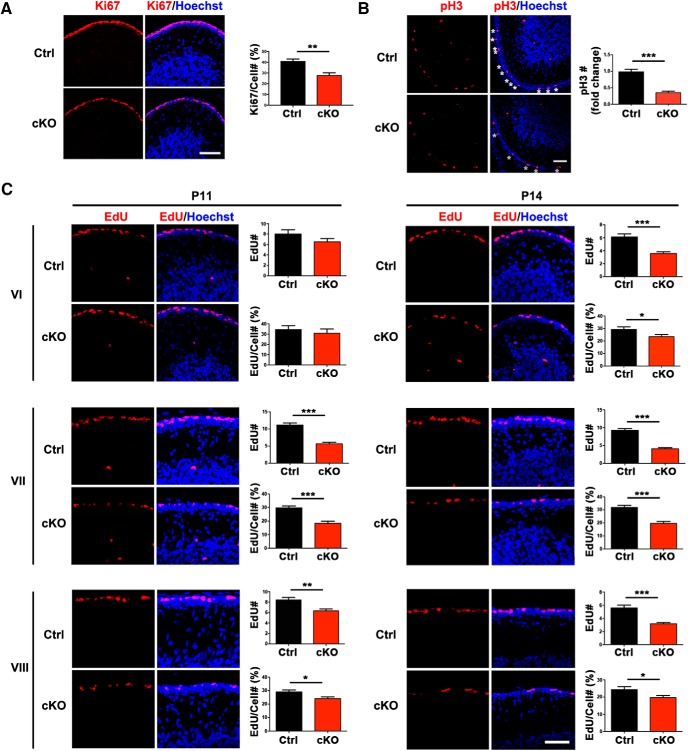
We next determined whether SnoN regulates the proliferation of granule neuron precursors in the postnatal cerebellum. We performed immunohistochemical analyses of the cerebellar cortex in control and conditional SnoN KO mice at P14 using an antibody to the protein antigen Ki67, which labels cycling cells, and an antibody to the phosphorylated histone H3 (pH3), which labels cells specifically during mitosis (Espinosa and Luo, 2008). Conditional KO of SnoN robustly reduced the number and percentage of Ki67- and pH3-positive granule neuron precursors in the oEGL (Fig. 4A,B), suggesting that the proliferation of granule neuron precursors is impaired upon SnoN KO.
Figure 4.
SnoN promotes proliferation of granule neuron precursors in the postnatal cerebellum. A, The cerebellum from P14 conditional SnoN KO and control littermate mice was subjected to immunohistochemical analyses using the Ki67 antibody and the DNA dye bisbenzimide (Hoechst). Substantially fewer Ki67-positive granule cell precursors were found in the EGL of conditional SnoN KO mice compared with control littermate animals. Scale bar, 50 μm. Graphical data are presented as mean ± SEM. **p < 0.01 (t test). n = 4 mice. B, The cerebellum from P14 conditional SnoN KO and control littermate mice was subjected to immunohistochemical analyses using an antibody to the pH3, which labels cells specifically during mitosis. Substantially fewer pH3-positive granule cell precursors were found in the EGL of conditional SnoN KO mice compared with control littermate animals. Scale bar, 50 μm. Graphical data are presented as mean ± SEM. ***p < 0.001 (t test). n = 4 mice. C, P11 and P14 conditional SnoN KO and littermate mice were injected intraperitoneally with the nucleotide derivative EdU. Thirty minutes after EdU injection, the cerebellum was subjected to histochemical analyses using an EdU detection reagent and the DNA dye bisbenzimide (Hoechst). The total number of EdU-labeled proliferating granule precursors and their proliferation index (percentage of EdU-labeled cells among all EGL cells) was significantly reduced in lobules VI, VII, and VIII at P14 and VII, VIII at P11 upon conditional KO of SnoN. Scale bar, 50 μm. The measured area equals 1000 μm2. All analyses were performed on three midline sections per brain. Graphical data are presented as mean ± SEM. ***p < 0.001 (t test). **p < 0.01 (t test). *p < 0.05 (t test). n = 4 mice.
To further elucidate the role of SnoN in the proliferation of granule neuron precursors in the oEGL, we performed analyses of the DNA synthesis phase of the cell cycle in these cells by injecting P11 or P14 control and conditional SnoN KO mice with the nucleotide derivative EdU. After a short pulse of EdU for 30 min, we subjected the cerebellar cortex from these mice to histochemical analyses using an EdU detection reagent and the DNA dye bisbenzimide (Hoechst), the latter to label cell nuclei. We measured the proliferation index of granule neuron precursors in the oEGL as the ratio of the number of cells in which EdU was incorporated to the total number of cells labeled with Hoechst in the EGL. We focused our analyses on the central lobules VI, VII, and VIII, the locale of maximal impairment of EGL development in conditional SnoN KO mice. The proliferation index of granule neuron precursors in the oEGL in lobules VI, VII, and VIII was significantly reduced in conditional SnoN KO mice (Fig. 4C; Table 4), suggesting a requirement for SnoN in the proliferation of granule neuron precursors in the oEGL.
Table 4.
Quantification of EdU-labeled EGL cells in different lobules of control and conditional SnoN knockout mice
| Age | Lobule | Marker | Genotype | No. or ratio (%) | No. of mice |
|---|---|---|---|---|---|
| P11 | VI | EdU# | Ctrl | 8 ± 0.8165 | 4 |
| cKO (mean ± SEM) | 6.5 ± 0.6455 | 4 | |||
| p | p = 0.1996 | ||||
| EdU#/cell# | Ctrl | 34.44 ± 3.684 | 4 | ||
| cKO (mean ± SEM) | 30.96 ± 4.081 | 4 | |||
| p | p = 0.5493 | ||||
| VII | EdU# | Ctrl | 11.17 ± 0.6009 | 4 | |
| cKO (mean ± SEM) | 5.5 ± 0.466 | 4 | |||
| p | p < 0.0001 | ||||
| EdU#/cell# | Ctrl | 29.76 ± 1.402 | 4 | ||
| cKO (mean ± SEM) | 18.29 ± 1.623 | 4 | |||
| p | p < 0.0001 | ||||
| VIII | EdU# | Ctrl | 8.417 ± 0.4994 | 4 | |
| cKO (mean ± SEM) | 6.333 ± 0.3553 | 4 | |||
| p | p = 0.0026 | ||||
| EdU#/cell# | Ctrl | 29.03 ± 1.431 | 4 | ||
| cKO (mean ± SEM) | 24.05 ± 1.278 | 4 | |||
| p | p = 0.0163 | ||||
| P14 | VI | EdU# | Ctrl | 6.125 ± 0.4596 | 4 |
| cKO (mean ± SEM) | 3.583 ± 0.2476 | 4 | |||
| p | p < 0.0001 | ||||
| EdU#/cell# | Ctrl | 29.27 ± 2.023 | 4 | ||
| cKO (mean ± SEM) | 23.44 ± 1.736 | 4 | |||
| p | p = 0.0338 | ||||
| VII | EdU# | Ctrl | 9.25 ± 0.4787 | 4 | |
| cKO (mean ± SEM) | 4.083 ± 0.3005 | 4 | |||
| p | p < 0.0001 | ||||
| EdU#/cell# | Ctrl | 31.8 ± 1.612 | 4 | ||
| cKO (mean ± SEM) | 19.55 ± 1.474 | 4 | |||
| p | p < 0.0001 | ||||
| VIII | EdU# | Ctrl | 5.583 ± 0.4337 | 4 | |
| cKO (mean ± SEM) | 3.167 ± 0.2056 | 4 | |||
| p | p < 0.0001 | ||||
| EdU#/cell# | Ctrl | 24.35 ± 1.704 | 4 | ||
| cKO (mean ± SEM) | 19.74 ± 1.203 | 4 | |||
| p | p = 0.0321 |
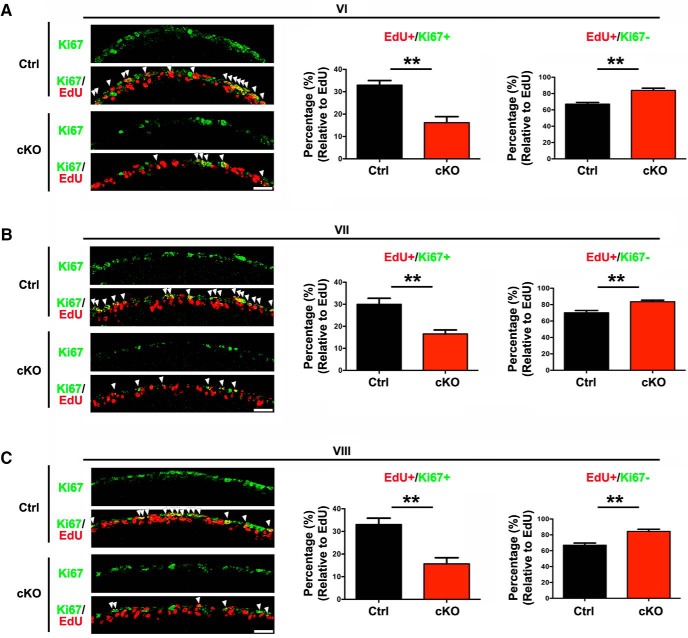
To determine whether SnoN regulates cell cycle exit of granule neuron precursors, we performed additional EdU incorporation analyses in which we injected P13 mice with EdU and subjected animals 24 h later at P14 to histochemical analyses using the EdU detection reagent and the proliferation marker Ki67. Cells that are positive for both EdU and Ki67 represent granule neuron precursors remaining in the cell cycle. However, EdU-positive, Ki67-negative cells represent granule neuron precursors that have exited the cell cycle. The number of EdU+/Ki67+ granule cells was significantly reduced in the EGL of lobules VI, VII, and VIII upon conditional SnoN KO (Fig. 5A–C; Table 5). Remarkably, the number of EdU+/Ki67− cells was significantly increased in the EGL of lobules VI, VII, and VIII upon conditional SnoN KO (Fig. 5A–C; Table 5). Together, these results suggest that SnoN promotes the proliferation of granule neuron precursors in the oEGL and concomitantly inhibits their exit from the cell cycle.
Figure 5.
Conditional SnoN KO impairs proliferation and promotes cell cycle exit of granule neuron precursors in the postnatal cerebellum. A–C, P13 conditional SnoN KO and control littermate mice were injected with EdU, and 24 h later, the cerebellum was subjected to histochemical analyses using the EdU detection reagent and Ki67 antibody. EdU+/Ki67+ cells represent granule neuron precursors, which remained in the cell cycle, whereas the EdU+/Ki67− cells represent granule neuron precursors that have exited the cell cycle and differentiated into postmitotic granule neurons within 24 h. The number of EdU+/Ki67+ granule neuron precursors was significantly reduced in the EGL of lobules VI (A), VII (B), and VIII (C) upon conditional KO of SnoN. By contrast, the number of EdU+/Ki67− postmitotic granule neurons was significantly increased in lobules VI (A), VII (B), and VIII (C) upon conditional KO of SnoN. Graphical data are presented as mean ± SEM. **p < 0.01 (t test). n = 4 mice.
Table 5.
Quantification of proliferation and cell cycle exit of granule neuron precursors
| Age | Lobule | Marker | Genotype | Ratio (%) | No. of mice |
|---|---|---|---|---|---|
| P13 To P14 | VI | EdU+/Ki67+ | Ctrl | 32.95 ± 2.064 | 4 |
| cKO (mean ± SEM) | 16.16 ± 2.729 | 4 | |||
| p | p = 0.0027 | ||||
| EdU+/Ki67− | Ctrl | 67.05 ± 2.064 | 4 | ||
| cKO (mean ± SEM) | 83.84 ± 2.729 | 4 | |||
| p | p = 0.0027 | ||||
| VII | EdU+/Ki67+ | Ctrl | 29.97 ± 2.761 | 4 | |
| cKO (mean ± SEM) | 16.50 ± 1.856 | 4 | |||
| p | p = 0.0067 | ||||
| EdU+/Ki67− | Ctrl | 70.03 ± 2.761 | 4 | ||
| cKO (mean ± SEM) | 83.50 ± 1.856 | 4 | |||
| p | p = 0.0067 | ||||
| VIII | EdU+/Ki67+ | Ctrl | 33.05 ± 2.796 | 4 | |
| cKO (mean ± SEM) | 15.65 ± 2.72 | 4 | |||
| p | p = 0.0043 | ||||
| EdU+/Ki67− | Ctrl | 66.95 ± 2.796 | 4 | ||
| cKO (mean ± SEM) | 84.35 ± 2.72 | 4 | |||
| p | p = 0.0043 |
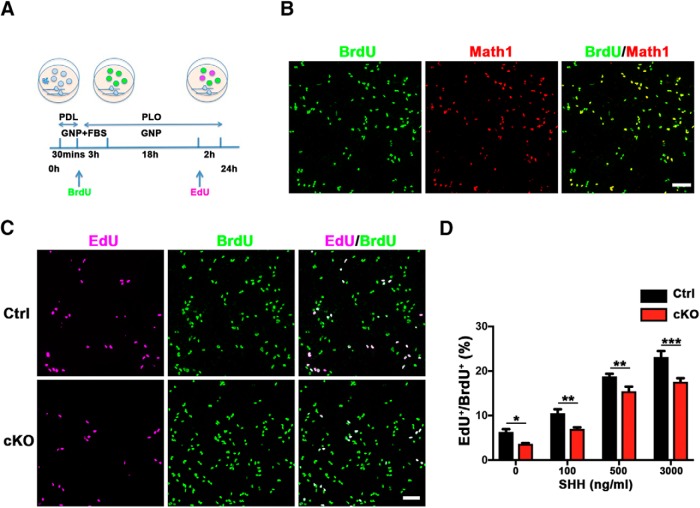
We next determined whether SnoN regulates granule neuron precursor proliferation in a cell-type autonomous manner. We therefore assessed the effect of conditional SnoN KO on the proliferation of primary granule neuron precursors cultured from the cerebellum of P6 mice. Proliferating granule neuron precursors were labeled with BrdU immediately after their isolation from the cerebellum, and 22 h later, they were pulsed with EdU for analysis of cell proliferation as described previously (Fig. 6A) (Kenney et al., 2003). Almost all BrdU-positive cells coexpressed the transcription factor Math1, confirming their identity of granule neuron precursors (Fig. 6B). Importantly, the rate of cell proliferation as measured by EdU incorporation was significantly reduced in conditional SnoN KO granule neuron precursors compared with the control (Fig. 6C). Notably, exposure of granule neuron precursors to the mitogen SHH robustly increased cell proliferation of both control and SnoN-deficient granule neuron precursors (Fig. 6D; Table 6). Conversely, SnoN depletion significantly reduced the rate of proliferation in granule neuron precursors in the presence or absence of SHH (Fig. 6D; Table 6). Collectively, our results suggest that SnoN promotes the proliferation of granule neuron precursors cell-autonomously and in a pathway that is parallel to SHH.
Figure 6.
SnoN promotes proliferation of granule neuron precursors cell-autonomously and independently of SHH. A, A schematic of preparation of primary granule neuron precursors from P6 cerebellum. BrdU was used to label proliferating granule neuron precursors immediately after isolation, and 22 h later, a 2 h pulse of EdU incorporation was used to trace granule neuron precursors that remained in the cell cycle or those that exited the cell cycle. PDL, Poly-d-lysine; PLO, poly-ornithine; GNP, granule neuron precursor culture medium. B, BrdU-labeled proliferating granule neuron precursors were subjected to Math1 immunofluorescence. BrdU-positive cells almost completely colabeled with Math1, indicating the identity of mitotic granule neuron precursors. Scale bar, 50 μm. C, BrdU-labeled proliferating granule neuron precursors were subjected to EdU counterstaining. The percentage of primary granule neuron precursors that remained in the cell cycle (EdU+/BrdU+) was reduced upon conditional SnoN KO. Scale bar, 50 μm. D, Quantification of the percentage of EdU+/BrdU+ cells in control and conditional SnoN KO primary granule neuron precursors showed SHH dosage-dependent expansion. Exposure to SHH robustly increased proliferation of both control and SnoN-deficient granule neuron precursors, and SnoN depletion significantly reduced the rate of proliferation of granule neuron precursors in the presence or absence of SHH. Graphical data are presented as mean ± SEM. ***p < 0.001 (ANOVA followed by Fisher's PLSD post hoc test). **p < 0.01 (ANOVA followed by Fisher's PLSD post hoc test). *p < 0.05 (ANOVA followed by Fisher's PLSD post hoc test). n = 3 biological replicates.
Table 6.
Quantification of EdU+/BrdU+ cells in control and conditional SnoN knockout primary granule neuron precursors
| SHH concentration (ng/ml) | Ctrl (mean ± SEM) | cKO (mean ± SEM) | N | p |
|---|---|---|---|---|
| 0 | 6.342 ± 0.5992 | 3.651 ± 0.1918 | 3 | 0.0310 |
| 100 | 10.52 ± 0.8816 | 7.003 ± 0.3561 | 3 | 0.0070 |
| 500 | 18.8 ± 0.5841 | 15.47 ± 1.02 | 3 | 0.0099 |
| 3000 | 23.14 ± 1.344 | 17.58 ± 0.8321 | 3 | 0.0002 |
SnoN regulates a program of cell proliferation gene expression during cerebellar development
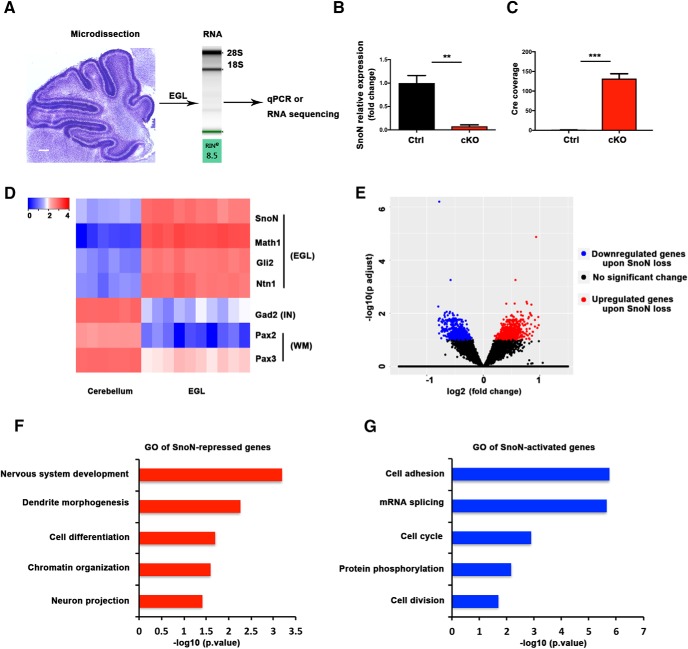
Having identified a function for SnoN in granule neuron precursor proliferation, we next characterized a mechanism underlying the novel function of SnoN in the mouse brain in vivo. Because granule neuron precursors proliferate and differentiate in the EGL, we deployed a laser capture microdissection (LCM) approach to focus our analyses of SnoN specifically on the EGL of the developing mouse cerebellum. Because SnoN is a transcriptional regulator, we subjected the EGL from conditional SnoN KO and control littermate mice to RNA-seq (LCMRNA-seq) analysis (Fig. 7A). We chose the postnatal age of P6, before the major effect of conditional SnoN KO on granule neuron precursor proliferation and EGL development. We confirmed that the recombinase Cre was expressed and that SnoN mRNA was dramatically downregulated in the EGL of conditional SnoN KO mice compared with control littermate mice (Fig. 7B,C). In addition, the microdissected EGL was enriched for genes that are highly expressed in the EGL and de-enriched for genes expressed in cells in other parts of the cerebellum (Fig. 7D) (Rosenberg et al., 2018), establishing the utility of LCMRNA-seq analyses to probe gene expression in a spatially restricted manner in the developing cerebellum.
Figure 7.
LCMRNA-seq of the EGL shows that SnoN activates expression of cell proliferation genes and represses neuronal differentiation genes. A, LCM was used to isolate the EGL from the central lobules of control and conditional SnoN KO animals at P6. After BioAnalyzer quality control (Agilent Technologies), intact RNA was used for quantitative PCR analysis or RNA sequencing. Scale bar, 200 μm. B, The EGL from conditional SnoN KO and control littermate mice was subjected to quantitative PCR analysis. SnoN mRNA levels were profoundly lower in the EGL of conditional SnoN KO mice compared with control mice. Graphical data are presented as mean ± SEM. **p < 0.01 (t test). n = 3 mice. C, The recombinase Cre was expressed in the LCM EGL of conditional SnoN KO but not control mice. Graphical data are presented as mean ± SEM. ***p < 0.001 (t test). n = 5 mice. D, RNA-seq analyses of the LCM EGL and whole cerebellum revealed that the EGL genes, Math1, Gli2, and Ntn1, were significantly higher in the EGL compared with whole cerebellum, and genes that are expressed in other region of the cerebellum, such as Gad2 (marker for interneuron [IN]), Pax2 and Pax3 (marker for white matter [WM]) were higher in the whole cerebellum than in the EGL. E, Volcano plot of gene expression changes. The transcriptome of the LCM EGL from conditional SnoN KO mice was compared with that of control mice using DESeq2. The x axis specifies the log2 fold-changes, and the y axis specifies the negative logarithm to the base 10 of the FDR adjusted p values. Red and blue dots represent genes expressed at significantly higher (n = 792) or lower (n = 600) levels upon SnoN loss, respectively (FDR-adjusted p < 0.1). F, Gene ontology analyses of SnoN-repressed genes (i.e., upregulated in the LCM EGL upon conditional SnoN KO) showed gene cluster functions related to neuronal development, cell differentiation, dendrite morphogenesis, and neuron projection. G, Gene ontology analyses of SnoN-activated genes (i.e., downregulated in the LCM EGL upon conditional SnoN KO) showed notable gene cluster functions in cell proliferation, cell adhesion, mRNA splicing, and protein phosphorylation.
Importantly, upon subjecting LCMRNA-seq datasets to DESeq2 analyses, using an FDR < 0.1, we identified a total of 1392 differentially expressed genes, with 600 genes downregulated and 792 genes upregulated in the EGL of conditional SnoN KO animals compared with littermate control animals (Fig. 7E). In gene ontology analyses, SnoN-repressed genes (i.e., upregulated in SnoN KO EGL) were related to neuronal development, cell differentiation, dendrite morphogenesis, and neuron projection (Fig. 7F). By contrast, SnoN-activated genes (i.e., downregulated in conditional SnoN KO mice) were associated most notably with the control of cell proliferation as well as with cell adhesion, mRNA splicing, and protein phosphorylation (Fig. 7G). These data suggest that SnoN regulates programs of genes that control cell proliferation and differentiation in granule neuron precursors in the postnatal cerebellum.
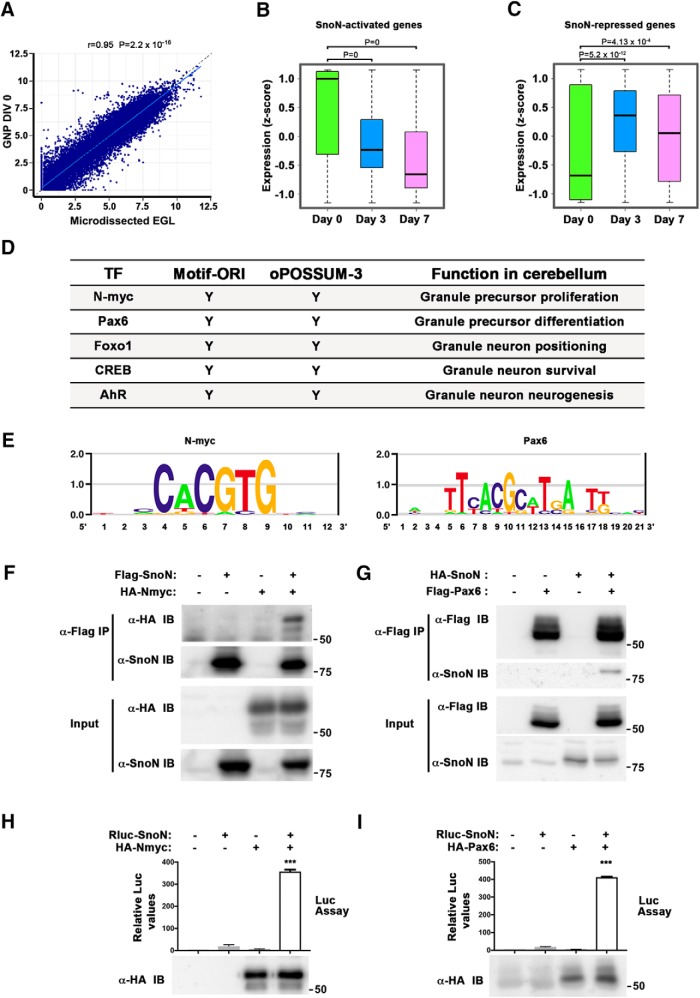
To further assess the role of the SnoN-regulated genes in EGL, we next characterized the temporal pattern of their expression in developing granule neuron precursors. The gene expression profile of the EGL was highly correlated with gene expression of acutely isolated granule neuron precursors (Fig. 8A) (Frank et al., 2015). Because the development of granule neuron precursors in the EGL is temporally mimicked by the developmental stages of proliferation and differentiation of primary granule cells (Frank et al., 2015), we next mapped the SnoN-activated and SnoN-repressed genes from LCMRNA-seq analyses of the EGL to profiling data of primary granule neuron precursors and granule neurons (Frank et al., 2015). These analyses showed that SnoN-activated genes are downregulated and SnoN-repressed genes are upregulated upon granule neuron precursor differentiation and granule neuron maturation (Fig. 8B,C). These data further strengthen the conclusion that SnoN regulates programs of gene expression in granule neuron precursors to promote their proliferation and inhibit their differentiation.
Figure 8.
Analysis of SnoN-regulated genes in the EGL uncovers novel interactions of SnoN with the EGL-enriched transcription factors N-myc and Pax6. A, The gene expression profile of the LCM EGL significantly correlated with primary granule neuron precursors, the latter described by Frank et al. (2015). B, C, Downregulated and upregulated genes in the LCM EGL upon SnoN depletion were mapped to the profiling data of primary granule neuron precursors at different days. SnoN-activated genes were downregulated, whereas SnoN-repressed genes were upregulated with differentiation of granule neuron precursors. Boxplots show the expression level distributions of downregulated (B) and upregulated (C) genes on days 0, 3, and 7 of granule neuron precursor cultures. The expression level was log2-transformed reads per kilobase of transcript per million mapped reads (RPKM) and was scaled by z score. Graphical data are presented as median (center line), 25th and 75th percentiles (bottom and top edges). ***p < 0.001 (Wilcoxon test). D, Motif enrichment analyses of promoter sequences of genes differentially expressed in the LCM EGL upon SnoN conditional KO revealed enrichment of binding sites for key transcription factors that regulate granule neuron precursor proliferation and differentiation. E, Position weight matrix logo for N-myc (left) and Pax6 (right) are shown. x axis shows the position of each nucleotide. The height of each base quantifies the bits of information content, which is related to the probability of its occurrence at that position. F, Lysates of HEK293T cells expressing HA-Nmyc, Flag-SnoN, or both were subjected to immunoprecipitation with the Flag antibody followed by immunoblotting with the HA or SnoN antibody. Total lysates were also subjected to immunoblotting with the HA or SnoN antibody. G, Lysates of HEK293T cells expressing HA-SnoN, Flag-Pax6, or both were subjected to immunoprecipitation with the Flag antibody followed by immunoblotting with the Flag or SnoN antibody. Total lysates were also subjected to immunoblotting with the Flag or SnoN antibody. H, Lysates of HEK293T cells expressing Renilla luciferase, Rluc (−), or Rluc-SnoN (+) alone or together with HA-Nmyc were subjected to immunoprecipitation with the HA antibody followed by luciferase assays. Aliquots of cell lysates were also assayed for luciferase activity as a measure of Rluc or Rluc-SnoN expression. The expression of HA-Nmyc in lysates (10%) was confirmed by immunoblotting using the HA antibody. Rluc or Rluc-SnoN values in the HA-Nmyc immunoprecipitates were normalized to respective input. Graphical data are presented as mean ± SEM. ***p < 0.001 (one-way ANOVA followed by Fisher's PLSD post hoc test). n = 4 biological replicates. I, Lysates of 293T cells expressing Rluc (−), Rluc-SnoN (+) alone or together with HA-Pax6 were subjected to immunoprecipitation with the HA antibody followed by luciferase assays. Aliquots of cell lysates were also assayed for luciferase activity. The expression of HA-Pax6 in aliquots of lysates (10%) was confirmed by immunoblotting using the HA antibody. Rluc or Rluc-SnoN values in the HA-Pax6 immunoprecipitation were normalized to respective input. Graphical data are presented as mean ± SEM. ***p < 0.001 (one-way ANOVA followed by Fisher's PLSD post hoc test). n = 4 biological replicates.
We next performed bioinformatics analyses to determine whether SnoN-regulated genes in granule neuron precursors share common regulatory elements. Because SnoN regulates transcription through interactions with transcription factors, we searched for enrichment of transcription factor binding motifs in the promoter sequences of the SnoN-regulated genes in granule neuron precursors using two independent algorithms. Motifs for five transcription factors were found in the promoter sequences of SnoN-regulated genes by both algorithms (Fig. 8D). Strikingly, two of these transcription factors, N-myc and Pax6, have been implicated in the control of granule neuron precursor proliferation and differentiation, respectively (Fig. 8E) (Kenney et al., 2003; Owa et al., 2018). We therefore asked whether SnoN might form a physical complex with these transcription factors. In coimmunoprecipitation analyses in cells, SnoN interacted with N-myc or Pax6 (Fig. 8F,G). In other analyses in which we used SnoN fused to luciferase (Rluc-SnoN), SnoN as measured by luciferase activity was robustly and specifically coprecipitated with N-myc and Pax6 (Fig. 8H,I). Together, these results reveal that SnoN forms physical complexes with N-myc and Pax6 and may thereby regulate programs of cell proliferation and cell differentiation gene expression in granule neuron precursors in the postnatal mammalian brain.
Role of granule neuron precursor SnoN in cerebellar-dependent learning
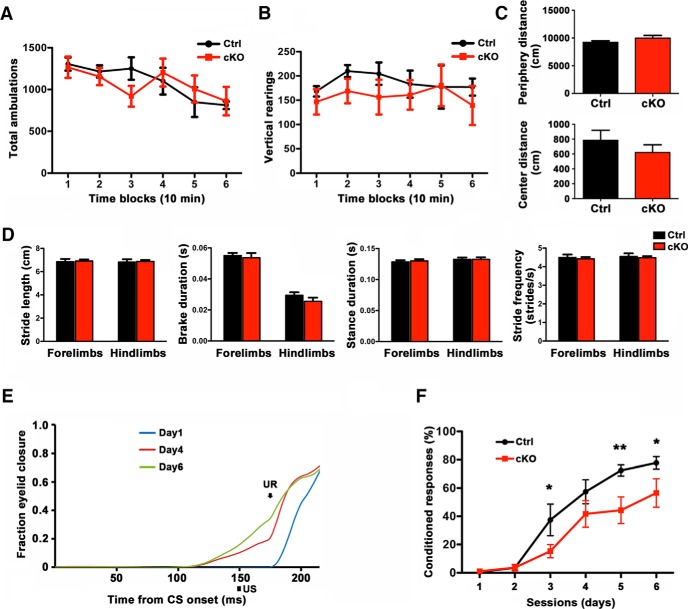
The identification of a function for SnoN in granule neuron precursor proliferation in the postnatal mammalian brain led us to ask whether the novel role of SnoN bears consequences in cerebellar function. The cerebellum is required for motor coordination and procedural motor learning (Heiney et al., 2014). We therefore assessed the effect of conditional KO of SnoN on behavioral paradigms designed to assess these cerebellar functions. We characterized the effect of conditional KO of SnoN in open field assay to measure effects on the whole-body movements, exploratory and anxiety behavior. Conditional KO of SnoN had little or no effect on total ambulations, levels of vertical rearing, distance traveled in a small peripheral zone surrounding the perimeter of the open field, or distance traveled in a small central zone within in the field (Fig. 9A–C), suggesting no obvious difference in general ambulations or exploratory behavior. Conditional SnoN KO did not have significant effects on the number of entries made or time spent in the center zone (data not shown). Although no significant differences were observed between groups for the three center-zone variables (distance traveled, time spent, or entries made into the center), there was a trend across all three for the SnoN conditional KO mice to exhibit lower levels (data not shown).
Figure 9.
Cerebellar-dependent learning is impaired in conditional SnoN KO mice. A, Conditional KO of SnoN had little or no effect on total ambulations in an open field assay (ANOVA followed by Fisher's PLSD post hoc test); n = 6 mice. B, Conditional SnoN KO and control littermate showed comparable levels of vertical rearing in the open field (ANOVA followed by Fisher's PLSD post hoc test); n = 6 mice. C, Conditional SnoN KO had little or no effect on distance traveled in a small peripheral zone surrounding the perimeter of the open field (top), distance traveled in a small central zone within in the field (bottom), or on the numbers of entries made or time spent in the center zone (data not shown) (t test, n = 6 mice). D, Gait dynamics were assessed in conditional SnoN KO and control littermate animals using ventral plane videography in which temporal and spatial characteristics of footprints were captured on a treadmill moving at 30 cm/s (DigiGait, Mouse Specifics). Conditional SnoN KO had little or no effect on gait-related variables, including stride length, brake duration, stance duration, or stride frequency (repeated-measures ANOVA, planned comparisons). E, Conditional SnoN KO and control littermate mice were subjected to eye-blink conditioning learning paradigm. Average eyelid CR position traces during the 150 ms CS-US (air puff) interval period throughout 6 d of conditioning to a light CS. Each trace is an average of the session-averaged traces for 100 trials. The CR amplitude gradually builds up. F, Impaired performance on the eye-blink conditioning task in conditional SnoN KO mice. **p < 0.01 (ANOVA followed by Fisher's PLSD post hoc test). *p < 0.05 (ANOVA followed by Fisher's PLSD post hoc test). n = 9 mice.
To assess motor coordination, we used the DigiGait paradigm. Gait dynamics were assessed in control and SnoN conditional KO mice using ventral plane videography, which involved quantifying temporal and spatial characteristics of digital images of footprints captured during ambulation on a transparent treadmill moving at 30 cm/s (DigiGait, Mouse Specifics). No convincing evidence was found for performance deficits in the conditional SnoN KO mice after analyzing many common gait-related variables, including stride length, brake duration, stance duration, or stride frequency (Fig. 9D). Together, our data suggest that conditional SnoN KO has little or no effect on motor coordination.
We next asked whether SnoN function in granule neuron precursor might influence the role of the cerebellum in adaptive responses. To assess cerebellar-dependent motor learning, we used the delay eye-blink conditioning paradigm. This assay permits quantification and accurate measurements and has been useful in determining the effect of genetic manipulations on cerebellar learning (Yang et al., 2016; Giovannucci et al., 2017; Valnegri et al., 2017). We characterized eye-blink responses to the blue LED light as the CS and air puffs presented to the cornea as the US. After 100 sessions of conditioning per day (150 ms CS-US interval; 100 trials per session), control animals displayed robust associative learning with acquisition of the CR (Fig. 9E). By contrast, conditional SnoN KO mice demonstrated significant deficit in learning compared with control littermate animals (Fig. 9F; Table 7). These data suggest that impairment of granule neuron precursor proliferation upon conditional KO of SnoN bears functional consequences at the organismal level for the role of the cerebellum-dependent learning.
Table 7.
Motor learning paradigm in control and conditional SnoN knockout mice in the delay eyeblink conditioning
| Sessions (days) | Genotype | Conditioned response (%) | No. of mice |
|---|---|---|---|
| Day 1 | Ctrl | 0.60 ± 0.30 | 9 |
| cKO (mean ± SEM) | 1.08 ± 0.66 | 9 | |
| p | p = 0.9598 | ||
| Day 2 | Ctrl | 3.25 ± 0.87 | 9 |
| cKO (mean ± SEM) | 3.6 ± 2.23 | 9 | |
| p | p = 0.9667 | ||
| Day 3 | Ctrl | 37.43 ± 11.23 | 9 |
| cKO (mean ± SEM) | 15.32 ± 4.65 | 9 | |
| p | p = 0.0218 | ||
| Day 4 | Ctrl | 57.39 ± 8.49 | 9 |
| cKO (mean ± SEM) | 41.7 ± 9.41 | 9 | |
| p | p = 0.1013 | ||
| Day 5 | Ctrl | 72.02 ± 4.01 | 9 |
| cKO (mean ± SEM) | 44.28 ± 9.43 | 9 | |
| p | p = 0.0043 | ||
| Day 6 | Ctrl | 77.54 ± 4.35 | 9 |
| cKO (mean ± SEM) | 56.51 ± 10.06 | 9 | |
| p | p = 0.0290 | ||
| Final | ANOVA | p = 0.0399 |
Discussion
In this study, we have discovered a requirement for the transcriptional regulator SnoN in the proliferation of cerebellar granule neuron precursors in the postnatal mammalian brain. Conditional KO of SnoN in granule neuron precursors impairs their proliferation, leading to structural defects in the EGL at later stages of cerebellar development. Strikingly, in LCMRNA-seq of the EGL in conditional SnoN KO and control littermate animals, we have found that SnoN activates a program of cell proliferation genes and represses cell differentiation genes in granule neuron precursors in vivo. Bioinformatics analyses have revealed that SnoN-regulated genes contain motifs for the transcription factors N-myc and Pax6. Accordingly, SnoN physically interacts with N-myc and Pax6 in cells, suggesting that SnoN may regulate cell proliferation and cell differentiation genes via interaction with these transcription factors in granule neuron precursors. In behavior analyses, conditional KO of SnoN in granule neuron precursors triggers impairment of cerebellar-dependent learning, suggesting that SnoN function in granule neuron precursors bears functional consequences at the organismal level. Our findings define a novel role and mechanism for SnoN in the control of cerebellar granule precursor cell proliferation in the mammalian brain.
The finding that SnoN promotes the proliferation of granule neuron precursors bears significant implications for our understanding of cerebellar development. The identification of SnoN control of granule neuron proliferation in a developmentally specific stage suggests that granule neuron precursors are comprised of heterogeneous subpopulations. Consistent with this interpretation, recent studies suggest that granule neuron precursors may be categorized into at least two distinct subpopulations based on expression of Math1 and the precursor cell marker Nestin (Li et al., 2013; Wojcinski et al., 2017). Our study suggests that Math1-positive precursors may be further categorized based on the requirement for SnoN in their proliferation. The pool of SnoN-regulated subpopulation of granule neuron precursors is substantial, giving rise to their contribution to the functions of the cerebellum, as evidenced by impairment of cerebellar-dependent motor plasticity in the delayed eye-blink conditioning associative learning paradigm in the conditional SnoN KO animals.
Aside from granule neuron precursors, the Math1-Cre driver used in our study to trigger SnoN KO is expressed in deep cerebellar nuclei neurons and in unipolar brush cells (Orvis et al., 2012). However, cerebellar nuclei neurons are generated during embryonic ages in mice (Hoshino et al., 2005; Machold and Fishell, 2005; Wang et al., 2005). As to unipolar brush cells, the overwhelming majority of these cells are present in the IGL in lobules IX and X (Englund et al., 2006), where the Math1-Cre driver line expresses Cre poorly (Orvis et al., 2012), and the effect of conditional SnoN KO is evident in the anterior and central lobules. These observations suggest that the effect of conditional SnoN KO on granule neuron precursors is likely to be cell-intrinsic rather than secondary to SnoN functions in the deep cerebellar nuclei or unipolar brush cells. Importantly, our primary culture data support the conclusion that SnoN operates as a cell-intrinsic regulator within granule neuron precursors to promote the proliferation of these cells.
The identification of a novel function for SnoN in the proliferation of granule neuron precursors in the postnatal brain advances our understanding of the role of this major transcriptional regulator in brain development. SnoN has been implicated in several aspects of the development of postmitotic granule neurons in the rodent cerebellum, including parallel fiber axon growth, neuron migration, and dendrite branching (Stegmüller et al., 2006; Ikeuchi et al., 2009; Huynh et al., 2011). SnoN function in postmitotic granule neurons is tightly regulated by the E3 ubiquitin ligase Cdh1-APC and the Smad signaling pathway (Stegmüller and Bonni, 2005; Stegmüller et al., 2006, 2008; Kim and Bonni, 2007; Yang et al., 2010). In future studies, it will be interesting to determine whether SnoN function in granule neuron precursors is subject to these regulatory pathways. Although SHH signaling is a pivotal regulator of granule neuron precursor proliferation (Lee et al., 2010), our findings suggest that SnoN and SHH operate in parallel pathways to promote granule neuron precursor proliferation. Therefore, it will be important in future studies to identify the extrinsic cues that regulate SnoN-dependent granule neuron precursor proliferation in the mammalian brain. In view of the major role of SnoN as a key regulator of TGF-β-mediated signaling (Stroschein et al., 1999; Sun et al., 1999; Bonni et al., 2001), it will be interesting to consider whether SnoN might mediate or control granule neuron proliferation in response to members of this family of growth factors.
To gain mechanistic insights into SnoN-dependent granule neuron precursor proliferation, we deployed LCMRNA-seq to profile changes in gene expression specifically in the EGL of the developing cerebellum. As predicted, LCMRNA-seq was much more sensitive than profiling the entire cerebellum in identification of SnoN-regulated genes in the EGL using Math1-Cre-induced KO of SnoN in the cerebellum (data not shown). In future studies, it will be interesting to determine the effect of granule neuron precursor-specific KO of SnoN on gene expression in all cell types in the developing cerebellum using a single-cell RNA-seq approach.
The identification of SnoN-regulated programs of gene expression in granule neuron precursors sheds light on the biological functions of SnoN in these cells. Notably, in granule neurons, SnoN promotes axon growth by activating transcription (Ikeuchi et al., 2009), whereas SnoN regulates neuron migration and branching by acting as a transcriptional corepressor in an isoform-specific manner (Huynh et al., 2011). In granule neuron precursors, our gene profiling studies suggest that SnoN may act by both activating and repressing transcription. Gene ontology analyses suggest that SnoN-activated genes may promote cell proliferation, whereas SnoN-repressed genes prevent premature cell cycle exit of these cells.
The identification of novel interactions between SnoN and the transcription factors N-myc and Pax6, inspired by our bioinformatics analyses of transcription factor binding sites within SnoN-regulated genes, further illuminates how SnoN may promote the proliferation of granule neuron precursors in the mammalian brain. N-myc and Pax6 promote the proliferation and differentiation of granule neuron precursors, respectively (Kenney et al., 2003; Owa et al., 2018). Hence, SnoN may act as a transcriptional coregulator for these major transcription factors in granule neuron precursors. Beyond granule neuron precursors, it will be interesting to determine whether the interaction of SnoN with N-myc and Pax6 might regulate biological functions in other regions of the brain and outside the nervous system.
Our study also bears important implications for understanding neurological diseases. Impairment of granule neuron function within cerebellar circuits may contribute to neurodevelopmental disorders, including autism spectrum disorders (Kloth et al., 2015; Stoodley et al., 2017). Thus, deregulation of SnoN function in granule neuron precursors may contribute to the pathogenesis of these disorders. In addition, granule cells represent the cells of origin of medulloblastoma, the most common childhood brain tumor (Yang et al., 2008; Roussel and Hatten, 2011). In view of the role of SnoN in promoting granule neuron proliferation, it will be important to determine whether deregulation of SnoN function contributes to the pathogenesis of a subset of these tumors.
Footnotes
This work was supported by National Institutes of Health Grant NS041021 to A.B., and Natural Sciences and Engineering Research Council of Canada Grant to S.B. Confocal microscopic analyses were performed at the Washington University Center for Cellular Imaging, supported by Washington University School of Medicine, the Children's Discovery Institute Washington University, and St. Louis Children's Hospital CDI-CORE-2015-505 and NS086741. Sequencing was performed at the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine. We thank the members of the Bonni laboratory for helpful discussion and critical reading of the manuscript; Alexandra L. Joyner and Anna Marie Kenney for helpful comments on the project; and the Washington University Animal Behavior Core for DigiGait and open field behavior assay and data analysis.
The authors declare no competing financial interests.
References
- Alcaraz WA, Gold DA, Raponi E, Gent PM, Concepcion D, Hamilton BA (2006) Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation. Proc Natl Acad Sci U S A 103:19424–19429. 10.1073/pnas.0609184103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J. (1997) Development of the cerebellar system: in relation to its evolution, structure, and functions. Boca Raton, FL: CRC. [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY (1997) Math1 is essential for genesis of cerebellar granule neurons. Nature 390:169–172. 10.1038/36579 [DOI] [PubMed] [Google Scholar]
- Bonni S, Bonni A (2012) SnoN signaling in proliferating cells and postmitotic neurons. FEBS Lett 586:1977–1983. 10.1016/j.febslet.2012.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL (2001) TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol 3:587–595. 10.1038/35078562 [DOI] [PubMed] [Google Scholar]
- Cassady JP, D'Alessio AC, Sarkar S, Dani VS, Fan ZP, Ganz K, Roessler R, Sur M, Young RA, Jaenisch R (2014) Direct lineage conversion of adult mouse liver cells and B lymphocytes to neural stem cells. Stem Cell Reports 3:948–956. 10.1016/j.stemcr.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang K, Zhou L, Gao X, Wang J, Yao Y, He F, Luo Y, Yu Y, Li S, Cheng L, Sun YE (2016) Coupled electrophysiological recording and single cell transcriptome analyses revealed molecular mechanisms underlying neuronal maturation. Protein Cell 7:175–186. 10.1007/s13238-016-0247-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Bonni A (2011) Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain. Neuron 72:22–40. 10.1016/j.neuron.2011.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Kennedy H (2007) Cell-cycle control and cortical development. Nat Rev Neurosci 8:438–450. 10.1038/nrn2097 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Kowalczyk T, Daza RA, Dagan A, Lau C, Rose MF, Hevner RF (2006) Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci 26:9184–9195. 10.1523/JNEUROSCI.1610-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JS, Luo L (2008) Timing neurogenesis and differentiation: insights from quantitative clonal analyses of cerebellar granule cells. J Neurosci 28:2301–2312. 10.1523/JNEUROSCI.5157-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora A, Klisch TJ, Schuster G, Zoghbi HY (2009) Deletion of Atoh1 disrupts sonic hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science 326:1424–1427. 10.1126/science.1181453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, Vockley CM, Safi A, Gersbach CA, Crawford GE, West AE (2015) Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci 18:647–656. 10.1038/nn.3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Badura A, Deverett B, Najafi F, Pereira TD, Gao Z, Ozden I, Kloth AD, Pnevmatikakis E, Paninski L, De Zeeuw CI, Medina JF, Wang SS (2017) Cerebellar granule cells acquire a widespread predictive feedback signal during motor learning. Nat Neurosci 20:727–734. 10.1038/nn.4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME, Heintz N (1995) Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci 18:385–408. 10.1146/annurev.ne.18.030195.002125 [DOI] [PubMed] [Google Scholar]
- Heiney SA, Wohl MP, Chettih SN, Ruffolo LI, Medina JF (2014) Cerebellar-dependent expression of motor learning during eyeblink conditioning in head-fixed mice. J Neurosci 34:14845–14853. 10.1523/JNEUROSCI.2820-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y (2005) Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47:201–213. 10.1016/j.neuron.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Hua R, Yu S, Liu M, Li H (2018) A PCR-based method for RNA probes and applications in neuroscience. Front Neurosci 12:266. 10.3389/fnins.2018.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MA, Ikeuchi Y, Netherton S, de la Torre-Ubieta L, Kanadia R, Stegmüller J, Cepko C, Bonni S, Bonni A (2011) An isoform-specific SnoN1-FOXO1 repressor complex controls neuronal morphogenesis and positioning in the mammalian brain. Neuron 69:930–944. 10.1016/j.neuron.2011.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y, Stegmüller J, Netherton S, Huynh MA, Masu M, Frank D, Bonni S, Bonni A (2009) A SnoN-Ccd1 pathway promotes axonal morphogenesis in the mammalian brain. J Neurosci 29:4312–4321. 10.1523/JNEUROSCI.0126-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y, Dadakhujaev S, Chandhoke AS, Huynh MA, Oldenborg A, Ikeuchi M, Deng L, Bennett EJ, Harper JW, Bonni A, Bonni S (2014) TIF1gamma protein regulates epithelial-mesenchymal transition by operating as a small ubiquitin-like modifier (SUMO) E3 ligase for the transcriptional regulator SnoN1. J Biol Chem 289:25067–25078. 10.1074/jbc.M114.575878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH (2003) Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130:15–28. 10.1242/dev.00182 [DOI] [PubMed] [Google Scholar]
- Kim AH, Bonni A (2007) Thinking within the D box: initial identification of Cdh1-APC substrates in the nervous system. Mol Cell Neurosci 34:281–287. 10.1016/j.mcn.2006.11.019 [DOI] [PubMed] [Google Scholar]
- Kloth AD, Badura A, Li A, Cherskov A, Connolly SG, Giovannucci A, Bangash MA, Grasselli G, Peñagarikano O, Piochon C, Tsai PT, Geschwind DH, Hansel C, Sahin M, Takumi T, Worley PF, Wang SS (2015) Cerebellar associative sensory learning defects in five mouse autism models. eLife 4:e06085. 10.7554/eLife.06085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Stegmüller J, Matsuda T, Bonni S, Bonni A (2004) Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science 303:1026–1030. 10.1126/science.1093712 [DOI] [PubMed] [Google Scholar]
- Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW (2012) oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-seq datasets. G3 (Bethesda) 2:987–1002. 10.1534/g3.112.003202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Ji H, Ouyang Z, Zhou B, Ma W, Vokes SA, McMahon AP, Wong WH, Scott MP (2010) Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc Natl Acad Sci U S A 107:9736–9741. 10.1073/pnas.1004602107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leto K, Arancillo M, Becker EB, Buffo A, Chiang C, Ding B, Dobyns WB, Dusart I, Haldipur P, Hatten ME, Hoshino M, Joyner AL, Kano M, Kilpatrick DL, Koibuchi N, Marino S, Martinez S, Millen KJ, Millner TO, Miyata T, et al. (2016) Consensus paper: cerebellar development. Cerebellum 15:789–828. 10.1007/s12311-015-0724-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Du F, Yuelling LW, Lin T, Muradimova RE, Tricarico R, Wang J, Enikolopov G, Bellacosa A, Wechsler-Reya RJ, Yang ZJ (2013) A population of nestin-expressing progenitors in the cerebellum exhibits increased tumorigenicity. Nat Neurosci 16:1737–1744. 10.1038/nn.3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G (2005) Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48:17–24. 10.1016/j.neuron.2005.08.028 [DOI] [PubMed] [Google Scholar]
- Netherton SJ, Bonni S (2010) Suppression of TGFbeta-induced epithelial-mesenchymal transition like phenotype by a PIAS1 regulated sumoylation pathway in NMuMG epithelial cells. PLoS One 5:e13971. 10.1371/journal.pone.0013971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvis GD, Hartzell AL, Smith JB, Barraza LH, Wilson SL, Szulc KU, Turnbull DH, Joyner AL (2012) The engrailed homeobox genes are required in multiple cell lineages to coordinate sequential formation of fissures and growth of the cerebellum. Dev Biol 367:25–39. 10.1016/j.ydbio.2012.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owa T, Taya S, Miyashita S, Yamashita M, Adachi T, Yamada K, Yokoyama M, Aida S, Nishioka T, Inoue YU, Goitsuka R, Nakamura T, Inoue T, Kaibuchi K, Hoshino M (2018) Meis1 coordinates cerebellar granule cell development by regulating Pax6 transcription, BMP signaling and Atoh1 degradation. J Neurosci 38:1277–1294. 10.1523/JNEUROSCI.1545-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot I, Patel S, Deng L, Chandhoke AS, Zhang C, Bonni A, Bonni S (2013) Identification of a novel link between the protein kinase NDR1 and TGFbeta signaling in epithelial cells. PLoS One 8:e67178. 10.1371/journal.pone.0067178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon y Cajal S. (1911) Histology of the nervous system of man and vertebrates. Oxford, UK: Oxford UP. [Google Scholar]
- Rios I, Alvarez-Rodríguez R, Martí E, Pons S (2004) Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development 131:3159–3168. 10.1242/dev.01188 [DOI] [PubMed] [Google Scholar]
- Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, Pun SH, Sellers DL, Tasic B, Seelig G (2018) Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360:176–182. 10.1126/science.aam8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel MF, Hatten ME (2011) Cerebellum development and medulloblastoma. Curr Top Dev Biol 94:235–282. 10.1016/B978-0-12-380916-2.00008-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker KP, Wilson SM, Bonni S (2005) SnoN is a cell type-specific mediator of transforming growth factor-beta responses. J Biol Chem 280:13037–13046. 10.1074/jbc.M409367200 [DOI] [PubMed] [Google Scholar]
- Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudillière B, Yuan Z, Stegmüller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A (2006) A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311:1012–1017. 10.1126/science.1122513 [DOI] [PubMed] [Google Scholar]
- Sillitoe RV, Joyner AL (2007) Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu Rev Cell Dev Biol 23:549–577. 10.1146/annurev.cellbio.23.090506.123237 [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME (2001) Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron 31:557–568. 10.1016/S0896-6273(01)00395-6 [DOI] [PubMed] [Google Scholar]
- Stegmüller J, Bonni A (2005) Moving past proliferation: new roles for Cdh1-APC in postmitotic neurons. Trends Neurosci 28:596–601. 10.1016/j.tins.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Stegmüller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A (2006) Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron 50:389–400. 10.1016/j.neuron.2006.03.034 [DOI] [PubMed] [Google Scholar]
- Stegmüller J, Huynh MA, Yuan Z, Konishi Y, Bonni A (2008) TGFbeta-Smad2 signaling regulates the Cdh1-APC/SnoN pathway of axonal morphogenesis. J Neurosci 28:1961–1969. 10.1523/JNEUROSCI.3061-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley CJ, D'Mello AM, Ellegood J, Jakkamsetti V, Liu P, Nebel MB, Gibson JM, Kelly E, Meng F, Cano CA, Pascual JM, Mostofsky SH, Lerch JP, Tsai PT (2017) Altered cerebellar connectivity in autism and cerebellar-mediated rescue of autism-related behaviors in mice. Nat Neurosci 20:1744–1751. 10.1038/s41593-017-0004-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K (1999) Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science 286:771–774. 10.1126/science.286.5440.771 [DOI] [PubMed] [Google Scholar]
- Stroschein SL, Bonni S, Wrana JL, Luo K (2001) Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev 15:2822–2836. 10.1101/gad.912901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA (1999) SnoN and ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc Natl Acad Sci U S A 96:12442–12447. 10.1073/pnas.96.22.12442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnegri P, Huang J, Yamada T, Yang Y, Mejia LA, Cho HY, Oldenborg A, Bonni A (2017) RNF8/UBC13 ubiquitin signaling suppresses synapse formation in the mammalian brain. Nat Commun 8:1271. 10.1038/s41467-017-01333-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo BT, Wolf E, Kawauchi D, Gebhardt A, Rehg JE, Finkelstein D, Walz S, Murphy BL, Youn YH, Han YG, Eilers M, Roussel MF (2016) The interaction of myc with Miz1 defines medulloblastoma subgroup identity. Cancer Cell 29:5–16. 10.1016/j.ccell.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28:2184–2185. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY (2005) Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron 48:31–43. 10.1016/j.neuron.2005.08.024 [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP (1999) Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron 22:103–114. 10.1016/S0896-6273(00)80682-0 [DOI] [PubMed] [Google Scholar]
- Whittaker DE, Riegman KL, Kasah S, Mohan C, Yu T, Sala BP, Hebaishi H, Caruso A, Marques AC, Michetti C, Smachetti ME, Shah A, Sabbioni M, Kulhanci O, Tee WW, Reinberg D, Scattoni ML, Volk H, McGonnell I, Wardle FC, et al. (2017) The chromatin remodeling factor CHD7 controls cerebellar development by regulating reelin expression. J Clin Invest 127:874–887. 10.1172/JCI83408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcinski A, Lawton AK, Bayin NS, Lao Z, Stephen DN, Joyner AL (2017) Cerebellar granule cell replenishment postinjury by adaptive reprogramming of Nestin(+) progenitors. Nat Neurosci 20:1361–1370. 10.1038/nn.4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Huang J, Coppola G, Geschwind DH, Bonni A (2013) Sumoylated MEF2A coordinately eliminates orphan presynaptic sites and promotes maturation of presynaptic boutons. J Neurosci 33:4726–4740. 10.1523/JNEUROSCI.4191-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Yamada T, Wu B, Bilimoria PM, Ikeuchi Y, de la Iglesia N, Shen J, Bonni A (2009) A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science 326:575–578. 10.1126/science.1177087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Bonni A (2010) The dynamic ubiquitin ligase duo: Cdh1-APC and Cdc20-APC regulate neuronal morphogenesis and connectivity. Curr Opin Neurobiol 20:92–99. 10.1016/j.conb.2009.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yamada T, Hill KK, Hemberg M, Reddy NC, Cho HY, Guthrie AN, Oldenborg A, Heiney SA, Ohmae S, Medina JF, Holy TE, Bonni A (2016) Chromatin remodeling inactivates activity genes and regulates neural coding. Science 353:300–305. 10.1126/science.aad4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schüller U, Machold R, Fishell G, Rowitch DH, Wainwright BJ, Wechsler-Reya RJ (2008) Medulloblastoma can be initiated by deletion of patched in lineage-restricted progenitors or stem cells. Cancer Cell 14:135–145. 10.1016/j.ccr.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Schriefer LA, Stormo GD (2007) Identification of muscle-specific regulatory modules in Caenorhabditis elegans. Genome Res 17:348–357. 10.1101/gr.5989907 [DOI] [PMC free article] [PubMed] [Google Scholar]