Abstract
Objective
The oxidized low density lipoprotein receptor 1 (OLR1) gene plays an important role in the degradation of oxidized low-density lipoprotein and adipocyte proliferation in mammals. For this reason, we aimed at investigating the association of OLR1 gene polymorphisms with carcass quality traits in Chinese Qinchuan cattle.
Methods
The single nucleotide polymorphism (SNP) was identified in the 3′ untranslated region of bovine OLR1 gene by DNA sequencing. In addition, the haplotype frequency and linkage disequilibrium estimates of three SNPs were evaluated in 520 individuals.
Results
Results indicated that the studied three SNPs were within the range of moderate genetic diversity (0.25< polymorphism information content<0.5). Haplotype analysis of three SNPs showed that ten different haplotypes were identified, but only five haplotypes were listed as those with a frequency of <0.05 were excluded. The Hap3 (-G1T2C3-) had the highest haplotype frequency (42.10%). Linkage disequilibrium analysis showed that the three SNPs had a low linkage (r2<0.001). The T10588C and C10647T were significantly associated with backfat thickness and intramuscular fat content in Qinchuan cattle.
Conclusion
Based on our results, we believe that the OLR1 gene could be a strong candidate gene for influencing carcass quality traits in Qinchuan cattle.
Keywords: Carcass Quality Traits, Qinchuan Cattle, Association Analysis, Oxidized Low-density Lipoprotein Receptor 1 Gene
INTRODUCTION
In China, Qinchuan cattle (Bos taurus) are generally considered as efficient converters of poor quality roughage to meat [1]. However, the carcass quality traits, i.e., back fat thickness, loin muscle area (LMA) and intramuscular fat (IMF), are noticeably lower than those of other exotic commercial cattle breeds (such as Wagyu cattle) [2]. In the process of livestock breeding, carcass traits are a good tool to assess the economic value of animals, which, at least in part, are affected by genetic variability [3]. Therefore, to identify polymorphisms of effective genes associated with carcass quality traits would be significant for marker assisted selection programs for breed improvement.
The oxidized low-density lipoprotein receptor 1 (OLR1), also known as LOX1, is a type-II membrane cell-surface protein [4], that plays an important role in the degradation of oxidized low-density lipoprotein (Ox-LDL) and adipocyte proliferation [5]. The OLR1 gene was first identified by Sawamura et al [6] and found to influence lipid metabolism in the liver and mammary glands [6,7]. In mouse adipose tissue, the over-expression of the OLR1 gene may improve cholesterol content, free fatty acid intake and lipid drops [8]. In pig adipose tissue, the expression of OLR1 was correlated with peroxisome proliferator-activated receptor gamma, fas cell surface death receptor, and sterol regulatory element binding transcription factor-1c, which demonstrated that OLR1 gene was highly associated with fat deposition and its transcription [9]. As a core surface receptor, OLR1 was reported to support bind, internalizes and proteolytic degrades Ox-LDL, thereby controlling glucose and lipid metabolism in the mammary gland [10,11].
Based on the role of OLR1 gene in lipid metabolism, the objectives of this study were to investigate the genetic variations within OLR1 gene in Chinese native breed, and to analyze the associations between genetic variations and carcass quality traits. Results gleaned from this study could possibly contribute to better breeding plan and policies.
MATERIALS AND METHODS
All animal procedures were performed according to guidelines laid down by the China Council on Animal Care, and the protocols were approved by the Experimental Animal Manage Committee (EAMC) of Qinghai University.
A total number of 520 cows (aged 18 to 24 months) were randomly collected from the experiment farm of national beef cattle improvement center (Yangling, Shaanxi, China). All cows were kept under similar dietary and environmental conditions. Genomic DNA was extracted from whole blood samples using a Blood DNA Kit (OMGAM Bio-Tek, Doraville, USA). Meanwhile, ultrasound measurements were available for each individual, including backfat thickness, LMA and IMF content.
Available sequence information from bovine OLR1 gene (Genbank accession no AC_000162) was used to design polymerase chain reaction (PCR) primers. Details of primer nucleotide sequences for all primers, PCR product lengths, primers locations and annealing temperatures are shown in Table 1. PCR was performed in a 20 μL reaction mixture containing 50 ng of genomic DNA, 10 pM of each upstream and downstream primer, 0.20 mM of dNTPs, 2.5 mM MgCl2 and 0.5 U Taq polymerase (TaKaRa, Dalian, China). The reaction conditions were as follows: initial denaturation of DNA for 5 min at 95°C, followed by 35 cycles at 94°C for 30 s, the annealing temperature 30 s, and 40 s at 72°C followed by a final extension of primer for 10 min at 72°C.
Table 1.
Primer sequences used in PCR assays with OLR1 gene fragments
| Primer name | Primer sequence (5′ to 3′) | Annealing temperature (°C) | Product length (bp) | Amplified region |
|---|---|---|---|---|
| L1 | ACTCCAGCAGGAACTCACA AATGATAAGCCAACTTGGT |
56.7 | 336 | Exon 1 and intron 1 |
| L2 | ATATCTGATATTGAATCCCA ATGAGGGCTTATAAACA |
60.0 | 381 | Intron 1, exon 2, and intron 2 |
| L3 | TTATTGGGAATTGGAATTGG CTCTATTTTTGTCATCCT |
57.2 | 448 | Intron 2, exon 3, and intron 2 |
| L4 | GTTAAGAATTGTAGAAATAT GACGCCCCACTTGTAAG |
63.5 | 653 | Intron 3, exon 4, intron 4, and exon 5 |
| L5 | AAGGCGAATCTATTGAGAGC CCTAGAAGAAAGCATAGGAC |
60.4 | 360 | Exon 6 and 3′UTR |
PCR, polymerase chain reaction; OLR1, oxidized low density lipoprotein receptor 1, UTR, untranslated region.
Gene frequencies, Hardy-Weinberg equilibrium (HWE), and polymorphism information content (PIC) were tested by POPGENE software package (Version 3.2). Linkage disequilibrium (LD) and haplotype construction was performed with the online version of SHEsis software [12]. Combinations with frequencies below 5.0% were not included, and the remaining combinations were selected for further analysis.
According to the pedigree information, the 520 daughters came from 6 different sires. In livestock species, this affinity may influence the genotype of offspring. So, the sire is the random effect in the model. Age factors were divided into four levels (18, 20, 22, and 24-month age periods, respectively) in the model, due to the longer fattening periods of Qinchuan cattle. The association of each single nucleotide polymorphism (SNP) marker genotype and carcass traits was studied using a general linear model (GLM) procedure implemented in SPSS 16.0 (IBM Company, NY, USA) software package. The mixed model equation used in the study was as follows:
Where Yijk is the trait measurement, μ is the overall mean, Gi is the fixed effect of genotype (i = 1, 2, and 3), Aj is the fixed covariate of age (j = 1, 2, 3, and 4), Sk is the random effect of sire (k = 1 through 6) and eijk is the random error.
For a more detailed review of the results, we also corrected p values via the Bonferroni correction [13]. This correction was used to account for multiple tests and yielded more robust results. Differences were considered significant if p<0.05. Data are expressed as the mean±standard error.
RESULTS
Investigation of polymorphic loci on the bovine OLR1 gene
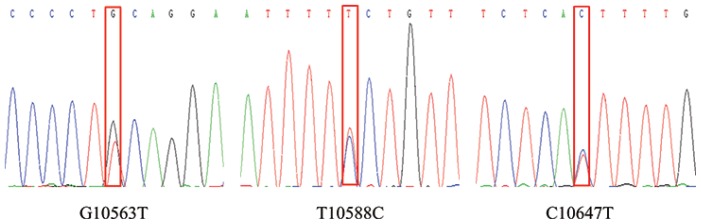
In this study, three SNPs (G10563T, T10588C, and C10647T) were identified by pool DNA sequencing in bovine OLR1 gene (Figure 1). Table 2 illustrates the genotypic and allelic frequencies at all loci of bovine OLR1 gene. The G10563T-A allele, T10588C-A allele, and C10647T-C allele were found to be predominant in the studied samples. In this study, the calculated PIC values for each locus ranged from 0.258 to 0.328. According to the convention for classification of PIC, three SNPs were within the range of moderate genetic diversity (0.25<p<0.50). Besides, the chi-square test showed that those SNPs were in HWE.
Figure 1.
The sequencing peak of three single nucleotide polymorphisms (SNPs) from oxidized low density lipoprotein receptor 1 (OLR1) gene.
Table 2.
Distribution of genotype and allele frequencies in Qinchuan cattle
| Site | Genotype (N) | Genotypic frequency (%) | Alleles | Allele frequency (%) | χ2 (HWE) | PIC |
|---|---|---|---|---|---|---|
| G10563T | GG (348) | 66.92 | G | 81.25 | p>0.05 | 0.2583 |
| rs722568839 | GT (149) | 28.65 | T | 18.75 | ||
| TT (23) | 4.43 | - | - | |||
| T10588C | TT (290) | 55.77 | T | 74.04 | p>0.05 | 0.3105 |
| rs132917098 | TC (190) | 36.54 | C | 25.96 | ||
| CC (40) | 7.69 | - | - | |||
| C10647T | CC (268) | 72.12 | C | 70.77 | p>0.05 | 0.3281 |
| rs45938133 | CT (200) | 24.23 | T | 29.23 | ||
| TT (52) | 3.65 | - | - |
χ2 (HWE), Hardy-Weinberg equilibrium χ2 value, Hard-Weinberg equilibrium (p>0.05), Hardy–Weinberg disequilibrium (p<0.05).
Linkage disequilibrium within bovine OLR1 gene
As shown in Table 3, the Hap3 (-G1T2C3-, 42.10%) occurred with greater frequency than the others in Qinchuan cattle. The values of D′ varied from 0.019 to 0.094, and the r2 values were from 0.000 to 0.001, indicating that those SNPs had low LD. It could be argued that recombination will be high, and LD will be low in genovariation-dense regions.
Table 3.
Haplotypes of OLR1 gene and their frequencies in Qinchuan cattle1)
| Haplotype | G10563T | T10588C | C10647T | Frequency (%) |
|---|---|---|---|---|
| Hap1 | G1 | C2 | C3 | 15.70 |
| Hap2 | G1 | C2 | T3 | 5.90 |
| Hap3 | G1 | T2 | C3 | 42.10 |
| Hap4 | G1 | T2 | T3 | 17.60 |
| Hap5 | T1 | T2 | C3 | 9.80 |
OLR1, oxidized low density lipoprotein receptor 1.
C1 and T1 were haplotypes of G10563T, C2 and T2 were haplotypes of T10588C, C3 and T3 were haplotypes of C10647T.
Effects of different genotypes of OLR1 gene SNP loci on carcass quality traits
The association results are presented in Table 4. At the T10588C locus, the influence of TT genotype resulted in the highest mean for backfat thickness compared to animals with genotype CC in Qinchuan cattle (p<0.05). At the C10647T locus, individuals carrying the TT genotype had significantly greater backfat thickness than those with CC genotype (p<0.05). At the G10563T locus, genotypes did not show any significant correlation with carcass quality traits (p>0.05).
Table 4.
Association of carcass traits with marker genotypes within OLR1 gene in Qinchuan cattle
| Site | Trait | Genotype (mean±standard error) | p value1) | ||
|---|---|---|---|---|---|
| GG | GT | TT | |||
| G10563T | BT (cm) | 0.981±0.017 | 0.963±0.026 | 1.031±0.066 | 0.902 |
| rs722568839 | LMA (cm2) | 64.195±0.467 | 64.367±0.714 | 61.753±1.817 | 0.543 |
| IMF (%) | 7.138±0.041 | 7.207±0.063 | 7.482±0.161 | 0.338 | |
| TT | TC | CC | |||
| T10588C | BT (cm) | 0.987±0.019a | 0.991±0.023a | 0.850±0.050b | 0.031d |
| rs132917098 | LMA (cm2) | 63.541±0.511 | 64.757±0.631 | 65.504±1.376 | 0.404 |
| IMF (%) | 7.264±0.045 | 7.146±0.055 | 6.639±0.120 | 0.177 | |
| CC | CT | TT | |||
| C10647T | BT (cm) | 0.907±0.019b | 1.005±0.021 | 1.240±0.042a | 0.013d |
| rs45938133 | LMA (cm2) | 64.720±0.532 | 63.608±0.616 | 63.159±1.207 | 0.517 |
| IMF (%) | 7.084±0.047 | 7.223±0.054 | 7.440±0.106 | 0.069 | |
OLR1, oxidized low density lipoprotein receptor 1; BT, backfat thickness; LMA, loin muscle area; IMF, intramuscular fat content.
Probability of the F-test for genotype effect.
Means with different superscripts are significantly different (p<0.05).
Significant effect (p<0.05) after modified Bonferroni correction for trait-wise multiple tests.
Table 5 shows the association of selected haplotypes (with frequencies of 5% or more) with carcass quality traits. When the combination results were compared no significant differences were detected between the combined haplotype of these SNPs and carcass quality traits in the Qinchuan cattle population (p>0.05).
Table 5.
Associations of carcass traits with diplotypes within OLR1 gene in Qinchuan cattle
| Haplotypes | Hap3/1 (67) | Hap3/2 (41) | Hap3/3 (94) | Hap3/4 (81) | Hap3/5 (48) | p-value1) |
|---|---|---|---|---|---|---|
| BT (cm) | 0.921±0.037 | 1.055±0.047 | 0.907±0.031 | 1.027±0.034 | 0.984±0.044 | 0.085 |
| LMA (cm2) | 64.347±1.084 | 63.905±1.385 | 64.476±0.915 | 63.347±0.986 | 64.904±1.280 | 0.773 |
| IMF (%) | 7.021±0.094 | 7.070±0.120 | 7.186±0.079 | 7.293±0.085 | 7.127±0.110 | 0.254 |
OLR1, oxidized low density lipoprotein receptor 1; BT, backfat thickness; LMA, loin muscle area; IMF, intramuscular fat content.
Probability of the F-test for genotype effect.
DISCUSSION
The OLR1 gene was reported to possess complex biological functions [14], especially regarding degradation of low-density lipoprotein and glucose metabolism in liver. Previous studies have shown that genetic polymorphisms in the OLR1 gene are associated with economic traits in livestock. Khatib et al [3] reported that the SNP A8232C in the 3′ untranslated region (UTR) of OLR1 gene was associated with OLR1 gene expression, and linked to significant effects on milk fat yield and fat percentage in Holstein dairy cattle. Similarly, one novel SNP (C223A) detected in OLR1 gene was associated with milk fat percentage in Polish Holstein-Friesian bulls [15]. Michael et al [16] showed that one novel SNP (c.-495T>C) in promoter region of OLR1 gene had a strong effect on rib-eye area in Angus steer population. The research of Fonseca et al [17] showed that a marker (rs109019599) on OLR1 gene was associated with rump fat thickness and weaning weight in Nelore cattle. Based on these outcomes, it is our belief that the OLR1 gene could be an excellent candidate gene for fat deposition-related traits in livestock.
The bovine OLR1 gene localizes on chromosome 18, has 6 exons [11], and is highly expressed in lung, liver and adipose tissue [9]. In the current study, we detected three SNPs in 3′ UTR, including G10563T, T10588C, and C10647T. Data analysis revealed that the individuals with genotype TT of T10588C and C10647T had significantly greater backfat thickness and IMF content than those with genotype CC, demonstrating that allele T might be associated with an increase in backfat thickness and IMF content in Qinchuan cattle. Therefore, we suggest that the selective breeding of the heterozygotes at both T10588C and C10647T loci of OLR1 in Qinchuan cattle may improve on the carcass quality traits.
The 3′ UTR miRNA-related SNPs may be within or at the vicinity of the miRNA binding site, which could impair regulatory functions of the associated miRNA [18,19]. This could result in variations in the level or timing of gene expression, ultimately affecting phenotypes [20,21], as has been previously demonstrated in Duroc pigs [22], Polish Holstein-Friesian [23], and Guanzhong dairy goats [24]. In the current paper, bioinformatics analysis was used to predict the effects of the two SNPs in the 3′ UTR of the OLR1 gene on the miRNA binding sites using the online version of Targetscan software (http://www.targetscan.org/vert). The data indicated that allele C in T10588C, and allele T in C10647T altered the base within binding sites of bta-miR-12047 and bta-miR-12021, respectively. Thus, it can be reasonably inferred that the mutations may alter OLR1 gene expression level by modifying miRNA binding sites within the 3′ UTR. As a result, the mutations could affect fatty acid metabolism in Qinchuan cattle.
In summary, three polymorphisms in the OLR1 were identified in Qinchuan cattle. The association analysis of single markers (T10588C and C10647T) revealed prominent effects on carcass quality traits. However, further research and validation of various allelic effects, functional mechanisms and bioactivity are needed in larger population to explore the usage of OLR1 gene in beef cattle breeding.
ACKNOWLEDGMENTS
The research was supported by the program for Weng Hongwu Original Research Fund of Peking University (WHW201509), National Natural Science Foundation of Qinghai Province (2018-ZJ-922Q and 2016-ZJ-935Q) and National Natural Science Foundation of China (No.31660662).
Footnotes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
REFERENCES
- 1.Sun Y, Lan X, Lei C, Zhang C, Chen H. Haplotype combination of the bovine CFL2 gene sequence variants and association with growth traits in Qinchuan cattle. Gene. 2015;563:136–41. doi: 10.1016/j.gene.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Gui L, Wang H, Wei S, Zhang Y, Zan L. Molecular characterization, expression profiles, and analysis of Qinchuan cattle SIRT1 gene association with meat quality and body measurement traits (Bos taurus) Mol Biol Rep. 2014;41:5237–46. doi: 10.1007/s11033-014-3393-1. [DOI] [PubMed] [Google Scholar]
- 3.Khatib H, Leonard SD, Schutzkus V, Luo W, Chang YM. Association of the OLR1 gene with milk composition in Holstein dairy cattle. J Dairy Sci. 2006;89:1753–60. doi: 10.3168/jds.S0022-0302(06)72243-3. [DOI] [PubMed] [Google Scholar]
- 4.Mei C, Wang H, Liao Q, et al. Genetic architecture and selection of Chinese cattle revealed by whole genome resequencing. Mol Biol Evol. 2017 Dec 19; doi: 10.1093/molbev/msx322. [Epub] [DOI] [PubMed] [Google Scholar]
- 5.Liao CH, Shaw HM, Chao PM. Impairment of glucose metabolism in mice induced by dietary oxidized frying oil is different from that induced by conjugated linoleic acid. Nutrition. 2008;24:744–52. doi: 10.1016/j.nut.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Sawamura T, Kume N, Aoyama T, et al. An endothelial receptor for oxidized low-density lipoprotein. Nature. 1997;386:73–7. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- 7.Ringseis R, Dathe C, Muschick A, Brandsch C, Eder K. Oxidized fat reduces milk triacylglycerol concentrations by inhibiting gene expression of lipoprotein lipase and fatty acid transporters in the mammary gland of rats. J Nutr. 2007;137:2056–61. doi: 10.1093/jn/137.9.2056. [DOI] [PubMed] [Google Scholar]
- 8.Chui PC, Guan HP, Lehrke M, Lazar MA. PPARgamma regulates adipocyte cholesterol metabolism via oxidized LDL receptor 1. J Clin Invest. 2005;115:2244–56. doi: 10.1172/JCI24130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun C, Liu C, Zhang Z, et al. Cloning of OLR1 gene in pig adipose tissue and preliminary study on its lipid-accumulating effect. Asian-Australas J Anim Sci. 2009;22:1420–8. [Google Scholar]
- 10.Murase T, Kume N, Kataoka H, et al. Identification of soluble forms of lectin-like oxidized LDL receptor-1. Arterioscler Thromb Vasc Biol. 2000;20:715–20. doi: 10.1161/01.atv.20.3.715. [DOI] [PubMed] [Google Scholar]
- 11.Khatib H, Rosa GJ, Weigel K, et al. Additional support for an association between OLR1 and milk fat traits in cattle. Anim Genet. 2007;38:308–10. doi: 10.1111/j.1365-2052.2007.01584.x. [DOI] [PubMed] [Google Scholar]
- 12.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–8. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 13.Tanner MS, Sharrard MJ, Rigby AS. Gene polymorphisms and the use of the bonferroni correction factor: when and when not to apply? Arch Dis Child. 1997;76:385. doi: 10.1136/adc.76.4.385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M, Qiu H, Lin X, et al. Lectin-like oxidized low-density lipoprotein receptor (LOX-1) in sickle cell disease vasculopathy. Blood Cells Mol Dis. 2016;60:44–8. doi: 10.1016/j.bcmd.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komisarek J, Dorynek Z. Effect of ABCG2, PPARGC1A, OLR1, and SCD1 gene polymorphism on estimated breeding values for functional and production traits in Polish Holstein-Friesian bulls. J Appl Genet. 2009;50:125–32. doi: 10.1007/BF03195663. [DOI] [PubMed] [Google Scholar]
- 16.Michael V, Khandker I, Chen L, Li C. SHORT COMMUNICATION: Association analyses of a single nucleotide polymorphism in the promoter of OLR1 with growth, feed efficiency, fat deposition, and carcass merit traits in hybrid, Angus and Charolais beef cattle. Can J Anim Sci. 2013;93:193–7. [Google Scholar]
- 17.Fonseca PD, de Souza FR, de Camargo GM, et al. Association of ADIPOQ, OLR1 and PPARGC1A gene polymorphisms with growth and carcass traits in Nelore cattle. Meta Gene. 2015;4:1–7. doi: 10.1016/j.mgene.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano M, Mohri T, Fukami T, et al. Single-nucleotide polymorphisms in cytochrome P450 2E1 (CYP2E1) 3′-untranslated region affect the regulation of CYP2E1 by miR-570. Drug Metab Dispos. 2015;43:1450–7. doi: 10.1124/dmd.115.065664. [DOI] [PubMed] [Google Scholar]
- 19.Yie SM, Li LH, Xiao R, Librach CL. A single base-pair mutation in the 3′-untranslated region of HLA-G mRNA is associated with preeclampsia. Mol Hum Reprod. 2008;14:649–53. doi: 10.1093/molehr/gan059. [DOI] [PubMed] [Google Scholar]
- 20.Schwerin M, Maak S, Hagendorf A, von Lengerken G, Seyfert HM. A 3′-UTR variant of the inducible porcine hsp70.2 gene affects mRNA stability. Biochim Biophys Acta. 2002;1578:90–4. doi: 10.1016/s0167-4781(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 21.Lee I, Ajay SS, Yook JI, et al. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19:1175–83. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartz M, Kociucka B, Mankowska M, Switonski M, Szydlowski M. Transcript level of the porcine ME1 gene is affected by SNP in its 3′UTR, which is also associated with subcutaneous fat thickness. J Anim Breed Genet. 2014;131:271–8. doi: 10.1111/jbg.12065. [DOI] [PubMed] [Google Scholar]
- 23.Juszczuk-Kubiak E, Bujko K, Grzes M, et al. Study of bovine Mef2B gene: the temporal-spatial expression patterns, polymorphism and association analysis with meat production traits. J Anim Sci. 2016;94:4536–48. doi: 10.2527/jas.2016-0741. [DOI] [PubMed] [Google Scholar]
- 24.Hou J, An X, Song Y, et al. Two mutations in the caprine MTHFR 3′UTR regulated by microRNAs are associated with milk production traits. PLoS One. 2015;10:e0133015. doi: 10.1371/journal.pone.0133015. [DOI] [PMC free article] [PubMed] [Google Scholar]