Abstract
Long noncoding RNAs (lncRNAs) have been illustrated as vital molecules in regulating human cancer by emerging evidence. LINC00163 is a novel lncRNA without functional definition. In this study, we investigated its role in the tumorigenesis of lung cancer. The results showed that LINC00163 level was significantly downregulated in lung cancer tissues and cell lines by bioinformatics and qRT-PCR analyses. Notably, we observed that LINC00163 expression was lower in metastatic tissues than in non-metastatic cases. Furthermore, higher expression of LINC00163 in patients with lung cancer predicted better prognosis. Gain-of-function assays illustrated that upregulation of LINC00163 dramatically suppressed the proliferation, migration and invasion of lung cancer cells in vitro, whereas promoting apoptosis. Consistently, LINC00163 overexpression impaired tumor propagation in vivo. Mechanical study revealed that LINC00163 recruited ARID1A to the promoter of TCF21 and initiated its expression. In conclusion, we concluded that LINC00163/ARID1A/TCF21 regulatory loop modulated the development of lung cancer, providing a new insight on the mechanism underlying lung cancer progression.
Keywords: Long noncoding RNA, lung cancer, LINC00163, ARID1A, TCF21
Introduction
Lung cancer is the second most popular tumor and remains the first leading risk for cancer-related deaths in both females and males around the world [1]. In USA, every year there are about one million newly diagnosed patients with lung cancer and about 500,000 deaths caused by lung cancer [1]. Despite great improvement achieved in surgery, radiotherapy and chemotherapy, the five-year survival rate of lung cancer patients still remains rather low [2,3]. Especially, high rates of recurrence and metastasis increase the malignance of lung cancer [4]. Thus, it is urgent to understand the molecular mechanism of lung cancer and develop novel therapeutic strategies.
Long noncoding RNAs (lncRNAs) are characterized by no protein-coding ability and over 200 nucleotides in length [5]. In the recent decade, emerging evidences have illustrated the vital regulatory functions of lncRNAs [6]. Many studies revealed that lncRNAs play crucial roles in cellular homeostasis, embryo development, immune system and tumorigenesis [7-10]. Nowadays, the importance of lncRNA in cancer has been widely acknowledged. For example, lncRNA MEG3 suppresses bladder cancer growth and metastasis through regulating EZH2 ubiquitination [11]. LncRNA LOXL1-AS1 promotes CCND1 expression to increase the proliferation of prostate cancer cells by inhibiting miR-541-3p [12]. LncRNA SOX2OT overexpression contributes to metastasis of non-small-cell lung cancer (NSCLC) [13]. Additionally, lncRNA PRAL has been reported to serve as a diagnostic biomarker in NSCLC patients and regulate tumor metastasis [14]. Hence, lncRNAs possess an important therapeutic potential and their function requires to be investigated.
The role of LINC00163 has not been explored before. In our study, we focused to elucidate the potential functions of LINC00163 and illustrate its molecular mechanism. Our findings demonstrated that LINC00163 serves as a tumor suppressor in lung cancer by transcriptionally increasing TCF21 expression.
Materials and methods
Tissue collection
59 lung cancer tissues (all patients were NSCLC) and their corresponding adjacent normal tissues were obtained from Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. None of them received radiotherapy or chemotherapy before surgery. These samples were divided into two groups according to the expression value (median value) of LINC00163 or TCF21. This work was approved by the Ethics Committee of our hospital. And patients signed the informed consent.
Cell culture and transfection
Lung cancer cell lines (H1975, SPC-A1, A549, HCC827 and H1299) and the human immortalized normal epithelial cell line (BEAS-2B) were from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured with RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS, Thermo Fisher Scientific), 100 U/mL penicillin, and 100 mg/mL streptomycin (Thermo Fisher Scientific) at 37°C in a 5% CO2 atmosphere.
siRNAs against LINC00163 (5’-GCGAGUGCUCGGAGAACAAGC-3’), ARID1a (5’-AGUUCUAGGUUCAGUUGAAGU-3’) or TCF21 (5’-AGUGCAAUAUGUAAUUAUAAA-3’) and negative control were from from Thermo Fisher Scientific. For LINC00163 overexpression, LINC00163 sequence was constructed into pcDNA3 vector. siRNAs (50 nM) or pcDNA3-LINC00163 (1 μg) was transfected into cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
qRT-PCR
Total RNA was obtained from tissues or cell lines using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Reverse transcription was performed using DNA Reverse Transcription kit (Takara, Dalian, China). qRT-PCR was completed using SYBR Green Kit (Thermo Fisher Scientific) on ABI 7500 real-time polymerase chain reaction system. Relative expression was normalized to GAPDH and calculated based on the 2-ΔΔCt method.
Cell proliferation assays
2000 cells per well were seeded into 96-well plates (Corning Incorporated, Corning, NY, USA) and cultured for indicated days. Then cell proliferation was measured using the Cell Counting Kit-8 (CCK-8) kit (MedChemExpress, Monmouth Junction, NJ, USA) according to the manufacturer’s instructions.
For colony formation assay, 1000 cells were seeded into the 6-well plates and cultured for 14 days. Then colonies were fixed with methanol and stained with 0.5% crystal violet. Colony numbers were counted under a light microscope (IX71, Olympus, Tokyo, Japan).
Transwell assay
Transwell assay were used to determine cell migration and invasion. In migration assay, 2 × 104 cells in serum-free medium were seeded into the upper chamber (8 μm pore, Corning). The lower chamber was fixed with 600 μl serum-containing medium. After cultured for 2 days, the migrated cell number in the lower chamber was fixed and with methanol and stained with 0.5% crystal violet. Then cell number was counted using a light microscope. For invasion assay, the upper chamber was pre-coated with Matrigel and other steps were the same as migration assay.
Flow cytometric analysis of apoptosis
Apoptosis was determined using fluorescein isothiocyanate-Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s instructions.
Chromatin isolation by RNA purification (CHiRP)
The probe for this assay was designed online (https://www.biosearchtech.com/). This experiment was carried out according to a previous study [8]. In brief, biotin-labeled anti-sense DNA probes for LINC00163 (#1: 5’-AACTGGTCTGGATGCCACAG-3’; #2: 5’-AGCACAGTTGGAGACATCAC-3’ and #3: 5’-GATTCTGTGGTGTGTCGGTG-3’) were synthesized by Sangon (Shanghai, China). 1 × 106 Cells were crossed with 1% formaldehyde and quenched with 0.125 M glycine buffer for 5 min. Then nuclei were lyzed using lysis buffer (50 mM Tris, pH 7.0, 10 mM EDTA, 1% SDS, DTT, PMSF, protease inhibitor and RNase inhibitor). Genomes were sonicated three times into fragments 300-500 bp in length. Then genome fragments were incubated with biotin-labeled probes and enriched using Streptavidin-magnetic C1 beads. Finally, the precipitated DNA fragments were eluted from beads and analyzed by qRT-PCR.
RNA pulldown
Biotin-labeled LINC00163 and control RNAs were transcribed using biotin RNA labeling mix (Roche) in vitro. Then the cell lysates were incubated with Biotin-labeled LINC00163 and control RNAs for 4 h at 4°C, followed by addition of Streptavidin-coupled Dynabeads (Thermo Fisher Scientific Logo). Then the precipitants were analyzed by western blotting.
Tumor xenografts in nude mice
4-weeks old female BALB/c nude mice were subcutaneously injected with 5 × 106 LINC00163-stable-overexpressing or control A549 cells in the flank. Tumor volume was measured every 4 days and tumor weight was determined at the endpoint. Tumor volume was calculated according to the formula: Volume = Length × Width2 × 0.5. This experiment was approved by the Ethics Committee of our hospital.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using an EZ ChIP Kit (Millipore, Bedford, MA) in accordance with the manufacturer’s instructions and described before [15].
RNA immunoprecipitation (RIP) assay
RIP assay was performed to detect the interaction between LINC00163 and ARID1A according to a previous study [15].
Statistical analysis
All statistical analysis was carried out using GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Each experiment was repeated three times. Results were displayed as mean ± SD. Statistically significant differences were calculated using Student’s t-test or one-way analysis of variance. Survival analysis was completed using Kaplan-Meier procedure, followed by the log-rank test. P<0.05 was defined statistically significant.
Results
LINC00163 level is downregulated in lung cancer
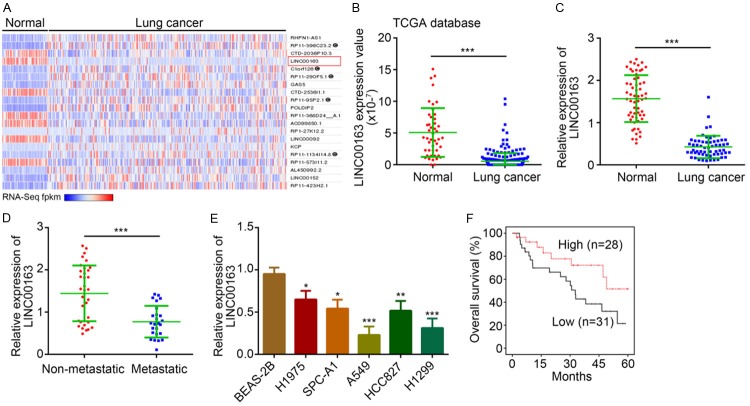
To explore how lncRNA regulates the development of lung cancer, we analyzed differentially expressed lncRNAs in lung cancer tissues and normal control tissues by bioinformatics (http://lncrnator.ewha.ac.kr/index.htm). As shown, LINC00163 was a highly downregulated lncRNAs in lung cancer tissues compared to normal tissues according to the TCGA database (Figure 1A and 1B). To validate it, we analyzed the expression of LINC00163 in 59 pairs of lung cancer tissues and matched normal controls. We observed a similar result that LINC00163 level was significantly downregulated in lung cancer tissues (Figure 1C). Moreover, in metastatic lung cancer tissues, the expression of LINC00163 was lower (Figure 1D). Similarly, the expression of LINC00163 was still reduced in lung cancer cell lines (Figure 1E). We further divided these tissues into two groups based on LINC00163 median value. Kaplan-Meier method was used to plot the survival rate and revealed that LINC00163 downregulation in lung cancer patients indicated a poor prognosis (Figure 1F). Taken together, LINC00163 downregulation might be involved in lung cancer development.
Figure 1.
LINC00163 level is downregulated in lung cancer. A. Expression heatmap of lncRNAs in lung cancer tissues (n=245) and normal tissues (n=46) according to the TCGA database was presented. B. According to the TCGA database, LINC00163 level was significantly reduced in lung cancer tissues (n=245) compared to normal tissues (n=46). C. Total RNAs were extracted from 59 pairs of lung cancer and normal tissues. And LINC00163 level was analyzed using qRT-PCR. D. LINC00163 level was analyzed using qRT-PCR in metastatic (n=34) and non-metastatic (n=25) lung cancer tissues. E. Total RNAs were extracted from described cell lines and LINC00163 expression was determined by qRT-PCR. F. Survival rate analysis was determined according to LINC00163 median expression level in the 59 lung cancer patients. *P<0.05, **P<0.01 and ***P<0.001.
LINC00163 overexpression suppresses proliferation, migration and invasion in lung cancer
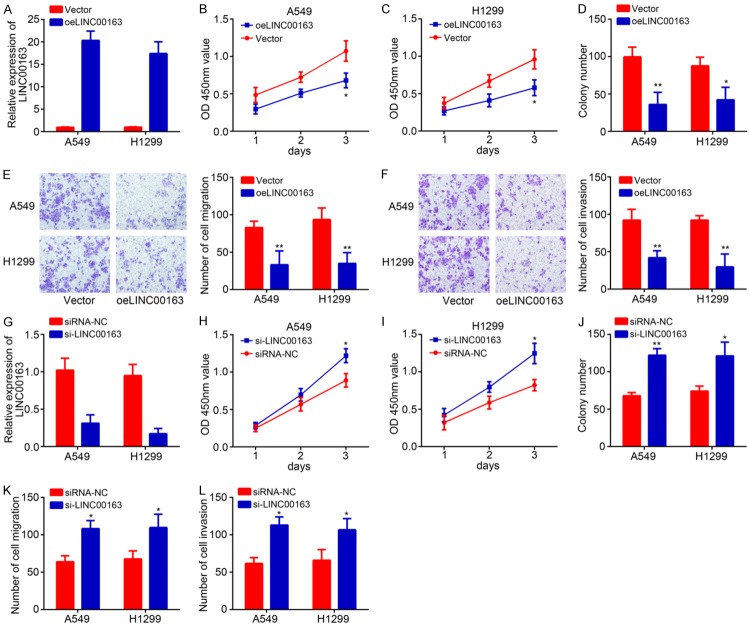
To further investigate the function of LINC00163, we overexpressed it in A549 and H1299 cells due to its lowest expression in these two cell lines. qRT-PCR results validated the successful transfection of pcDNA3-LINC00163 in A549 and H1299 cells (Figure 2A). CCK8 assay was further carried out to evaluate cell proliferation. Results indicated that LINC00163 overexpression markedly suppressed the proliferation of A549 and H1299 cells (Figure 2B and 2C), which was further validated by colony formation assay (Figure 2D). We have observed a negative correlation between LINC00163 level and metastasis. Thus, we then conducted Transwell assay. The results showed that LINC00163 overexpression led to reduced numbers of migrated and invaded A549 and H1299 cells (Figure 2E and 2F), indicating LINC00163 suppresses metastasis. Moreover, we knocked down LINC00163 in A549 and H1299 cells (Figure 2G), followed by CCK8, colony formation and transwell assasy. We found that LINC00163 silencing significantly promoted proliferation, colony formation, migration and invasion of A549 and H1299 cells (Figure 2H-L). In sum, our results demonstrated that LINC00163 inhibits lung cancer cell growth, migration and invasion.
Figure 2.
LINC00163 overexpression suppresses proliferation, migration and invasion in lung cancer. A. LINC00163 was overexpressed in A549 and H1299 through transfection with pcDNA3-LINC00163. And the efficiency of LINC00163 overexpression was analyzed by qRT-PCR. B and C. A549 and H1299 cells were transfected with pcDNA3-LINC00163. And CCK8 assay was used to clarify cell proliferation at different time points including 1 day, 2 days and 3 days. oeLINC00163 indicates overexpression of LINC00163. D. A549 and H1299 cells were transfected with pcDNA3-LINC00163. Colony formation assay was used to analyze cell proliferation. E and F. After transfection, images and quantification of cell migration and invasion of A549 and H1299 were determined by transwell assay. G. LINC00163 was knocked down in A549 and H1299 cells by transfection with si-LINC00163. The efficiency of knockdown was analyzed by qRT-PCR. siRNA-NC: siRNA negative control. H and I. After infection, CCK8 assay was used to analyze cell proliferation in A549 and H1299 cells. J. Colony formation assay indicated that LINC00163 silencing increased colony number. K and L. Cell migration and invasion was determined by Transwell assay after LINC00163 silencing. *P<0.05 and **P<0.01.
LINC00163 promotes lung cancer cell apoptosis and impairs tumor growth in vivo
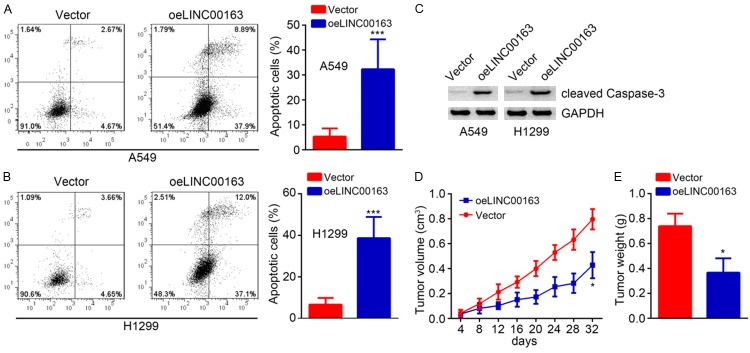
Afterwards, the apoptosis of A549 and H1299 were analyzed. Results showed that LINC00163 upregulation significantly increased the apoptotic cell rates (Figure 3A and 3B). Consistently, LINC00163 overexpression promoted the expression of cleaved Caspase-3 (Figure 3C). Moreover, through in vivo xenograft assay using stably LINC01296-overexpressing A549 cells, we found that LINC00163 upregulation inhibited tumor volumes (Figure 3D) and decreased tumor weights (Figure 3E). Collectively, these data suggest that LINC00163 promotes apoptosis and prevent lung cancer growth in vivo.
Figure 3.
LINC00163 promotes lung cancer cell apoptosis and impairs tumor growth in vivo. A and B. LINC00163 overexpression gave rise to increased apoptotic A549 and H1299 cells. C. Western blotting was used to detect the expression of cleaved Caspase-3. D. Tumor volumes were plotted. E. Tumor weight was determined. LINC00163 overexpression suppressed the tumor weight. *P<0.05 and ***P<0.001.
LINC00163 promotes TCF21 expression
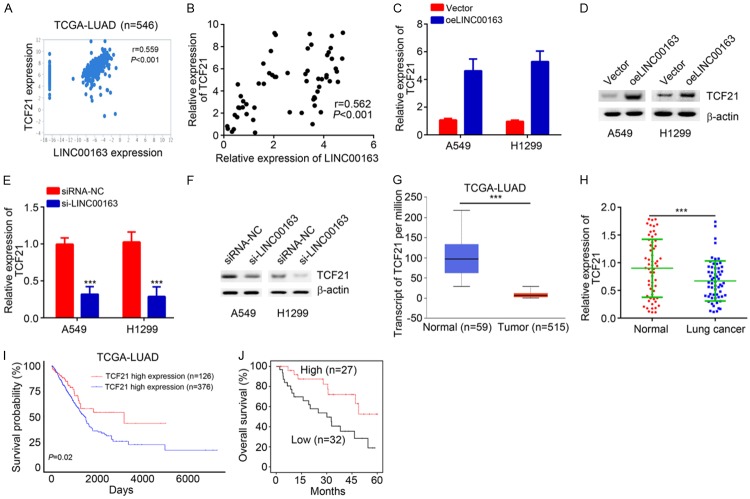
Then we sought to determine the mechanism of LINC00163. We analyzed the co-expressed mRNAs with LINC00163 according to the TCGA database. Results indicated that LINC00163 was positively correlated with TCF21 in lung cancer tissues (Figure 4A). qRT-PCR also confirmed that LINC00163 expression was positively correlated with TCF21 expression in the 59 lung cancer tissues (Figure 4B). Furthermore, through qRT-PCR and western blotting analysis, upregulation of LINC00163 promoted the expression of TCF21 in A549 and H1299 cells (Figure 4C and 4D), and vice versa (Figure 4E and 4F). Interestingly, the TCGA database revealed that TCF21 was downregulated in lung cancer tissues (Figure 4G). qRT-PCR analysis also indicated LINC00163 was reduced in lung cancer tissues (Figure 4H), suggesting TCF21 might also suppress lung cancer development. Finally, analysis based on TCGA database and Kaplan-Meier method revealed that TCF21 downregulation in lung cancer tissues correlates with a poor prognosis (Figure 4I and 4J). These data demonstrated that LINC00163 promotes TCF21 expression and TCF21 might also suppress lung cancer progression.
Figure 4.
LINC00163 promotes TCF21 expression. A. A positive correlation between LINC00163 and TCF21 expression in lung cancer tissues according to TCGA database. B. qRT-PCR analysis showed that there was a positive correlation between LINC00163 and TCF21 expression in lung cancer tissues. C and D. After LINC00163 overexpression, the expression of TCF21 was analyzed by qRT-PCR and western blot in lung cancer cells. E and F. After transfection with si-LINC00163, the expression of TCF21 was determined by qRT-PCR and western blot in lung cancer cells. G. TCF21 expression was downregulated in lung cancer tissues according to TCGA database. H. TCF21 expression in lung cancer tissues and normal tissues was measured by qRT-PCR. I. Survival rate analysis based on TCF21 expression according to TCGA database. J. Higher TCF21 expression indicated higher survival rate by Kaplan-Meier analysis. ***P<0.001.
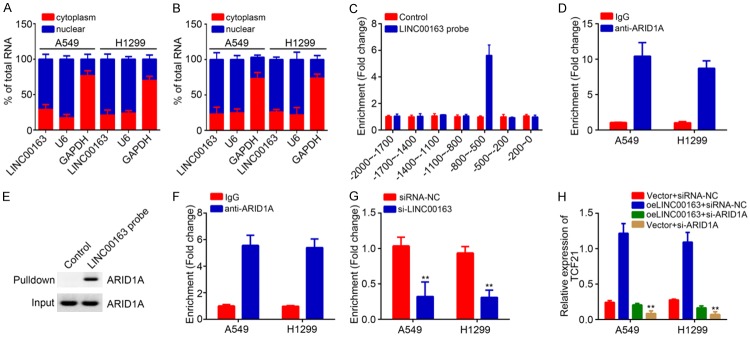
LINC00163 recruits ARID1A to initiate TCF21 expression
In order to further explore how LINC00163 regulates TCF21 expression, we first analyzed the localization of LINC00163 in lung cancer cells. qRT-PCR results showed that LINC00163 was mainly located in the nucleus of A549 and H1299 cells (Figure 5A). Moreover, in LINC00163 ectopically expressing A549 and H1299 cells, LINC00163 was also mainly located in the nucleus (Figure 5B). A previous study has revealed that lncRNA could regulate gene expression transcriptionally [16]. Thus, we speculated LINC00163 might also utilize a similar mechanism to promote TCF21 expression. Notably, CHIRP assay indicated that LINC00163 could enrich on the promoter region of TCF21 promoter in A549 cells (Figure 5C). Through RIP and RNA pulldown assay, we found that LINC00163 interacted with the chromatin remodeler ARID1A (Figure 5D and 5E) [15], impelling us to guess that LINC00163 cooperates with ARID1A to regulate TCF21 expression. Surprisingly, ChIP results indicated that ARID1A also enriched on the same region of TCF21 promoter as LINC00163 (Figure 5F). And LINC00163 silencing led to reduced enrichment of ARID1A on TCF21 promoter (Figure 5G). Moreover, knockdown of ARID1A also significantly suppressed the expression of TCF21 in lung cancer cells (Figure 5H). Taken together, above data suggest that LINC00163 interacts with ARID1A to initiate TCF21 expression in lung cancer.
Figure 5.
LINC00163 recruits ARID1A to initiate TCF21 expression. A. The subcellular localization of LINC00163 in cytoplasm and nuclear was analyzed by qRT-PCR. B. The subcellular localization of LINC00163 in LINC00163-overexpressing A549 and H1299 cells was analyzed by qRT-PCR. C. Chromatin isolation by RNA purification (ChIRP) assay using LINC00163 biotin probes showed that LINC00163 was enriched on the promoter of TCF21 in A549 cells. The biotin-labeled probes against LINC00163 were incubated with the ultrasonically treated cell lysates. Then the precipitated DNA fragment was isolated and analyzed by qPCR. D. RIP assay showed that LINC00163 interacts with ARID1A. E. The interaction between LINC00163and ARID1A in lung cancer was analyzed by RNA pulldown assay. A549 cell lysates were used. F. ChIP assay showed that ARID1A was enriched on the promoter of TCF21. G. ChIP assay showed that LINC00163 silencing inhibited the enrichment of ARID1A on TCF21 promoter. H. qRT-PCR analysis for TCF21 expression in described cells. **P<0.01.
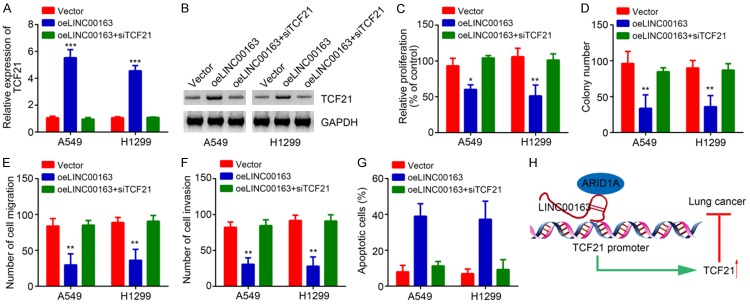
Knockdown of TCF21 rescues the proliferation, migration and invasion of lung cancer cells
Finally, we sought to determine whether TCF21 works downstream of LINC00163. We knocked down TCF21 in LINC00163-overexpressing A549 and H1299 cells to restore its level (Figure 6A and 6B). CCK8 and colony formation assays showed that LINC00163 overexpression suppressed lung cancer cell proliferation (Figure 6C and 6D). However, knockdown of TCF21 reversed the effect of LINC00163 overexpression (Figure 6C and 6D). Similarly, Transwell assay also demonstrated that LINC00163 overexpression-induced inhibition of migration and invasion was reversed by TCF21 knockdown (Figure 6E and 6F). Finally, we also observed that knockdown of TCF21 decreased the apoptotic A549 and H1299 cells (Figure 6G). In sum, our results demonstrated that LINC00163 cooperates with ARID1A to initiate TCF21 expression and suppress lung cancer development (Figure 6H).
Figure 6.
Knockdown of TCF21 rescues the proliferation, migration and invasion of lung cancer cells. A. After transfection with described plasmids, qRT-PCR was used to analyze TCF21 expression. B. Western blotting was utilized to test TCF21 expression in described cells. C. CCK8 assay was used to analyze cell proliferation in described cells. D. Colony formation assay was utilized to detect proliferation potential. E and F. Transwell assay was used to analyze cell migration and invasion. G. Apoptosis was determined after transfection with indicated plasmids in A549 and H1299 cells. H. Work model. LINC00163 recruits ARID1A to promote TCF21 expression, leading to inhibition of lung cancer cell proliferation, migration and invasion. *P<0.05, **P<0.01 and ***P<0.001.
Discussion
In the past years, the important functions of lncRNAs in human cancer have drawn increasing attentions [17]. In our study, we illustrated the function of LINC00163 in lung cancer. We showed that LINC00163 expression was significantly downregulated in lung cancer tissues and cell lines. Moreover, LINC00163 expression was reversely correlated with lung cancer metastasis. Importantly, our results suggested that LINC00163 downregulation in lung cancer patients predicts poor prognosis. Functional results indicated that LINC00163 suppresses the proliferation, migration and invasion of lung cancer whereas promoting apoptosis. Thus, our research demonstrated the correlation between LINC00163 expression and lung cancer progression.
In lung cancer, numerous lncRNAs are reported to work as oncogenes or tumor suppressors [18,19]. For example, lncRNA SNHG15 promotes NSCLC cell proliferation and invasion through upregulating CDK14 expression [20]. LINC00707 is shown to enhance proliferation of lung adenocarcinoma cells via regulating Cdc42 expression [21]. In addition, lncRNA HOTAIR silencing increases the drug resistance of NSCLC cells by activating autophagy [22]. The functions of lncRNAs in lung cancer still remain further investigation. In our study, we found that many undefined lncRNAs were differentially expressed in lung cancer tissues according to bioinformatics analysis. We focused on the study of LINC00163 function. Through CCK8 and colony formation assay, we found that LINC00163 suppresses the proliferation of lung cancer cells. Transwell assay demonstrated that LINC00163 ectopic expression inhibits the migration and invasion of lung cancer cells while promoting apoptosis in vitro. Additionally, in vivo assay also demonstrated that LINC00163 prevents lung cancer growth in nude mice. Therefore, we concluded that LINC00163 inhibits lung cancer tumorigenesis. We for the first time validated that LINC00163 acts as a tumor suppressor in lung cancer.
Subsequently, we researched the target gene of LINC00163. We found that LINC00163 expression was positively correlated with TCF21 in lung cancer tissues. TCF21 is a transcription factor and reported to suppress progression of various cancers, including lung cancer [23]. For instance, upregulation of TCF21 inhibits the metastasis of esophageal squamous cell carcinoma [24]. TCF21 also suppresses colorectal cancer development by inhibiting PI3K/AKT pathway [25,26]. Additionally, downregulation of TCF21 predicts poor prognosis in clear cell renal cell carcinoma [27]. Nevertheless, the mechanism regulating TCF21 expression remains poorly understood. In our work, we showed that LINC00163 promotes TCF21 expression transcriptionally. We demonstrated that LINC00163 directly enriched on the promoter of TCF21. Moreover, we demonstrated that LINC00163 recruits ARID1A to TCF21 promoter. ARID1A is a chromatin remodeler and also reported to suppress progression of bladder cancer [19], breast cancer [28] and gastric cancer [29]. Our results showed that deposition of ARID1A by LINC00163 led to increased expression of TCF21 in lung cancer. Furthermore, in accordance with previous work [23], our study also indicated that TCF21 is downregulated in lung cancer and predicts poor prognosis. Finally, we performed rescue assays, we demonstrated that TCF21 inhibition reversed the suppression of lung cancer cell proliferation, migration and invasion by LINC00163 overexpression.
Taken together, the data in our study confirm that LINC00163 is downregulated in lung cancer and suppresses tumor progression through ARID1A-dependent TCF21 expression, indicating a valuable treatment strategy for lung cancer.
Acknowledgements
We thank all patients involved in this study.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Evans M. Lung cancer: needs assessment, treatment and therapies. Br J Nurs. 2013;22:S15–16. S18, S20–22. doi: 10.12968/bjon.2013.22.Sup17.S15. [DOI] [PubMed] [Google Scholar]
- 3.Anagnostou VK, Brahmer JR. Cancer immunotherapy: a future paradigm shift in the treatment of non-small cell lung cancer. Clin Cancer Res. 2015;21:976–984. doi: 10.1158/1078-0432.CCR-14-1187. [DOI] [PubMed] [Google Scholar]
- 4.Tang LX, Chen GH, Li H, He P, Zhang Y, Xu XW. Long non-coding RNA OGFRP1 regulates LYPD3 expression by sponging miR-124-3p and promotes non-small cell lung cancer progression. Biochem Biophys Res Commun. 2018;505:578–585. doi: 10.1016/j.bbrc.2018.09.146. [DOI] [PubMed] [Google Scholar]
- 5.Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, Du Y, Gao G, Tian Y, He L, Fan Z. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu P, Du Y, Wu J, Qin X, Chen R, Tian Y, Fan Z. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 9.Ye B, Liu B, Yang L, Zhu X, Zhang D, Wu W, Zhu P, Wang Y, Wang S, Xia P, Du Y, Meng S, Huang G, Wu J, Chen R, Tian Y, Fan Z. LncKdm2b controls self-renewal of embryonic stem cells via activating expression of transcription factor Zbtb3. EMBO J. 2018;37 doi: 10.15252/embj.201797174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L, Wei B, Hui H, Sun Y, Liu Y, Yu X, Dai J. Positive feedback loop of lncRNA LINC01296/miR-598/Twist1 promotes non-small cell lung cancer tumorigenesis. J Cell Physiol. 2018 doi: 10.1002/jcp.27235. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Jin L, Cai Q, Wang S, Wang S, Mondal T, Wang J, Quan Z. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018;9:1017. doi: 10.1038/s41419-018-1064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long B, Li N, Xu XX, Li XX, Xu XJ, Liu JY, Wu ZH. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun. 2018;505:561–568. doi: 10.1016/j.bbrc.2018.09.160. [DOI] [PubMed] [Google Scholar]
- 13.Zhang K, Li Y, Qu L, Ma X, Zhao H, Tang Y. Long noncoding RNA Sox2 overlapping transcript (SOX2OT) promotes non-small-cell lung cancer migration and invasion via sponging microRNA 132 (miR-132) Onco Targets Ther. 2018;11:5269–5278. doi: 10.2147/OTT.S168654. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Wang H, Wang J, Liang CF, Zhou T. Expression of long non-coding RNA PRAL as a potential biomarker for diagnosis in non-small-cell lung cancer patients is associated with the inhibition of cell proliferation and metastasis. Clin Lab. 2018;64:1341–1348. doi: 10.7754/Clin.Lab.2018.171237. [DOI] [PubMed] [Google Scholar]
- 15.Fang C, He W, Xu T, Dai J, Xu L, Sun F. Upregulation of lncRNA DGCR5 correlates with better prognosis and inhibits bladder cancer progression via transcriptionally facilitating P21 expression. J Cell Physiol. 2018 doi: 10.1002/jcp.27356. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Xu YC, Liang CJ, Zhang DX, Li GQ, Gao X, Fu JZ, Xia F, Ji JJ, Zhang LJ, Li GM, Wu JX. LncSHRG promotes hepatocellular carcinoma progression by activating HES6. Oncotarget. 2017;8:70630–70641. doi: 10.18632/oncotarget.19906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong X, Chen R, Lin H, Lin T, Pan S. lncRNA BG981369 inhibits cell proliferation, migration, and invasion, and promotes cell apoptosis by SRY-related high-mobility group box 4 (SOX4) signaling pathway in human gastric cancer. Med Sci Monit. 2018;24:718–726. doi: 10.12659/MSM.905965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan S, Liu Q, Hu Z, Zhou Z, Wang G, Li C, Xie W, Meng G, Xiang Y, Wu N, Wu L, Yu Z, Bai L, Li Y. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis. 2018;9:450. doi: 10.1038/s41419-018-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Dong HX, Zeng J, Pan J, Jin XY. LncRNA DGCR5 contributes to CSC-like properties via modulating miR-330-5p/CD44 in NSCLC. J Cell Physiol. 2018;233:7447–7456. doi: 10.1002/jcp.26590. [DOI] [PubMed] [Google Scholar]
- 20.Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to accelerate non-small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233:7164–7172. doi: 10.1002/jcp.26543. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Ma T, Ma H, Zou Z, He X, Liu Y, Shuai Y, Xie M, Zhang Z. The long intergenic noncoding RNA 00707 promotes lung adenocarcinoma cell proliferation and migration by regulating Cdc42. Cell Physiol Biochem. 2018;45:1566–1580. doi: 10.1159/000487693. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Jiang C, Yang Y, Guo L, Huang J, Liu X, Wu C, Zou J. Silencing of LncRNA-HOTAIR decreases drug resistance of Non-Small Cell Lung Cancer cells by inactivating autophagy via suppressing the phosphorylation of ULK1. Biochem Biophys Res Commun. 2018;497:1003–1010. doi: 10.1016/j.bbrc.2018.02.141. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17:2080–2090. doi: 10.1080/15384101.2018.1515553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Zhang C, Chen J, Zhang B, Zhang H, Yang X, Liu J, Wu Q. Expression of transcription factor 21 (TCF21) and upregulation its level inhibits invasion and metastasis in esophageal squamous cell carcinoma. Med Sci Monit. 2018;24:4128–4136. doi: 10.12659/MSM.909138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai Y, Duan H, Duan C, Zhu H, Zhou R, Pei H, Shen L. TCF21 functions as a tumor suppressor in colorectal cancer through inactivation of PI3K/AKT signaling. Onco Targets Ther. 2017;10:1603–1611. doi: 10.2147/OTT.S118151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai Y, Duan H, Duan C, Zhou R, He Y, Tu Q, Shen L. Down-regulation of TCF21 by hypermethylation induces cell proliferation, migration and invasion in colorectal cancer. Biochem Biophys Res Commun. 2016;469:430–436. doi: 10.1016/j.bbrc.2015.09.109. [DOI] [PubMed] [Google Scholar]
- 27.Ye YW, Jiang ZM, Li WH, Li ZS, Han YH, Sun L, Wang Y, Xie J, Liu YC, Zhao J, Tang AF, Li XX, Guan ZC, Gui YT, Cai ZM. Down-regulation of TCF21 is associated with poor survival in clear cell renal cell carcinoma. Neoplasma. 2012;59:599–605. doi: 10.4149/neo_2012_076. [DOI] [PubMed] [Google Scholar]
- 28.Guo X, Zhang Y, Mayakonda A, Madan V, Ding LW, Lin LH, Zia S, Gery S, Tyner JW, Zhou W, Yin D, Lin DC, Koeffler HP. ARID1A and CEBPalpha cooperatively inhibit UCA1 transcription in breast cancer. Oncogene. 2018;37:5939–5951. doi: 10.1038/s41388-018-0371-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhu YP, Sheng LL, Wu J, Yang M, Cheng XF, Wu NN, Ye XB, Cai J, Wang L, Shen Q, Wu JQ. Loss of ARID1A expression is associated with poor prognosis in patients with gastric cancer. Hum Pathol. 2018;78:28–35. doi: 10.1016/j.humpath.2018.04.003. [DOI] [PubMed] [Google Scholar]