Abstract
YEATS domain containing 4 (YEATS4) has functions of chromatin modification and transcriptional regulation and is in a gene-amplified region (12q13) in various human cancers. In this study, we tested whether YEATS4 acts as a cancer-promoting gene through its activation/overexpression in gastric cancer (GC). We analyzed 5 GC cell lines and 135 primary tumor samples of GC, which were curatively resected in our hospital. Overexpression of the YEATS4 protein was frequently detected in GC cell lines (5/5 cell lines, 100%) and primary GC tumor tissues (32/135 cases, 23.7%). Knockdown of YEATS4 inhibited proliferation, migration and invasion of GC cells through NOTCH2 down-regulation in a TP53 mutation-independent manner, and induced apoptosis in wild-type TP53 GC cells. Moreover, knockdown of YEATS4 improved chemosensitivity for CDDP and L-OHP. Overexpression of YEATS4 protein significantly correlated with more aggressive lymphatic invasion, larger tumor size, deeper tumor depth, positive lymph node metastasis and recurrence. Patients with YEATS4-overexpressing tumors had a lower overall survival rate than those with non-expressing tumors. Multivariate analysis demonstrated that YEATS4 was independently associated with poor outcomes. These findings suggest that YEATS4 plays a pivotal role in tumor malignant potential through its overexpression and highlight its usefulness as a prognostic factor and potential therapeutic target in GC.
Keywords: YEATS4, overexpression, gastric cancer, TP53, chemosensitivity
Introduction
Gastric cancer (GC) is one of the most common causes of death from cancer worldwide [1]. Recent advances in diagnostic technologies and perioperative management have increased early detection of GC and decreased morbidity and mortality rates [2,3]. However, patients with advanced GC still frequently experience recurrence despite extended radical resections, which results in an extremely poor survival rate [4,5]. Numerous genes have been analyzed to understand the molecular mechanisms of carcinogenesis and improve clinical outcomes for human GCs; however, only a few genes with frequent alterations have been identified [6,7], such as the gene amplifications of MET and ERBB2 [8,9]; hypermethylation of p16 [10,11]; mutations of TP53, APC and E-cadherin [12-14]; oncogenic activation of beta-catenin and K-ras [15,16]; and inactivation of the mismatch repair gene hMLH1, which is associated with microsatellite instability [17]. However, in clinical settings, only a few genes have been used as diagnostic biomarkers and/or therapeutic targets [18-20]. Therefore, we aimed to identify novel genes associated with the progression of GC.
YEATS domain containing 4 (YEATS4) was first reported in glioma by Fischer U in 1997 [21], and is involved in chromatin modification and transcriptional regulation [22]. Oncogenic functions of YEATS4 have recently been reported in various types of cancer, such as non-small cell lung cancer [23], glioblastoma [24], liposarcoma [25], ovarian cancer [26], colorectal cancer [27,28], pancreatic cancer [29] and GC [30]. In addition, YEATS4 is located in chromosome 12q13-15, in which genomic high-copy amplification has been reported to contribute to the progression of several kinds of cancers including liposarcoma [31], sarcoma [32], leukemia [33], lymphoma [34], malignant tumor of salivary gland [35], osteosarcoma [36], glioblastoma [37], nasopharyngeal carcinoma [38] and bladder cancer [39]. However, to date, there has been only one report on YEATS4 molecular function in GC [30]. These findings prompted us to investigate the effects of YEATS4 overexpression and activation in primary GC.
In this study, we attempted to investigate the effects of YEATS4 overexpression and activation in GC. Consequently, we demonstrated that YEATS4 was frequently overexpressed in GC cell lines and primary GC tissues, and the overexpression of YEATS4 was identified as an independent factor predicting a poor prognosis. We also demonstrated that knockdown of YEATS4 expression in YEATS4-overexpressing GC cells suppressed the cell proliferation, migration and invasion and induced apoptosis in a p53-dependent manner. Our results provide evidence that YEATS4 could be an important molecular marker for determining the malignant properties of tumors and a promising therapeutic target in patients with GC.
Materials and methods
Patients and primary samples
All experimental methods were carried out in accordance with relevant guidelines (such as REMARK guidelines and regulations). Written informed consent was obtained from all patients to use their tissue specimens and blood samples. This study was approved by the institutional review board of Kyoto Prefectural University of Medicine. No patients underwent endoscopic mucosal resection, palliative resection, preoperative chemotherapy or radiation procedures. Patients with synchronous or metachronous multiple cancers were excluded.
Paraffin-embedded primary GC tissue samples were collected from 135 consecutive GC patients who underwent curative gastrectomy at Kyoto Prefectural University of Medicine (Kyoto, Japan). Tumor stages were assessed according to the Union for International Cancer Control classification system [40]. In all cases, at least two pathologists agreed with the pathological observations and confirmed the diagnosis. Relevant clinical and survival data were available for all patients (Supplementary Table 1).
Cell lines
Five GC cell lines (NUGC4, HGC27, MKN7, MKN28, MKN45), a wild-type p53 osteosarcoma cell line, a p53-null osteosarcoma cell line and a fibroblast cell line were used in this study. All cell lines were purchased from RIKEN Cell Bank (Tsukuba, Japan), the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan) and Cell Lines Service (Eppelheim, Germany). All cell lines were identified by Short Tandem Repeat profiling. No mycoplasma contamination was detected in any of the cultures. HGC27 and U2OS cells were cultured in Dulbecco’s Minimum Essential Medium (Sigma, St. Louis, MO). SaOS2 cells were cultured in HyClone McCoy’s 5A Medium (GE Healthcare Life Sciences, Amersham, UK). The other cells were cultured in Roswell Park Memorial Institute 1640 medium (Sigma, St. Louis, MO). All mediums were supplemented with 10% fetal bovine serum (Trace Scientific, Melbourne, Australia). All cells were cultured in 5% carbon dioxide at 37°C in a humidified chamber.
Quantification of mRNA by qRT-PCR
Total RNA was also extracted from four 15-μm-thick slices of formalin-fixed and paraffin-embedded tissue samples (60 μm thickness) and cell lines using a Recover All Total Nucleic Acid Isolation Kit (Ambion, Austin, TX) and an RNeasy Mini Kit (Qiagen, Valencia, CA), respectively. The total RNA of normal organs was purchased from Takara Bio Inc., Shiga, Japan (Human Total RNA Master Panel II (Cat. No. 636643) and Human Liver Total RNA (Cat. No. 636531)) and BioChain Institute Inc., CA, USA (Human Oesophagus Total RNA (Cat. No. R1234106-50)). Single-stranded complementary DNA generated from total RNA was amplified with primers specific for each gene, as described below. The abundance of mRNAs was quantified by qRT-PCR using a Human TaqMan Gene Expression Assays (Hs00232423_m1 for YEATS4; Applied Biosystems, Foster City, CA). The reverse transcription reaction was conducted with a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). qPCR was run on a StepOnePlus PCR system (Applied Biosystems). Cycle threshold (CT) values were calculated with StepOne Software v2.0 (Applied Biosystems). The results of gene expression were calculated as a ratio between YEATS4 and an internal reference gene (Hs01060665_g1 for β-actin; Applied Biosystems) that provides a normalization factor for the amount of RNA isolated from specimen. This assay was performed in triplicate for each sample.
Western blot analysis
Anti-YEATS4 mouse monoclonal antibody (Cat. No. sc-393708) was purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology, CA). Anti-ACTB, p21, AKT, phospho-AKT, Notch2, Caspase-3, Cleaved Caspase-3, and PARP were purchased from Cell Signaling Technology (Cat. No. 4970, 2946, 4691, 4060, 5732, 9662, 9664 and 9532, respectively; Cell Signaling Technology, USA). Cells were lysed in Tris buffer (50 mmol/l, pH 7.5) containing 150 mmol/L NaCl, 1 mmol/L EDTA, 0.5% NP-40, 10% glycerol, 100 mmol/L NaF, 10 mmol/L sodium pyrophosphate, 2 mmol/L Na2VO3 and a protease inhibitor cocktail (Roche, Tokyo, Japan), and their proteins were extracted using M-PER Mammalian® Protein Extraction Reagent (Thermo Scientific, USA). Twenty micrograms of proteins per lane were loaded for electrophoresis.
Immunofluorescence staining
For immunofluorescence staining, cells were subsequently fixed with 4% paraformaldehyde at room temperature for 20 min, permeabilized in 0.25% Triton X-100 in PBS, and incubated in blocking buffer containing 1% bovine serum albumin. Cells were then incubated with the anti-YEATS4 and anti-Notch2 antibody at room temperature for 1 h. After three washes in PBS, cells were incubated with Alexa Fluor 488-labeled goat anti-mouse and Alexa Fluor 594-labeled goat anti-rabbit secondary antibodies at room temperature for 1 h. After three washes in PBS, cells were incubated with rhodamine phalloidin and 40,6-diamidino-2-phenylindole (DAPI) for 30 min. DAPI staining was performed and slides were mounted with Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA, USA). The distribution of YEATS4 and Notch2 proteins was examined using BZ-X700 (Keyence, Tokyo, Japan).
Loss-of-function by siRNA and cell growth analysis
For loss-of-function analysis by the knocking down of endogenous gene expression, each of the siRNAs targeting YEATS4 (custom made SiRNA, 5’-CCUGUAACCCUGUAUCAUUUGCUAA-3’ for SiRNA-YEATS4 1, 5’-GCAACAAUUAUUGACAACAUCUCGU-3’ for SiRNA-YEATS4 2, and 5’-CAGAAUUUGCAGAGCUUGAAGUGAA-3’ for SiRNA-YEATS4 3; Sigma Genosys, Tokyo, Japan) or Luciferase (Luc) 5’-CGUACGCGGAAUACUUCGA-3’ (Sigma Genosys) was transfected into cells (10 nmol/l) using Lipofectamine RNAiMAX (Invitrogen Corporation, Carlsbad, CA) according to the manufacturer’s instructions. The knockdown of a target gene was confirmed by qRT-PCR and Western blotting.
To measure cell growth, the number of viable cells at various time points after transfection was assessed by a colorimetric water-soluble tetrazolium salt assay (Cell Counting Kit 8; Dojindo Laboratories, Kumamoto, Japan). Cell viability was determined by reading the optical density at 450 nm. The cell cycle was evaluated 72 h after transfection by fluorescence-activated cell sorting (FACS), as described elsewhere [41,42]. For the FACS analysis, harvested cells were fixed in 70% cold ethanol and treated with RNase A and propidium iodide. Samples were analyzed on a Becton Dickinson AccuriTM C6 Flow Cytometer (Becton Dickinson, San Jose, CA).
Apoptotic cell analysis
At 72 h after transfection, the SiRNA-transfected cells were harvested and stained with fluorescein isothiocyanate-conjugated annexin V and phosphatidylinositol using an Annexin V Kit (Beckman Coulter, Brea, CA). A Becton Dickinson AccuriTM C6 flow cytometer was used to analyze the proportion of apoptotic cells [43].
To assess the chemoresistance of GC cell lines to CDDP and L-OHP, NUGC4 (wild-type TP53) and HGC27 (mutant-type TP53) that were transfected with SiRNA-YEATS4 and its control were plated onto a 6-well plate (6 × 104 cells per well) and incubated overnight under normal culture conditions. The cells were then incubated with CDDP (2 μM) and L-OHP (5 μM). At 48 h after the addition of anticancer drug, apoptosis cell analysis was performed as mentioned above.
Trans-well migration and invasion assays
Transwell migration and invasion assays were conducted in 24-well modified Boyden chambers (Transwell chambers, BD Transduction, Franklin Lakes, NJ). The upper surface of 6.4-mm-diameter filters with 8-µm pores was precoated with (invasion assay) or without (migration assay) Matrigel (BD Transduction). The SiRNA transfectants (5 × 105 cells per well) were transferred into the upper chamber. Following 22 h of incubation, the migrated or invasive cells on the lower surface of the filters were fixed and stained with Diff-Quik stain (Sysmex, Kobe, Japan), and stained cell nuclei were counted directly in triplicate [44]. We assessed the ability of the cells to move through extracellular matrices by calculating the number of cells, which is the ratio of the percentage invasion through the matrigel-coated filters relative to migration through the uncoated filters of test cells over that in the control cells.
Immunohistochemistry
Tumor samples were fixed with 10% formaldehyde in PBS, embedded in paraffin, sectioned into 5 μm-thick slices and subjected to immunohistochemical staining of YEATS4 protein using the avidin-biotin-peroxidase method [45,46]. In brief, after deparaffinization, endogenous peroxidases were quenched by incubating the sections for 20 min in 3% H2O2. Antigen retrieval was performed by heating the samples in 10 mmol/L citrate buffer (pH 6.0) at 95°C for 60 min. After treatment with Block Ace (Dainippon Sumitomo Pharmaceutical, Osaka, Japan) for 30 min at room temperature, the sections were incubated at room temperature for 60 min and at 4°C overnight with anti-YEATS4 (1:500) antibody. The avidin-biotin-peroxidase complex system (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA) was used for color development with diaminobenzidine tetrahydrochloride. The slides were counterstained with Mayer’s hematoxylin. A formalin-fixed GC cell lines overexpressing YEATS4 (NUGC4), in which > 50% of cells showed staining of YEATS4 protein, was used as a positive control, whereas NUGC4 cells included without the YEATS4 antibody were used as a negative control.
For scoring YEATS4 expression, the intensity (intensity score: 0 = negative, 1 = weak, 2 = moderate, 3 = strong) was evaluated for each case. Expression of YEATS4 was graded as high expression (Intensity score ≥ 2), or low expression (Intensity score ≤ 1) using high powered (× 200) microscopy. For the scoring of p53 expression, the intensity (intensity score; 0 = negative, 1 = weak, 2 = moderate, 3 = strong) and the percentage of the total cell population (proportion score; 0% ≤ 0 < 30%, 30% ≤ 1 ≤ 100%) that expressed p53 were evaluated for each case. A distinct nuclear immunoreaction in both 3 and ≥ 30% of the cancer cells was judged positive. The remaining cases were judged negative.
Statistical analysis
Clinicopathological categorical variables pertaining to the corresponding patients were analyzed for significance by the Chi-square test or Fisher’s exact test. The differences of non-categorical variables between subgroups were tested with the non-parametric Mann-Whitney U test. For the analysis of survival, Kaplan-Meier survival curves were constructed for groups based on univariate predictors, and differences between the groups were tested with the log-rank test. Univariate and multivariate survival analyses were performed using the likelihood ratio test of the stratified Cox proportional hazards model. Differences were assessed with a two-sided test and were considered significant at the P < 0.05 level.
Results
Overexpression of YEATS4 in GC cell lines
Quantitative RT-PCR analysis was performed to test whether YEATS4 is overexpressed in GC cell lines compared with the normal organs and the fibroblast cell line WI-38 (Figure 1A). YEATS4 mRNA overexpression was observed in all GC cell lines (5 out of 5 lines, 100.0%) and testis compared with normal stomachs, other normal organs and the fibroblast cell line WI-38, suggesting that this gene is a cancer-testis antigen and a target for activation in GC. Expression of YEATS4 protein detected by the YEATS4-specific antibody (5 out of 5 lines, 100.0%) seemed to be correlated with that of YEATS4 mRNA in GC cell lines (Figure 1B). We used representative osteosarcoma cell lines (SaOS2; TP53-null, U2OS; wild-type TP53) and examined the status of a TP53 mutation in the GC cell lines, including these osteosarcoma cell lines by western blotting. These statuses of TP53 mutation in various cell lines are positively associated with their reported status of TP53 mutation in the database (http://p53.free.fr/index.html; W: wild-type TP53, M: mutant TP53). Note that, among TP53-mutated GC cell lines, HGC27 cells have a frameshift mutation.
Figure 1.
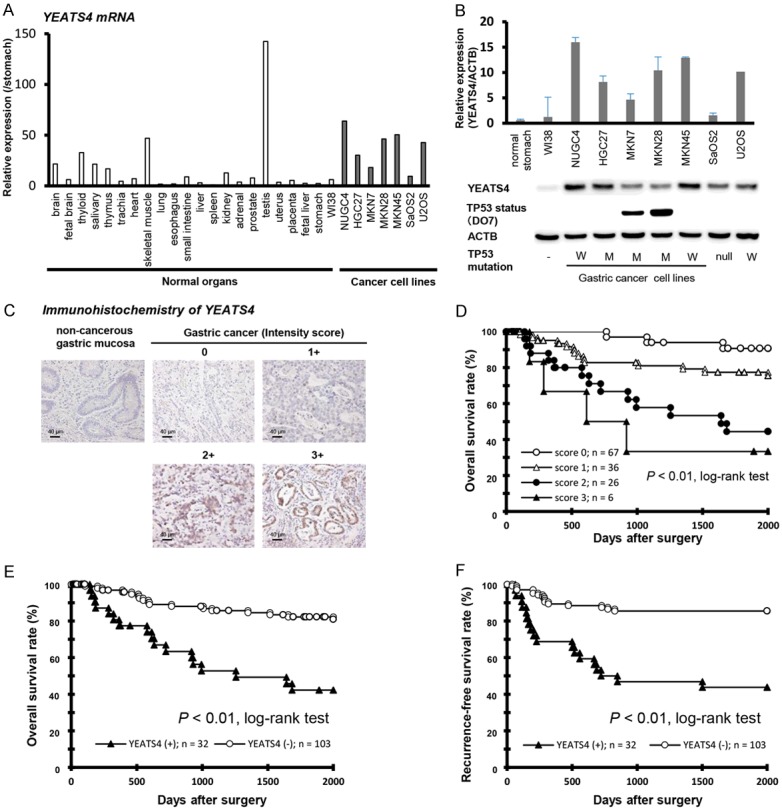
Overexpression of YEATS4 in GC. (A) Expression of YEATS4 mRNA in five GC cell lines compared with the normal organs, fibroblast cell line WI-38, TP53 wild osteosarcoma cells, U2OS and all TP53 null osteosarcoma cell lines, SaOS2. (B) Expression of YEATS4 in five GC cell lines and osteosarcoma cell lines compared with that in a normal stomach (n = 10) and fibroblast cell line WI-38. The level of YEATS4 mRNA was determined by RT-PCR in normal gastric mucosae and a panel of GC cell lines. The status of the TP53 mutation in each cancer cell line was evaluated by western blotting. The status of TP53 was positively associated with the reported status of a TP53 mutation in the database (http://p53.free.fr/index.html, W: wild-type TP53, M: mutant-type TP53). Note that, among TP53-mutated GC cell lines, KATO-III and HGC27 cells have a p53 gene deletion and a frameshift mutation, respectively. (C) Specific immunostaining of YEATS4 in representative primary tumor samples. Based on this result, the intensity scores for YEATS4 staining were determined as follows: 0 = negative, 1 = weak, 2 = medium, 3 = strong. (D) Kaplan-Meier plots depending on the intensity scores of specific immunostaining of YEATS4. The log-rank test was used for statistical analysis. P < 0.05 was considered statistically significant. Overall survival (E) and recurrence-free survival (F) rate of GC patients depended on the intensity scores in YEATS4 expression. Expression of YEATS4 was graded as high expression (intensity scores ≥ 2 for tumor cells showing immunopositivity) or low expression (intensity scores ≤ 1 for tumor cells showing immunopositivity).
Immunohistochemical analysis of YEATS4 expression in primary tumors of GC
Because the YEATS4 protein was overexpressed in all GC cell lines, it was hypothesized that YEATS4 was also highly expressed in GC tissues and would be associated with carcinogenesis and malignant outcomes. We examined the prognostic and clinicopathological significance of YEATS4 expression in primary tumor samples of GC based on the immunohistochemical staining pattern of this protein. YETS4 was mainly observed in the nucleus of cancer cells (Figure 1C). We classified 135 GC samples into positive and negative groups according to the intensity of YEATS4 staining among tumor cells, as described in materials and methods. In primary cases, YEATS4 protein expression was negative in the non-tumorous gastric mucosal cell population (Figure 1C). Kaplan-Meier survival estimates showed that YEATS4 immunoreactivity in tumor cells was significantly associated with a worse overall survival according to the intensity score (Figure 1D). Supplementary Table 1 shows the distribution of patients based on the intensity scores of YEATS4 immunoreactivity of tumor samples. In the score of intensity, the YEATS4 high expression group, with scores of ≥ 2 for tumor cells showing immunopositivity, presented a significantly poorer overall survival (P < 0.01, log-rank test) (Figure 1E) and recurrence-free survival rate (P < 0.01, log-rank test) (Figure 1F) than the low expression group.
Association between YEATS4 protein abundance and clinicopathological characteristics in primary cases of GC
The relationship between the expression of the YEATS4 protein and clinicopathological characteristics is summarized in Table 1. Protein expression of YEATS4 was significantly associated with more aggressive lymphatic invasion, larger tumor size, deeper tumor depth and a higher rate of lymph node metastasis and recurrence, whereas other characteristics were not. Patients with YEATS4-overexpressing tumors had a worse overall rate of survival than those with non-expressing tumors (P = 0.024, log-rank test) in an intensity-dependent manner. When data were stratified for multivariate analysis using both the forward and backward stepwise Cox regression procedures, YEATS4 immunoreactivity in tumor cells as an independent factor predicting a worse overall survival rate (hazard ratio 2.30; 95% confidence interval: 1.053-5.126) along with advanced pathological stage and venous invasion (Table 2).
Table 1.
Association between clinicopathological characteristics and YEATS4 expression in GC patients
| Variables | Intensity | Univariatea | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| High (2, 3) | (n = 32) | Low (0, 1) | (n = 103) | P-value | ||
| Gender | Female | 7 | 17% | 34 | 83% | 0.276 |
| Male | 25 | 27% | 69 | 73% | ||
| Age (60 years old) | < 60 | 12 | 23% | 40 | 77% | 0.937 |
| ≥ 60 | 20 | 24% | 63 | 76% | ||
| Tumor size | < 60 mm | 14 | 16% | 76 | 84% | < 0.001 |
| ≥ 60 mm | 17 | 46% | 20 | 54% | ||
| Lymphatic invasion | ly0, ly1 | 15 | 15% | 87 | 85% | < 0.001 |
| ly2, ly3 | 17 | 53% | 15 | 47% | ||
| Venous invasion | v0, v1 | 25 | 21% | 93 | 79% | 0.122 |
| v2, v3 | 7 | 41% | 10 | 59% | ||
| T factor | T1, T2 | 9 | 11% | 75 | 89% | < 0.001 |
| T3, T4 | 23 | 45% | 28 | 55% | ||
| N factor | N0, N1 | 13 | 13% | 88 | 87% | < 0.001 |
| N2, N3 | 19 | 56% | 15 | 44% | ||
| p Stage | p Stage I, II | 13 | 14% | 81 | 86% | < 0.001 |
| p Stage III | 19 | 46% | 22 | 54% | ||
| Recurrence | Absent | 14 | 13% | 92 | 87% | < 0.001 |
| Present | 18 | 62% | 11 | 38% | ||
Univariate analysis was assessed using the Chi squared test.
NOTE: significant values are in bold.
Table 2.
Univariate and multivariate analyses for survival of GC patients following gastrectomy using the Cox’s proportional hazards model
| Variable | n | Univariatea | Multivariateb | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| P-value | HRc | 95% CId | p-value | |||
| Gender | Male vs. female | 94 vs. 41 | 0.977 | |||
| Age (60 years old) | ≥ 60 vs. < 60 | 83 vs. 52 | 0.056 | |||
| Lymphatic invasion (ly) | Ly (+) vs. ly (-) | 66 vs. 69 | < 0.001 | |||
| Venous invasion (v) | v (+) vs. v (-) | 40 vs. 95 | 0.008 | 7.57 | 1.47-30.8 | 0.007 |
| p Stage (TNM classification) | Stage II, III vs. Stage I | 94 vs. 41 | < 0.001 | 30.3 | 9.84-133 | < 0.001 |
| YEATS4 intensity score | High vs. low | 38 vs. 97 | 0.012 | 2.30 | 1.05-5.12 | 0.037 |
Kaplan-Meier method; significance was determined by a log-rank test.
Multivariate survival analyses were performed using Cox’s proportional hazards model.
HR: Hazard ratio.
CI: Confidence interval.
NOTE: significant values are in bold.
Suppression of cell proliferation by knockdown of YEATS4 and the knockdown effect according to TP53 mutation status
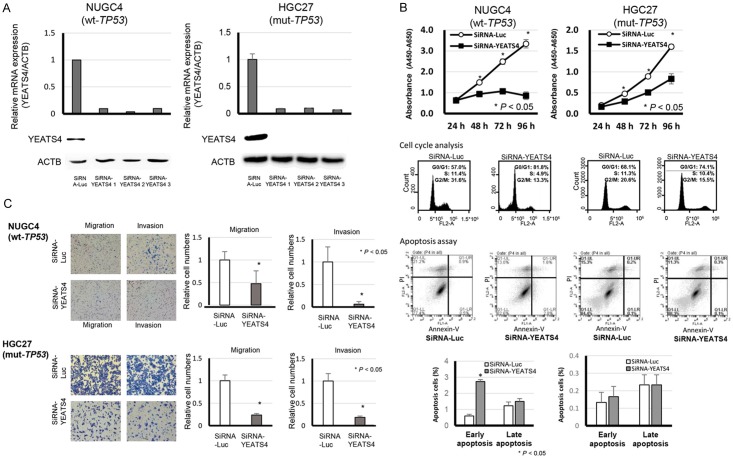
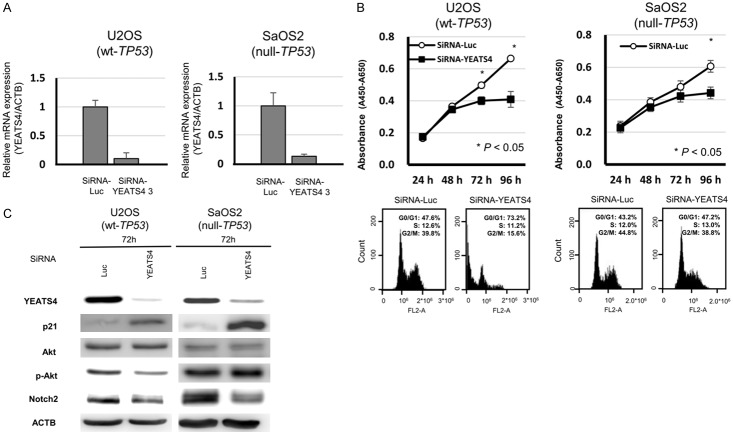
To gain insight into the potential role of YEATS4 as an oncogene whose overexpression could be associated with gastric carcinogenesis, we first performed a cell proliferation assay using siRNAs specific to YEATS4 (SiRNA-YEATS4) to investigate whether knockdown of YEATS4 would suppress the proliferation of GC cells that overexpress YEATS4. In both TP53 wild-type and TP53 mutant-type cell lines, such as NUGC4 (Figure 2A), HGC27 (Figure 2A), MKN45 (Figure 3A), MKN7 (Figure 3A), SaOS2 (Figure 4A) and U2OS (Figure 4A), expression of the YEATS4 protein was more efficiently knocked down 24-72 h after the transient introduction of a YEATS4-specific siRNA (siRNA-YEATS4) compared to a Luciferase-specific siRNA (siRNA-Luc) as a negative control. The proliferation of all these cell lines was significantly lower than with controls after the knockdown of endogenous YEATS4 expression (Figures 2B, 3B and 4B).
Figure 2.
Suppression of malignant behaviors in GC cells by knockdown of YEATS4 and the knockdown effect according to TP53 mutation status. A. Effects of YEATS4 knockdown by siRNA (siRNA-YEATS4) compared with those of the control siRNA in NUGC4 (wild-type TP53) and HGC27 (mutant-type TP53) cell lines. B. Effects of knocking down endogenous YEATS4 on cell proliferation at the indicated times. The results are the means ± SD (bars) for quadruplicate experiments. The Mann-Whitney U-test was used for the statistical analysis: *P < 0.05. Representative results of the population in each phase of the cell cycle in GC cells assessed by FACS at 72 h after treatment with siRNA (upper middle). Representative results of the apoptosis assay in GC cells at 72 h after treatment with siRNA (lower middle). C. Migration and invasion of cells transfected with siRNA targeting YEATS4 in NUGC4 and HGC27. The graphs on the bottom show the means ± SD (bars) of n = 4. The Mann-Whitney U-test test was used for statistical analysis. P < 0.05 was considered statistically significant.
Figure 3.
Suppression of malignant behaviors in GC cells by knockdown of YEATS4 and the knockdown effect according to TP53 mutation status. A. Effects of YEATS4 knockdown by siRNA (siRNA-YEATS4) compared with those of the control siRNA in MKN45 (wild-type TP53) and MKN7 (mutant-type TP53) cell lines. B. Effects of knocking down endogenous YEATS4 on cell proliferation at the indicated times. The results are the means ± SD (bars) for quadruplicate experiments. The Mann-Whitney U-test was used for the statistical analysis: *P < 0.05. Representative results of the population in each phase of the cell cycle in GC cells assessed by FACS at 72 h after treatment with siRNA. Representative results of the apoptosis assay in GC cells at 72 h after treatment with siRNA. C. Western blotting analyses showed that knockdown of YEATS4 by transfection with siRNA-YEATS4 induced the production of p21 and suppressed Notch2 expression. In NUGC4 (wild-type TP53) cells, knockdown of YEATS4 also induced phosphorylation activation of AKT.
Figure 4.
Suppression of malignant behaviors in osteosarcoma cells by knockdown of YEATS4 and the knockdown effect according to TP53 mutation status. A. Effects of YEATS4 knockdown by siRNA (siRNA-YEATS4) compared with those of the control siRNA in U2OS (wild-type TP53) and SaOS2 (null TP53) cell lines. B. Effects of knocking down endogenous YEATS4 on cell proliferation at the indicated times. The results are the means ± SD (bars) for quadruplicate experiments. The Mann-Whitney U-test was used for the statistical analysis: *P < 0.05. Representative results of the population in each phase of the cell cycle in osteosarcoma cells assessed by FACS at 72 h after treatment with siRNA. Representative results of the apoptosis assay in GC cells at 72 h after treatment with siRNA. C. Western blotting analyses showed that knockdown of YEATS4 by transfection with siRNA-YEATS4 induced the production of p21 and suppressed the Notch2 expression in U2OS and SaOS2 cells. In U2OS (wild-type TP53) cells only, knockdown of YEATS4 also induced phosphorylation activation of AKT.
Cell cycle analysis and apoptosis assay by silencing of YEATS4 expression using fluorescence-activated cell sorting
To investigate the reasons for the suppression of cell proliferation by knockdown of YEATS4, we performed a cell cycle analysis and an apoptosis assay. Fluorescence-activated cell sorting (FACS) analysis demonstrated that transfection of TP53 wild-type NUGC4 (Figure 2B), MKN45 (Figure 3B) and U2OS (Figure 4B) with siRNA-YEATS4 resulted in an accumulation of cells in the sub-G1 phase compared with transfection with control siRNA. In the TP53 mutant HGC27 (Figure 2B) and MKN7 (Figure 3B) and SaOS2 (Figure 4B), transfection with siRNA-YEATS4 resulted in an accumulation of cells in the G0/G1 phase compared with transfection with control siRNA.
The apoptotic cell analysis showed that transfection of TP53 wild-type NUGC4 with siRNA-YEATS4 increased early apoptosis (annexin V-positive/PI-negative) and late apoptosis (annexin V/PI-double positive) at 72 h after transfection compared with transfection with control siRNA (Figure 2B). Whereas, there was no difference by transfection with siRNA-YEATS4 In the TP53 mutant HGC27 (Figure 2B).
Western blotting showed that transfection of TP53 wild-type NUGC4 (Figure 5A) with siRNA-YEATS4 induced the cleavage of caspase 3 and PARP. In the TP53 mutant, HGC27 (Figure 5A) transfected with siRNA-YEATS4, the cleavage of caspase 3 and PARP was not found. These findings suggested that the knockdown of YEATS4 overexpression in TP53 wild-type cells induces cell apoptosis through caspase activation. However, in TP53 mutant cells, knockdown of YEATS4 overexpression did not induce cell apoptosis but did induce cell cycle arrest. Namely, YEATS4 might be related to cell apoptosis and the cell cycle in a TP53 mutation-dependent manner.
Figure 5.
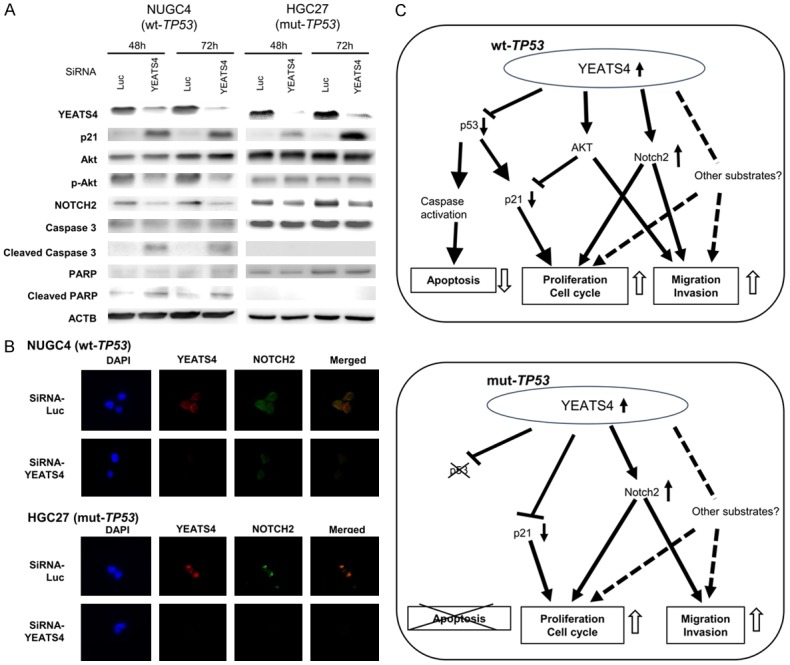
Molecular mechanisms by which overexpression of YEATS4 contributes to malignant potential in GC cells. A. Knockdown of YEATS4 by transfection with siRNA-YEATS4 induced the production of p21 and suppressed Notch2 expression. In NUGC4 (wild-type TP53) cells, knockdown of YEATS4 also induced phosphorylation activation of AKT, the cleavage of caspase 3 and PARP. B. Knockdown of YEATS4 suppressed Notch2 expression in both NUGC4 (wild-type TP53) and HGC27 (mutant-type TP53) GC cells. Moreover, the distribution of YEATS4 in GC cells was similar to that of Notch2. C. A hypothetical model of the overexpression/activation of YEATS4 in GC cells.
Suppression of cell migration and invasion by downregulation of YEATS4 expression
As shown in Table 1, protein expression of YEATS4 was significantly associated with lymphatic invasion in clinical samples. To confirm the relationship between YEATS4 and cell migration and invasion in vitro, transwell migration and invasion assays were performed. We examined the ability of TP53 wild-type NUGC4 and TP53 mutant HGC27 cells transfected with siRNA-YEATS4 to move through pores under different conditions. An uncoated membrane was used for the migration assays, whereas a Matrigel-coated membrane was used for the invasion assays. In Figure 2C, the numbers of NUGC4 and HGC27 cells that migrated into the lower chamber were significantly lower for siRNA-YEATS4-transfected cells than for siRNA-control-transfected cells under both conditions. The results suggest that the overexpression of YEATS4 can enhance the ability of GC cells to migrate and invade in both the TP53 wild-type and the TP53 mutant-type cell lines.
Molecular mechanisms by which overexpression of YEATS4 contributes to malignant potential in GC cells
The PI3K/AKT pathway is frequently activated in cancers and is important for tumor cell growth and survival [47]. Knockdown of YEATS4 expression by transfection with siRNA-YEATS4 suppressed the phosphorylation activation of AKT and induced production of p21 in TP53 wild-type NUGC4 (Figure 5A), MKN45 (Figure 3C) and U2OS (Figure 4C), whereas it induced production of p21 without suppressing the phosphorylation activation of AKT in TP53 mutant-type HGC27 (Figure 5A), MKN7 (Figure 3C) and TP53 null-type SaOS2 (Figure 4C).
Recently, YEATS4 has been reported as an activator of Notch2 [48], which is a key molecule on proliferation, migration and invasion of cancer cells [49]. We confirmed that knockdown of YEATS4 suppressed Notch2 expression in both TP53 wild-type and TP53 mutant-type GC cells (Figures 5B, 3C and 4C). The distribution of YEATS4 and Notch2 in TP53 wild-type NUGC4 and TP53 mutant-type HGC27 treated with siRNA-Luc and SiRNA-YEATS4 was examined using immunofluorescence staining. The distribution of YEATS4 in GC cells was similar to that of Notch2 (Figure 5B). These results suggested that overexpression of YEATS4 induced cell proliferation, migration and invasion through the activation of Notch2 in both TP53 wild-type and mutant-type GC cells. In addition, the overexpression of YEATS4 could induce cell migration and invasion through the activation of AKT in TP53 wild-type cells. Figure 5C shows a hypothetical model of the overexpression/activation of YEATS4 in GC cells.
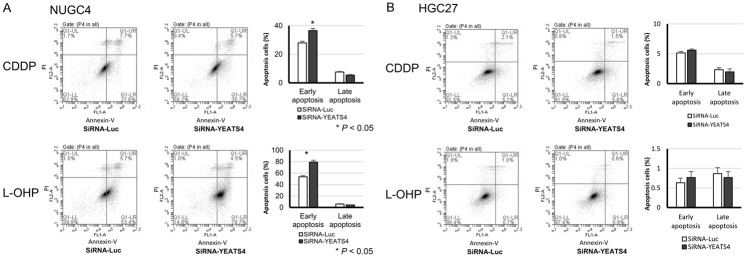
Association between high expression of YEATS4 and chemoresistance
Recently, it has been reported that the high expression of YEATS4 is associated with chemoresistance for L-OHP in colorectal cancer [28]. Thus, we examined whether YEATS4 was also associated with chemoresistance in GC. When treated with CDDP or L-OHP, transfection of TP53 wild-type NUGC4 with siRNA-YEATS4 increased early apoptosis at 72 h after transfection compared with transfection with control siRNA (Figure 6A). Whereas, the significant improvement of chemosensitivity was not observed by transfection with siRNA-YEATS4 in the TP53 mutant HGC27 (Figure 6B).
Figure 6.
Improvement of chemosensitivity by knockdown of YEATS4 in GC cells. When treated with CDDP or L-OHP, transfection of TP53 wild-type NUGC4 with siRNA-YEATS4 increased early apoptosis at 72 h after transfection compared with transfection of the control siRNA (A). The significant improvement of chemosensitivity was not observed by transfection with siRNA-YEATS4 in the TP53 mutant HGC27 (B).
Discussion
In this study, we demonstrated that YEATS4 was frequently overexpressed in GC cell lines and primary GC, and the overexpression was significantly associated with malignant tumor features and poor outcomes. Knockdown of YEATS4 using specific siRNA inhibited proliferation, migration and invasion of YEATS4-overexpressing GC cells through the NOTCH2 down-regulation in a TP53 mutation-independent manner, and induced apoptosis in wild-type TP53 GC cells. Moreover, knockdown of YEATS4 improved chemosensitivity for CDDP and L-OHP. These findings suggest that YEATS4 plays a crucial role in tumor malignant potential through its overexpression and highlight its usefulness as a prognostic factor and potential therapeutic target in GC.
YEATS4 is a member of a protein family characterized by the presence of an N-terminal YEATS domain [22]. As with other YEATS domain-containing proteins, YEATS4 is reported to be associated with chromatin modification and transcriptional regulation, which may involve recognition of chromatin targets and recruiting modification complexes to affect transcription. Moreover, previous studies have identified that YEATS4 is frequently gene-amplified and has various oncogenic functions in human cancers [23,50,51]. In our study, the immunoreactivity to the YEATS4 protein in each GC sample was significantly associated with a worse clinical outcome even after stratification with other clinicopathological characteristics in a multivariate analysis. In combination with p53, there was no relationship between the expression levels of YEATS4 and p53 in primary tumors, although some previous reports suggested these positive correlations in various cancers, such as glioma [52], lung cancer [23] and ovarian cancer [26]. These results suggest that the immunoreactivity to YEATS4 may be useful as an independent prognosticator in patients with GC.
In our in vitro analyses, knockdown of YEATS4 over-expression induced cell apoptosis in wild-type TP53 GC cells and G0/G1 cell cycle arrest in mutant-type TP53 cells. Furthermore, we investigated the molecular mechanism affecting the malignant potential in tumour cells with both overexpressions of YEATS4. Transwell migration and an invasion assay revealed that the knockdown of YEATS4 suppressed cell migration and invasion in both wild- and mutant-type TP53 cells. Recently, interaction between YEATS4 and Notch2 was suggested in hepatocellular carcinoma [48], so we explored whether there was similar positive association between YEATS4 and Notch2 in GC. The distribution of YEATS4 in GC cells was similar to that of Notch2 and knockdown of YEATS4 suppressed Notch2 expression in both wild- and mutant-type TP53 cells. These findings suggested that knockdown of YEATS4 suppressed cell migration and invasion through down-regulation of Notsh2 in GC cells. These YEATS4 functions, which were partly dependent on the TP53 mutation status, could be validated by experiments using TP53 wild sarcoma cells, U2OS and all TP53 null sarcoma cell lines, SaOS2. These findings strongly suggested that YEATS4 has crucial oncogenic roles and functions as a cancer-promoting gene in GC.
A recent fascinating study regarding chemotherapy identified that the downregulation of YEATS4 in colorectal cancer cells presented oxaliplatin (L-OHP)-induced cell apoptosis by inhibiting cytoprotective autophagy [28]. Cisplatin (CDDP) and oxaliplatin (L-OHP) are key drugs of chemotherapy for locally advanced or metastatic carcinoma of esophagus, gastroesophageal junction or stomach [53,54]. Therefore, we verified the significance of YEATS4 for CDDP or L-OHP chemoresistance in GC. As a result, knockdown of YEATS4 was strongly associated with chemosensitivity for CDDP and L-OHP in the TP53 wild NUGC4 cells, whereas, weakly in the TP53 mutant HGC27 cells. Therefore, these findings suggested that YEATS4 may be a key molecule for predicting chemoresistance and improving chemosensitivity of CDDP and L-OHP in prospective GC patients with overexpression of YEATS4. The detailed analyses are currently being evaluated.
In conclusion, we believe that this is the first report to show that YEATS4 has a crucial oncogenic role and is a potential therapeutic target in GC. We demonstrated the frequent overexpression of YEATS4 protein and its prognostic value in patients with GC. Although studies of larger cohorts are needed to validate these findings before moving to clinical settings, our results provide evidence that YEATS4 could be a pivotal molecular marker to determine the malignant properties of GC cells and a target for molecular therapy in patients with this cancer.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Jemal A, Wender RC, Gansler T, Ma J, Brawley OW. An assessment of progress in cancer control. CA Cancer J Clin. 2018;68:329–339. doi: 10.3322/caac.21460. [DOI] [PubMed] [Google Scholar]
- 3.Kanaji S, Suzuki S, Matsuda Y, Hasegawa H, Yamamoto M, Yamashita K, Oshikiri T, Matsuda T, Nakamura T, Sumi Y, Kakeji Y. Recent updates in perioperative chemotherapy and recurrence pattern of gastric cancer. Ann Gastroenterol Surg. 2018;2:400–405. doi: 10.1002/ags3.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin RC 2nd, Jaques DP, Brennan MF, Karpeh M. Extended local resection for advanced gastric cancer: increased survival versus increased morbidity. Ann Surg. 2002;236:159–165. doi: 10.1097/00000658-200208000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 6.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell. 2004;5:121–125. doi: 10.1016/s1535-6108(04)00033-9. [DOI] [PubMed] [Google Scholar]
- 8.Fukushige S, Matsubara K, Yoshida M, Sasaki M, Suzuki T, Semba K, Toyoshima K, Yamamoto T. Localization of a novel v-erbB-related gene, c-erbB-2, on human chromosome 17 and its amplification in a gastric cancer cell line. Mol Cell Biol. 1986;6:955–958. doi: 10.1128/mcb.6.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuniyasu H, Yasui W, Kitadai Y, Yokozaki H, Ito H, Tahara E. Frequent amplification of the c-met gene in scirrhous type stomach cancer. Biochem Biophys Res Commun. 1992;189:227–232. doi: 10.1016/0006-291x(92)91548-5. [DOI] [PubMed] [Google Scholar]
- 10.Oue N, Motoshita J, Yokozaki H, Hayashi K, Tahara E, Taniyama K, Matsusaki K, Yasui W. Distinct promoter hypermethylation of p16INK4a, CDH1, and RAR-beta in intestinal, diffuse-adherent, and diffuse-scattered type gastric carcinomas. J Pathol. 2002;198:55–59. doi: 10.1002/path.1170. [DOI] [PubMed] [Google Scholar]
- 11.Ding Y, Le XP, Zhang QX, Du P. Methylation and mutation analysis of p16 gene in gastric cancer. World J Gastroenterol. 2003;9:423–426. doi: 10.3748/wjg.v9.i3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Hofler H. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845–3852. [PubMed] [Google Scholar]
- 13.Maesawa C, Tamura G, Suzuki Y, Ogasawara S, Sakata K, Kashiwaba M, Satodate R. The sequential accumulation of genetic alterations characteristic of the colorectal adenoma-carcinoma sequence does not occur between gastric adenoma and adenocarcinoma. J Pathol. 1995;176:249–258. doi: 10.1002/path.1711760307. [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Abraham SC, Kim HS, Nam JH, Choi C, Lee MC, Park CS, Juhng SW, Rashid A, Hamilton SR, Wu TT. Inverse relationship between APC gene mutation in gastric adenomas and development of adenocarcinoma. Am J Pathol. 2002;161:611–618. doi: 10.1016/S0002-9440(10)64216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita K, Ohuchi N, Yao T, Okumura M, Fukushima Y, Kanakura Y, Kitamura Y, Fujita J. Frequent overexpression, but not activation by point mutation, of ras genes in primary human gastric cancers. Gastroenterology. 1987;93:1339–1345. doi: 10.1016/0016-5085(87)90264-2. [DOI] [PubMed] [Google Scholar]
- 16.Park WS, Oh RR, Park JY, Lee SH, Shin MS, Kim YS, Kim SY, Lee HK, Kim PJ, Oh ST, Yoo NJ, Lee JY. Frequent somatic mutations of the beta-catenin gene in intestinal-type gastric cancer. Cancer Res. 1999;59:4257–4260. [PubMed] [Google Scholar]
- 17.Fang DC, Wang RQ, Yang SM, Yang JM, Liu HF, Peng GY, Xiao TL, Luo YH. Mutation and methylation of hMLH1 in gastric carcinomas with microsatellite instability. World J Gastroenterol. 2003;9:655–659. doi: 10.3748/wjg.v9.i4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Ruschoff J, Kang YK. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 19.Wilke H, Muro K, Van Cutsem E, Oh SC, Bodoky G, Shimada Y, Hironaka S, Sugimoto N, Lipatov O, Kim TY, Cunningham D, Rougier P, Komatsu Y, Ajani J, Emig M, Carlesi R, Ferry D, Chandrawansa K, Schwartz JD, Ohtsu A. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15:1224–1235. doi: 10.1016/S1470-2045(14)70420-6. [DOI] [PubMed] [Google Scholar]
- 20.Kang YK, Boku N, Satoh T, Ryu MH, Chao Y, Kato K, Chung HC, Chen JS, Muro K, Kang WK, Yeh KH, Yoshikawa T, Oh SC, Bai LY, Tamura T, Lee KW, Hamamoto Y, Kim JG, Chin K, Oh DY, Minashi K, Cho JY, Tsuda M, Chen LT. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–2471. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 21.Fischer U, Heckel D, Michel A, Janka M, Hulsebos T, Meese E. Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Hum Mol Genet. 1997;6:1817–1822. doi: 10.1093/hmg/6.11.1817. [DOI] [PubMed] [Google Scholar]
- 22.Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem Cell Biol. 2009;87:65–75. doi: 10.1139/O08-111. [DOI] [PubMed] [Google Scholar]
- 23.Pikor LA, Lockwood WW, Thu KL, Vucic EA, Chari R, Gazdar AF, Lam S, Lam WL. YEATS4 is a novel oncogene amplified in non-small cell lung cancer that regulates the p53 pathway. Cancer Res. 2013;73:7301–7312. doi: 10.1158/0008-5472.CAN-13-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piccinni E, Chelstowska A, Hanus J, Widlak P, Loreti S, Tata AM, Augusti-Tocco G, Bianchi MM, Negri R. Direct interaction of Gas41 and Myc encoded by amplified genes in nervous system tumours. Acta Biochim Pol. 2011;58:529–534. [PubMed] [Google Scholar]
- 25.Persson F, Olofsson A, Sjogren H, Chebbo N, Nilsson B, Stenman G, Aman P. Characterization of the 12q amplicons by high-resolution, oligonucleotide array CGH and expression analyses of a novel liposarcoma cell line. Cancer Lett. 2008;260:37–47. doi: 10.1016/j.canlet.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Kim YR, Park MS, Eum KH, Kim J, Lee JW, Bae T, Lee DH, Choi JW. Transcriptome analysis indicates TFEB1 and YEATS4 as regulatory transcription factors for drug resistance of ovarian cancer. Oncotarget. 2015;6:31030–31038. doi: 10.18632/oncotarget.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao K, Yang J, Hu Y, Deng A. Knockdown of YEATS4 inhibits colorectal cancer cell proliferation and induces apoptosis. Am J Transl Res. 2015;7:616–623. [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Q, Cheng J, Zhang J, Zhang Y, Chen X, Xie J, Luo S. Downregulation of YEATS4 by miR-218 sensitizes colorectal cancer cells to L-OHP-induced cell apoptosis by inhibiting cytoprotective autophagy. Oncol Rep. 2016;36:3682–3690. doi: 10.3892/or.2016.5195. [DOI] [PubMed] [Google Scholar]
- 29.Jixiang C, Shengchun D, Jianguo Q, Zhengfa M, Xin F, Xuqing W, Jianxin Z, Lei C. YEATS4 promotes the tumorigenesis of pancreatic cancer by activating beta-catenin/TCF signaling. Oncotarget. 2017;8:25200–25210. doi: 10.18632/oncotarget.15633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji S, Zhang Y, Yang B. YEATS domain containing 4 promotes gastric cancer cell proliferation and mediates tumor progression via activating the wnt/beta-catenin signaling pathway. Oncol Res. 2017;25:1633–1641. doi: 10.3727/096504017X14878528144150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Arheden K, Ronne M, Mandahl N, Heim S, Kinzler KW, Vogelstein B, Mitelman F. In situ hybridization localizes the human putative oncogene GLI to chromosome subbands 12q13.3-14.1. Hum Genet. 1989;82:1–2. doi: 10.1007/BF00288260. [DOI] [PubMed] [Google Scholar]
- 32.Berner JM, Forus A, Elkahloun A, Meltzer PS, Fodstad O, Myklebost O. Separate amplified regions encompassing CDK4 and MDM2 in human sarcomas. Genes Chromosomes Cancer. 1996;17:254–259. doi: 10.1002/(SICI)1098-2264(199612)17:4<254::AID-GCC7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Merup M, Juliusson G, Wu X, Jansson M, Stellan B, Rasool O, Roijer E, Stenman G, Gahrton G, Einhorn S. Amplification of multiple regions of chromosome 12, including 12q13-15, in chronic lymphocytic leukaemia. Eur J Haematol. 1997;58:174–180. doi: 10.1111/j.1600-0609.1997.tb00944.x. [DOI] [PubMed] [Google Scholar]
- 34.Dierlamm J, Wlodarska I, Michaux L, Vermeesch JR, Meeus P, Stul M, Criel A, Verhoef G, Thomas J, Delannoy A, Louwagie A, Cassiman JJ, Mecucci C, Hagemeijer A, Van den Berghe H. FISH identifies different types of duplications with 12q13-15 as the commonly involved segment in B-cell lymphoproliferative malignancies characterized by partial trisomy 12. Genes Chromosomes Cancer. 1997;20:155–166. [PubMed] [Google Scholar]
- 35.Rao PH, Murty VV, Louie DC, Chaganti RS. Nonsyntenic amplification of MYC with CDK4 and MDM2 in a malignant mixed tumor of salivary gland. Cancer Genet Cytogenet. 1998;105:160–163. doi: 10.1016/s0165-4608(98)00013-2. [DOI] [PubMed] [Google Scholar]
- 36.Wunder JS, Eppert K, Burrow SR, Gokgoz N, Bell RS, Andrulis IL. Co-amplification and overexpression of CDK4, SAS and MDM2 occurs frequently in human parosteal osteosarcomas. Oncogene. 1999;18:783–788. doi: 10.1038/sj.onc.1202346. [DOI] [PubMed] [Google Scholar]
- 37.Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, Cloughesy TF, Nelson SF. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 38.Huang Z, Desper R, Schaffer AA, Yin Z, Li X, Yao K. Construction of tree models for pathogenesis of nasopharyngeal carcinoma. Genes Chromosomes Cancer. 2004;40:307–315. doi: 10.1002/gcc.20036. [DOI] [PubMed] [Google Scholar]
- 39.Marra L, Cantile M, Scognamiglio G, Perdona S, La Mantia E, Cerrone M, Gigantino V, Cillo C, Caraglia M, Pignata S, Facchini G, Botti G, Chieffi S, Chieffi P, Franco R. Deregulation of HOX B13 expression in urinary bladder cancer progression. Curr Med Chem. 2013;20:833–839. [PubMed] [Google Scholar]
- 40.Rice TW, Ishwaran H, Hofstetter WL, Kelsen DP, Apperson-Hansen C, Blackstone EH. Recommendations for pathologic staging (pTNM) of cancer of the esophagus and esophagogastric junction for the 8th edition AJCC/UICC staging manuals. Dis Esophagus. 2016;29:897–905. doi: 10.1111/dote.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura Y, Komatsu S, Ichikawa D, Nagata H, Hirajima S, Takeshita H, Kawaguchi T, Arita T, Konishi H, Kashimoto K, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br J Cancer. 2013;108:1324–1331. doi: 10.1038/bjc.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohashi T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Imamura T, Kiuchi J, Kosuga T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of PBK/TOPK relates to tumour malignant potential and poor outcome of gastric carcinoma. Br J Cancer. 2017;116:218–226. doi: 10.1038/bjc.2016.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imamura T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Ohashi T, Kiuchi J, Nishibeppu K, Kosuga T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of ZRF1 is related to tumor malignant potential and a poor outcome of gastric carcinoma. Carcinogenesis. 2018;39:263–271. doi: 10.1093/carcin/bgx139. [DOI] [PubMed] [Google Scholar]
- 44.Komatsu S, Ichikawa D, Hirajima S, Nagata H, Nishimura Y, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Imoto I, Inazawa J, Otsuji E. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br J Cancer. 2015;112:357–364. doi: 10.1038/bjc.2014.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naoi Y, Miyoshi Y, Taguchi T, Kim SJ, Arai T, Maruyama N, Tamaki Y, Noguchi S. Connexin26 expression is associated with aggressive phenotype in human papillary and follicular thyroid cancers. Cancer Lett. 2008;262:248–256. doi: 10.1016/j.canlet.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, Aiko S, Yoshizumi Y, Ichikawa D, Otsuji E, Inazawa J. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30:1139–1146. doi: 10.1093/carcin/bgp116. [DOI] [PubMed] [Google Scholar]
- 47.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 48.Huang G, Jiang H, Lin Y, Wu Y, Cai W, Shi B, Luo Y, Jian Z, Zhou X. lncAKHE enhances cell growth and migration in hepatocellular carcinoma via activation of NOTCH2 signaling. Cell Death Dis. 2018;9:487. doi: 10.1038/s41419-018-0554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivasankaran B, Degen M, Ghaffari A, Hegi ME, Hamou MF, Ionescu MC, Zweifel C, Tolnay M, Wasner M, Mergenthaler S, Miserez AR, Kiss R, Lino MM, Merlo A, Chiquet-Ehrismann R, Boulay JL. Tenascin-C is a novel RBPJkappa-induced target gene for notch signaling in gliomas. Cancer Res. 2009;69:458–465. doi: 10.1158/0008-5472.CAN-08-2610. [DOI] [PubMed] [Google Scholar]
- 50.Debernardi S, Bassini A, Jones LK, Chaplin T, Linder B, de Bruijn DR, Meese E, Young BD. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002;99:275–281. doi: 10.1182/blood.v99.1.275. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt J, Fischer U, Heisel S, Strickfaden H, Backes C, Ruggieri A, Keller A, Chang P, Meese E. GAS41 amplification results in overexpression of a new spindle pole protein. Genes Chromosomes Cancer. 2012;51:868–880. doi: 10.1002/gcc.21971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park JH, Smith RJ, Shieh SY, Roeder RG. The GAS41-PP2Cbeta complex dephosphorylates p53 at serine 366 and regulates its stability. J Biol Chem. 2011;286:10911–10917. doi: 10.1074/jbc.C110.210211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koizumi W, Narahara H, Hara T, Takagane A, Akiya T, Takagi M, Miyashita K, Nishizaki T, Kobayashi O, Takiyama W, Toh Y, Nagaie T, Takagi S, Yamamura Y, Yanaoka K, Orita H, Takeuchi M. S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): a phase III trial. Lancet Oncol. 2008;9:215–221. doi: 10.1016/S1470-2045(08)70035-4. [DOI] [PubMed] [Google Scholar]
- 54.Cunningham D, Starling N, Rao S, Iveson T, Nicolson M, Coxon F, Middleton G, Daniel F, Oates J, Norman AR. Capecitabine and oxaliplatin for advanced esophagogastric cancer. N Engl J Med. 2008;358:36–46. doi: 10.1056/NEJMoa073149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.