Abstract
Colorectal cancer (CRC) is a major worldwide health problem due to its high prevalence and mortality rate. microRNA has been reported playing an important role in a variety of cancers including colorectal cancer. miR-203a-3p has been found up-regulated in CRC tissues compare with the adjacent normal tissues. But, how miR-203a-3p regulates CRC development remains to be elucidated. In this study, gain and loss-of-function assays showed that miR-203a-3p promotes colorectal cancer cell proliferation, colony formation and migration and invasion by targeting PDE4D. And miR-203a-3p/β-catenin/Cyclin D1/c-Myc signaling pathway is involved in the CRC. In summary, this study highlights an onco-miRNA role for miR-203a-3p by regulating PDE4D in CRC and suggests that miR-203a-3p may be a novel molecular therapeutic target for CRC.
Keywords: Colorectal cancer, micro-RNA, miR-203a-3p, PDE4D
Introduction
Colorectal cancer (CRC) is one of the most common cancers and a leading cause of death worldwide. Most diagnoses of CRC occur in an advanced stage of the disease, leading to a poor prognosis [1,2]. Uncover the molecular basis of individual susceptibility to colorectal cancer and determining the factors that initiate the development of the tumor, drive its progression are the most important tasks in the study of CRC [3].
MicroRNAs (miRNAs) are a large class of endogenous tiny regulatory RNAs 21-25 nucleotides in length that extensively regulate gene expression by binding to the 3’-untranslated region (UTR) of mRNAs [4,5]. This imperfect base pairing with specific sequences lead to the degradation or translational inhibition of the corresponding mRNAs [6,7]. It has been reported that Many miRNAs playing an important role in the regulation of cancer cellular proliferation [8], differentiation [9], apoptosis [10], migration and invasion [11,12] by a variety of pathways. MiRNAs may be potential biomarkers for the diagnosis and prognosis of tumors as well as their therapy [13].
miR-203a-3p has been reported playing an important role in a variety of cancers, such as hepatocellular carcinoma [14], esophageal cancer [15], bladder cancer [16] and nasopharyngeal cancer [17]. In hepatocellular carcinoma, it promotes HCC cell proliferation and metastasis. In Barrett’s esophagus cells, up-regulating miR-203a-3p can inhibit cell proliferation and induces G0/G1 cell cycle arrest. In bladder tumors, downregulation of miR-203a-3p was significantly linked to progression in non-muscle invasive bladder tumors. In nasopharyngeal cancer, miR-203a-3p suppresses cell proliferation and metastasis. And There was a report that lncRNA HOTAIR could regulate the progression and chemoresistance of CRC via modulating the expression levels of miR-203a-3p [18].
miR-203a-3p has been found up-regulated in CRC tissues compare with the adjacent normal tissues [19]. miR-203a-3p is a member of the miR-203 family. In the present study, we have clarified demonstrated that miR-203a-3p promoted proliferation, colony formation, apoptosis, invasion and migration by suppressing the expression of PDE4D in CRC. We further identified that the β-catenin signal pathway plays an important role in this process. miR-203a-3p could potentially be utilized as a research and therapeutic target on proliferation and migration features in RCC.
Materials and methods
Meta-analysis of miRNA expression in CRC
We used an online database YM500 to analyze and compare the miRNA expression in colon cancer tissues and normal solid tissues. This database compared the miRNA expression profiles between 8 normal solid colon tissues and 429 primary solid colon tumors [19]. We found miR-203a was significantly upregulated.
Clinical samples and patient information
89 pairs of paraffin-embedded archived CRC and adjacent non-tumor colorectal mucosal tissues (ANT) were collected from our Department between January 2000 and December 2013. 8 pairs of fresh CRC and ANT were collected in 2015. No patients had received chemotherapy and/or radiotherapy before operation. The histopathology of the disease was determined by two pathologists according to the criteria of the World Health Organization. For the research purposes, these clinical materials, prior patient consents and approval from the Institutional Research Ethics Committee were obtained.
Cell lines
The human CRC cell lines SW480 and SW620 were maintained in Leibovitz’s L-15 Medium (Invitrogen, Carlsbad, CA). HCT116 was grown in McCoy’s 5A, Medium (Invitrogen). LOVO and SW1116 were cultured in RPMI-1640 medium (Invitrogen). HT29 was maintained in Dulbecco’s modified Eagle’s medium (Invitrogen). The human colonic epithelial cell line NCM460 was cultured in RPMI-1640 medium. All lines were purchased from Cell bank of Chinese Academy of Science (Shanghai, China) and authenticated by their karyotypes, and detailed gene expression in 2015. All media were supplemented with 10% (v/v) fetal bovine serum (Invitrogen), 1 × antibiotic/antimycotic (100 units/mL streptomycin, 100 units/mL, penicillin, and 0.25 mg/ ml amphotericin B). All cell lines were cultured in a humidified incubator at 37°C with 5% CO2.
Small interfering RNA (siRNA) sequences
The small interfering RNA (siRNA) specifically for PDE4D was chemically synthesized and purified from Ribbon Inc. (Guangzhou, China). The PDE4D siRNA sequences were: sense 5’-GCAGGGTCAAACTGAGAAAdTdT-3’. The siRNA was transfected using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer’s instructions. Scrambled siRNA were used as the negative control group. The human miR-203a-3p expression virus and miR-203a-3p-sponge virus was purchased from Hanheng (Shanghai, China).
miR-203a-3p overexpression and blocking
miR-203a-3p over-expression and blocking were achieved by infecting CRC cells with miR-203a-3p virus or miR-203a-3p-sp virus, respectively. miR-203a-3p virus and miR-203a-3p-sp virus and negative control virus were purchased from Han heng (Shanghai, China). SW480, HCT116 and SW1116 cells were seeded in 6-well plates and infected with the virus on the following day when the cells were approximately 30% confluent to get the stable miR-203a-3p over-expression and blocking cell lines. All the miR-203a-3p over-expression and blocking related experiments use these stable cell lines.
Cell proliferation assay
HCT116 and SW480 cells and stable miR-203a-3p blocking cell lines (2 × 103) were plated onto 96-well plates with medium containing 10% FBS and incubated overnight. After transfection with PDE4D-siRNA, cell proliferation was determined at 0, 24, 48, 72, and 96 h using the Cell Counting Kit-8 (CCK8) (Keygene, China). The absorbance (OD) was measured at a wavelength of 450 nm using a micro-plate Auto-reader (BioTek Instruments, USA). This experiment was performed in triplicate.
Analysis of apoptosis
5 × 105 cells were seeded in 6-well plates and incubated overnight until 50-60% confluence. The cells were transfected with 100 nM PDE4D-siRNA and harvested at 48 h, washed in cold PBS, fixed with 80% ethanol for 8 h at 4°C, then stained with propidium iodide buffer (50 mg/ml propidium iodide, 0.1% sodium citrate and 0.1% Triton X-100) for 3 h at 4°C. Scrambled-siRNA was used as the control group. 2 × 104 cells were analyzed for apoptosis using a Becton Dickinson FACScan (Becton Dickinson Immunocytometry Systems, San Jose, CA). The percentage of cells in each phase of the apoptotic cells was quantified using Cell Quest software. This experiment was performed in triplicate.
Colony formation assay
After 48 h transfection with PDE4D-siRNA, the cells at 2 × 102 per well were plated in 6-well plates and grown for 2 weeks, respectively. Then, the cells were washed twice with phosphate buffer saline (PBS), fixed with 4% paraformaldehyde and stained with 0.5% crystal violet for 15 min. The number of colonies in 10 random view fields was counted under a microscope and the average number of colonies was achieved. The experiment was triplicated independently.
Transwell migration and Boyden invasion assays
Migration and invasion assays were carried out in Transwell chambers containing polycarbonate filters (8 μm pore size; Corning Incorporated, Life Sciences, NY, USA). After transfected with PDE4D-siRNA for 48 h, cells (migration/2 × 104 cells; invasion/1 × 105 cells) in a 100 μl volume of serum free medium were placed in the upper chambers and incubated at 37°C with 5% CO2 for 18 hours, while a 500 μl volume of medium containing 10% FBS was added to the lower chamber as chemoattractant. Cells were allowed to invade through the matrigel (BD Biosciences) or migrate for 24 hours at 37°C with 5% CO2. Following invasion or migration, cells were fixed with 4% formaldehyde and stained with 1% crystal violet. Cells on the upper surface of the filters were removed by wiping with a cotton swab. Cell counts were the mean number of cells per fields of view. Three independent experiments were performed and the data were presented as mean ± standard deviation (SD).
Quantitative real time PCR
Reverse transcription was performed using One step Prime Script miRNA and mRNA cDNA Synthesis Kit (Takara Biotechnology Co. Ltd, Dalian, China), and quantitative real-time PCR were performed using SYBR Premix Ex Taq II (Takara Biotechnology). RNAU6B snRNA and GAPDH was used for sample loading normalization. The specific forward primer of miR-203a-3p was 5’-CCGGTGAAATGTTTAGGACCACTAG-3’. Reverse primer for miR-203a-3p was 5’GCCGCGTGAAATGTTTAGG3’, U6B snRNA was Uni-miR qPCR primer (TakaRa code D350A). The primer sequences used for β-catenin were followed: forward: 5’-TTG AAAATCCAGCGTGGACA-3’; reverse: 5’-TCGAGTCATTGCATACTGTC-3’. The primer sequences used for PDE4D were followed: forward: 5’-GGGCGCTCCAAGAAGTAAAG-3’; reverse: 5’-AAGGGAGGTGCTGGTTAAGG-3’. The quantity of miR-203a-3p in each CRC tissue relative to its paired ANT was calculated using the equation [RQ = 2-ΔΔCT, ΔΔCT = (CTmiRNA-CTU6) T - (CTmiRNA-CTU6) N]. The expression level of miR-203a-3p was classified into low expression and high expression group compared with RQ ratio = RQ (CRC)/RQ (ANT). The geometric mean of housekeeping gene GAPDH was used to normalize the variability in mRNA expression levels. All experiments were performed in triplicate.
Western blot analysis
Protein isolation and western blot lysis buffer (Beyotime, Shanghai, China) with freshly added PMSF (Beyotime, Shanghai, China) was used to isolate the protein from cells or tissues. Proteins were separated by SDS-PAGE (Bio-Rad). Antibodies against PDE4D, E-cadherin, and GAPDH were purchased from Santa Cruz Biotechnology ((sc-365349, sc-40, sc-130545 and sc-25778) respectively; Santa Cruz, CA, USA).
Luciferase reporter assay
The wild-type 3’-UTR sequence of PDE4D predicted to interact with miR-203a-3p or a mutated sequence within the target sites was synthesized and inserted into psiCHECK2 vector.
Cells were seeded in 48-well plates and cultured for 48 h. The psiCHECK2-PDE4D-3’-UTR-wt or psiCHECK2-PDE4D-3’-UTR-mut plasmid was co-transfected into HCT116 and SW480 with the miR-203a-3p mimics, mimics control using lipofectamine 2000 reagent (Invitrogen), respectively. Firefly luciferase and Renilla luciferase activities were evaluated 48 h post transfection using the Dual Luciferase Reporter Assay Kit (keygentec) according to the manufacturer’s instructions. All experiments were performed independently in triplicate.
Statistical analyses
Groups from cell culture experiments were compared using an unpaired, two-tailed Student’s tests and results were presented as mean ± SD. For CCK8 assay, comparisons were made by univariate variance analysis (two-way ANOVA). Statistical analyses were performed using the SPSS 16.0 statistical software. P < 0.05 was considered to be statistically significant.
Results
miR-203a-3p was up-regulated in most of the CRC tissues and cells
We examined the expression of miR-203a-3p in human CRC tissues, we found that miR-203a-3p levels were significantly increased in the 89 pairs of Paraffin CRC tissues compared with their matched adjacent normal tissues (Figure 1A). And high miR-203a-3p expression was found in 5 of 8 CRC fresh tissues compared with adjacent non-tumor tissues (Figure 1B). High miR-203a-3p expression was found in 4 of 6 CRC cell lines including HCT116, HT29, Lovo and SW1116 cell lines compared with human colonic epithelial cell line NCM460 (Figure 1C).
Figure 1.
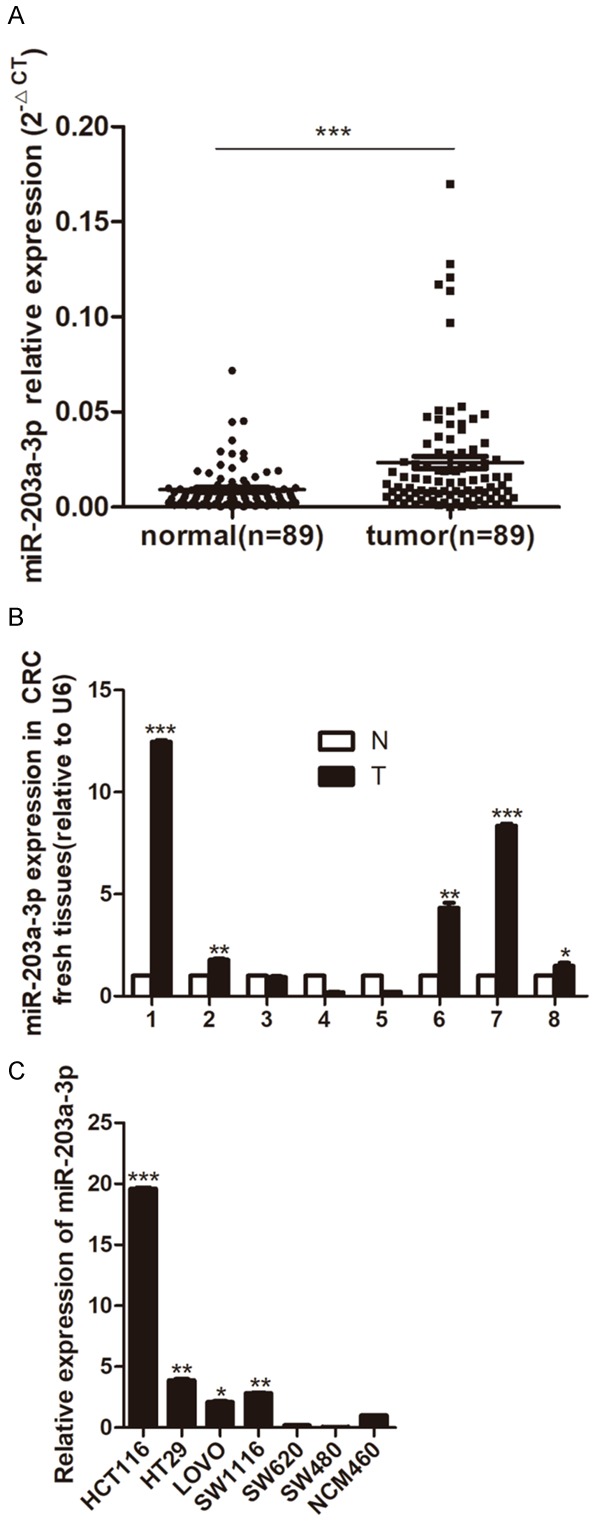
miR-203a-3p expression in CRC tissues and cell lines. A. miR-203a-3p expression level in 89 paired of Paraffin CRC tissues (tumor) and adjacent non-tumor colorectal mucosa tissues (normal) by quantitative real-time PCR analysis. B. miR-203a-3p expression level in 8 paired of fresh CRC samples (T) and adjacent non-tumor colorectal mucosa tissues (N). C. miR-203a-3p expression level in CRC cell lines. Data were normalized to the miR-203a-3p expression level in NCM460 cells. qPCR data represent mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
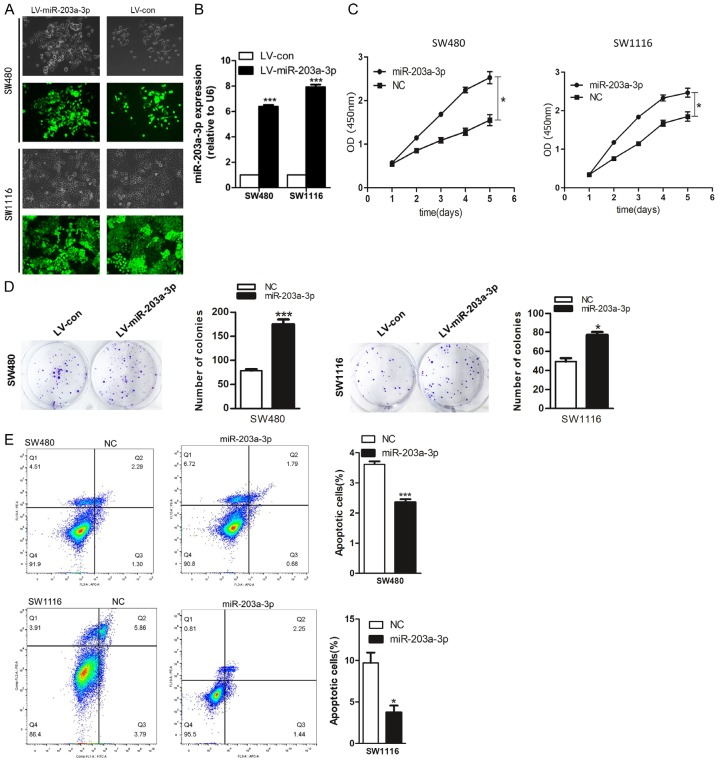
Over-expression of miR-203a-3p increases CRC cell proliferation, colony formation and inhibits cell apoptosis
To determine whether miR-203a-3p over-expression modulates CRC tumorigenesis, CRC cell lines SW480 and SW1116 were transduced with recombinant lentivirus carrying the miR-203a-3p gene (Figure 2A) and over-expression confirmed via qRT-PCR (Figure 2B). As evident from the CCK8 assay, miR-203a-3p over-expression induced CRC cell viability (Figure 2C). The promote effect of miR-203a-3p on CRC cell proliferation was further confirmed using the colony formation assay. The number of colonies was significantly increased in cells over-expressing miR-203a-3p, compared with the negative control group (Figure 2D). The flow cytometry assay was further employed to measure apoptosis. Apoptotic rate was significantly decreased upon over-expression of miR-203a-3p in the CRC cells (Figure 2E). Our data strongly suggest that miR-203a-3p exerts growth promotion effects on CRC.
Figure 2.
miR-203a-3p promoted the CRC cell proliferation and colony formation, inhibited apoptosis. (A, B) SW480 and SW1116 cells were infected with miR-203a-3p or negative control lenti-virus and Quantitative RT-PCR used to determine miR-203a-3p expression. (C, D) miR-203a-3p significantly promoted cell proliferation (C) and colony formation (D) in SW480 and SW1116 cells. (E) miR-203a-3p significantly reduced cell apoptosis in SW480 and SW1116 cells. Each assay was repeated three times. The data represent means ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
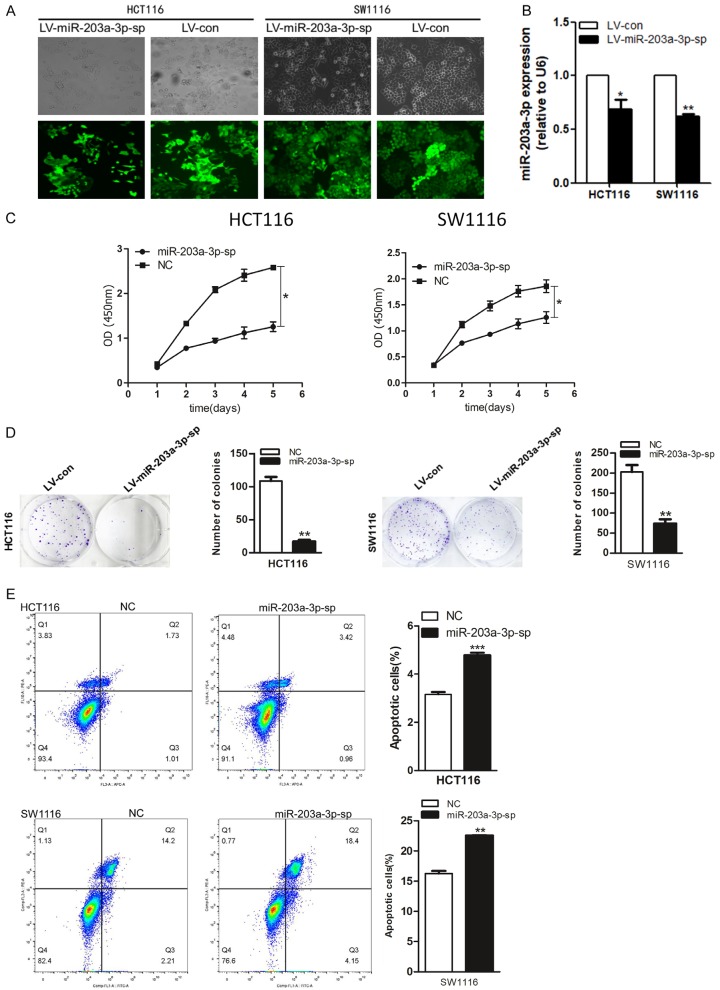
The silence of miR-203a-3p by sponges significantly reduces CRC cell viability and proliferation, promotes CRC cell apoptosis
To determine whether silence of miR-203a-3p could modulates CRC tumorigenesis, we use MiR-203a-3p sponges. MiR-203a-3p sponges can bind to sites in the 3’ untranslated region (UTR) of mRNA to derepress microRNA targets at least as strongly as chemically modified antisense oligo nucleotides (MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells). CRC cell lines HCT116 and SW1116 were infected with recombinant lentivirus carrying the miR-203a-3p-sp gene (Figure 3A) and the effects on expression of miR-203a-3p were confirmed via qRT-PCR (Figure 3B). As evident from the CCK8 assay, silence of endogenous miR-203a-3p decreases CRC cell viability (Figure 3C). The inhibit effect of miR-203a-3p-sp on CRC cell proliferation was further confirmed using the colony formation assay. The number of colonies was significantly decreased in cells silence of endogenous miR-203a-3p, compared with the negative control group (Figure 3D). The flow cytometry assay was further employed to measure apoptosis. Apoptotic rate was significantly increased upon silence of endogenous miR-203a-3p in the CRC cells (Figure 3E).
Figure 3.
Silence of miR-203a-3p by sponges significantly reduced CRC cell proliferation and colony formation, promoted apoptosis. (A, B) HCT116 and SW1116 cells were infected with miR-203a-3p-sp or negative control lenti-virus and Quantitative RT-PCR used to determine miR-203a-3p expression. (C, D) miR-203a-3p-sp significantly reduced cell proliferation (C) and clone formation (D) in HCT116 and SW1116 cells. (E) miR-203a-3p-sp significantly promoted cell apoptosis in HCT116 and SW1116 cells. Each assay was repeated three times. The data represent means ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Alterations in miR-203a-3p expression influence the migration and invasion of CRC cells
We found that over-expression of miR-203a-3p in SW480 and SW1116 cells significantly increased cell migration and invasion ability compared with control cells (Figure 4A). Silence of endogenous miR-203a-3p decreased cell migration and invasion ability compared with control cells (Figure 4B). These results suggest that miR-203a-3p can promote cell migration and invasion in CRC cells.
Figure 4.
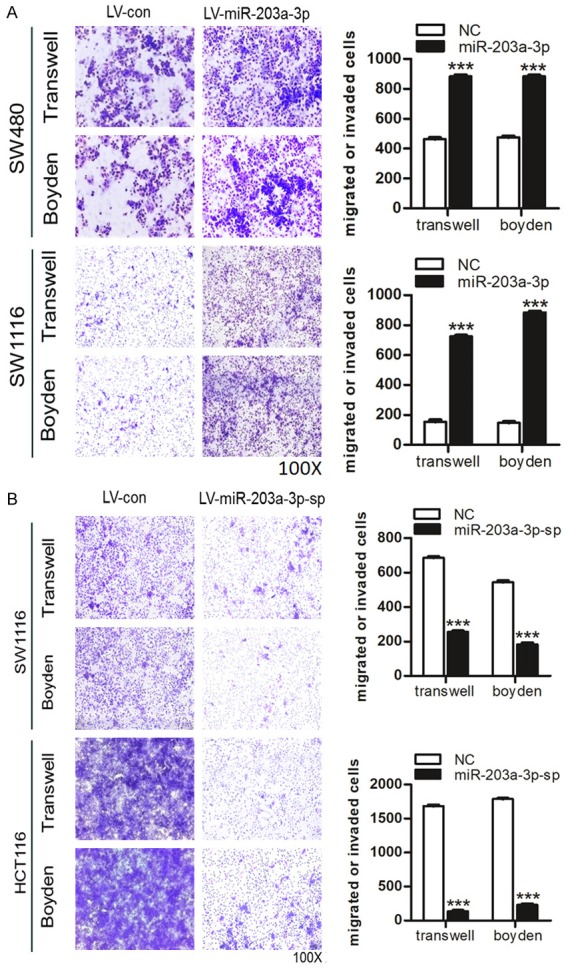
Effects of miR-203a-3p on CRC cells in migration and invasion. A. Over-expression of miR-203a-3p significantly promoted cell migration and invasion in SW480 and SW1116 cells. B. The Silence of miR-203a-3p significantly inhibited cell migration and invasion in HCT116 and SW1116 cells. Each assay was repeated three times. The data represent means ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
PDE4D is a direct target of miR-203a-3p
To investigate the mechanism by which miR-203a-3p exerts promote effects on CRC cells, we searched for target genes using Pictar, miRDB and TargetScan databases. These databases revealed that PDE4D was a potential target of miR-203a-3p.
To determine whether miR-203a-3p regulates PDE4D expression in CRC cancer cells, we evaluated the expression of PDE4D in CRC fresh tissues, CRC cancer cell lines and CRC cell lines with miR-203a-3p overexpression or silence of endogenous miR-203a-3p by sponges. An inverse correlation between miR-203a-3p and PDE4D expression was observed in these CRC tissues and cell lines (Figure 5A). Overexpression of miR-203a-3p significantly suppressed PDE4D at both the mRNA and protein levels in SW480 and SW1116 cells (Figure 5B). Silence of endogenous miR-203a-3p significantly increased PDE4D at both the mRNA and protein levels in HCT116 and SW1116 cells (Figure 5C).
Figure 5.
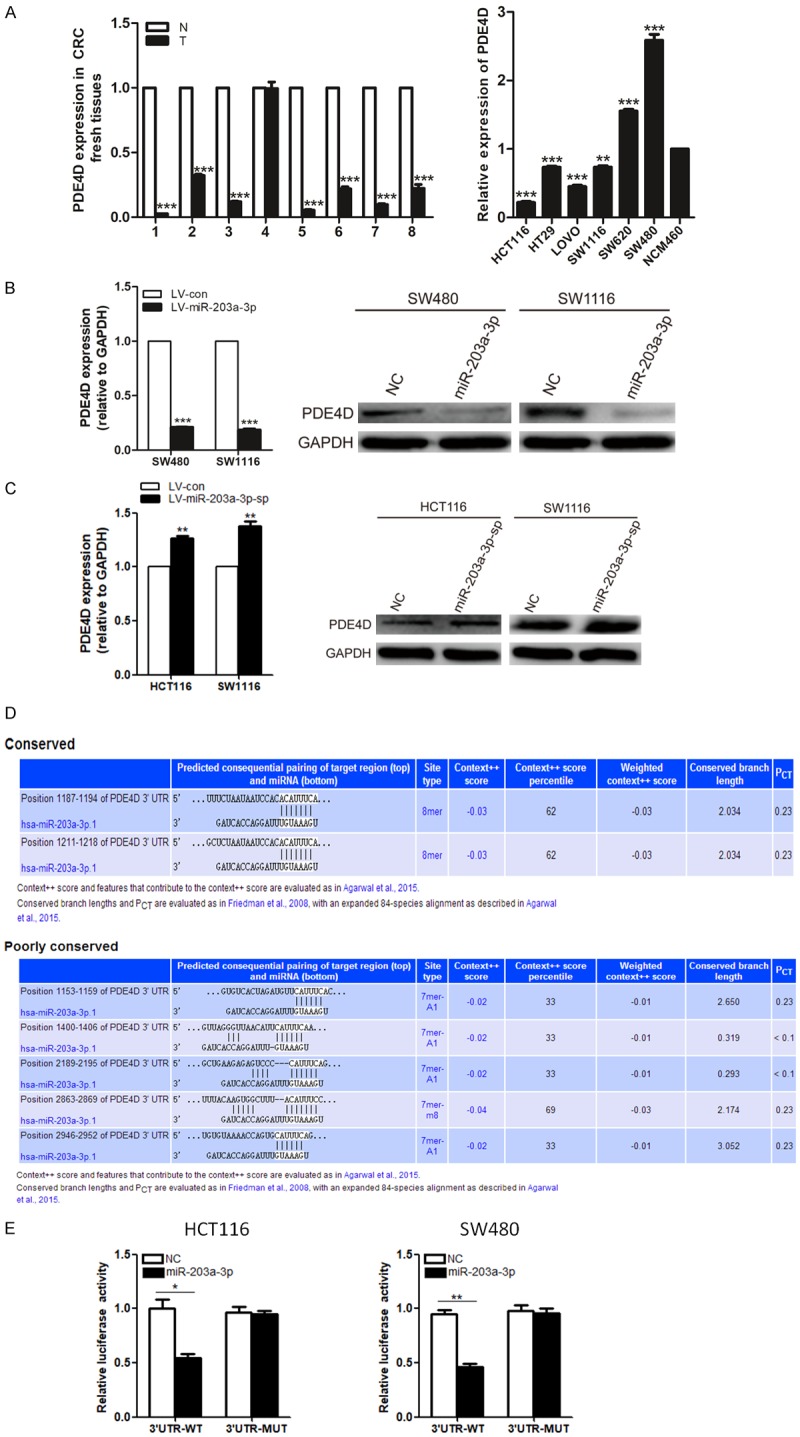
Identification of PDE4D as a miR-203a-3p target. A. Quantitative RT-PCR analysis of PDE4D expression levels in the same 8 pairs of CRC and normal tissue samples and in the same CRC cell lines. B. Over-expression of miR-203a-3p significantly suppressed PDE4D at both the mRNA and protein levels in SW480 and SW1116 cells. C. Silence of endogenous miR-203a-3p significantly increased PDE4D at both the mRNA and protein levels in HCT116 and SW1116 cells. D. Binding sites of miR-203a-3p at the 3’-UTR of PDE4D mRNA. E. PDE4D is a target gene of miR-203a-3p in HCT116 and SW480 cells based on the luciferase reporter assay. Each assay was repeated three times. The data represent means ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
To ascertain whether PDE4D is regulated via direct binding of miR-203a-3p to its 3’-UTR region, we did Luciferase Reporter Assay. The 3’-UTR of PDE4D mRNA contains 7 complementary sites in the seed region of miR-203a-3p (Figure 5D). Compared with the control group, luciferase activity was significantly suppressed in psiCHECK2-PDE4D-3’-UTR-wt transfected but not psiCHECK2-PDE4D-3’-UTR-mut transfected cells (Figure 5E). The results indicate that miR-203a-3p binds directly to PDE4D 3’-UTR to repress its expression.
The PDE4D mRNA is down-regulated in colorectal tumors and PDE4D lower expression group had a poorer overall survival rate
Down-regulation of PDE4D was found in 11 of 20 cancer types, especially cancers of digestive system, such as colorectal cancer and gastric cancer, through oncomine data-mining analysis (Figure 6A). In kaiser’s dataset, we found that level of PDE4D mRNA was significantly decreased from colon tissues to colorectal tumors with probe (204491_at) (Figure 6B). The expression level of PDE4D mRNA was significantly decreased from colon, rectum to colorectal tumors in probe (A_24_P419353) in TCGA dataset. And we found that PDE4D lower expression group had a poorer overall survival rate (P < 0.05) in COAD (Colon adenocarcinoma) and READ (Rectum adenocarcinoma) datasets on gepia.cancer websites (Figure 6C).
Figure 6.
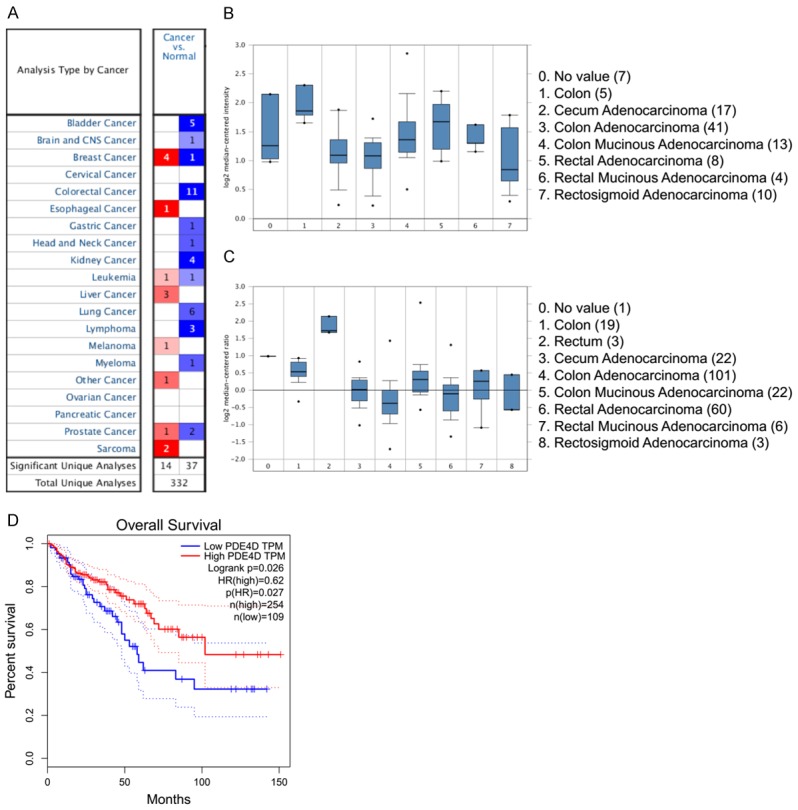
The PDE4D mRNA is down-regulated in colorectal cancer compared with colon as revealed by oncomine data-mining analysis and PDE4D lower expression group had a poorer overall survival rate. A. Down-regulation of PDE4D was found in 11 of 20 cancer types. B. Level of PDE4D mRNA was significantly decreased from colon to CRC with probe (204491_at) in the kaiser’s dataset. C. The expression level of PDE4D mRNA was significantly decreased from colon, rectum to CRC in probe (A_24_P419353) in TCGA dataset. D. PDE4D lower expression group had a poorer overall survival rate (P < 0.05) in COAD (Colon adenocarcinoma) and READ (Rectum adenocarcinoma) datasets on gepia.cancer websites.
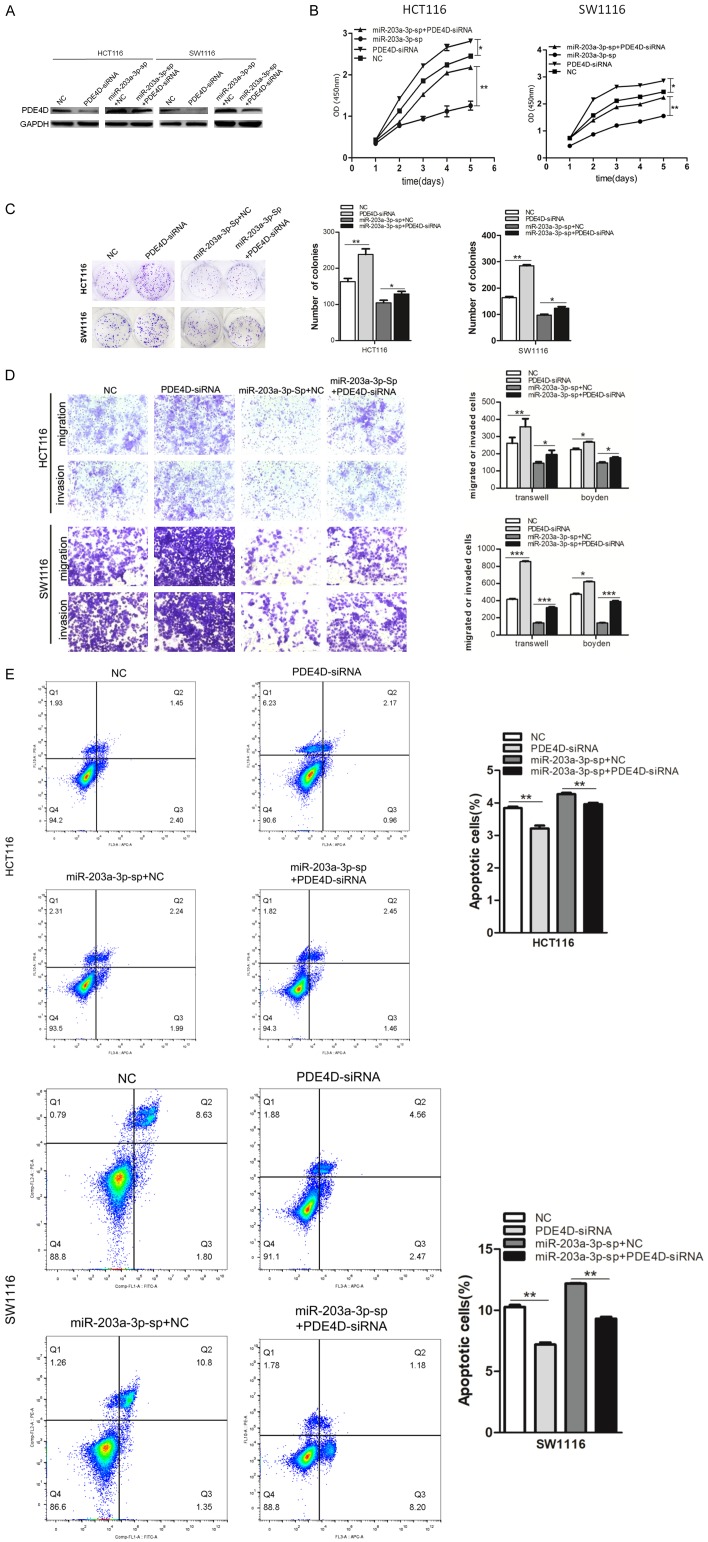
Alterations in PDE4D expression influence the effects of miR-203a -3p on CRC cells
We knocked down PDE4D in HCT116 and SW1116 cells and HCT116 and SW1116 miR-203a-3p-sp cells using specific siRNAs (Figure 7A). Functional assays disclosed that knockdown of PDE4D increases HCT116 and SW1116 cell proliferation (Figure 7B) colony formation (Figure 7C), migration and invasion (Figure 7D) and significantly inhibited cell apoptosis (Figure 7E). These effects were similar to the effects of over-expression of miR-203a-3p. PDE4D knockdown reversed these effects induced by silence of miR-203a-3p. All the results above provided further evidence of the PDE4D as a downstream mediator of miR-203a-3p.
Figure 7.
miR-203a-3p promoted CRC cell proliferation, colony formation and inhibited apoptosis by targeting PDE4D. (A) HCT116 and SW1116 cells were transfected with PDE4D siRNA or negative control. After 48 h, the PDE4D protein level was examined via western blot analysis. (B-E) knockdown of PDE4D increased HCT116 and SW1116 cell proliferation (B), colony formation (C), migration and invasion (D) and significantly inhibited cell apoptosis (E). In addition, PDE4D knockdown reversed of these functions induced by the silence of miR-203a-3p. Each assay was repeated three times. The data represent means ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
MiR-203a-3p/β-catenin/c-Myc/cyclin D1 signaling pathway is involved in CRC
The protein expression levels of of β-catenin, c-Myc and Cyclin D1 were increased after over-expression of miR-203a-3p and the result was similar with suppresse the expression of PDE4D in SW480 and SW1116 cells (Figure 8A). After silence of endogenous miR-203a-3p in HCT116 and SW1116 cells, the protein expression levels of of β-catenin, c-Myc and Cyclin D1 were decreased. And knock down of PDE4D can rescue this Phenotype (Figure 8B). Together, these results demonstrated that miR-203a-3p suppressed the expression of PDE4D, thus inducing Wnt/β-catenin signaling activation and CRC cell proliferation and migration.
Figure 8.
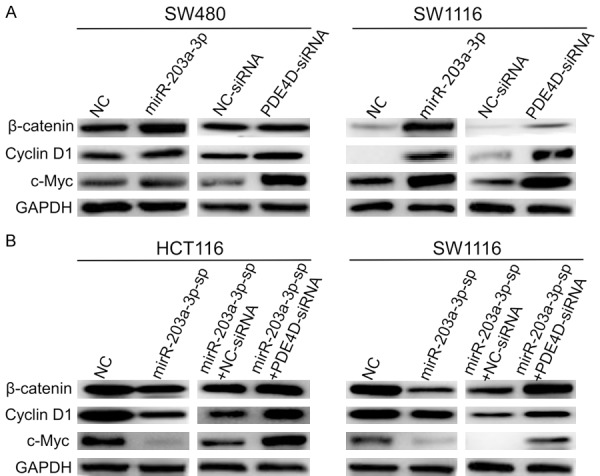
MiR-203a-3p/β-catenin/Cyclin D1/c-Myc signaling pathway is involved in CRC. A. The protein expression levels of of β-catenin, Cyclin D1 and c-Myc were increased after over-expression of miR-203a-3p and the effect was same as suppresses the expression of PDE4D in SW480 and SW1116 cells. B. The protein expression levels of of β-catenin, Cyclin D1 and c-Myc were decreased after the silence of endogenous miR-203a-3p in HCT116 and SW1116 cells. And knock down of PDE4D can rescue this Phenotype. Each assay was repeated three times.
Discussion
In this study, we used an online database YM500 to analyze and compare the miRNA expression profiles between 429 primary solid colon tumors and 8 normal solid colon tissues. MiR-203a was significantly up-regulated in the primary solid colon tumors compare to the normal solid colon tissues. And then we detected the expression of miR-203a-3p in 89 pairs of clinical primary solid colorectal tumors Paraffin tissues, 8 pairs of clinical primary solid colorectal fresh tissues. The results showed that miR-203a-3p is up-regulated in CRC tissue samples.
There were multiple evidences that demonstrated the expression of miR-203a-3p was increased in many kinds of cancers, but how miR-203a-3p regulates CRC development remains to be elucidated. We found that miR-203a-3p promotes colorectal cancer proliferation, colony formation, migration and invasion. MicroRNAs can regulate gene expression by binding to the 3’-untranslated regions (3’-UTR) of target genes for translational repression or degradation. Based on sequence complementarity, plus by using miRNA target prediction software, we inferred that PDE4D was the target of miR-203a-3p. And Luciferase Reporter Assay further confirmed that MiR-203a-3p binds directly to the 3’-UTR of PDE4D. In order to investigate the roles of miR-203a-3p and PDE4D in the development of CRC, we did gain and loss-of-function assays. The results showed that miR-203a-3p promoted colorectal cancer proliferation, colony formation and migration and invasion by targeting PDE4D, which was consistent with previous studies in hepatocellular carcinoma of miR-203a-3p [14]. We also found that PDE4D knockdown induced the same phenotype as miR-203a-3p over-expression, which were consistent with the negative regulation of miR-203a-3p on PDE4D.
Phosphodiesterase 4D (PDE4D) is a subtype of metallohydrolases. PDE4D gene has many variants attributed to alternative splicing and the use of multiple promoters. PDE4D enzyme degrades cAMP. The distribution pattern of PDE4D variants varies within both cells and tissues [20]. There is one paper said that Expression of PDE4D isoforms is consistently altered in primary human prostate cancer compared to benign tissue, with PDE4D7 being up-regulated while PDE4D5 and PDE4D9 are down-regulated. Disease progression is marked by an overall down-regulation of long PDE4D isoforms, while short isoforms (PDE4D1/2) appear to be relatively unaffected [21]. It has been reported that PDE4D functions as a proliferation promoting factor in some types of tumors, including prostate cancer [22], lung cancer [23], nasopharyngeal carcinoma [24] and medulloblastoma [25]. In our study, an inverse correlation between micoRNA-203a-3p and PDE4D expression was observed in CRC tissues and cell lines. And the PDE4D mRNA is down-regulated in colorectal cancer compared with normal colon as revealed by oncomine data-mining analysis (Figure 6B and 6C) and PDE4D lower expression group had a poorer overall survival rate (P < 0.05) on gepia.cancer websites (Figure 6D). micoRNA-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Maybe our data showed the long PDE4D isoforms altered in primary human colorectal cancer compared to benign tissue. Since microRNAs often have more than one target, future studies could explore if micoRNA-203a-3p regulates the production of other cancer-related proteins as well.
Growing evidences have focused on the role of miR-203a-3p in cancer progression, but the results were not consistent. In hepatocellular carcinoma, miR-203a-3p.1 acted as an onco-miRNA by targeting IL-24. Inhibition of miR-203a-3p.1 in cells could lead to the reversal of HCC cell proliferation and metastasis.
In Barrett’s esophageal cancer, Omeprazole Inhibits Cell Proliferation and Induces G0/G1 Cell Cycle Arrest through Up-regulating miR-203a-3p Expression. In nasopharyngeal carcinoma, miR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1.
Xiao et al. have reported both HOTAIR knockdown and miR-203a-3p overexpression in CRC cell lines led to inhibit cell proliferation [18]. They studied them in Colo205 and SW620 cells. Our results suggested that miR-203a-3p was an onco-miR in colorectal cancer. We found that miR-203a-3p promotes colorectal cancer proliferation, colony formation, migration and invasion. As we Know, Colo205 and SW620 derived from CRC metastatic sites (ascites and lymph node). We studied miR-203a-3p in SW480, HCT116 and SW11160 cells, which derived from the CRC primary tumor. We know that EMT program results in suppression of cancer cell proliferation [26]. And EMT program can involve in tumor metastasis [27]. So there may be some differences between the metastatic sites cell lines (Colo205 and SW620) and CRC primary tumor cell lines (SW480, HCT116 and SW11160). And epigenetic events can regulate the EMT process [27] and affect tumor proliferation and migration. miRNA may play different roles in different tumor cells original from different tumor sites and status. The mechanism of this is complicated.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant NO.: 81472251, 81272636) and Natural Science Foundation of Guangdong Province (2017A030313552).
Disclosure of conflict of interest
None.
References
- 1.Li Y, Li L, Chen M, Yu X, Gu Z, Qiu H, Qin G, Long Q, Fu X, Liu T, Li W, Huang W, Shi D, Kang T, Luo M, Wu X, Deng W. MAD2L2 inhibits colorectal cancer growth by promoting NCOA3 ubiquitination and degradation. Mol Oncol. 2018;12:391–405. doi: 10.1002/1878-0261.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6:391–401. [PMC free article] [PubMed] [Google Scholar]
- 3.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–63. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 5.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–8. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Hede K. Studies define role of microRNA in cancer. J Natl Cancer Inst. 2005;97:1114–5. doi: 10.1093/jnci/dji260. [DOI] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 9.Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6:8474–90. doi: 10.18632/oncotarget.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 12.Khella HW, White NM, Faragalla H, Gabril M, Boazak M, Dorian D, Khalil B, Antonios H, Bao TT, Pasic MD, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA, Yousef GM. Exploring the role of miRNAs in renal cell carcinoma progression and metastasis through bioinformatic and experimental analyses. Tumour Biol. 2012;33:131–40. doi: 10.1007/s13277-011-0255-5. [DOI] [PubMed] [Google Scholar]
- 13.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, Cheng JQ. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–89. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huo W, Du M, Pan X, Zhu X, Gao Y, Li Z. miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma cell growth and metastasis. FEBS Open Bio. 2017;7:1085–91. doi: 10.1002/2211-5463.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou Y, Hu Q, Huang J, Xiong H. Omeprazole inhibits cell proliferation and induces G0/G1 cell cycle arrest through up-regulating miR-203a-3p expression in barrett’s esophagus cells. Front Pharmacol. 2017;8:968. doi: 10.3389/fphar.2017.00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenherr SM, Tsai S, Silva Neto B, Sullivan TB, Cimmino CB, Logvinenko T, Gee J, Huang W, Libertino JA, Summerhayes IC, Rieger-Christ KM. MicroRNA expression profile identifies high grade, non-muscle-invasive bladder tumors at elevated risk to progress to an invasive phenotype. Genes (Basel) 2017:8. doi: 10.3390/genes8020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang N, Jiang X, Chen Z, Song X, Wu L, Zong D, Song D, Yin L, Wang D, Chen C, Bian X, He X. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2017;36:138. doi: 10.1186/s13046-017-0604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-mediated Wnt/ss-Catenin signaling pathway. Cell Physiol Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui S, Gu Y, Sun W, You C, Liu Z, Sun F, Wang Y, Fu Z, Ye C, Zhang C, Li J, Chen X. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16:53. doi: 10.1186/s12943-017-0625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powers GL, Hammer KD, Domenech M, Frantskevich K, Malinowski RL, Bushman W, Beebe DJ, Marker PC. Phosphodiesterase 4D inhibitors limit prostate cancer growth potential. Mol Cancer Res. 2015;13:149–60. doi: 10.1158/1541-7786.MCR-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Böttcher R, Dulla K, van Strijp D, Dits N, Verhoef EI, Baillie GS, van Leenders GJ, Houslay MD, Jenster G, Hoffmann R. Human PDE4D isoform composition is deregulated in primary prostate cancer and indicative for disease progression and development of distant metastases. Oncotarget. 2016;7:70669–70684. doi: 10.18632/oncotarget.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahrmann EP, Collier LS, Knutson TP, Doyal ME, Kuslak SL, Green LE, Malinowski RL, Roethe L, Akagi K, Waknitz M, Huang W, Largaespada DA, Marker PC. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a Sleeping Beauty transposon-based somatic mutagenesis screen. Cancer Res. 2009;69:4388–97. doi: 10.1158/0008-5472.CAN-08-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pullamsetti SS, Banat GA, Schmall A, Szibor M, Pomagruk D, Hänze J, Kolosionek E, Wilhelm J, Braun T, Grimminger F, Seeger W, Schermuly RT, Savai R. Phosphodiesterase-4 promotes proliferation and angiogenesis of lung cancer by crosstalk with HIF. Oncogene. 2013;32:1121–34. doi: 10.1038/onc.2012.136. [DOI] [PubMed] [Google Scholar]
- 24.Xu T, Wu S, Yuan Y, Yan G, Xiao D. Knockdown of phosphodiesterase 4D inhibits nasopharyngeal carcinoma proliferation via the epidermal growth factor receptor signaling pathway. Oncol Lett. 2014;8:2110–2116. doi: 10.3892/ol.2014.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge X, Milenkovic L, Suyama K, Hartl T, Purzner T, Winans A, Meyer T, Scott MP. Phosphodiesterase 4D acts downstream of Neuropilin to control Hedgehog signal transduction and the growth of medulloblastoma. Elife. 2015:4. doi: 10.7554/eLife.07068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng X, Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heerboth S, Housman G, Leary M, Longacre M, Byler S, Lapinska K, Willbanks A, Sarkar S. EMT and tumor metastasis. Clin Transl Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]