Abstract
Colorectal cancer (CRC) is one of the most common malignant tumors in the world. Invasion and metastasis are the main cause of mortality in most CRC patients. Polypeptide N-acetylgalactosaminyltransferase 6 (GALNT6) regulated glycosylation, which is frequently altered in cancers, and play an important role in cancer development. However, the role of GALNT6 in CRC remains unknown. To investigate the role of GALNT6 in CRC, first we studied correlation of GALNT6 expression levels with outcomes of CRC patients and found CRC patients with higher expression of GALNT6 had a better overall survival than those with lower expression. In addition, GALNT6 expression were significantly associated with tumor size, histological differentiation and lymph node metastasis. In vitro, GALNT6 overexpression dramatically inhibited cellular colony formation, migration, and invasion, and promoted the apoptosis of CRC cells. In vivo, CRC with GALNT6 overexpression showed reduced pulmonary metastasis in recipient mice compared with the controls. GALNT6 expression was significantly increased in SW480 and SW1116 cells cultured in hypoxic condition, and decreased in HT29 and LOVO cells with oxidative stress. Affimetrix microarray analysis showed that GALNT6 overexpression induced 279 genes up-regulated and 215 genes down-regulated in CRC. GALNT6 overexpression dramatically suppressed AKT and activated CD28 signaling pathway in CRC. AKT rescue experiment found that AKT was involved in GALNT6-induced CRC cell migration and invasion. In conclusion, our results first suggest that GALNT6 plays an important role in development and progression of CRC as a tumor suppressor gene.
Keywords: GALNT6, colorectal cancer, tumor suppressor gene
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors in the world. The incidence of CRC is also increasing over the past few decades in China. For most patients with CRC, metastasis is the main cause of mortality. The high mortality is related to the difficulty of CRC early diagnosis, a lack of effective chemotherapeutic treatment, and a tendency of CRC to infiltrate locally and metastasize into other organs, rendering surgical resection ineffective. Therefore, it is critical to understand the mechanisms underlying the invasive phenotype of CRC.
Glycosylation is frequently altered in cancers and may contribute to the progression of malignant tumors [1]. Some of the most common and striking characteristics of malignant tumors are perturbations to the O-glycosylation pathway and the altered expression of short O-glycans [2]. For instance, there is a marked increase in polypeptide alpha-linked terminal GalNAc in about 90% of solid breast cancers [3]. This single sugar glycan forms the Tn antigen [4], which can be detected by lectins, such as Vicia villosa lectin (VVL) and Helix pomatia lectin [5]. Tn is produced by the polypeptide N-acetylgalactosaminyltransferases (GALNTs) family of enzymes, transmembrane Golgi-resident proteins that catalyze the addition of GalNAc to Ser or Thr residues.
GALNT6 is derived from the polypeptide N-acetyltransferase family, which play an important role in the human tumors (need ref). GALNT6 is a new prognostic indicator for breast cancer and endometrial cancer [6-9]. Overexpression of GALNT6 might contribute to mammary carcinogenesis through aberrant glycosylation and stabilization of mucin-1 (MUC1) [10]. At the same time, fibronectin was O-glycosylated in vivo and thereby stabilized by GALNT6. GALNT6-fibronectin pathway could be a critical component for breast cancer development and progression [11]. Recently, GALNT6 overexpression has also been found in pancreatic cancer [12], ovarian carcinoma and associates with prognosis of these patients [13]. However, the role of GALNT6 in CRC remains unknown. Our current study is to investigate the role of GALNT6 in CRC and the mechanism underlying its function.
Material and method
Patient information and tissue specimens
A total of 84 samples of paraffin-embedded CRC tissues and 77 samples of paraffin-embedded adjacent non-tumor mucosa (ANM) tissues collected between 1990 and 2009, and 8 pairs of fresh CRC tissues and their respective ANM samples collected in 2016 were all from Department of Pathology, the first Affiliated Hospital, Sun Yat-sen University, Guangzhou, China. Prior patient consent and approval from the Institutional Research Ethics Committee were obtained. No patients had received chemotherapy or radiotherapy before operation. The histopathology of the disease was determined according to the criteria of WHO. Clinical staging was evaluated according to Dukes classification.
Cell lines and small interfering RNA (siRNA) sequences
The human CRC cell lines SW480 and SW620 were maintained in Leibovitz’s L-15 Medium (Invitrogen, Carlsbad, CA), HCT116 was grown in McCoy’s 5A Medium (Invitrogen). LOVO and SW1116 were cultured in RPMI-1640 medium (Invitrogen). HT29 was maintained in Dulbecco’s modified Eagle’s medium (Invitrogen). The human colonic epithelial cell line NCM460 was cultured in RPMI-1640 medium. All medium were supplemented with 10% (v/v) fetal bovine serum (Invitrogen), 1× antibiotic/antimycotic (100 units/mL streptomycin, 100 units/mL penicillin, and 0.25 mg/mL amphotericin B). All cell lines were cultured in humidified incubator at 37°C with 5% CO2.
The sequences of GALNT6 siRNA were: sense 5’-GAG AAA UCC UUC GGU GAC ATT-3’, antisense 5’-UGU CAC CGA AGG AUU UCU CTT-3’. The siRNA duplexes were chemically synthesized and purified by Sangon Biotech (Shanghai, China) Co., Ltd. The siRNA was transfected using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer’s instructions. NC-siRNA (Sangon Biotech) were used as negative control.
Quantitative real-time PCR analysis
Quantitative real-time PCR was run on ABI Step One Plus PCR system (Applied Biosystems). The primer sequences used for GALNT6 were followed. Forward primer: 5’-GAC AAG ACA GTG GTG GTG AG. Reverse primer: 5’ GAA GGT CAG GCT CCA GTC A. The geometric mean of housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Takara bio, Dalian, China) was used as internal controls. In hypoxia and oxidative stress assay, β-actin (Sangon biotech, Shanghai, China) was used as internal controls. All experiments were performed in triplicate.
Immunohistochemistry staining and evaluation
The working concentrations of primary antibody for the detection of GALNT6 (Abcam) were 1:500. The immunostaining of GALNT6 was defined as the proportion score multiplied by the staining intensity score. The percentage of GALNT6-positive tumor cells was calculated by counting at least 1,000 carcinoma cells. Staining intensity was graded according to the following criteria: 0 (no staining); 1 (weak staining = light yellow), 2 (moderate staining = yellow brown), and 3 (strong staining = brown). Each tissue section was evalusated in 5 different areas and an average score was used for subsequent analyses. Based on mean staining score of GALNT6 expression, the samples were further divided into two groups: GALNT6high and GALNT6low.
Western blot analysis
20 μg of total proteins were loaded onto 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerico, MA) that were subsequently blocked in 5% non-fat milk in TBST (20 mmol/L Tris, pH 7.6, 137 mmol/L NaCl, 0.1% Tween-20). The membranes were incubated with primary antibodies including rabbit GALNT6 (Abcam, dilution 1:1000) at 4°C overnight. Rabbit anti-glyceraldehyde-3-Phosphate dehydrogenase (GAPDH) (Cell Signaling Technology, dilution 1:1000) was used as the loading control. After washing, the membranes were incubated with HRP-conjugated goat anti-rabbit (Cell Signalling Technology, dilution 1:4000) secondary antibody for 1 h at room temperature and visualized by enhanced chemiluminescence detection kit (Millipore). All experiments were performed three times using 3 individual samples.
Migration and invasion assays
Migration and invasion assays were carried out in Transwell chambers containing polycarbonate filters (8 μm pore size; Corning Incorporated, Life Sciences, NY). 1×105 cells were diluted in a 200 μl volume of serum-free medium and placed in the upper chambers of a 24-well plate, while a 600 μl volume of medium containing 10% FBS was added to the lower chamber as chemoattractant. For invasion assay, the upper chambers were pre-loaded with matrigel. Cells could migrate or invade through the matrigel (BD Biosciences) for 48 to 72 h at 37°C with 5% CO2. Following invasion or migration, cells in the upper chamber were fixed with 4% formaldehyde and stained with 1% crystal violet. Cells on the upper surface of the filters were removed by wiping with a cotton swab. We counted the cells on the lower surface of the filters and get the mean number of cells in three fields of the view. Three independent experiments were performed and the data were presented as mean ± standard deviation (SD).
Scratch wound assay
Cells were plated in 6-well plates and incubated overnight until 70%-90% confluent, then vertical scratches were then made using a 100 μl plastic filter tip to create a ‘wound’ of approximately 100 μm in diameter. To eliminate dislodged cells, culture medium was removed and wells were washed with phosphate buffered saline (PBS). ‘Wound closure’ was observed at 0, 12, 24, 48 hours and digital images were taken under an inverted microscope.
CCK8 assay
Cells (5×103) were plated onto 96-well plates with medium containing 10% FBS and incubated overnight. Cell proliferation was determined at 0, 24, 48, 72, and 96 hours using CCK8 (Jiangsu KeyGEN BioTECH Corp., Ltd, China). The absorbance (OD) was measured at a wavelength of 450 nm using Spectrum Max M5 (Molecular Devices). This experiment was performed in triplicate.
Colony formation assay
For SW1116, HT29, LOVO cells, a total of 100 cells were plated onto 6-well plates, as for SW480 cells, the cell number was 400, and cells were incubated at 37°C in a 5% CO2 incubator for 2 weeks. At the end-point, the cells were washed with cold PBS twice, fixed with 4% paraformaldehyde for 30 minutes and stained with 1% crystal violet solution for 20 minutes at room temperature. The visible colony numbers were counted. This experiment was performed in triplicate.
Cell cycle and apoptosis assay
Stably transfected cells were seeded in 6-well plates and incubated overnight until 50%-60% confluent. The cells were digested from the plates and washed with PBS, collected and adjusted to a cell concentration of 1×106 /ml, taking 1 ml single cell suspension. After centrifugation, the supernatant was removed and the cells were added to the volume fraction of 70% cold ethanol 500 ul fixed 2 hours, washed with PBS to remove the fixative before staining, added 500 μL propidium iodide (PI)/RNase A (Jiangsu KeyGEN BioTECH Corp, Ltd, Cell Cycle Detection Kit, China) staining, protected from light 30-60 min at room temperature, detected on the Cytoflex machine (Beckman).
As for apoptosis assay, the cells were digested from the plates with Trypsin without EDTA and washed with PBS, collected for 1×105, then stained with Annexin V-APC (Jiangsu KeyGEN BioTECH Corp., Ltd, China) binding buffer and PI buffer for 15 minutes at room temperature protected from light.
3×104 cells were analyzed for cell cycle and apoptosis using Flow cytometry analysis (CYTOFLEX, BECHMAN COULTER). The percentage of cells in each phase of the cell cycle and apoptotic cells were quantified using Modfit and Flow jow software, respectively. This experiment was performed in triplicate.
Hypoxia and oxidative stress assay
2×106 cells were plated onto a 3.5 cm2 diameter dish and cultured in 1% oxygen and 37°C incubator. Cells were collected at 12 h, 24 h, 36 h, and 48 h and stored in Trizol at -80°C.
Oxidative stress assay was performed using a complete medium containing 100 μmol/L H2O2 to culture the cells. Cells were collected at 12 h, 24 h, 36 h, and 48 h after cultured in complete medium containing 100 μmol/L H2O2, and stored in trizol at -80°C.
Xenograft tumor model
We used female BALB/c-nude mice (4-5 weeks old and weighing 15-16 g) which were housed under pathogen-free conditions. The mice were randomly assigned into two groups. GALNT6-transfected SW1116 cell lines were injected into one group and empty vector-transfected (controls) SW1116 cell lines were injected into the other BALB/c-nude mice group by tail vein (5×106/mouse). The mice were treated with xx or vehicle for 4 weeks. At the end of the experiment, all mice were euthanized and xx were analyzed. All the experiments were performed following the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication).
Microarray hybridization and gene expression analysis
After performing RNA quality tests with the Thermo NanoDrop2000 and Agilent 2100, aRNA (amplified RNA) was prepared using the GeneChip 3’IVT Express Kit. aRNAs were purified and then fragmented and hybridized to a chip probe (Affymetrix GeneChip Hybridization Wash and Stain Kit). After hybridization, the chips were dyed and finally the images and raw data were scanned using a GeneChip Scanner 3000 (Affymetrix).
The Affymetrix GeneChip® PrimeViewTM Human Gene Expression Arrays were used to measure the gene expression profile of cancer cell lines after extraction. This assays used 49,395 probes, therefore covering 36,000 transcripts and their variants. GALNT6 overexpression and Vector control groups were tested. Three technical replicates were analysed per extract per cell line, resulting in a total of 6 samples submitted for Affymetrix analysis.
Statistical analyses
The Kaplan-Meier survival curves were used to estimate overall survival (OS). The significance of predictor variables for OS was evaluated by the long-rank test. Prognostic factors associated with OS were investigated according to the Cox proportional hazards regression model in a stepwise manner. Only those factors that were statistically significant (P < 0.05) in the univariate survival analysis were included in the multivariate analyses. Statistical analyses were performed using SPSS 16.0 statistical software. P < 0.05 was considered to be statistically significant. Results from cell culture and PCR were compared using an unpaired, two-tailed Student’s tests and results were presented as mean ± SD.
Result
GALNT6 expression in CRC cell lines and fresh CRC tissues
GALNT6 mRNA and protein expressions were higher in CRC cell lines including LOVO, HT29, and SW620, but lower in SW480 and SW1116 compared with human colonic epithelial cell line NCM460 by Q-RT PCR and western blot analysis (Figure 1A, 1B). In addition, GALNT6 expression were significantly higher in all 8 samples of fresh CRC tissues compared with their respective ANM tissues by western blot analysis (Figure 1C).
Figure 1.
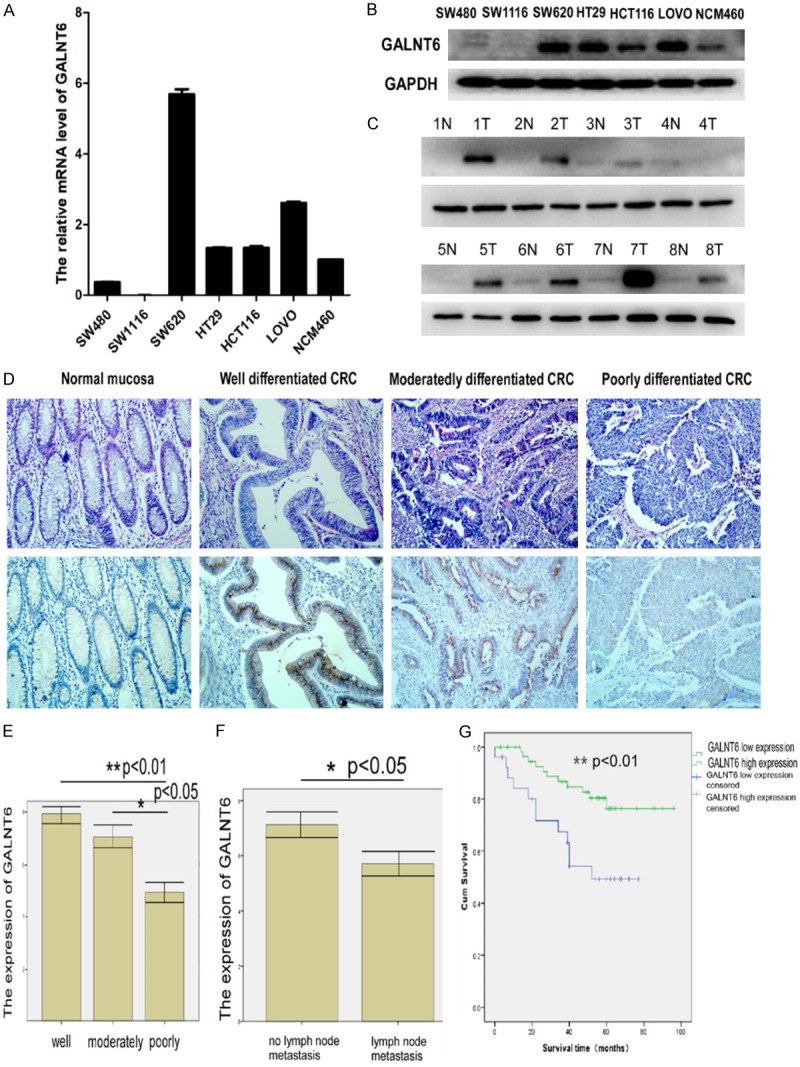
(A, B) GALNT6 expression in CRC cell lines and normal colonic mucosa epithelial cell (NCM460) by real-time PCR (A) and western blot analysis (B); (C) GALNT6 expression in 8 pairs of fresh CRC and adjacent non-tumor colorectal mucosa (ANM) tissues by western blot analysis; (D) GALNT6 expression in normal colorectal mucosa and different differentiated CRC tissues by immunohistochemistry staining ×200; (E) GALNT6 expression decreased with the increased histological grade of CRC; (F) GALNT6 expression was significantly higher in CRC without lymph node metastasis than that with lymph node metastasis; (G) CRC patients with high GALNT6 expression had a significantly better OS rate compared with that with low GALNT6 expression.
GALNT6 expression in paraffin-embedded CRC tissues and its relationship with clinicopathological features of CRC
In our studies, GALNT6 positive signals which present as “sand dots” were mostly located in the cytoplasm of CRC cells by immunohistochemistry staining (Figure 1D). 69% (58 out of 84 samples of paraffin-embedded CRC tissues) versus 14% (11 out of 77 samples of ANM tissues) were GALNT6high (P < 0.001). In addition, GALNT6 expression were significantly associated with tumor size (P = 0.041, Table 1), histological differentiation (P = 0.002, Table 1) and lymph node metastasis (P = 0.013, Figure 3B). GALNT6 expression was significantly higher in well differentiated CRC than that in moderately or poorly differentiated CRC (Figure 1E). In addition, GALNT6 expression was higher in CRC without lymph node metastasis than that in CRC with lymph node metastasis (P < 0.05) (Figure 1F). However, there is no significant correlation between GALNT6 expression and patients’ gender, age, tumor clinical stage, the depth of cancer invasion and tumor metastasis after operation (Table 1).
Table 1.
The relationship between GALNT6 expression and the clinicopathological features of CRC
| Characteristics | GALNT6 expression | |||
|---|---|---|---|---|
|
| ||||
| Low (%) | High (%) | p value | ||
| Gender | Male | 13 (27.7) | 34 (72.3) | 0.138 |
| Female | 13 (35.1) | 24 (64.9) | ||
| Age (years) | ≥ 60 | 17 (34.7) | 32 (65.3) | 0.187 |
| < 60 | 9 (25.7) | 26 (74.3) | ||
| Tumor size (cm) | ≥ 5 | 15 (37.5) | 25 (62.5) | 0.041 |
| < 5 | 11 (25.0) | 33 (75.0) | ||
| Histological differentiation | Well | 3 (16.7) | 15 (83.3) | 0.002 |
| Moderately | 8 (21.1) | 30 (78.9) | ||
| Poorly | 15 (53.6) | 13 (46.4) | ||
| Dukes Staging | A | 0 (0.0) | 5 (100.0) | 0.126 |
| B | 8 (20.5) | 31 (79.5) | ||
| C | 14 (46.7) | 16 (53.3) | ||
| D | 4 (40.0) | 6 (60.0) | ||
| T classification | T1 | 0 (0.0) | 2 (100.0) | 0.430 |
| T2 | 3 (23.1) | 10 (76.9) | ||
| T3+T4 | 23 (33.3) | 46 (66.7) | ||
| N classification | No | 16 (45.7) | 19 (54.3) | 0.013 |
| N1+N2 | 10 (20.4) | 39 (79.6) | ||
| M classification | M0 | 4 (33.3) | 8 (66.7) | 0.917 |
| M1 | 22 (30.6) | 50 (69.4) | ||
Figure 3.
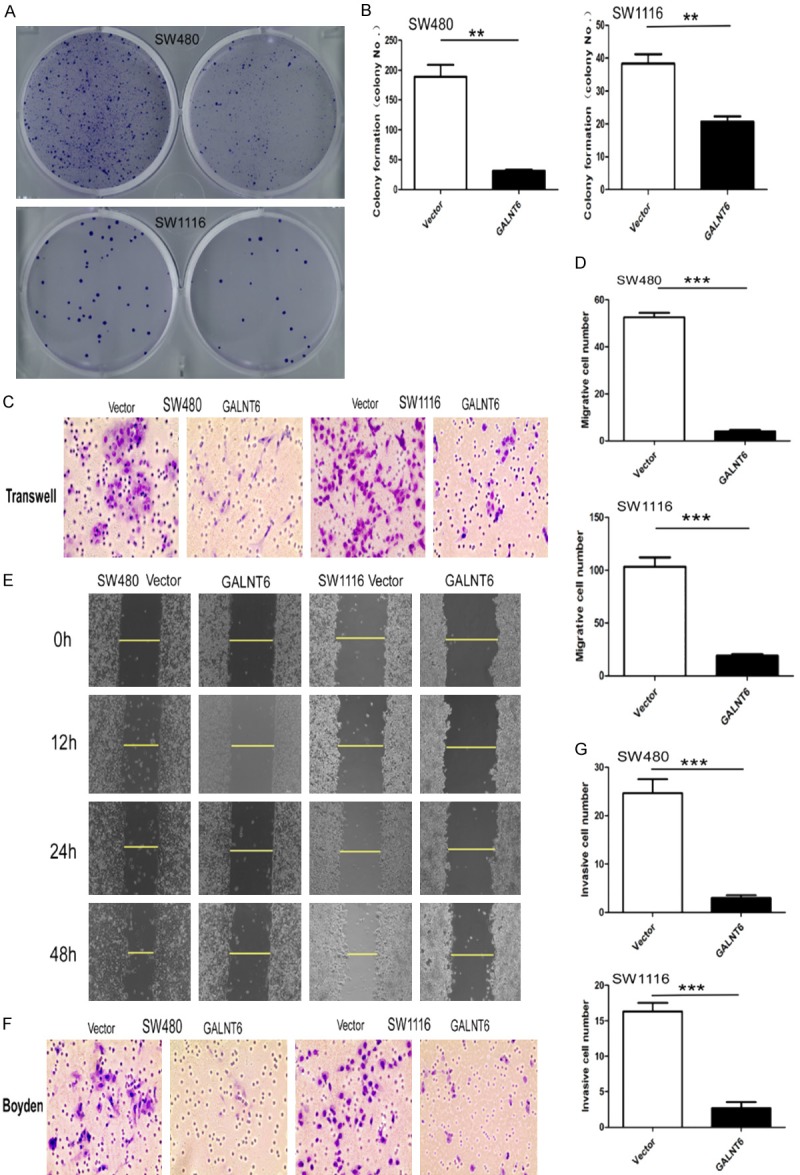
A, B. GALNT6 overexpression dramatically suppressed colony formation of SW480 and SW1116 cells compared with the control group, respectively; C, D. GALNT6 overexpression dramatically inhibited the migration of SW480 and SW1116 cells by transwell analysis; E. GALNT6 overexpression dramatically inhibited the migration of SW480 and SW1116 by wound healing experiment; F, G. GALNT6 overexpression dramatically inhibited the invasion of SW480 and SW1116 cells by Boyden experiment. **P < 0.01, ***P < 0.001.
Prognostic significance of GALNT6 expression in CRC
Kaplan-Meier analysis showed that high GALNT6 expression CRC patients had a significantly higher OS rate compared with low GALNT6 expression patients (LOG RANK P = 0.007, BRESLOW P = 0.004) (Figure 1G). The OS rate for high GALNT6 expression patients at 5-year follow up was 76.4% versus 49.3% for low GALNT6 expression patients. univariate Cox regression analysis indicated that high GALNT6 expression was significantly associated with increased OS (P = 0.023) in CRC patients, indicating GALNT6 expression was an independent prognostic factor for CRC patients. In addition, other clinical parameters including tumor size, histological differentiation, clinical stage and metastasis after operation were also significant prognostic indicators for CRC patients’ OS (Table 2). Furthermore, multivariate Cox regression analysis demonstrated that high GALNT6 expression and clinical stage was an independent prognostic indicator for CRC patients’ OS, respectively (P = 0.047 and P = 0.044) (Table 3).
Table 2.
Univariate Cox regression analysis for the prognostic value of GALNT6 expression and clinicopathological features in CRC
| Characteristics | GALNT6 expression | Hazard ratio | 95% CI | P value | ||
|---|---|---|---|---|---|---|
|
| ||||||
| Low (%) | High (%) | |||||
| Gender | Male | 13 (27.7) | 34 (72.3) | 0.623 | ||
| Female | 13 (35.1) | 24 (64.9) | ||||
| Age (years) | ≥ 60 | 17 (34.7) | 32 (65.3) | 0.822 | ||
| < 60 | 9 (25.7) | 26 (74.3) | ||||
| Tumor size (cm) | ≥ 5 | 15 (37.5) | 25 (62.5) | 1.317 | 1.115~1.555 | 0.001 |
| < 5 | 11 (25.0) | 33 (75.0) | ||||
| Histological differentiation | Well | 3 (16.7) | 15 (83.3) | 0.384 | 0.201~0.735 | 0.004 |
| Moderately | 8 (21.1) | 30 (78.9) | ||||
| Poorly | 15 (53.6) | 13 (46.4) | ||||
| Dukes Staging | A | 0 (0.0) | 5 (100.0) | 3.722 | 2.114~6.553 | 0.000 |
| B | 8 (20.5) | 31 (79.5) | ||||
| C | 14 (46.7) | 16 (53.3) | ||||
| D | 4 (40.0) | 6 (60.0) | ||||
| T classification | T1 | 0 (0.0) | 2 (100.0) | 0.227 | ||
| T2 | 3 (23.1) | 10 (76.9) | ||||
| T3+T4 | 23 (33.3) | 46 (66.7) | ||||
| N classification | No | 16 (45.7) | 19 (54.3) | 2.372 | 1.026 ~5.484 | 0.043 |
| N1+N2 | 10 (20.4) | 39 (79.6) | ||||
| M classification | M0 | 4 (33.3) | 8 (66.7) | 6.971 | 2.941~16.519 | 0.000 |
| M1 | 22 (30.6) | 50 (69.4) | ||||
| GALNT6 expression | Low | 0.846 | 0.733~0.977 | 0.023 | ||
| High | ||||||
Table 3.
Multivariate Cox regression analysis for the prognostic value of GALNT6 expression and clinicopathological feature of CRC
| Characteristics | GALNT6 expression | Hazard ratio | 95% CI | P value | ||
|---|---|---|---|---|---|---|
|
| ||||||
| Low (%) | High (%) | |||||
| Dukes staging | A | 0 (0.0) | 5 (100.0) | 2.230 | 1.023~4.860 | 0.044 |
| B | 8 (20.5) | 31 (79.5) | ||||
| C | 14 (46.7) | 16 (53.3) | ||||
| D | 4 (40.0) | 6 (60.0) | ||||
| M classification | M0 | 4 (33.3) | 8 (66.7) | 2.963 | 0.814~10.783 | 0.099 |
| M1 | 22 (30.6) | 50 (69.4) | ||||
| GALNT6 expression | Low | 0.844 | 0.713~0.998 | 0.047 | ||
| High | ||||||
GALNT6 had no effect on proliferation and cell cycle but induced apoptosis and inhibited cell colony formation in CRC cells
GALNT6 mRNA and protein levels in SW480 and SW1116 cells transfected with GALNT6 overexpressing vector were significantly higher than those in the untreated and vector-transfected control cells using Q-RT PCR and western blot analysis, respectively (Figure 2A, 2B). GALNT6 mRNA and protein levels in LOVO and HT29 transfected with GALNT6 siRNA were significantly lower than those in the untreated and control siRNA-transfected cells using quantitative real-time PCR and western blot analysis, respectively (Figure 2C, 2D).
Figure 2.
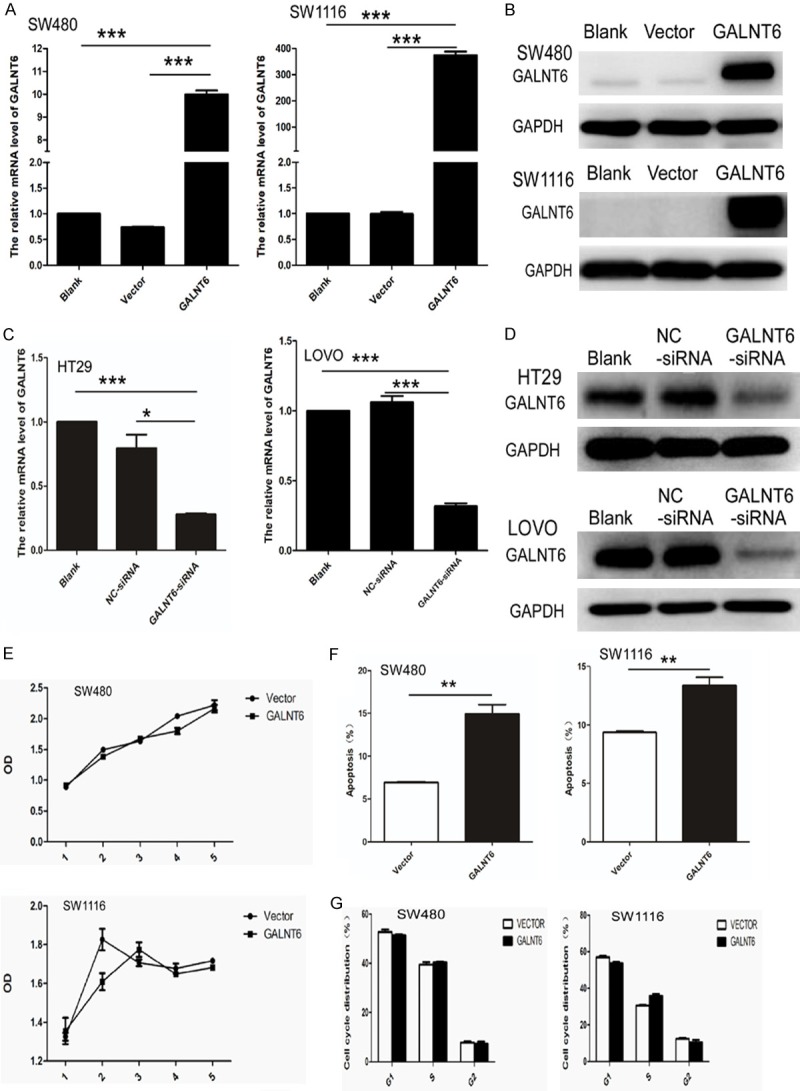
(A, B) GALNT6 mRNA and protein expression increased in SW480 and SW1116 cells stably transfected with GALNT6 expression virus by real-time PCR (A) and western blot analysis (B), respectively; (C, D) GALNT6 mRNA and protein expression decreased in HT29 and LOVO cells transfected with GALNT6-siRNA by real-time PCR (C) and western blot analysis (D), respectively. GAPDH was used as loading control; (E, F) GALNT6 overexpression had no effect on cell proliferation (E) and cell cycle (F) of SW480 and SW1116, respectively; (G) Cell apoptosis was significantly increased in GALNT6 over-expression group compared with the vector-transfected control group. **P < 0.01.
CCK8 assay showed that there was no significant difference on cell proliferation between GALNT6-overexpressing SW480 and SW1116 cells and the vector-transfected control counterparts (Figure 2E). In addition, no significant difference on cell proliferation was observed between GALNT6 knockdown (KD) HT29 and LOVO cells compared with control cells. Flow cytometry analysis showed that there was no significant difference on cell cycling rate between GALNT6 overexpressing and control SW480 and SW1116 cells (Figure 2F) and between GALNT6 KD and control HT29 and LOVO cells. However, cell apoptosis was significantly increased in GALNT6-overexpressing SW480 (P = 0.0020) and SW1116 (P = 0.0047) cells compared with the control counterparts (Figure 2G). GALNT6 overexpression (OE) significantly suppressed the cell colony formation capacity in SW480 and SW1116 cells (Figure 3A, 3B).
GALNT6 expression inhibited migration and invasion capacity of CRC cells
Two different experiments were carried out to clarify the effect of GALNT6 on migration of CRC cells. Transwell migration assay showed that the mean number of migrated cells per field of view (SW480: mean number = 4, SW1116: mean number = 19) was significantly lower in GALNT6 OE cells than that (SW480: mean number = 52, SW1116: mean number = 103) in control cells (SW480: P < 0.0001, SW1116: P = 0.0007) (Figure 3C, 3D). Scratch wound assay showed that cell migration measured at 12, 24, and 48 hours was inhibited in SW480 and SW1116 cells with GALNT6 OE compared with the control cells (Figure 3E).
Transwell matrix penetration assay (Boyden) showed that the mean number of migrated cells per field of view (SW480 mean number = 3, SW1116 mean number = 3) was significantly lower in GALNT6 overexpression cells than that (SW480 mean number = 17, SW1116 mean number = 25) in the control cells (SW480 P < 0.001, SW1116 P < 0.001), (Figure 3F, 3G). The mean number of migrated cells per field of view (HT29 mean number = 22, LOVO mean number = 66) was significantly higher in GALNT6 KD cells than that (HT29 mean number = 7, LOVO mean number = 39) in control cells (HT29 P < 0.001, LOVO P < 0.001, Figure 4A). The mean number of invasive cells (HT29 mean number = 12, LOVO mean number = 74) was significantly higher in GALNT6 KD cells than that (HT29 mean number = 4, LOVO mean number = 15) in control cells (HT29 P < 0.001, LOVO P < 0.001), respectively (Figure 4B).
Figure 4.
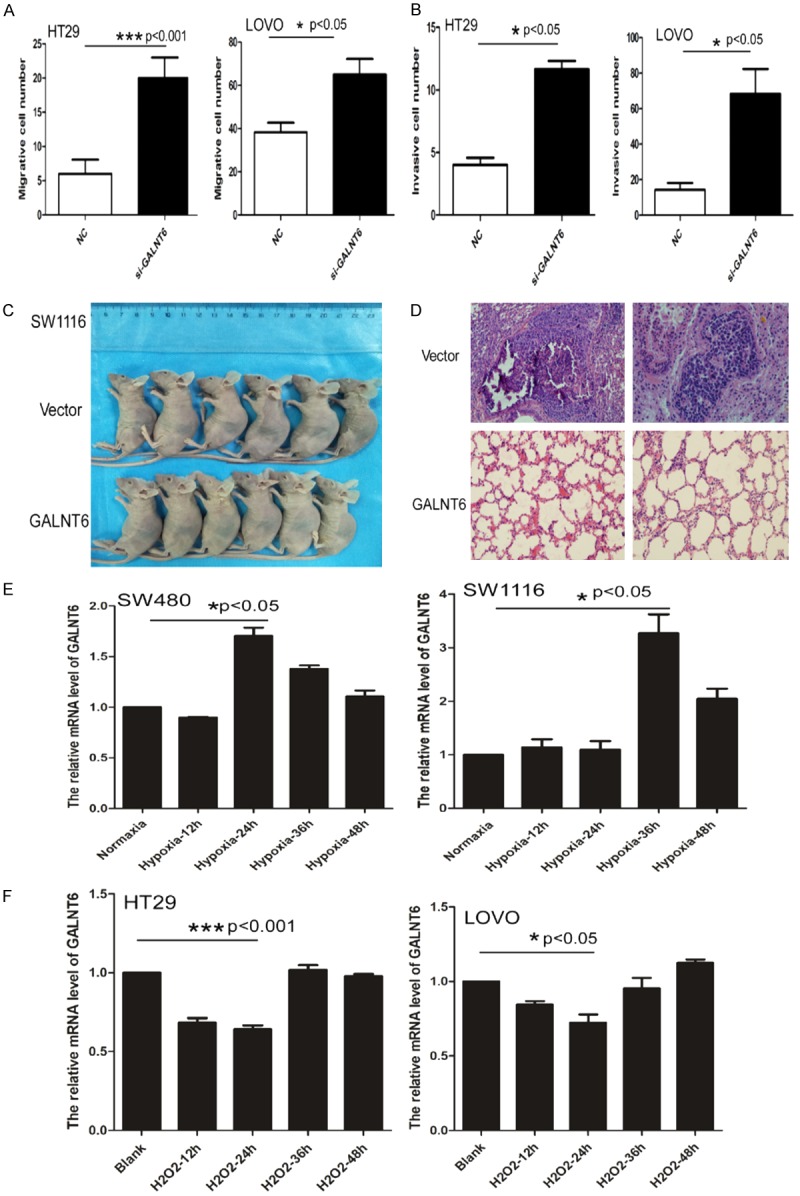
(A, B) GALNT6 knockdown dramatically promoted the migration (A) and invasion (B) of HT29 and LOVO, respectively; (C, D) Nude mice with GALNT6 expression group and vector group (C); Histological staining showed the pulmonary metastatic CRC in the vector group but no CRC metastasis in GALNT6 over-expression group. Haematoxylin & eosin staining ×200. (E) GALNT6 expression significantly increased under hypoxia culture condition for SW480 at 24 hours and SW1116 cells at 36 hours compared with the control group, respectively by real-time PCR. (F) H2O2 treatment dramatically inhibited GALNT6 expression in HT29 and LOVO cells at 12 and 24 hours compared with the control group, respectively by real-time PCR.
GALNT6 inhibit the metastasis of CRC cells in vivo
To determine the role of GALNT6 levels in CRC metastasis in vivo, we transplanted SW1116 cells with or without GALNT6 OE into nude mice. After 4 weeks’ follow-up, the mice were euthanized and autopsy was performed. The result showed that metastatic CRC tumor was found in the lung (2/6 mice) in the recipient mice transplanted with control cells, but not in the recipient mice transplanted with GALNT6 OE cells (Figure 4C, 4D).
Hypoxia increased GALNT6 expression and oxidative stress inhibited GALNT6 expression in CRC cells
To determine the effect of hypoxia on GALNT6 expression in CRC cells, SW480 and SW1116 cells were cultured in hypoxia (1% oxygen concentration) for 12, 24, 36, and 48 hours and then GALNT6 expression was detected by real-time PCR analysis. The result showed that GALNT6 mRNA expression was significantly increased at 24 h in SW480 cells (P = 0.0133) and at 36 h in SW1116 cells compared with the control cells (P = 0.0228) (Figure 4E).
To further determine the effect of oxidative stress on GALNT6 expression in CRC cells, we cultured HT29 and LOVO cells which is high in GALNT6 expression with 100 umol/L H2O2 to cause oxidative stress. We found expression level of GALNT6 mRNA was significantly decreased in HT29 and LOVO cells at 12 h and 24 h with oxidative stress compared with the control cells without oxidative stress (HT29 P=0.0048, LOVO P=0.0377) (Figure 4F).
GeneChip analysis and ingenuity pathway analysis (IPA)
As the cluster analysis map showed in Figure 5A, the Affymetrix GeneChip analysis using SW480 cells with or without GALNT6 overexpression revealed that GALNT6 overexpression induced up-regulation of 279 genes and down-regulation of 215 genes (fold change > 1.5, P < 0.05). IPA analysis showed that GALNT6 overexpression resulted in some disease and function change dramatically (Figure 5B). Overexpression of GALNT6 significantly increased apoptosis (Z-score = 2.426) and cell death related genes (Z-score = 2.266), and significantly suppressed tumor invasion related pathways in CRC cell lines (Z-score = -3.121).
Figure 5.
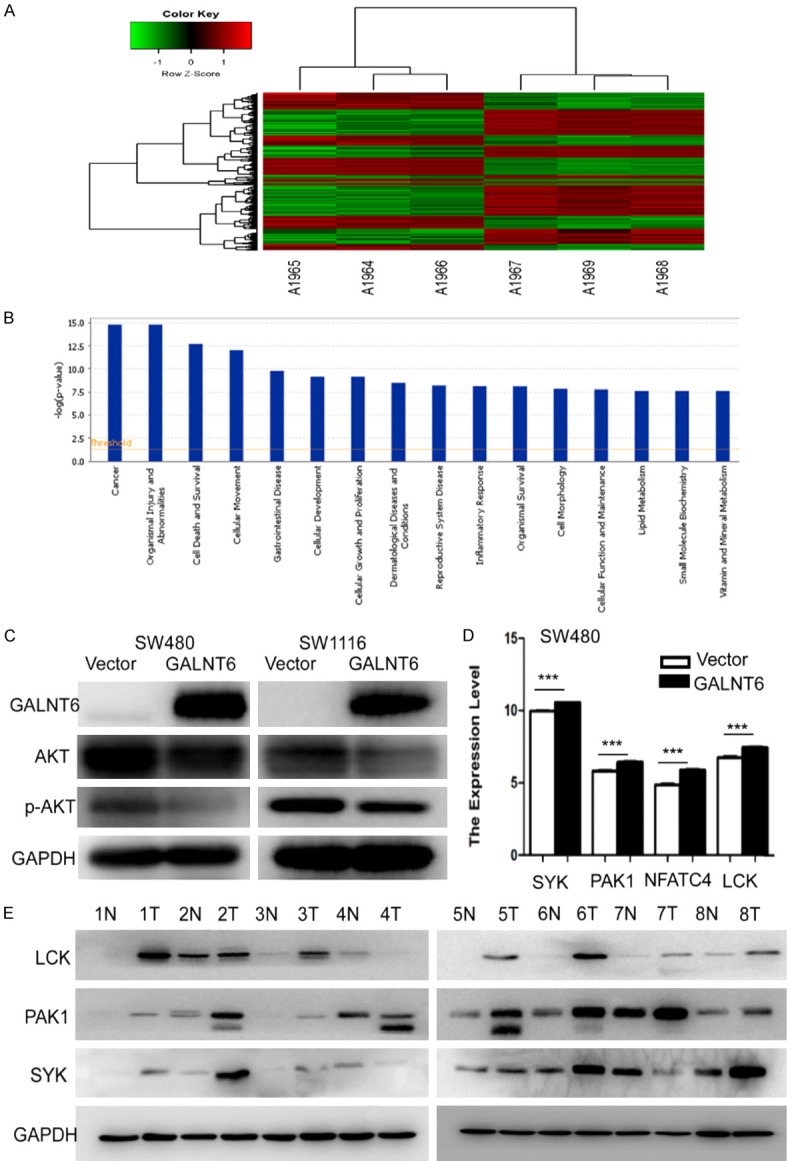
A. The graph of GALNT6 overexpression group and Vector control group which were clustered by using the expression profile of differential genes screened with Fold Change > 1.5 and FDR < 0.05 as standard. In the cluster analysis graph, each column represented a sample (A1964-1966 represented the three replicate groups of vector, A1967-1969 represented the three replicate groups of GALNT6 overexpression), each row represented a differential gene; the upper tree structure was based on the expression profile of the differential genes, aggregation or classification of all samples; the left tree structure represented the difference of genes. Red color indicates the relative increase of gene expression, green color indicates that gene expression is relatively down-regulated, black color indicates no significant change of gene expression, and gray color indicates that the signal intensity of the gene is not detected; B. The significant enrichment of the differentially expressed genes in disease and function. The abscissa is the path name and the ordinate is the level of significance of enrichment (base 10 negative logarithmic transformation); C. GALNT6 overexpression inhibited AKT and p-AKT expression in SW480 and SW1116 cells by western blot analysis; D. SYK, PAK1, NFAT4, and LCK expression were significantly upregulated in SW480 stably transfected with GALNT6 overexpression virus compared with the vector control group by real-time PCR analysis; E. LCK, PAK1, and SYK expression were elevated in 8 pairs of fresh CRC tissues compared with their respective ANM tissues by western blot analysis.
The regulatory network in the regulatory effect analysis suggested that Akt might inhibit the migration, invasion of carcinoma, and vasculogenesis through regulating ALOX5AP, AMOT, etc (data not shown). We verified the changes of AKT and p-AKT in SW480 and SW1116 cells with GALNT6 overexpression and found that AKT and p-AKT expression were significantly decreased in CRC cells with GALNT6 overexpression by western blot analysis (Figure 5C). Using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analysis, we found that the CD28 signaling pathway was activated. Furthermore, we found that CD28 signaling pathway related genes including PAK1, LCK, SYK, and NFAT4 were significantly upregulated in SW480 cells with GALNT6 overexpression compared with the control cells by Genechip analysis (Figure 5D). We also verified that PAK1, LCK and SYK protein expression were elevated in 8 pairs of fresh CRC tissues compared with their respective ANM tissues using western blot analysis (Figure 5E).
The AKT agonist reversed the inhibitory effect of GALNT6 on AKT in CRC cells
In order to detect whether AKT activator could rescue inhibition of GALNT6 on p-AKT in CRC cells, SW480 and SW1116 with GALNT6 OE were treated with SC79 (AKT agonist, Selleckchem No. S7863, TX, USA) at the concentrations of 4 ug/ml and 8 ug/ml for 30 minutes and 5 hours, respectively. Western blot showed that SC79 treatment significantly increased p-AKT expression in GALNT6 OE SW480 and SW1116 cells at 30 minutes and 5 h. No significant difference on p-AKT expression level was observed in SW480 and SW1116 cells, regardless of SC79 treatment (Figure 6A, 6B).
Figure 6.
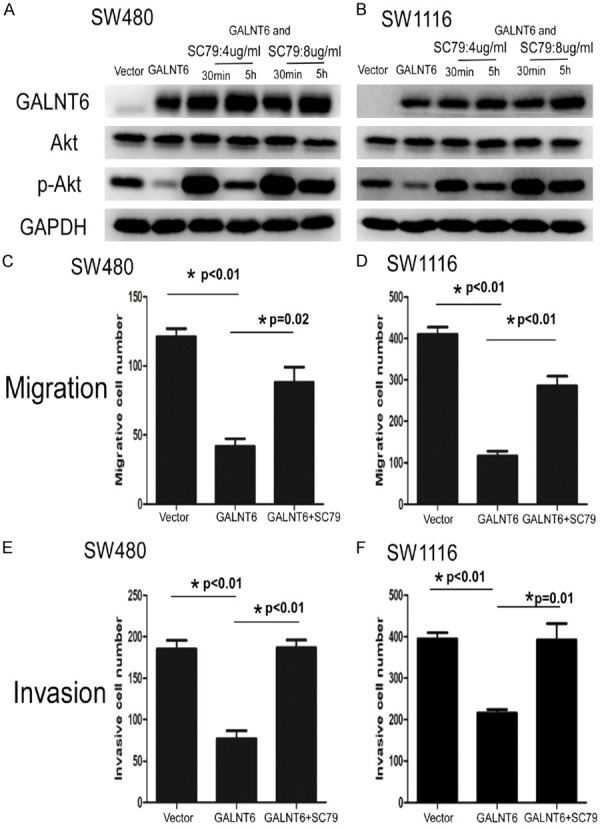
A, B. p-AKT expression was significantly reversed the suppression in SW480 and SW1116 with GALNT6 overexpression by SC79 (AKT agonist) treatment at the concentration of 4 ug/ml and 8 ug/ml for 30 minutes and 5 hours using western blot ananlysis, respectively. C, D. SC79 treatment at the concentration of 4 ug/ml for 30 minutes dramatically reversed migration suppression in SW480 and SW1116 with GALNT6 overexpression by transwell analysis, respectively. E, F. SC79 treatment at the concentration of 4 ug/ml for 30 minutes dramatically reversed invasion suppression in SW480 and SW1116 with GALNT6 overexpression by transwell matrix penetration assay, respectively.
To clarify the involvement of AKT on GALNT6-induced decreased migration and invasion of CRC cells, we performed transwell migration analysis and showed that SC79 treatment at the concentration of 4 ug/ml for 30 minutes dramatically rescued migration suppression in SW480 and SW1116 cells caused by GALNT6 overexpression (SW480 P = 0.02; SW1116 P = 0.003) (Figure 6C, 6D). Invasion assay also showed that SC79 treatment (4 ug/ml) for 30 minutes dramatically reversed invasion suppression in SW480 and SW1116 cells resulted from GALNT6 overexpression (SW480 P = 0.001; SW1116 P = 0.013) (Figure 6E, 6F). However, SC79 treatment fail to reverse the increased apoptosis of SW480 and SW1116 induced by GALNT6 overexpression by flow cytometry analysis (data not shown).
Discussion
In this study, our results showed that GALNT6 was highly expressed in CRC. GALNT6 expression was closely related to histological differentiation, tumor size, and lymph node metastasis. Emerging evidence have demonstrated that GALNT6 is highly expressed in various type of cancers. The positive expression of GALNT6 is associated with low-grade breast cancer (6). Additionally, GALNT6 expression is associated with recurrence of ovarian cancer, lymph node metastasis, and increased resistance to chemotherapy. The higher GALNT6 level is significantly associated with poorer patient survival (13). However, GALNT6-positive pancreatic cancer patients have a good prognosis [14], and GALNT6-positive endometrial cancer patients also have a higher survival rate (8). These contradictory findings suggest that the role of GALNT6 is related to the type of cancer. Our data showed that GALNT6 expression is associated with histological differentiation of CRC, with higher levels seen in the less differentiated CRC. CRC Patients with higher GALNT6 expression had a better prognosis than those with lower GALNT6 expression, suggesting GALNT6 expression is an independent prognostic factor for CRC patients.
As for the biological role of GALNT6 in cancers, overexpression of GALNT6 may be accompanied by abnormal glycosylation of mucin-1 (MUC1), which stabilize it and promote the development of breast cancer. Fibronectin can also be stabilized by O-glycosylation, affecting the evolution of breast cancer (10). Overexpression of GALNT6 reduces cell adhesion and destroys mammary acinar (11). Upregulation of GALNT6 expression in pancreatic cancer results in a high O-glycosylation of mucins. Knockdown of GALNT6 expression in pancreatic cancer cells by siRNA reduces the amount of Mucin 4 and decreases the levels of human epidermal growth factor receptor 2 (EGFR2) and extracellular signal-regulated kinase, and significantly reduces the proliferation of pancreatic cancer cells. GALNT6 knock-down also causes changes in the morphology of pancreatic cells, accompanied by a shift from P-cadherin to E-cadherin (12). GALNT6 knock-down significantly inhibited the viability, migration, and invasiveness of ovarian cancer cells while reducing the phosphorylation of EGFR. Experiments with Vicia villosa lectin (VVA) have shown that GALNT6 can modify O-glycans on EGFR. The EGFR inhibitor erlotinib significantly reversed the high invasiveness of GALNT6-induced ovarian cancer cells (13). GALNT6 is highly expressed in up to 79% of gastric cancer tissues, and its expression level is significantly correlated with the venous invasiveness of cancer cells [15]. The results suggest that GALNT6 as an oncogene promotes the development of these malignancies and is associated with a poor prognosis in patients. However, studies have shown that GALNT6 expression is low in endometrial cancer cell lines, and there is a significant correlation with well-differentiated endometrial cancer (P = 0.002) and non-muscle infiltration (P = 0.017). Overexpression of GALNT6 significantly reduced the invasiveness of endometrial cancer cells (8). The results suggest that GALNT6 may play a tumor suppressor gene role in endometrial cancer. The role of GALNT6 in CRC remains unknown. Lavrsen et al. have reported that GALNT6 inhibits the differentiation and proliferation of colon cancer cells [16]. Their study used the well-differentiated human LS174T colon adenocarcinoma cell line to evaluate colon cell growth. Our study has found that up-regulation of GALNT6 inhibits the ability of CRC cell colony formation, migration and invasion, but promotes the apoptosis of CRC cells, although the effect of GALNT6 on the proliferation and cell cycle of CRC cells was not significant. O-GlcNAc level was elevated in response to numerous forms of cell stress and tissue injury including heat stress, oxidative stress, ethanolic stress, genotoxic stress, reductive stress, ER stress, hypoxia reoxygenation, osmotic stress, ATP depletion, ischemia reperfusion injury, and trauma hemorrhage [17]. Studies have shown that oxidative stress can lead to genetic changes in cells and changes in the level of DNA methylation eventually lead to carcinogenesis. Genetic changes caused by reactive oxygen species include DNA base modifications, DNA strand damage, and DNA-protein cross-linking, all of which increase gene mutation rates. In addition, changes in DNA methylation induced by reactive oxygen species can lead to silencing of tumor suppressor genes or activation of oncogenes, thereby promoting malignant transformation of cells [18]. Therefore, the change of GALNT6 expression may be related to the regulation of promoter methylation or demethylation. Gene chip results suggest that DNA demethylation can upregulate GALNT6 expression in colorectal cancer cells, and our result showed that GALNT6 expression significantly increased under hypoxia culture condition for SW480 at 24 hours and SW1116 cells at 36 hours. H2O2 treatment dramatically inhibited GALNT6 expression in HT29 and LOVO cells at 12 and 24 hours. Then we hypothesized that the GALNT6 promoter may be demethylated and result in up-regulation of GALNT6 expression due to a decrease in intracellular oxygen levels during hypoxia. Based on the above data, GALNT6 plays an important role in CRC as tumor suppressor gene. Hypoxia could induce GALNT6 expression in CRC.
Through the microarray IPA, we have found that overexpression of GALNT6 inhibits tumor-associated genes, regulates differential genes by inhibiting AKT expression, and activates CD28 signaling pathways to affect the function of colorectal cancer cells. We verified the changes of AKT and p-AKT in SW480 and SW1116 cells stably transfected with GALNT6 overexpression virus, and found that AKT and p-AKT expression were significantly decreased upon GALNT6 up-regulation by Western blot analysis. Activation of PI3K/Akt signaling pathway leads to decreased apoptosis and stimulates cell growth and proliferation (or your own data?). Under normal conditions, the activation of PI3K/Akt is tightly controlled and depends on extracellular growth signals as well as the supply of amino acids and glucose. The PI3K/Akt signaling pathway is continuously activated in the development of most tumors, including lung cancer [19-23], gastric cancer [24-27], and colorectal cancer (10, 15, 16). Studies have shown that phosphorylation and glycosylation may be mutually antagonistic at some protein modification sites [28,29]. Moreover, up-regulation of GALNT6 significantly activates the CD28 signaling pathway, we found that PAK1, LCK, SYK, and NFAT4 were significantly upregulated in SW480 cells with GALNT6 OE. We also verified that expression of PAK1, LCK and SYK were elevated in fresh CRC tissues compared to their respective ANM tissues. CD28 signaling pathway promotes the maturation of naive CD8+ T cells and kills tumor cells [30]. Furthermore, AKT rescue experiment showed that AKT mediated GALNT6-induced reduction of CRC cell migration and invasion. The mechanism of AKT regulating CRC metastasis through GALNT6 needs further study.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81472251, 81272636) and Natural Science Foundation of Guangdong Province (2017A030313552).
Disclosure of conflict of interest
None.
References
- 1.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 2.Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim Biophys Acta. 1999;1473:67–95. doi: 10.1016/s0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- 3.Gill DJ, Tham KM, Chia J, Wang SC, Steentoft C, Clausen H, Bard-Chapeau EA, Bard FA. Initiation of GalNAc-type O-glycosylation in the endoplasmic reticulum promotes cancer cell invasiveness. Proc Natl Acad Sci U S A. 2013;110:E3152–E3161. doi: 10.1073/pnas.1305269110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira JA, Magalhães A, Gomes J, Peixoto A, Gaiteiro C, Fernandes E, Santos LL, Reis CA. Protein glycosylation in gastric and colorectal cancers: toward cancer detection and targeted therapeutics. Cancer Lett. 2017;387:32–45. doi: 10.1016/j.canlet.2016.01.044. [DOI] [PubMed] [Google Scholar]
- 5.Ju T, Otto VI, Cummings RD. The Tn antigen-structural simplicity and biological complexity. Angew Chem Int Ed Engl. 2011;50:1770–1791. doi: 10.1002/anie.201002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen TT, Kurita T, Koi C, Murakami M, Kagami S, Hachisuga T, Masanori H, Morimoto Y, Izumi H. GalNAc-T6 in the relationship with invasion ability of endometrial carcinomas and prognostic significance. Am J Cancer Res. 2017;7:1188–1197. [PMC free article] [PubMed] [Google Scholar]
- 7.Yang R, Zhang H, Ma Y, Gong S, Niu J, Ma J, Zhong A. The role of ppGalNAc-T family in breast cancer development and progression. Indian J Cancer. 2015;52(Suppl 3):E144–E147. doi: 10.4103/0019-509X.186556. [DOI] [PubMed] [Google Scholar]
- 8.Berois N, Mazal D, Ubillos L, Trajtenberg F, Nicolas A, Sastre-Garau X, Magdelenat H, Osinaga E. UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-6 as a new immunohistochemical breast cancer marker. J Histochem Cytochem. 2006;54:317–328. doi: 10.1369/jhc.5A6783.2005. [DOI] [PubMed] [Google Scholar]
- 9.Freire T, Berois N, Sonora C, Varangot M, Barrios E, Osinaga E. UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 6 (ppGalNAc-T6) mRNA as a potential new marker for detection of bone marrow-disseminated breast cancer cells. Int J Cancer. 2006;119:1383–1388. doi: 10.1002/ijc.21959. [DOI] [PubMed] [Google Scholar]
- 10.Park JH, Nishidate T, Kijima K, Ohashi T, Takegawa K, Fujikane T, Hirata K, Nakamura Y, Katagiri T. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70:2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. [DOI] [PubMed] [Google Scholar]
- 11.Park JH, Katagiri T, Chung S, Kijima K, Nakamura Y. Polypeptide N-acetylgalactosaminyltransferase 6 disrupts mammary acinar morphogenesis through O-glycosylation of fibronectin. Neoplasia. 2011;13:320–326. doi: 10.1593/neo.101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarhan YE, Kato T, Jang M, Haga Y, Ueda K, Nakamura Y, Park JH. Morphological changes, cadherin switching, and growth suppression in pancreatic cancer by GALNT6 knockdown. Neoplasia. 2016;18:265–272. doi: 10.1016/j.neo.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin TC, Chen ST, Huang MC, Huang J, Hsu CL, Juan HF, Lin HH, Chen CH. GALNT6 expression enhances aggressive phenotypes of ovarian cancer cells by regulating EGFR activity. Oncotarget. 2017;8:42588–42601. doi: 10.18632/oncotarget.16585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Yamada S, Inenaga S, Imamura T, Wu Y, Wang KY, Shimajiri S, Nakano R, Izumi H, Kohno K, Sasaguri Y. Polypeptide N-acetylgalactosaminyltransferase 6 expression in pancreatic cancer is an independent prognostic factor indicating better overall survival. Br J Cancer. 2011;104:1882–1889. doi: 10.1038/bjc.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes J, Marcos NT, Berois N, Osinaga E, Magalhães A, Pinto-de-Sousa J, Almeida R, Gärtner F, Reis CA. Expression of UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-6 in gastric mucosa, intestinal metaplasia, and gastric carcinoma. J Histochem Cytochem. 2009;57:79–86. doi: 10.1369/jhc.2008.952283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavrsen K, Dabelsteen S, Vakhrushev SY, Levann AMR, Haue AD, Dylander A, Mandel U, Hansen L, Frödin M, Bennett EP, Wandall HH. De novo expression of human polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) in colon adenocarcinoma inhibits the differentiation of colonic epithelium. J Biol Chem. 2018;293:1298–1314. doi: 10.1074/jbc.M117.812826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groves JA, Lee A, Yildirir G, Zachara NE. Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones. 2013;18:535–558. doi: 10.1007/s12192-013-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yara S, Lavoie JC, Levy E. Oxidative stress and DNA methylation regulation in the metabolic syndrome. Epigenomics. 2015;7:283–300. doi: 10.2217/epi.14.84. [DOI] [PubMed] [Google Scholar]
- 19.Sheta R, Woo CM, Roux-Dalvai F, Fournier F, Bourassa S, Droit A, Bertozzi CR, Bachvarov D. A metabolic labeling approach for glycoproteomic analysis reveals altered glycoprotein expression upon GALNT3 knockdown in ovarian cancer cells. J Proteomics. 2016;145:91–102. doi: 10.1016/j.jprot.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyahara N, Shoda J, Kawamoto T, Furukawa M, Ueda T, Todoroki T, Tanaka N, Matsuo K, Yamada Y, Kohno K, Irimura T. Expression of UDP-N-acetyl-alpha-D-galactosamine-polypeptide N-acetylgalactosaminyltransferase isozyme 3 in the subserosal layer correlates with postsurgical survival of pathological tumor stage 2 carcinoma of the gallbladder. Clin Cancer Res. 2004;10:2090–2099. doi: 10.1158/1078-0432.ccr-1024-03. [DOI] [PubMed] [Google Scholar]
- 21.Sheta R, Roux-Dalvai F, Woo CM, Fournier F, Bourassa S, Bertozzi CR, Droit A, Bachvarov D. Proteomic dataset for altered glycoprotein expression upon GALNT3 knockdown in ovarian cancer cells. Data Brief. 2016;8:342–349. doi: 10.1016/j.dib.2016.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito N, Wijenayaka AR, Prideaux M, Kogawa M, Ormsby RT, Evdokiou A, Bonewald LF, Findlay DM, Atkins GJ. Regulation of FGF23 expression in IDG-SW3 osteocytes and human bone by pro-inflammatory stimuli. Mol Cell Endocrinol. 2015;399:208–218. doi: 10.1016/j.mce.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Harada Y, Izumi H, Noguchi H, Kuma A, Kawatsu Y, Kimura T, Kitada S, Uramoto H, Wang KY, Sasaguri Y, Hijioka H, Miyawaki A, Oya R, Nakayama T, Kohno K, Yamada S. Strong expression of polypeptide N-acetylgalactosaminyltransferase 3 independently predicts shortened disease-free survival in patients with early stage oral squamous cell carcinoma. Tumour Biol. 2016;37:1357–1368. doi: 10.1007/s13277-015-3928-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibao K, Izumi H, Nakayama Y, Ohta R, Nagata N, Nomoto M, Matsuo K, Yamada Y, Kitazato K, Itoh H, Kohno K. Expression of UDP-N-acetyl-alpha-D-galactosamine-polypeptide galNAc N-acetylgalactosaminyl transferase-3 in relation to differentiation and prognosis in patients with colorectal carcinoma. Cancer. 2002;94:1939–1946. doi: 10.1002/cncr.10423. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa M, Kitayama J, Nariko H, Kohno K, Nagawa H. The expression pattern of UDP-N-acetyl-alpha-d-galactosamine: polypeptide N-acetylgalactosaminyl transferase-3 in early gastric carcinoma. J Surg Oncol. 2004;86:28–33. doi: 10.1002/jso.20042. [DOI] [PubMed] [Google Scholar]
- 26.Kitada S, Yamada S, Kuma A, Ouchi S, Tasaki T, Nabeshima A, Noguchi H, Wang KY, Shimajiri S, Nakano R, Izumi H, Kohno K, Matsumoto T, Sasaguri Y. Polypeptide N-acetylgalactosaminyl transferase 3 independently predicts high-grade tumours and poor prognosis in patients with renal cell carcinomas. Br J Cancer. 2013;109:472–481. doi: 10.1038/bjc.2013.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang ZQ, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, Sebastianelli A, Bachvarov D. Role of the polypeptide N-acetylgalactosaminyltransferase 3 in ovarian cancer progression: possible implications in abnormal mucin O-glycosylation. Oncotarget. 2014;5:544–560. doi: 10.18632/oncotarget.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Jiang K, Li Y, Gao D, Sun L, Zhang S, Liu T, Guo K, Liu Y. Histidine-rich glycoprotein function in hepatocellular carcinoma depends on its N-glycosylation status, and it regulates cell proliferation by inhibiting Erk1/2 phosphorylation. Oncotarget. 2015;6:30222–30231. doi: 10.18632/oncotarget.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu SY, Shun CT, Hung KY, Juan HF, Hsu CL, Huang MC, Lai IR. Mucin glycosylating enzyme GALNT2 suppresses malignancy in gastric adenocarcinoma by reducing MET phosphorylation. Oncotarget. 2016;7:11251–11262. doi: 10.18632/oncotarget.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki T, Ogawa S, Tanabe K, Tahara H, Abe R, Kishimoto H. Induction of antitumor immune response by homeostatic proliferation and CD28 signaling. J Immunol. 2008;180:4596–4605. doi: 10.4049/jimmunol.180.7.4596. [DOI] [PubMed] [Google Scholar]


