Abstract
LncRNAs play significant roles in various cell biological processes. In the present study, we demonstrated that PICART1 expression was down-regulated in non-small cell lung cancer (NSCLC) tissues. Lower expression level of PICART1 was associated with advanced stage. In addition, PICART1 expression was down-regulated in NSCLC cell lines. Overexpression of PICART1 inhibited NSCLC cell growth and induced cell cycle arrest at G2/M phase. Elevated expression of PICART1 suppressed NSCLC cell colony formation and cell invasion. Ectopic expression of PICART1 promoted the expression of epithelial marker E-cadherin while suppressed the mesenchymal marker expression such as N-cadherin and Snail and Vimentin. Furthermore, PICART1 overexpression suppressed AKT phosphorylation and c-Myc expression while inhibited the p21 expression in NSCLC cell. AKT phosphorylation was involved in PICART1 mediated suppression of cell growth and invasion. These results suggested that overexpression of PICART1 suppressed cell growth and invasion partly through regulating AKT signaling pathway in NSCLC.
Keywords: Non-small cell lung cancer, long non-coding RNAs, PICART1, AKT
Introduction
Lung cancer ranks the leading cause of tumor-related death worldwide [1-5]. This tumor consists of two groups: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) [6-8]. NSCLC is further divided into subtypes of large cell carcinoma, adenocarcinoma and squamous cell carcinoma [6,9-11]. The predominant subtype of lung cancer is adenocarcinoma [12-14]. The causes of NSCLC are attributed to the combination of genetic factors, air pollution, radon gas, tobacco smoke and other factors [15-17]. It is challenging to diagnose lung cancer at early stage and most cases are diagnosed at advanced stage [7,18,19]. Therefore, it is important to study the mechanisms underlying NSCLC metastasis and invasion and find novel therapeutic strategies for NSCLC.
Long non-coding RNAs (lncRNAs) are longer than 200 nucleotides in length with no or limited protein-coding capacity [20-23]. Accumulating studies proved that lncRNAs were involved in various biological processes including cell growth, development, differentiation, invasion and fate decision [24-27]. Moreover, lncRNAs were deregulated in many tumors such as osteosarcoma, hepatocellular carcinoma, gastric cancer, colorectal cancer, gallbladder cancer and bladder cancer [28-33]. Recently, a new lncRNA p53-inducible cancer-associated RNA transcript 1 (PICART1) was identified by using transcriptome sequencing analysis [34]. Previous evidences showed that PICART1 expression was down-regulated in colorectal and breast cancer. However, the expression and role of PICART1 are still unknown in NSCLC.
The present study tried to determine the expression and role of PICART1 in NSCLC. We demonstrated that PICART1 expression level was down-regulated in NSCLC tissues compared to the paired adjacent non-cancerous tissues. Moreover, overexpression of PICART1 inhibited NSCLC cell proliferation and induced cell cycle arrest at G2/M phase. Elevated expression of PICART1 suppressed NSCLC cell colony formation and cell invasion.
Materials and methods
Patient samples and cell cultured and transfected
A total of 40 NSCLC samples and paired adjacent non-cancerous tissues were gathered from The First Affiliated Hospital of Harbin Medical University. None of these cases underwent radiotherapy or chemotherapy before surgery. All samples were collected and immediately stored at the -80°C until the protein and RNA were extracted. Our study was approved by the ethics committee of The First Affiliated Hospital of Harbin Medical University. Written consent from each patient was obtained. Human lung adenocarcinoma cell lines (A549, H23, H1299 and SPC-A1) and one bronchial epithelial cell line (16HBE) were purchased from Cell Biology of Shanghai Institute (Shanghai, China) and were kelpt in the RPMI-1640 (Hyclone, USA) and Dulbecco’s Modified Eagle Medium (DMEM) supplemented with penicillin-streptomycin (Sigma, USA) and 10% FBS (Invitrogen-GIBCO). pcDNA-PICART1 vector and the control vector were obtained from the GenePharma company (Shanghai, China). Cell tranfection was carried out by using the Lipofectamine 2000 (Invitrogen, USA) following to the manufacturer’s information.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNAs from samples or tissues were extracted by using TRizol reagent (Invitrogen, USA) and cDNA was synthesized following to the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) was carried out to determine the expression of PICART1 and cyclin D1, E-cadherin, N-cadherin, Snail and Vimentin by using SYBR Green qPCR Mixes (Invitrogen, CA) following to the manufacturer’s information. qRT-PCR was conducted on the IQ5 real-time PCR System machine. The primers were shown in the Table 1.
Table 1.
Primer sequence
| Name | Sequence (5’-3’) |
|---|---|
| Real-time PCR primer sequence | |
| U6 snRNA | CTCGCTTCGGCAGCACATATACT |
| ACGCTTCACGAATTTGCGTGTC | |
| Cyclin D1 | AACTACCTGGACCGCTTCCT |
| CCACTTGAGCTTGTTCACCA | |
| GAPDH | AATGGGCAGCCGTTAGGAAA |
| TGAAGGGGTCATTGATGGCA | |
| Vimentin | GAAGAGGTTAGTGGAGTGA |
| TGCTGTTCCTGAATCTGA |
Western blot analysis
Total protein lysates were extracted from cells or tissues. Equal amounts of protein were separated by using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (PVDF, Bio-Rad, Hercules, CA). After blocking with non-fat milk, the membrane was incubated with GAPDH and p-AKT antibodies (Bioworld Biotechnology) at 1:3000 dilutions at 4°C overnight. After being washed in TBST, he membrane was incubated was incubated with secondary antibody (Pierce, IL, USA). The protein band was detected by using ECL reagents (Pierce, IL, USA).
Cell growth, cell cycle and colony formation and invasion
For cell growth assay, cells were incubated in the 96-well plate and stained with sterile MTT dye (Invitrogen) at differenct time points. The absorbance at the 490 nm was determined by using the multilabel plate reader (PerkinElmer, MA). For cell cycle, cell was trypsinized, then washed with PBS and fixed with 75% cold ethanol overnight. Then, the cell was centrifuged and resuspended in PBS and treated with propidium iodide (PI) and ribonuclease A. The data were analyzed by flow cytometric analysis on the flow cytometer (FACSort, San Jose, CA, USA). For cell colony formation, cells were cultured in the six-well and continued to culture for 2 weeks. The colonies were treated with 10% formaldehyde and stained with 1% crystal violet. Cell invasion was measured by using matrigel coated transwell chambers (Millipore, MA, USA). Cells were cultured on the upper chamber with no serum. The medium with 10% FBS was added in the bottom well as the chemoattractant. The invasive cells were fixed with 1% crystal violet and counted.
Statistical analysis
All results were shown as mean ± standard deviation (SD). All these experiments were performed in triplicate. Statistical analysis was carried out by using SPSS statistical (SPSS, Chicago, USA). The statistical difference was measured by Student’s test or one-way analysis of variance. P<0.05 was considered statistically significant.
Result
PICART1 expression was decreased in NSCLC samples
We firstly determined PICART1 expression in NSCLC samples and the paired adjacent non-cancerous tissues. PICART1 expression was down-regulated in NSCLC tissues (28/40, 70%) compared to the paired adjacent non-cancerous tissues (Figure 1A). Moreover, PICART1 expression level was lower in NSCLC sample than in the adjacent non-cancerous tissues (Figure 1B). Moreover, lower expression levels of PICART1 were associated with advanced stage in NSCLC patients (Figure 1C).
Figure 1.

PICART1 expression was decreased in the NSCLC samples. A. The expression of PICART1 in NSCLC tissues and paired adjacent non-cancerous tissues was detected by qRT-PCR. B. The expression level of PICART1 was lower in the NSCLC tissues compared to paired adjacent non-cancerous tissues. C. The PICART1 expression level in the late-stage (III+IV) NSCLC patients was lower than in the early-stage (I+II) NSCLC patients. Statistically significant difference was determined using Student’s t test.
Overexpression of PICART1 suppressed NSCLC cell proliferation and cell cylce
We furtherly determined PICART1 expression in NSCLC cell lines. We demonstrated that the expression level of PICART1 was lower in lung adenocarcinoma cell lines (A549, H23, H1299 and SPC-A1) than in one bronchial epithelial cell line (16HBE) (Figure 2A). To determine the functional effect of PICART1 on NSCLC cell, PICART1 overexpression models were established by transfecting with pcDNA-PICART1 in NSCLC cell lines A549 (Figure 2B). Overexpression of PICART1 suppressed A549 cell proliferation (Figure 2C). In addition, ectopic expression of PICART1 inhibited cyclin D1 expression in A549 cell (Figure 2D). Moreover, elevated expression of PICART1 decreased A549 cell cycle (Figure 2E).
Figure 2.
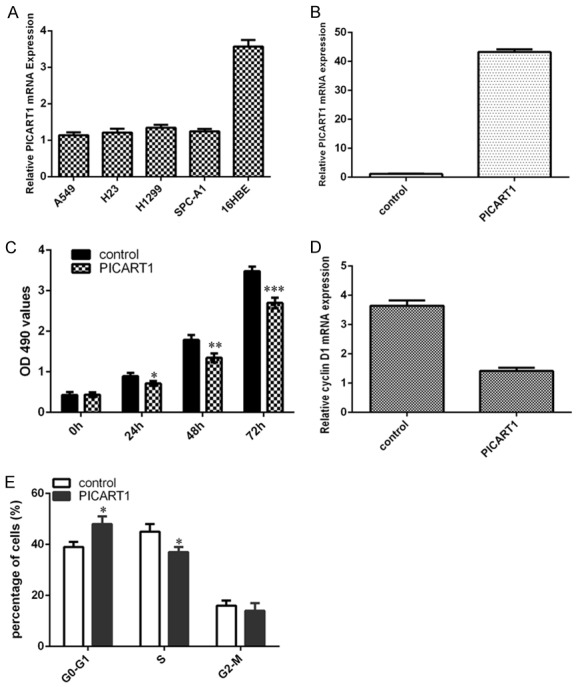
Overexpression of PICART1 suppressed NSCLC cell proliferation and cell cylce. A. The expression level of PICART1 in the lung adenocarcinoma cell lines (A549, H23, H1299 and SPC-A1) and one bronchial epithelial cell line (16HBE) was measured by qRT-PCR. B. The PICART1 expression was measured in the A549 cell after treated with pcDNA-PICART1 using qRT-PCR. C. CCK-8 assay was performed to detect the A549 cell proliferation. Ectopic expression of PICART1 decreased the A549 cell proliferation. D. Overexpression of PICART1 suppressed the expression of cyclin D1 in the A549 cell. E. Elevated expression of PICART1 suppressed the A549 cell cycle. *P<0.05, **P<0.01 and ***P<0.001. Statistically significant difference was determined using Student’s t test.
Overexpression of PICART1 suppressed NSCLC cell colony formation and invasion
Furthermore, overexpression of PICART1 decreased A549 cell colony formation (Figure 3A). In addition, we performed the transwell assays with Matrigel to determine cell invasion. We showed that restored expression of PICART1 suppressed A549 cell invasion (Figure 3B).
Figure 3.
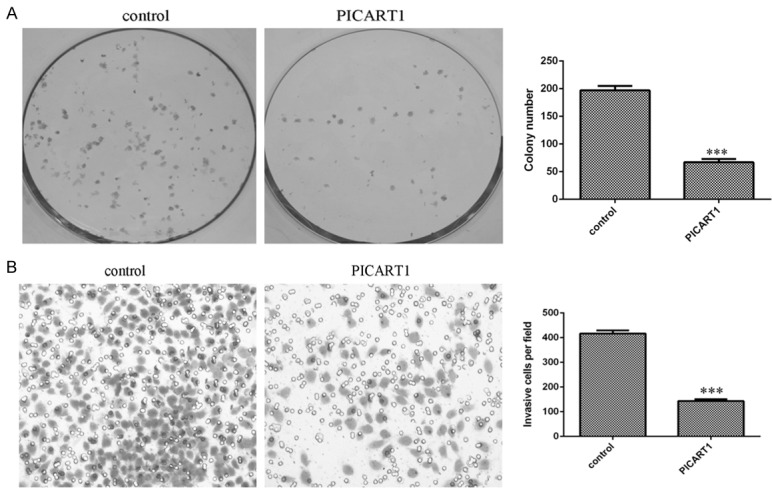
Ectopic expression of PICART1 inhibited NSCLC cell colony formation and invasion. A. Overexpression of PICART1 suppressed the A549 cell colony formation. The relative colony numbers were shown. B. Restored expression of PICART1 decreased the A549 cell invasion. ***P<0.001. Statistically significant difference was determined using Student’s t test.
PICART1 inhibited epithelial-mesenchymal transition (EMT) progression
Overexpression of PICART1 promoted the expression of epithelial marker E-cadherin in A549 cell (Figure 4A). Ovexpression of PICART1 inhibited the expression of mesenchymal marker N-cadherin (Figure 4B). Furthermore, increased expression of PICART1 decreased the expression of Snail (Figure 4C) and Vimentin (Figure 4D).
Figure 4.
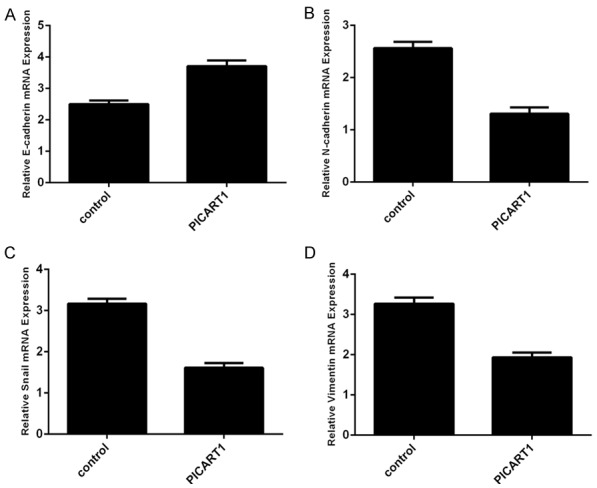
Overexpression of PICART1 suppressed epithelial-mesenchymal transition (EMT) progression. A. The expression of E-cadherin was measured by qRT-PCR. Ectopic expression of PICART1 enhanced the E-cadherin expression in the A549 cell. B. Ovexpression of PICART1 decreased the N-cadherin expression in the A549 cell. C. Elevated expression of PICART1 suppressed the Snail expression in the A549 cell. D. The expression of Vimentin was determined by qRT-PCR.
PICART1 decreased AKT phosphorylation and c-Myc expression
We further studied the underlying mechanism through which PICART1 functions in NSCLC. Overexpression of PICART1 decreased p-AKT expression but not t-AKT expression in A549 cell (Figure 5A). In line with previous data, PICART1 overexpression suppressed c-Myc expression in A549 cell (Figure 5B). PICART1 overexpression promoted p21 expression in A549 cell (Figure 5C).
Figure 5.
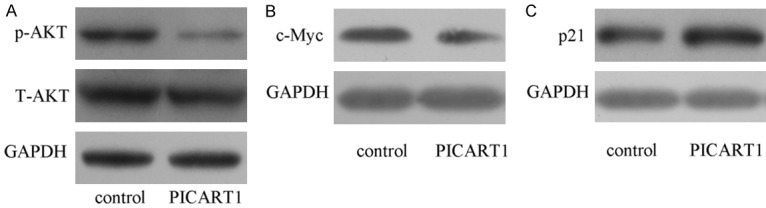
PICART1 decreased AKT phosphorylation and c-Myc expression. A. The protein expression of p-AKT, AKT and GAPDH was determined by western blot. GAPDH was used as the internal control. B. Elevated expression of PICART1 suppressed the c-Myc expression in the A549 cell. C. Overexpression of PICART1 enhanced the p21 expression in the A549 cell.
AKT was involved in PICART1 mediated suppression of cell growth and invasion
To determine the role of AKT in PICART1 mediated cell growth and invasion suppression in NSCLC cell, we used AKT inhibitor MK-2206 (Selleck, Houston, TX) to determine the functional relevance of PICART1/AKT interation in A549 cell. We showed that the expression of p-AKT was down-regulated in A549 cell after treated with AKT inhibitor MK-2206 (Figure 6A). Our data suggested that exposure to the MK-2206 abolished the stimulation of PICART1 silencing on cell growth in A549 cell (Figure 6B). The expression of cyclin D1 was decreased in the si-PICART1-overexpressing A549 cell after treated with MK-2206 (Figure 6C). Furthermore, we showed that exposure to the MK-2206 decreased the stimulation of PICART1 silencing on colony formation (Figure 6D) and cell invasion (Figure 6E) in A549 cell.
Figure 6.

AKT was involved in PICART1 mediated suppression of cell proliferation and invasion. A. The protein expression of p-AKT, AKT and GAPDH was determined by western blot. GAPDH was used as the internal control. B. The AKT inhibitor MK-2206 abolished the stimulation of PICART1 silencing on cell growth in the A549 cell. C. The expression of cyclin D1 was decreased in the si-PICART1-overexpressing A549 cell after treated with MK-2206. D. Exposure to the MK-2206 decreased the stimulation of PICART1 silencing on colony formation. The relative colony numbers were shown. E. Exposure to the MK-2206 decreased the stimulation of PICART1 silencing on colony invasion. The relative invasive numbers were shown. *P<0.05, **P<0.01 and ***P<0.001. Statistically significant difference was determined using Student’s t test.
Discussion
Recently, several pieces of evidences demonstrated that a lot of lncRNAs were deregulated and acted crucial roles in tumor development and pathogenesis, suggesting that these lncRNAs might provide novel insights into the treatment of these diseases [35-39]. Our research showed that PICART1 expression was down-regulated in NSCLC tissues compared to paired adjacent non-cancerous tissues. Moreover, lower expression levels of PICART1 were associated with advanced stage. In addition, PICART1 expression was reduced in NSCLC cell lines. Overexpression of PICART1 inhibited NSCLC cell growth and induced cell cycle arrest at G2/M phase. Elevated expression of PICART1 decreased NSCLC cell colony formation and cell invasion. Overexpression of PICART1 promoted the expression of epithelial marker E-cadherin and suppressed the expression of mesenchymal marker. Furthermore, PICART1 overexpression suppressed AKT phosphorylation and c-Myc expression while inhibited p21 expression in NSCLC. AKT phosphorylation was involved in PICART1 mediated suppression of cell growth and invasion. These results suggested that overexpression of PICART1 inhibited growth and invasion by regulating AKT signaling pathway in NSCLC.
PICART1 was located at 17q21.33 and consists of 3 exons and 2533 bp in length [34]. Previous study proved that PICART1 was down-regulated in colon and breast cancer tissues and cell lines. In addition, p53 increased the expression of PICART1. Overexpression of PICART1 decreased cell invasion, migration and proliferation through regulation of AKT/GSK3β/β-catenin pathway [34]. However, the role of PICART1 is still unknown in NSCLC. Firstly, we explored PICART1 expression in NSCLC tissues and cell lines. PICART1 expression was down-regulated in NSCLC tissues and cell lines. Lower expression level of PICART1 was associated with advanced tumor stage. Moreover, ectopic expression of PICART1 decreased NSCLC cell growth, cell cycle, colony formation and invasion. In conclusion, our data suggested that PICART1 might act as a tumor suppressor gene in NSCLC.
To explore the molecular mechanisms of PICART1-regulated tumor suppression in NSCLC cell, our study further researched the underlying cellular signaling pathway. In line with previous results, we showed that overexpression of PICART1 inhibited AKT phosphorylation and c-Myc expression while inhibited the p21 expression in NSCLC. AKT is one central component regulating signals from phosphatidylinositol 3-kinase (PI3K) and receptor tyrosine kinases [40-42]. As one crucial component of this pathway, AKT medicate signaling of several biological and pathophysiological processes such as development, angiogenesis, cell proliferation, apoptosis, migration, survival and invasion [43-46]. AKT pathway was proved to play important roles in tumor development [47,48]. In this study, we demonstrated that overexpression of PICART1 decreased p-AKT expression but not t-AKT expression in NSCLC cell. In line with previous research, we showed that elevated expression of PICART1 suppressed c-Myc expression in NSCLC cell. PICART1 overexpression promoted p21 expression in NSCLC cell. MK-2206 was a selective AKT inhibitor and our results suggested that exposure to the MK-2206 decreased the stimulation of PICART1 silencing on colony formation and cell invasion in A549 cell. These results suggested that AKT was involved in PICART1 mediated suppression of cell growth and invasion.
In conclusion, PICART1 expression were down-regulated in NSCLC cell lines and tissues and PICART1 overexpression suppressed cell growth, cell colony formation and cell invasion partly though modulating the AKT signaling pathway in NSCLC. Our results suggested that PICART1 might act as a tumor suppressor gene in NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Tejero R, Navarro A, Campayo M, Vinolas N, Marrades RM, Cordeiro A, Ruiz-Martinez M, Santasusagna S, Molins L, Ramirez J, Monzo M. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One. 2014;9:e101899. doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ai C, Jiang R, Fu L, Chen Y. MicroRNA-495 mimics delivery inhibits lung tumor progression. Tumour Biol. 2015;36:729–735. doi: 10.1007/s13277-014-2687-1. [DOI] [PubMed] [Google Scholar]
- 3.Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M, De Maria R. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ. 2014;21:774–82. doi: 10.1038/cdd.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye XW, Yu H, Jin YK, Jing XT, Xu M, Wan ZF, Zhang XY. miR-138 inhibits proliferation by targeting 3-phosphoinositide-dependent protein kinase-1 in non-small cell lung cancer cells. Clin Respir J. 2015;9:27–33. doi: 10.1111/crj.12100. [DOI] [PubMed] [Google Scholar]
- 5.Shi WY, Liu KD, Xu SG, Zhang JT, Yu LL, Xu KQ, Zhang TF. Gene expression analysis of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:217–228. [PubMed] [Google Scholar]
- 6.Chen C, Zhao Z, Liu Y, Mu D. microRNA-99a is downregulated and promotes proliferation, migration and invasion in non-small cell lung cancer A549 and H1299 cells. Oncol Lett. 2015;9:1128–1134. doi: 10.3892/ol.2015.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu W, He J, Chen D, Zhang B, Xu L, Ma H, Liu X, Zhang Y, Le H. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS One. 2014;9:e87780. doi: 10.1371/journal.pone.0087780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Li P, Chen T, Gao G, Chen X, Du Y, Zhang R, Yang R, Zhao W, Dun S, Gao F, Zhang G. Expression of microRNA-96 and its potential functions by targeting FOXO3 in non-small cell lung cancer. Tumour Biol. 2015;36:685–692. doi: 10.1007/s13277-014-2698-y. [DOI] [PubMed] [Google Scholar]
- 9.Xie X, Liu H, Wang M, Ding F, Xiao H, Hu F, Hu R, Mei J. miR-342-3p targets RAP2B to suppress proliferation and invasion of non-small cell lung cancer cells. Tumour Biol. 2015;36:5031–8. doi: 10.1007/s13277-015-3154-3. [DOI] [PubMed] [Google Scholar]
- 10.Rothschild SI, Tschan MP, Federzoni EA, Jaggi R, Fey MF, Gugger M, Gautschi O. MicroRNA-29b is involved in the Src-ID1 signaling pathway and is dysregulated in human lung adenocarcinoma. Oncogene. 2012;31:4221–4232. doi: 10.1038/onc.2011.578. [DOI] [PubMed] [Google Scholar]
- 11.Ge X, Zheng L, Huang M, Wang Y, Bi F. MicroRNA expression profiles associated with acquired gefitinib-resistance in human lung adenocarcinoma cells. Mol Med Rep. 2015;11:333–340. doi: 10.3892/mmr.2014.2757. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Shi Y, Yin Z, Xue X, Zhou B. An eight-miRNA signature as a potential biomarker for predicting survival in lung adenocarcinoma. J Transl Med. 2014;12:159. doi: 10.1186/1479-5876-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patnaik S, Mallick R, Kannisto E, Sharma R, Bshara W, Yendamuri S, Dhillon SS. MiR-205 and MiR-375 MicroRNA assays to distinguish squamous cell carcinoma from adenocarcinoma in lung cancer biopsies. J Thorac Oncol. 2015;10:446–453. doi: 10.1097/JTO.0000000000000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiou GY, Cherng JY, Hsu HS, Wang ML, Tsai CM, Lu KH, Chien Y, Hung SC, Chen YW, Wong CI, Tseng LM, Huang PI, Yu CC, Hsu WH, Chiou SH. Cationic polyurethanes-short branch PEI-mediated delivery of Mir145 inhibited epithelial-mesenchymal transdifferentiation and cancer stem-like properties and in lung adenocarcinoma. J Control Release. 2012;159:240–250. doi: 10.1016/j.jconrel.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45:2197–2206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 16.Vucic EA, Thu KL, Pikor LA, Enfield KS, Yee J, English JC, MacAulay CE, Lam S, Jurisica I, Lam WL. Smoking status impacts microRNA mediated prognosis and lung adenocarcinoma biology. BMC Cancer. 2014;14:778. doi: 10.1186/1471-2407-14-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Zhang T, Ti X, Shi J, Wu C, Ren X, Yin H. Curcumin promotes apoptosis in A549/DDP multidrug-resistant human lung adenocarcinoma cells through an miRNA signaling pathway. Biochem Biophys Res Commun. 2010;399:1–6. doi: 10.1016/j.bbrc.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Wu S, Zhao L, Zhao J, Liu J, Wang Z. Clinical use of microRNAs as potential non-invasive biomarkers for detecting non-small cell lung cancer: a meta-analysis. Respirology. 2015;20:56–65. doi: 10.1111/resp.12444. [DOI] [PubMed] [Google Scholar]
- 19.Wu C, Cao Y, He Z, He J, Hu C, Duan H, Jiang J. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med. 2014;232:85–95. doi: 10.1620/tjem.232.85. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Li Z. Long non-coding RNA HOTAIR: a novel oncogene (Review) Mol Med Rep. 2015;12:5611–5618. doi: 10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett. 2015;10:1953–1958. doi: 10.3892/ol.2015.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Yu X, Shen JX. Long non-coding RNAs: emerging players in osteosarcoma. Tumour Biol. 2016;37:2811–2816. doi: 10.1007/s13277-015-4749-4. [DOI] [PubMed] [Google Scholar]
- 23.Sun L, Sun P, Zhou QY, Gao XC, Han Q. Long noncoding RNA MALAT1 promotes uveal melanoma cell growth and invasion by silencing of miR-140. Am J Transl Res. 2016;8:3939–3946. [PMC free article] [PubMed] [Google Scholar]
- 24.Kitagawa M, Kotake Y, Ohhata T. Long non-coding RNAs involved in cancer development and cell fate determination. Curr Drug Targets. 2012;13:1616–1621. doi: 10.2174/138945012803530026. [DOI] [PubMed] [Google Scholar]
- 25.Yu QY, Zhou XF, Xia Q, Shen J, Yan J, Zhu JT, Li X, Shu M. Long non-coding RNA CCAT1 that can be activated by c-Myc promotes pancreatic cancer cell proliferation and migration. Am J Transl Res. 2016;8:5444–5454. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YH, Fu J, Zhang ZJ, Ge CC, Yi Y. LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8:5286–5297. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao HX, Hou WG, Tao JG, Zhao YL, Wan G, Ma C, Xu HB. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/beta-catenin signaling pathway. Am J Transl Res. 2016;8:3503–3512. [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CB, Lin JJ. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res. 2016;8:4095–4105. [PMC free article] [PubMed] [Google Scholar]
- 29.Dou J, Ni YY, He XF, Wu D, Li M, Wu SY, Zhang R, Guo M, Zhao FS. Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am J Transl Res. 2016;8:98–108. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YY, Yang R, Lian JC, Xu HY. LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res. 2016;8:5035–5043. [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H, Li X, Song Y, Zhang P, Xiao Y, Xing Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. 2015;467:223–228. doi: 10.1016/j.bbrc.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ, Ma WL. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:3076–3082. [PMC free article] [PubMed] [Google Scholar]
- 33.Wan X, Ding X, Chen S, Song H, Jiang H, Fang Y, Li P, Guo J. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumour Biol. 2015;36:521–532. doi: 10.1007/s13277-015-3136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Y, Lin ML, Bu YW, Ling HY, He YC, Huang CF, Shen Y, Song B, Cao DL. p53-inducible long non-coding RNA PICART1 mediates cancer cell proliferation and migration. Int J Oncol. 2017;50:1671–1682. doi: 10.3892/ijo.2017.3918. [DOI] [PubMed] [Google Scholar]
- 35.Ding YC, Yu W, Ma C, Wang Q, Huang CS, Huang T. Expression of long non-coding RNA LOC285194 and its prognostic significance in human pancreatic ductal adenocarcinoma. Int J Clin Exp Pathol. 2014;7:8065–8070. [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Bai HS, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci. 2015;19:3187–3193. [PubMed] [Google Scholar]
- 37.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye Z, Zhou M, Tian B, Wu B, Li J. Expression of lncRNA-CCAT1, E-cadherin and N-cadherin in colorectal cancer and its clinical significance. Int J Clin Exp Med. 2015;8:3707–3715. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang XF, Liu T, Li Y, Li S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 2015;8:9440–9445. [PMC free article] [PubMed] [Google Scholar]
- 40.Emamian ES. AKT/GSK3 signaling pathway and schizophrenia. Front Mol Neurosci. 2012;5:33. doi: 10.3389/fnmol.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Kong X, Wang Y, Yang Q. BRCC2 inhibits breast cancer cell growth and metastasis in vitro and in vivo via downregulating AKT pathway. Cell Death Dis. 2013;4:e757. doi: 10.1038/cddis.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun G, Zhou Y, Li H, Guo Y, Shan J, Xia M, Li Y, Li S, Long D, Feng L. Over-expression of microRNA-494 up-regulates hypoxia-inducible factor-1 alpha expression via PI3K/Akt pathway and protects against hypoxia-induced apoptosis. J Biomed Sci. 2013;20:100. doi: 10.1186/1423-0127-20-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Huang X, Liu X, Xiao S, Zhang Y, Xiang T, Shen X, Wang G, Sheng B. miR-21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int J Mol Sci. 2014;15:4007–4018. doi: 10.3390/ijms15034007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, You T, Jing J. MiR-125b inhibits cell biological progression of Ewing’s sarcoma by suppressing the PI3K/Akt signalling pathway. Cell Prolif. 2014;47:152–160. doi: 10.1111/cpr.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao C, Chen W, Fan X, Jiang X, Qiu L, Chen C, Zhu Y, Wang H. MicroRNA-200c inhibits apoptosis in pituitary adenoma cells by targeting the PTEN/Akt signaling pathway. Oncol Res. 2014;21:129–136. doi: 10.3727/096504013X13832473329999. [DOI] [PubMed] [Google Scholar]
- 47.Saxena NK, Sharma D, Ding X, Lin S, Marra F, Merlin D, Anania FA. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007;67:2497–2507. doi: 10.1158/0008-5472.CAN-06-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu L, Zhang J, Guo X, Li Z, Zhang P. MicroRNA-224 upregulation and AKT activation synergistically predict poor prognosis in patients with hepatocellular carcinoma. Cancer Epidemiol. 2014;38:408–413. doi: 10.1016/j.canep.2014.05.001. [DOI] [PubMed] [Google Scholar]


