Abstract
Colorectal cancer (CRC) is a heterogeneous disease in which unique subtypes are characterized by distinct genetic and epigenetic alterations. DNA methylation, a well-documented epigenetic modification, is a promising biomarker for the diagnosis and prognosis of cancers, including CRC. WNT10A is a member of the Wnt family. It belongs to the Wnt signaling pathway and is involved in CRC. However, studies regarding the methylation and expression of WNT10A in CRC are limited. In the current study, we analyzed the methylation status of WNT10A in 146 patients with CRC and normal controls. These samples were classified into two groups. The first group was an initial discovery set (i.e., fresh tissue samples from 40 patients with CRC and adjacent normal control samples). The second group was an independent validation set (i.e., formalin-fixed and paraffin-embeded [FFPE] samples from 106 patients with CRC and cutting edge tissues). The results showed a higher level of WNT10A hypermethylation of in CRC samples than in controls (Fresh tissue cohort: P = 2.8E-5; FFPE cohort: P = 3.6E-4).This finding was verified by WNT10A methylation data from The Cancer Genome Atlas portal (TCGA) (P = 1.9E-83). Subgroup analysis of clinical characteristics showed a higher WNT10A methylation level in elder patients (aged > 60 y) (P = 0.037) and, patients with distant metastasis (P = 0.033), rectal cancer (P = 0.03), and mucinous adenocarcinoma (P = 0.02). Furthermore, TCGA RNAseq data demonstrated lower WNT10A expression in patients with CRC than in controls (P = 4.0E-3) and showed a negative correlation between expression and methylation (r = -0.37, P = 5.7E-13). Moreover, the efficiency of WNT10A methylation for CRC diagnosis was analyzed in both cohorts of the present study and the TCGA cohorts, which indicated the potential use of WNT10A methylation as a tool for diagnosis of CRC.
Keywords: DNA methylation, WNT10A, CRC, TCGA, diagnosis
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer among both males and females in the United States of America (USA). The American Cancer Society provides (https://www.cancer.org/research.html) annual comprehensive overviews of the incidence of human cancers, (including CRC), reporting the numbers of new cases and mortality in the USA. Although, the reports show a decreased trend in the incidence of CRC from 1975 to 2014, it is estimated that 140,250 new cases and 50,630 new deaths will occur in 2018 in the USA [1]. Moreover, the incidence in individuals aged < 55 years increased by approximately 2% per year from 1990 to 2014 [2]. Nevertheless, the burden of CRC cases and deaths in China is much higher than that reported in the USA [3].
The etiology and pathology of CRC are well documented. Diet is a major etiological factor. A range of putative dietary carcinogens or daily behaviors, such as Western diet, a low level of physical activity, high consumption of red meat and processed food, are significantly correlated with the development of CRC [4,5]. However, multiple theories have emerged following the rising incidence of CRC in younger individuals. One theory hypothesized that older generations were exposed to a lower-risk environment than the younger generation [6]. Hessami and his colleagues comprehensively analyzed the association of CRC incidence in younger and elderly patients with dietary risks. They concluded that the unique genetic pattern of different populations cannot be ignored [7]. In addition, previous studies demonstrated strong evidence for genetic susceptibility to CRC through, interaction with the environment, including diet [8,9].
The multistage nature of carcinogenesis is defined by an accumulation of genetics and/or epigenetic events. Genome wide association studies (GWAS) revealed the significance of genes and pathways hitherto unsuspected of playing a role in CRC [10]. Genetic alterations involve mutations in oncogenes and tumor suppressor genes, such as the adenomatous polyposis coli (APC), Kirsten-ras (K-ras), and p53 [11-13]. It has been largely demonstrated that defects in these genes affect certain basic biological processes, including cell fate, cell survival, and genome maintenance [13,14]. However, only 10% of CRC cases are characterized by the “classic” genotype [15,16]. In additional, the initiation and promotion of cancer may be caused by epigenetic mechanisms, which are defined as heritable changes in gene function that are not due to changes in DNA sequence [15]; in particular, aberrant epigenetic modifications can result in transcriptional silencing [17,18]. Epigenetic alterations-specifically DNA methylation-are an emerging biomarker that may be potential useful in diagnosis, prognosis and treatment, of various disease including cancer, type 1 diabetes [19,20].
Aberrant methylation of tumor suppressor genes (TSGs) involved in multiple biological possesses, such as DNA repair and apoptosis, such as MLH1, MGMT, CDKN2A and RAFF1A [21-23], may be detrimental to health of patients and lead to progression of cancer [24-27]. Although the development of human CRCs is considered a consequence of the accumulation of multiple genetic and epigenetic alterations/interactions, the Wnt signaling pathway appears to play a critical role in the etiology of this disease [14,28]. Wnts in vertebrates are divided into canonical signaling and noncanonical members [29]. The most extensively characterized Wnt signaling pathway-the canonical Wnt/β-catenin pathway-regulates the stability of transcription co-activator β-catenin. Consequently, this activates the expression of a set of target genes, regulating cell proliferation, behavior, and survival [30,31]. As a member of the Wnt family, WNT10A is located on 2q35. Notably, the methylation and expression level of WNT10A in CRC is inconsistent [32-34].
The primary objective of the present study was to analyze the methylation status of WNT10A in 146 paired samples obtained from patients and corresponding normal controls, and investigate the associations between methylated WNT10A and clinical features. Moreover, the diagnostic strength of WNT10A hypermethylation in recognizing of CRC was analyzed. Subsequently, methylation and expression data along with clinical characteristics from a public database included large numbers of patients were accessed. This analysis was performed to verify the results and analyze the expression status of WNT10A in patients with CRC.
Materials and methods
Patient characteristics and collection of tissue specimens
An initial discovery set of 40 CRC fresh tissues and paired adjacent normal tissues were collected from CRC surgical specimens at the Department of Gastrointestinal Surgery in Ningbo First Hospital between April 2011 and April 2015. None of the patients underwent chemotherapy or radiotherapy prior to surgery. Following surgical excision, all specimens were immediately preserved in RNA-fixer reagent and stored in liquid nitrogen. Pathological diagnosis was performed by two experienced pathologists. Among the samples, there were 30 moderately differentiated cases, and 10 poorly differentiated cases. Of note, there were seven Stage I cases, 12 Stage II cases, 13 Stage III cases and eight Stage IV cases. All patients provided written informed consent for the collection of tissue samples prior to their participation in this study. The study was approved by the Human Research Ethical Committee of Ningbo First Hospital.
An independent validation set of 106 paired formalin-fixed and paraffin-embeded (FFPE) CRC and cutting edge tissues were collected from Diagnostic Pathology Central of Ningbo. Among those, there were two well differentiated, 70 moderately differentiated cases, and 34 poorly differentiated cases. Of note, there were nine Stage I cases, 43 Stage II cases, 50 Stage III cases and four Stage IV cases. The clinical characteristics of all the patients were available. All of tumors were staged by experienced surgeons according to the eighth edition (2016) of the tumor-node-metastasis staging system established by the American Joint Committee on Cancer (AJCC).
DNA extraction
Genomic DNA of fresh samples and FFPE samples were separately extracted using the QIAamp DNA Mini Kit and DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) as recommended by the manufacturer. Subsequently, the concentration and quality of the DNA was measured using the ultramicro nucleic acid ultraviolet tester (NANODROP 2000c, Thermo, USA).
Bisulfite treatment
Extracted genomic DNA was bisulphite-converted with ZYMO EZ DNA Methylation-Gold Kit according to the manufacturer’s protocol (Zymo Research, Orange, CA, USA). The bisulphite-modified DNA was used for the following methylation analysis.
Quantitative methylation-specific polymerase chain reaction (qMSP)
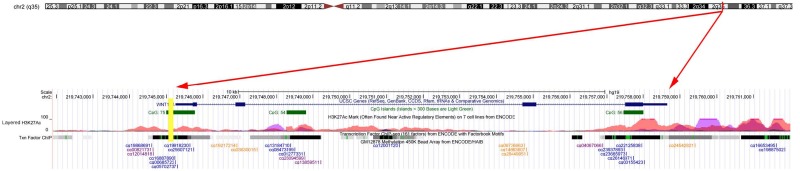
Bisulphite modified DNA was amplified through fluorescence-based real-time quantitative methylation-specific PCR (qMSP) using SYBR Green Master Mix (Roche Diagnosis) with Roche 480II RT-PCR instrument (9 -well plates, Roche Diagnostics). The qMSP primers for WNT10A were designed by MethPrimer (http://www.urogene.org/cgi-bin/methprimer2/Meth-Primer.cgi) [35]. Briefly, we initially determined the gene description, such as position, CpG islands, histone modification status, and the position of each CpG sits, from UCSC Genome Browser (http://genome.ucsc.edu/). Subsequently, we identified and downloaded the sequences of target fragment of the WNT10A promoter region and input them into MethPrimer to design the qMSP primers (Figure 1). One pair of WNT10A primers that could amplify a specific product was used for the methylation analysis. Primers for ACTB gene were simultaneously amplified in each sample and used as internal control [36]. The primers used for the WNT10A were as follows: sense, 5’-GCGGGTATAGGTAGGTAA-3’; antisense, 5’-CACACAACAAATTAACGAAA-3’. The primers used for ACTB were as follows: sense, 5’-TGGTGATGGAGGAGGTTTAGTAAGT-3’; antisense, 5’-AACCAATAAAACCTACTCCTCCCTTAA-3’. The values obtained for the WNT10A gene were compared with these of the internal reference gene to calculated a ratio that was then multiplied by 1000 for simple tabulation (WNT10A/ACTB×1000) [36]. This calculation determined the relative levels of methylated WNT10A in each sample.
Figure 1.
The detail information of the WNT10A gene. Each of the cg loci of WNT10A is labeled in the last layer according to its position; the yellow bar is the amplified segment of WNT10A in the current study; the green bands in the second layer indicate the CpG islands of WNT10A; the information of the layered H3K27Ac and the Txn factor chip are shown in the third and fourth layer. This information was downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/).
Validation dataset from the cancer genome atlas portal (TCGA) portal
To ascertain our results, we downloaded the Illumina HumanMethylation 450K methylation data and RNA-seq expression of WNT10A, combined with clinical characteristics from TCGA (https://cancergenome.nih.gov/). Collectively, a total of 443 patients’ methylation frequencies and expression data of WNT10A along with the clinical characteristics were obtained, including 398 CRCs and 45 normal controls.
Regarding the methylation data of WNT10A, we abstracted the level 3 methylation of 49 CpGs sites. There were eight CpG sites (including cg19868691, cg00821731, cg12014818, cg16887890, cg00685723, cg05702737, cg19918230, and cg25607121) entered into further analysis, based on the following three criteria: 1) significantly correlated CpG sites according to Pearson’s correlation; 2) located on the CpG island of the WNT10A promoter region annotated by Illumina HumanMethylation 450K annotation file downloaded from the Illumina official website (https://support.illumina.com/array/array_kits/infinium_humanmethylation-450_beadchip_kit/downloads.html); and 3) neighbored the targeted fragment in our study. The mean methylation levels of included eight CpG sites were computed to represent the methylation level of the promoter region of WNT10A.
Regarding the expression data of WNT10A, we also obtained the level 3 of RNASeq (Version 2). A total of 394 samples (including 373 CRC patients and 21 normal controls) simultaneously available of WNT10A methylation and expression data were used to investigate the expression of WNT10A and its correlation with WNT10A methylation in CRC.
Statistical analyses
All the statistical analyses were performed using the SPSS Version 18.0 (SPSS Inc., Chicago, IL, USA). Pearson’s correlation was performed to identify the significant correlated CpG sites, and to analyze the correlation between WNT10A methylation and expression. Differences in continuous data between pairs of groups were detected by Student’s t test. Namely, an independent t test was performed for unpaired subjects. Paired t test was utilized for paired subjects, and one-way analysis of variance (ANOVA) test was applied between multiple groups (i.e., more than two groups). The diagnostic value of WNT10A methylation was assessed using the receiver operating characteristic (ROC) curve and the area under the curve (AUC). In addition, the positive predict value (PPV) and negative predict value (NPV) were also computed. A two-tailed P value of less than 0.05 was considered statistically significant. All the figures were drawn using the GraphPad Prism 6 software (GraphPad Software, La Jolla, CA, USA) or R 3.1.2 software (http://www.r-project.org/).
Results
Hypermethylation of WNT10A in the study cohorts
The fragment of the CpG island in the promoter region was amplified using qMSP to verify the methylation level of WNT10A (Figure 1). In the fresh-tissue cohort, bisulphite modified DNA from 40 CRC and corresponding normal samples were analyzed. The results showed higher methylation level of WNT10A in CRCs was higher than that observed in controls (P = 2.8E-5, Figure 2A). In an independent validation set, the methylation frequencies of WNT10A in 106 bisulphite-converted DNA from samples from FFPE CRC and cutting-edge tissues were studied. These results corroborated the aforementioned findings, indicating a higher methylation level of WNT10A in patients with CRCs than in normal controls (P = 3.6E-4, Figure 2B).
Figure 2.
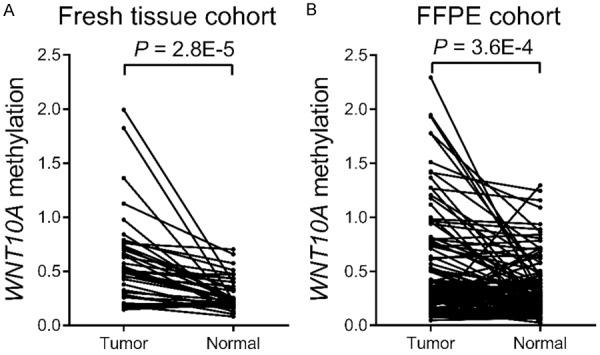
Comparison of WNT10A methylation between CRCs and controls in the cohorts of the present study. A: Comparison of WNT10A methylation between 40 paired CRCs and control samples in the fresh tissue cohort. B: Comparison of WNT10A methylation between 106 paired CRCs and control samples in the FFPE cohort.
Subsequently, a methylation difference analysis based on clinical characteristics was performed. Although, the observed differences in the study cohorts of the present study was not statistically significant (Table 1), an upward trend for the mean methylation frequency of WNT10A related to the advanced stage of CRC was still notable. Furthermore, the mean methylation level was increased in patients with lymph metastasis or distant metastasis versus those without metastasis. These results indicate the potential important role of WNT10A methylation in the progression of CRC.
Table 1.
Association of WNT10A methylation with clinical characteristics of CRC in the cohorts of the present study
| Characteristics | N | Mean methylation | P | |
|---|---|---|---|---|
| Gender | Male | 86 | 55% | 0.67 |
| Female | 60 | 59% | ||
| Age (year) | ≤ 60 | 58 | 55% | 0.73 |
| > 60 | 88 | 59% | ||
| Stage | I | 16 | 30% | 0.24 |
| II | 55 | 55% | ||
| III | 63 | 61% | ||
| IV | 12 | 64% | ||
| Lymph metastasis | No | 73 | 52% | 0.41 |
| Yes | 73 | 61% | ||
| Distant metastasis | No | 133 | 55% | 0.35 |
| Yes | 13 | 71% | ||
| Tumor location | Colon | 75 | 52% | 0.35 |
| Rectum | 71 | 61% | ||
| Differentiation | Well | 2 | 48% | 0.21 |
| Moderate | 100 | 44% | ||
| Poor | 44 | 85% | ||
| Tumor size | < 5 cm | 83 | 56% | 0.98 |
| ≥ 5 cm | 63 | 56% | ||
| Histological classification | Adenocarcinoma | 142 | 56% | 0.56 |
| Mucinous adenocarcinoma | 4 | 74% | ||
Diagnostic value of methylated WNT10A in CRC
Subsequently, an ROC analysis was conducted to determine the diagnostic power of methylation WNT10A in identification of CRC from normal control in both the fresh-tissue cohort and FFPE cohorts. In the fresh-tissue cohort, the under area of ROC curve (AUC) was 0.76 with 85% specificity and 63% sensitivity. And the PPV and NPV were 0.81 and 0.69, respectively (Figure 3A). In the FFPE cohort, the value of AUC was 0.59 with 91% specificity and 26% sensitivity; the PPV is 0.73 and NPV were 0.55, respectively (Figure 3B).
Figure 3.
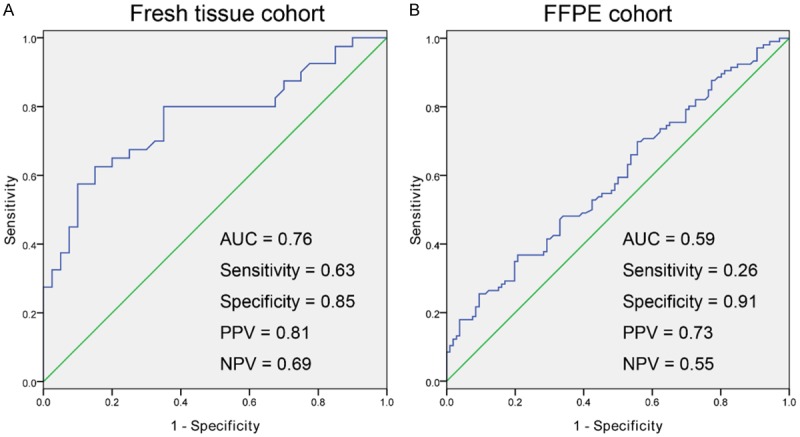
Diagnostic power of methylated WNT10A in distinguishing of CRC from controls in the cohorts of the present study. A: Diagnostic power of detection of methylated WNT10A in distinguishing of CRC from controls in the fresh tissue cohort. B: Diagnostic power of detection of methylated WNT10A in distinguishing of CRC from controls in the FFPE cohort.
Validation datasets from TCGA
To ascertain our results, methylation data of WNT10A from 398 CRC samples and 45 normal controls were download from TCGA portal. The results demonstrated elevated methylation frequency in CRC samples than in controls (P = 1.9E-83, Figure 4A).
Figure 4.
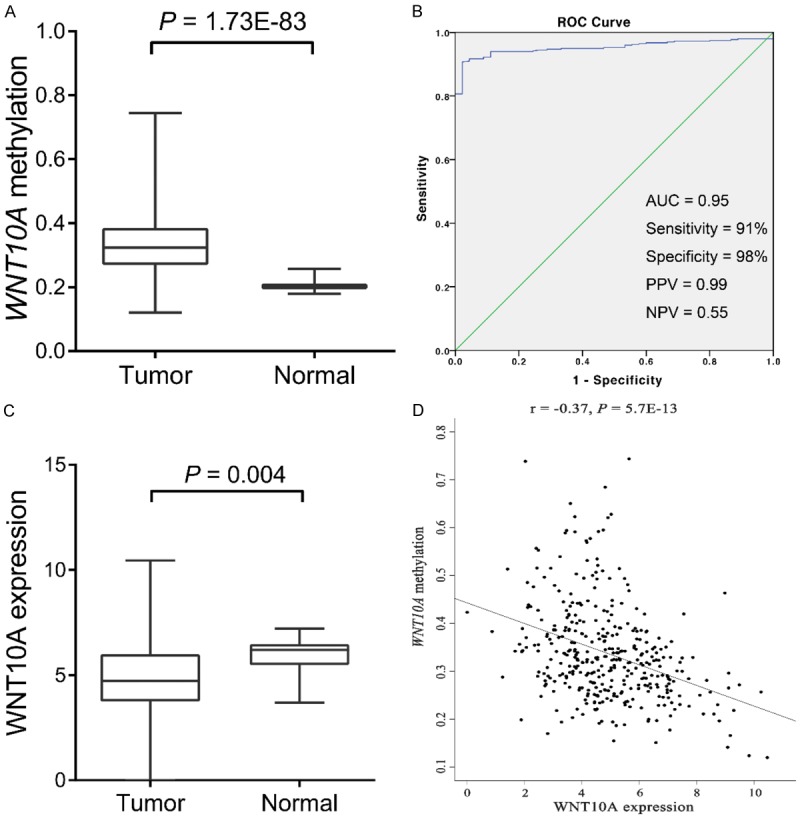
Validation of WNT10A methylation from TCGA dataset. A: Comparison of WNT10A methylation between patients with CRC and controls in TCGA cohorts. B: Diagnostic power of methylated WNT10A in distinguishing of patients with CRC from controls in TCGA cohorts. C: Comparison of WNT10A expression between patients with CRC and controls from TCGA portal. D: Negative correlation of WNT10A methylation and expression from TCGA portal.
Then, 394 CRC samples with available data for clinical characteristics were used for subgroup comparison analyses based on gender, age, different stage of disease, lymph invasive status, as well as distant metastasis status. In TCGA cohorts, a higher methylation level of WNT10A was observed with statistical significance in elderly patients (aged > 60 years) (P = 0.037), patients with distant metastasis (P = 0.033), and those diagnosed with rectal cancer (P = 0.03) and mucinous adenocarcinoma (P = 0.02) (Table 2). However, there were no associations between WNT10A methylation and the clinical stage of disease, lymph metastasis status or gender.
Table 2.
Association of WNT10A methylation with clinical characteristics of CRC in TCGA cohorts
| Characteristics | N | Mean methylation | P | |
|---|---|---|---|---|
| Gender | Male | 212 | 33% | 0.11 |
| Female | 182 | 35% | ||
| Age (year) | ≤ 60 | 153 | 32% | 0.037* |
| > 60 | 241 | 35% | ||
| Stage | I | 58 | 31% | 0.12 |
| II | 153 | 34% | ||
| III | 127 | 34% | ||
| IV | 56 | 35% | ||
| Lymph metastasis | No | 217 | 33% | 0.31 |
| Yes | 177 | 34% | ||
| Distant metastasis | No | 341 | 31% | 0.033* |
| Yes | 53 | 34% | ||
| Tumor location | Colon | 296 | 32% | 0.03* |
| Rectum | 98 | 35% | ||
| Histological classification | Adenocarcinoma | 344 | 33% | 0.02* |
| Mucinous adenocarcinoma | 50 | 37% | ||
P < 0.05.
Moreover, the diagnostic power of methylated WNT10A in TCGA cohort was studied. In this cohort, the AUC was 0.95 with 98% specificity and 91% sensitivity, and the PPV and NPV were 0.99 and 0.55, respectively (Figure 4B). These results represented the that WNT10A have potential to be served as useful biomarker for identification of CRC from normal control, which is still needed further prospective study to ascertain our results.
Expression data from TCGA
Numerous studies revealed a negative correlation between methylation and expression of WNT10A. The expression level of WNT10A was also studied in present study. Due to the absence of an adequate number of samples, we applied the expression data available at TCGA database. Firstly, the expression of WNT10A in CRC and controls was compared. This comparison revealed a reduced expression of WNT10A in patients with CRC was observed than in controls (P = 4.0E-3, Figure 4C). Secondly, the Pearson’s analysis showed an inverse correlation between WNT10A expression and methylation (r = -0.37, P = 5.7E-13, Figure 4D).
Discussion
Although the overall incidence of CRC is gradually decreased, the rejuvenation of CRC is observed and the number of individual who is diagnosed as CRC is still large in China [3]. Despite the introduction and improvement of endoscopy and biopsies technology, the number of diagnosed CRC is just the tip of the iceberg for untypical clinical symptom of most of early stage of this disease. Approximately 35% of patients are diagnosed as localized stage CRC, for which the 5-year survival is 90% [2]. However, the survival rate declines to 71% and 14% for patients diagnosed with regional and advanced stage disease, respectively [2,37]. Therefore, early detection is of crucial importance to improve the survival rate of patients with CRC, in whom curative surgical resection or endoscopic treatment is possible. The application of molecular markers specific to CRC offers new possibilities for early detection [38]. Although proteins or other small molecular markers derived from cancers proved useful in certain cases, markers based on changes in the genetic material offer the powerful advantage of allowing signal amplification using PCR, hence increasing sensitivity [39]. Thus far, studies strongly suggest that the analysis of DNA methylation patterns may become a powerful tool for the accurate and early diagnosis of CRC, with unparalleled specificity and sensitivity [40-42].
The Wnt-β-catenin-TCF signaling pathway consists of WNTs, Frizzled genes (FZD), intracellular signaling molecules, and TCF transcription factors, which are implicated in carcinogenesis and embryogenesis [43]. Wnts are secreted-type glycoproteins with conserved cysteine residues [44,45]. They bind to seven-transmembrane-type WNT receptors containing a cysteine-rich domain, encoded by the FZD genes [44,46]. Genetic alterations in the members of the canonical Wnt/β-catenin pathway moleculars, such as APC, AXIN1, β-catenin and TCF are involved in carcinogenesis through transcriptional activation of the Wnt target genes [47,48]. The human WNT gene family consists of at least 19 members. A previous study revealed that WNT10A is clustered with WNT6 on chromosome 2q35 in a head-to-tail manner [49]. The WNT10A gene encodes a 417-amino-acid polypeptide with a Wnt core domain, which is 64% identical to WNT10B [49,50]. Developmentally, WNT10A is extensively studied in the context of ectodermal lineages, primarily in the deregulation of ectodermal tissues resulting in a variety of disorders [51,52]. Furthermore, WNT10A was previously implicated in a variety of cancers and has been shown to promote proliferation, migration, and chemo-resistance in renal cell carcinoma cell lines via regulation of β-catenin [32,53-55]. Besides, Kurasawa and his colleagues reported that WNT10A was associated with epithelial-mesenchymal transition (EMT) and progression of cancer through mesenchymal-specific DNA hypermethylation [34]. Other reports indicate that WNT10A is upregulated in esophageal cancer, gastric, and colon cancer cells and tumors [32]. However, hypermethylated of the WNT10A promoter region resulting in lower expression has been identified in head and neck squamous cell carcinoma and oligodendroglioma cell lines [33,34]. Numerous studies have addressed the causal relationship between promoter hypermethylation and abated transcriptional.
The current data regarding the methylation and expression of WNT10A in human cancers are inconsistenct. Thus, in the present study, we analyzed its methylation and expression in CRC. Herein, the results demonstrated that WNT10A is frequently methylated in CRC primary tumors, but less frequently in normal colon tissues. Kurasawa and his colleagues have demonstrated the hypermethylation level of WNT10A in head and neck carcinomas and the associations of methylation and clinicopathology; specifically, the elevated level of WNT10A methylation in the advanced stages of the disease [34]. Although significant associations between WNT10A methylation and clinical features were not found in this study, an upward tendency of methylation in advanced of CRC was observed in the present cohort. Public data were also accessed/analyzed to verify our results, showing increased methylation level in patients with lymph metastasis and distant metastasis. This finding in accordance with previous study results. Furthermore, in TCGA dataset, a higher level of WNT10A methylation was observed in patients diagnosed with rectal cancer than in those with colon cancer. Andrew and his colleagues have conducted an assessment of loci methylation level throughout the genome of ascending colon and rectal normal colon epithelial cells, revealing multiple differentially methylated genes [56]. An explanation for this observation may be that different sublocations arise from different embryological sources and different methylation levels are associated with particular types tissues and cell lineages [57,58]. Previous studies also reported multiple differentially methylated genes among four different sublocations of CRC [59,60]. These findings suggest that assessing the diagnostic value of gene methylation may be useful in distinguishing different sublocations of CRC. Hypermethylation of the gene promoter region is accompanied with dysregulation of expression [61,62]. However, up-regulation of WNT10A was reported in numerous human cancers, such as gastric cancer, esophageal cancer or other cancers [32]. Nevertheless, in the current study, we analyzed the expression level of WNT10A in CRC using public data. The results showed lower expression of WNT10A in CRC tissues and a negative correlation between gene expression and methylation in CRC.
An indication that DNA methylation may be used as biomarkers provided by the use of hypermethylation events at the genes O6-methylguanine-DNA methyltransferase (MGMT) in glioma [63] and the glutathione S-transferase pi 1 (GSTP1) in prostate cancer [64]. These hypermethylation events were shown to be effective in determining an appropriate treatment choice for glioma and in the diagnosis of prostate cancer, respectively. Therefore, the detection of epigenetic alterations is a promising tool for the diagnosis and prognosis of disease. In the current study, the diagnostic power of methylated WNT10A was also analyzed in each of cohort. In our study cohorts A and cohort B, the AUC were 0.76 and 0.59 with high specificity and middle/low sensitivity, respectively. The PPV for CRC was more than 70%, whereas the NPV was not quite satisfactory. The disparity in AUC due to the different types of samples included in these two cohorts. The FFPE samples of cohort B were embedded in formalin, which induced the fragmentation of DNA. Furthermore, the public data from TCGA were used to verify the results. This analysis provided a consolidated conclusion, indicating that the detection of WNT10A methylation is potentially useful in distinguishing CRC from normal controls.
In summary, the present findings revealed the hypermethylation of WNT10A in CRC, which was negatively correlated with the expression of WNT10A. An upward tendency of WNT10A hypermethylation was observed in CRC. Further studies with large sample-size are warranted to confirm these results. The methylation status of WNT10A is a potentially useful biomarker for the diagnosis of CRC.
Acknowledgements
This work is supported by Ningbo Natural Science Foundation [NO. 2017A610157].
Disclosure of conflict of interest
None.
References
- 1.Society AC. Cancer Facts & Figures 2018. Available from: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-2018.pdf Accessed January 05, 2018.
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Baena R, Salinas P. Diet and colorectal cancer. Maturitas. 2015;80:258–264. doi: 10.1016/j.maturitas.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Vulcan A, Manjer J, Ericson U, Ohlsson B. Intake of different types of red meat, poultry, and fish and incident colorectal cancer in women and men: results from the malmo diet and cancer study. Food Nutr Res. 2017;61:1341810. doi: 10.1080/16546628.2017.1341810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haghighi P, Nasr K, Mohallatee EA, Ghassemi H, Sadri S, Nabizadeh I, Sheikholeslami MH, Mostafavi N. Colorectal polyps and carcinoma in Southern Iran. Cancer. 1977;39:274–278. doi: 10.1002/1097-0142(197701)39:1<274::aid-cncr2820390142>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 7.Hessami Arani S, Kerachian MA. Rising rates of colorectal cancer among younger Iranians: is diet to blame? Curr Oncol. 2017;24:e131–e137. doi: 10.3747/co.23.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson LR. Recent advances in understanding of interactions between genes and diet in the etiology of colorectal cancer. World J Gastrointest Oncol. 2010;2:125–129. doi: 10.4251/wjgo.v2.i3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YS, Milner JA. Dietary modulation of colon cancer risk. J Nutr. 2007;137:2576S–2579S. doi: 10.1093/jn/137.11.2576S. [DOI] [PubMed] [Google Scholar]
- 10.Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149:1177–1190.e3. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li XL, Zhou J, Chen ZR, Chng WJ. P53 mutations in colorectal cancer - molecular pathogenesis and pharmacological reactivation. World J Gastroenterol. 2015;21:84–93. doi: 10.3748/wjg.v21.i1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon BS, Jeong WJ, Park J, Kim TI, Min do S, Choi KY. Role of oncogenic K-Ras in cancer stem cell activation by aberrant Wnt/beta-catenin signaling. J Natl Cancer Inst. 2014;106:djt373. doi: 10.1093/jnci/djt373. [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 16.Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR. Mutations in APC, Kirsten-ras, and p53--alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci U S A. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 19.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33. doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, Daunay A, Busato F, Mein CA, Manfras B, Dias KR, Bell CG, Tost J, Boehm BO, Beck S, Leslie RD. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7:e1002300. doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 22.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1997 update of recommendations for the use of tumor markers in breast and colorectal cancer. Adopted on November 7, 1997 by the american society of clinical oncology. J. Clin. Oncol. 1998;16:793–795. doi: 10.1200/JCO.1998.16.2.793. [DOI] [PubMed] [Google Scholar]
- 25.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 26.Shin SK, Nagasaka T, Jung BH, Matsubara N, Kim WH, Carethers JM, Boland CR, Goel A. Epigenetic and genetic alterations in Netrin-1 receptors UNC5C and DCC in human colon cancer. Gastroenterology. 2007;133:1849–1857. doi: 10.1053/j.gastro.2007.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, Markowitz SD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baig-Lewis S, Peterson-Nedry W, Wehrli M. Wingless/wnt signal transduction requires distinct initiation and amplification steps that both depend on arrow/LRP. Dev Biol. 2007;306:94–111. doi: 10.1016/j.ydbio.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong GT, Gavin BJ, McMahon AP. Differential transformation of mammary epithelial cells by Wnt genes. Mol Cell Biol. 1994;14:6278–6286. doi: 10.1128/mcb.14.9.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 31.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 32.Kirikoshi H, Inoue S, Sekihara H, Katoh M. Expression of WNT10A in human cancer. Int J Oncol. 2001;19:997–1001. doi: 10.3892/ijo.19.5.997. [DOI] [PubMed] [Google Scholar]
- 33.Ordway JM, Bedell JA, Citek RW, Nunberg A, Garrido A, Kendall R, Stevens JR, Cao D, Doerge RW, Korshunova Y, Holemon H, McPherson JD, Lakey N, Leon J, Martienssen RA, Jeddeloh JA. Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis. 2006;27:2409–2423. doi: 10.1093/carcin/bgl161. [DOI] [PubMed] [Google Scholar]
- 34.Kurasawa Y, Kozaki K, Pimkhaokham A, Muramatsu T, Ono H, Ishihara T, Uzawa N, Imoto I, Amagasa T, Inazawa J. Stabilization of phenotypic plasticity through mesenchymal-specific DNA hypermethylation in cancer cells. Oncogene. 2012;31:1963–1974. doi: 10.1038/onc.2011.373. [DOI] [PubMed] [Google Scholar]
- 35.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 36.Jeronimo C, Henrique R, Hoque MO, Ribeiro FR, Oliveira J, Fonseca D, Teixeira MR, Lopes C, Sidransky D. Quantitative RARbeta2 hypermethylation: a promising prostate cancer marker. Clin Cancer Res. 2004;10:4010–4014. doi: 10.1158/1078-0432.CCR-03-0643. [DOI] [PubMed] [Google Scholar]
- 37.Lee MM, MacKinlay A, Semira C, Schieber C, Jimeno Yepes AJ, Lee B, Wong R, Hettia-rachchige CKH, Gunn N, Tie J, Wong HL, Skinner I, Jones IT, Keck J, Kosmider S, Tran B, Field K, Gibbs P. Stage-based variation in the effect of primary tumor side on all stages of colorectal cancer recurrence and survival. Clin Colorectal Cancer. 2018;17:e569–e577. doi: 10.1016/j.clcc.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Duffy MJ, Lamerz R, Haglund C, Nicolini A, Kalousova M, Holubec L, Sturgeon C. Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int J Cancer. 2014;134:2513–2522. doi: 10.1002/ijc.28384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sidransky D. Nucleic acid-based methods for the detection of cancer. Science. 1997;278:1054–1059. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- 40.Lange CP, Campan M, Hinoue T, Schmitz RF, van der Meulen-de Jong AE, Slingerland H, Kok PJ, van Dijk CM, Weisenberger DJ, Shen H, Tollenaar RA, Laird PW. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One. 2012;7:e50266. doi: 10.1371/journal.pone.0050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summers T, Langan RC, Nissan A, Brucher BL, Bilchik AJ, Protic M, Daumer M, Avital I, Stojadinovic A. Serum-based DNA methylation biomarkers in colorectal cancer: potential for screening and early detection. J Cancer. 2013;4:210–216. doi: 10.7150/jca.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gyparaki MT, Basdra EK, Papavassiliou AG. DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. J Mol Med (Berl) 2013;91:1249–1256. doi: 10.1007/s00109-013-1088-z. [DOI] [PubMed] [Google Scholar]
- 43.Sagara N, Katoh M. Mitomycin C resistance induced by TCF-3 overexpression in gastric cancer cell line MKN28 is associated with DT-diaphorase down-regulation. Cancer Res. 2000;60:5959–5962. [PubMed] [Google Scholar]
- 44.Miller JR. The Wnts. Genome Biol. 2002;3:REVIEWS3001. doi: 10.1186/gb-2001-3-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, Donigan L, Fischer M, Chaudhari A, Ji M, Kapoun AM, Lam A, Lazetic S, Ma S, Mitra S, Park IK, Pickell K, Sato A, Satyal S, Stroud M, Tran H, Yen WC, Lewicki J, Hoey T. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A. 2012;109:11717–11722. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dijksterhuis JP, Petersen J, Schulte G. WNT/frizzled signalling: receptor-ligand selectivity with focus on FZD-G protein signalling and its physiological relevance: IUPHAR review 3. Br J Pharmacol. 2014;171:1195–1209. doi: 10.1111/bph.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P, McKechnie D, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 48.Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, Sasaki Y, Imaoka S, Murata M, Shimano T, Yamaoka Y, Nakamura Y. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 49.Kirikoshi H, Sekihara H, Katoh M. WNT10A and WNT6, clustered in human chromosome 2q35 region with head-to-tail manner, are strongly coexpressed in SW480 cells. Biochem Biophys Res Commun. 2001;283:798–805. doi: 10.1006/bbrc.2001.4855. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Shackleford GM. Murine Wnt10a and Wnt10b: cloning and expression in developing limbs, face and skin of embryos and in adults. Oncogene. 1996;13:1537–1544. [PubMed] [Google Scholar]
- 51.Adaimy L, Chouery E, Megarbane H, Mroueh S, Delague V, Nicolas E, Belguith H, de Mazancourt P, Megarbane A. Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. Am J Hum Genet. 2007;81:821–828. doi: 10.1086/520064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bohring A, Stamm T, Spaich C, Haase C, Spree K, Hehr U, Hoffmann M, Ledig S, Sel S, Wieacker P, Ropke A. WNT10A mutations are a frequent cause of a broad spectrum of ectodermal dysplasias with sex-biased manifestation pattern in heterozygotes. Am J Hum Genet. 2009;85:97–105. doi: 10.1016/j.ajhg.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Peet GW, Balzarano D, Li X, Massa P, Barton RW, Marcu KB. Novel NEMO/IkappaB kinase and NF-kappa B target genes at the pre-B to immature B cell transition. J Biol Chem. 2001;276:18579–18590. doi: 10.1074/jbc.M100846200. [DOI] [PubMed] [Google Scholar]
- 54.Kirikoshi H, Sekihara H, Katoh M. Up-regulation of WNT10A by tumor necrosis factor alpha and Helicobacter pylori in gastric cancer. Int J Oncol. 2001;19:533–536. [PubMed] [Google Scholar]
- 55.Hsu RJ, Ho JY, Cha TL, Yu DS, Wu CL, Huang WP, Chu P, Chen YH, Chen JT, Yu CP. WNT10A plays an oncogenic role in renal cell carcinoma by activating WNT/beta-catenin pathway. PLoS One. 2012;7:e47649. doi: 10.1371/journal.pone.0047649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaz AM, Wong CJ, Dzieciatkowski S, Luo Y, Schoen RE, Grady WM. Patterns of DNA methylation in the normal colon vary by anatomical location, gender, and age. Epigenetics. 2014;9:492–502. doi: 10.4161/epi.27650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidl C, Klug M, Boeld TJ, Andreesen R, Hoffmann P, Edinger M, Rehli M. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19:1165–1174. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Langford C, Bird A. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Chen C, Bi X, Zhou C, Huang T, Ni C, Yang P, Chen S, Ye M, Duan S. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1–7. doi: 10.1016/j.gene.2017.07.082. [DOI] [PubMed] [Google Scholar]
- 60.Draht MX, Smits KM, Tournier B, Jooste V, Chapusot C, Carvalho B, Cleven AH, Derks S, Wouters KA, Belt EJ, Stockmann HB, Bril H, Weijenberg MP, van den Brandt PA, de Bruine AP, Herman JG, Meijer GA, Piard F, Melotte V, van Engeland M. Promoter CpG island methylation of RET predicts poor prognosis in stage II colorectal cancer patients. Mol Oncol. 2014;8:679–688. doi: 10.1016/j.molonc.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 62.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 64.Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res. 1998;58:4515–4518. [PubMed] [Google Scholar]