Abstract
Lung cancer is the most common cancer in both the developing and developed countries, which has an unoptimistic outcome. As a result of high rates in mortality and considerable difficulties in treatment, early detection of lung cancer is thought as one of the potential solutions for this stigma. Tissue biopsy has been widely used for cancer diagnosis, but the invasive nature limits their application, especially when repeated biopsies are needed. Liquid biopsy, a minimally invasive procedure aiming to primarily analyze circulating tumor cells (CTCs) and/or circulating tumor DNA (ctDNA) for diagnosing and profiling cancer, has gained interest from oncologists and basic researchers. A great number of achievements in the field of liquid biopsy has been developed, thus liquid biopsy is more feasible in clinical practice than before. More importantly, liquid biopsy is being used, in addition to the diagnosis of lung cancer, to predict prognosis according to genetic alterations and monitor disease based on signature molecular markers. In this review, we briefly summarize techniques in liquid biopsy and focus on its applications in disease diagnosis, prognosis prediction, and condition monitoring of lung cancer.
Keywords: Lung cancer, liquid biopsy, prognosis, treatment responses
Introduction
Lung cancer is the most common cancer in the developing and developed worlds, and the leading cause of cancer-related death. According to the report of GLOBOCAN 2012 [1,2], it has been estimated that there are 1.8 million (12.9% of all cancer cases) new cases of lung cancer, and nearly 1.6 million deaths related to lung cancer (19.4% of all death cases) in the single year of 2012. The 5-year survival rate of lung cancer patients is between 4 and 17%, depending on different stages and regions [3]. These data indicate a cold fact that lung cancer heavily affects the population health. The major reasons for the poor prognosis of lung cancer are a large fraction of patients are in advanced stages when diagnosed and many of them are lack of therapeutic responses [3]. Symptoms of lung cancer are usually covert and unspecific, which allows few patients to seek medical attention due to symptoms. Currently, the only screening method in effect for lung cancer is low-dose computed tomography [4] recommended by the US Preventive Services Task Force [5], which also requires traditional tissue biopsy to confirm the characteristics of nodules observed by CT scan.
Compared to tissue biopsy, liquid biopsy has many advancements. Liquid biopsy is a collection of methods that are used to enrich, detect, and analyze circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) in cancer patients [6] (Figure 1; Tables 1 and 2). CTCs migrate from solid tumors and enter the bloodstream due to tumor invasion and shedding, or in response to a mechanical stress such as surgery. It is generally accepted that CTCs are a group of heterogeneous tumor cells derived from primary and/or metastatic tumors. CTCs, presenting at a low level in the circulation, are difficult to isolate and distinguish from the peripheral blood. Current technologies to isolate CTCs focus on the development of high-efficiency methods to capture pure tumor cell populations across many stages and types of tumor cells [7]. After tumor cell death, DNA released into the blood is referred to as cell-free tumor DNA (cfDNA), among which ctDNA is a component. Circulating DNA levels tend to be higher in patients with metastatic cancers compared to healthy individuals and non-metastatic patients [8]. Levels of cfDNA in the blood are positively correlated with the severity of cancer. The significance of CTCs and ctDNA in predicting prognosis has been demonstrated in some tumors, as CTCs and ctDNA can be used to detect drug-resistant mutations, mainly of which are acquired. ctDNA contains tumor-specific mutations that can be used to determine mutation conditions in tumor DNA. To be succinct, we summarize techniques used in liquid biopsy in Tables 1 and 2, and concentrate the values of these methods in disease diagnosis, prognosis prediction, and treatment monitoring.
Figure 1.
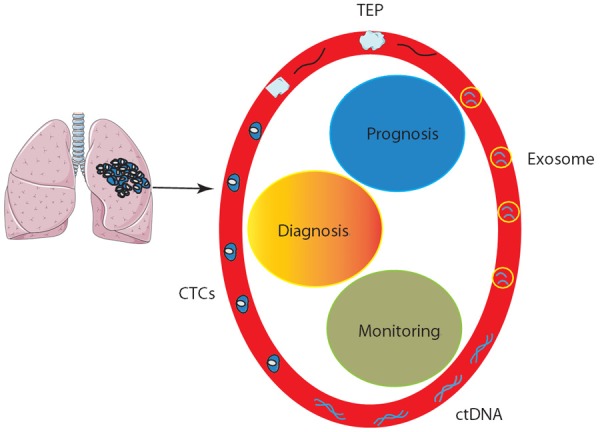
Overview of liquid biopsy in lung cancer. Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), exosomes, and tumor educated platelets (TEP) can be detected from the blood stream and utilized for diagnosis, prognosis prediction, and disease monitoring of patients with lung cancer.
Table 1.
Methods utilized for analyzing CTCs and ctDNA
| Methods | Mechanisms | Applications | Available products or markers | |
|---|---|---|---|---|
| CTC isolation | Positive selection | Selection of epithelial adhesion molecule (EpCAM) positive cells [64,65] | Magnetic beads, columns, cartridges, magnetic nanoparticles (MNPs) [66,67], nanoclusters [68], immunomagnetic nanospheres (IMNs) [69,70], iron oxide magnetic particles [71], lipid microbubbles [72], and microchips [64] | CellSearch® system [73] |
| Negative selection | Selection of CD45 negative cells [74,75] | |||
| Physical isolation | Selection by the density [76], size [77,78], and deformability [79,80] by centrifugation [81], microporous filters [82], microfluidic technology [83], and adhesion-based methods [84] | The microfluid platform [85]. | ||
| CTC analysis | Nucleic acid analysis | PCR, qRT-PCR, sequencing, array-comparative genomic hybridization [86] | KRAS, PIK3CA,and APC [86] | |
| Cell count | To count cell numbers and those that express certain markers. | Cytology, immunocytochemistry, flow cytometry | ||
| ctDNA analysis | To measure levels of ctDNA, and analyze sequences. | Real time PCR [88], droplet digital PCR (ddPCR) [89]; whole genome sequencing (WGS) [90], whole exome sequencing(WES) [91] and targeted region sequencing(TRS) [90,91] | Loss of heterozygosity, gene amplification, cancer-related viral sequences, hypermethylation in promoters, single-nucleotide mutations | |
Table 2.
Comparisons between CTCs and ctDNA
| CTCs | ctDNA | |
|---|---|---|
| Concentration in blood | Rare | Rich |
| Mechanism of release into the blood | Unclear | Unclear |
| Quantification | Yes | Difficult to do |
| Detection specificity | Higher than ctDNA | Lower than CTCs |
| Mutation detection | Yes | Yes |
| Predict prognosis | Yes | Yes |
| Monitor disease status | Yes | Yes |
| Predict treatment response | Yes | Yes |
| Predict relapse | Yes | Yes |
| Predict drug resistance | Yes | Yes |
| Detect molecular markers | Yes | Yes |
| Detection in CSF | Difficult to do | Yes |
| Indication of tumor burden and kinetics | Difficult to do | Yes |
| In vitro culture | Yes | No |
Applications of liquid biopsy in diagnosis of lung cancer
Liquid biopsy can be used to diagnose lung cancer in patients with high risks. One of advantages of liquid biopsy, compared to tissue biopsy, is that liquid biopsy can be performed repeatedly, especially when a confirmed diagnosis fails to be initially achieved. Ilie et al reported that CTCs were detected in patients with COPD patients when lung cancer was suspected [9]. In this study, patients with COPD were screened annually by low-dose spiral CT, and approximately 3% of these patients (5 of 168 COPD patients) had positive CTCs but normal CT results. The annual monitor of these five patients revealed lung nodules 1 to 4 years after the initial detection of CTCs, leading to surgical resection and histological diagnosis of early lung cancer. More importantly, CTCs failed to be detected in control individuals regardless of smoking or not. These indicate CTCs, at least combined with CT scans, might serve to detect early lung cancer.
cfDNA and circulating miRNA profiles have been tested in values of diagnosing lung cancer. A study involving 60 non-small cell lung cancer (NSCLC) patients and 40 COPD individuals analyzed the diagnostic values of cfDNA (both the short and long fragment) [10], which indicated that levels of cfDNA have an accuracy rate of 92.1% (the long fragment) and 83.6% (the short fragment) in lung cancer diagnosis, respectively. Levels of plasma miRNA expression can be used to predict lung cancer. Wozniak et al [11] analyzed plasma miRNA profiles in patients with stage I-IIIA lung cancer with the comparison with the control individuals, which detected 24 miRNA patterns different between the two groups. In another study [12], levels of miR944 and miR3662 were reported to be at least a fourfold increase in NSCLC patients with respect to healthy control. Moreover, in addition to the overall sensitivity (86%) and specificity (97%), a combination of four miRNAs (miR21, miR126, miR210, and miR485-5p) in the plasma is more sensitive in adenocarcinoma than squamous cell carcinoma (91% v.s. 82%) [13]. These data raise the possibility that circulating miRNA profiles can be combined with CT scans to identify lung cancer patients, which has been tested in small cohorts [14]. In the future, values of liquid biopsy in diagnosis of lung cancer, especially during the screening for early lung cancer in the high-risk population, requires well-designed prospective studies involving large cohorts.
Prognosis prediction of CTCs in lung cancer
Values of CTCs in predicting prognosis in lung cancer patients have been appreciated by clinical oncologists (Table 3).
Table 3.
Values of liquid biopsy in lung cancer diagnosis, prognosis prediction, and disease monitoring
| Diagnosis | Prognosis | Disease monitoring | |
|---|---|---|---|
| CTCs | Lung cancer patients bear more CTCs | High number indicates poor prognosis | Higher number indicates high risks for recurrence |
| KRAS and P53 reemerging indicates recurrence | |||
| Apoptotic CTCs | Positive results indicate poor prognosis | ||
| PD-1 on CTCs | Positive results indicate poor prognosis | ||
| ctDNA | SNV detected in early NSCLC patients | High concentration indicates poor prognosis | Determination of mutation patterns for targeted therapy decision making |
| High concentration during follow-up predicts recurrence | |||
| Exosome-derived miRNA | Potential for early cancer detection | ||
| Secretome | Differential diagnosis of pleural effusion | ||
| TEP-derived mRNA | mRNA profiles for diagnosis | Detection of KRAS mutation, EGFR mutation, MET overexpression |
Baseline CTC counts
Accurately predicting the prognosis of cancer patients using CTCs has been achieved in several types of tumors. Baseline CTC counts are an important prognostic factor, as patients with CTCs at baseline indicates a poor prognosis. Dedicated studies demonstrating significant correlations between CTC counts and prognosis have been observed in many tumor types, including small cell lung cancer (SCLC) [15] and NSCLC [16]. In a study, the correlation of CTC counts and prognosis in 51 patients with newly diagnosed SCLC was analyzed, where baseline CTCs were detected in 40 patients with a median of 4 CTCs [15]. At baseline, after treatment, and at relapse, patients with a CTC count < 9 (the favorable population), or conversion from a CTC count > 9 (the unfavorable population) to a favorable CTC-count, had better survival rates than those with a CTC count > 9 [15,17]. Similarly, in NSCLC patients, detection of CTCs at baseline may predict a poorer prognosis when compared to patients without detectable CTCs at baseline [17]. The presence of CTCs in metastatic NSCLC patients and SCLC is associated with poor outcome [18,19]. Krebs and colleagues [18] also reported that the progression-free survival (PFS) was 6.8 months in patients bearing fewer than 5 CTCs and 2.4 months in those having more than 5 CTCs before chemotherapy (the overall survival: 8.1 v.s. 4.3 months). Similar results were observed in lung cancer patients with radiotherapy [20] and surgery [21].
As disease progress, the positive detection rate of CTCs increases. Thus, CTCs may be used as a factor in staging advanced diseases. A study showed that the positive rates and the range of CTC counts at baseline were 13% and 0-1 among patients with stage IIIA, 20% and 0-16 among patients with stage IIIB, 38% and 0-4 among patients with stage IV, and 60% and 0-67 among naive NSCLC patients, respectively [17].
These results are from relatively small cohorts, but suggest important values of CTCs in disease staging and prognosis prediction. It is also imperative to compare the values of CTCs and the current TNM system in selected lung cancer patients to emphasize CTCs’ significance.
Apoptotic CTCs
Apoptotic CTCs are also an independent prognostic factor, as the presence of CTCs with apoptotic nuclear morphology appears to indicate a poorer prognosis [22]. Positive detection of CTC apoptosis was associated with a worse prognosis in SCLC patients with a shorter PFS and OS [23], indicating the importance of CTC apoptosis in predicting the outcome. It is worth noting that the biological functions of apoptotic CTCs have not been elucidated, which warrants investigations in the future.
Programmed death 1 expressed on CTCs
Programmed death 1 (PD-1) is an immune checkpoint that regulates T cell-mediated responses. PD-1 and its ligand, programmed death ligand l (PD-L1), can also be used as a biomarker for prognosis of cancer patients. It has been established that lung cancer patients with high PD-L1 expression had a poor prognosis as tumor cells escaped from immune surveillance by the interaction between PD-L1 expressed by lung cancer cells and PD-1 expressed by tumor-infiltrating dendritic cells [24]. In a study that recruited 24 metastatic NSCLC patients receiving anti-PD-1 antibody Nivolumab, CTCs and surface PD-L1 on CTCs were examined by the CellSearch system. Patients who had detected CTCs and surface PD-L1 expression at baseline and 3 months after treatment had an unfavorable outcome [25]. Controversially, high expression of PD-L1 (more than 50% tumor cells expressed PD-L1) also predicted a good response upon PD-1 blockade (Pembrolizumab) in lung cancer patients [26]. The seeming difference between the above two trials [25,26] might result from administration of two different types of antibodies, or tumors cells possibly have varied ability to leaving the tumor bed and entering the circulation because of different levels of PD-L1 expression. The further understanding activation of the PD-1-PD-L1-axis will help to unfold the underlying mechanisms.
ctDNA in diagnosis lung cancer
The cellular source of ctDNA can be both healthy and tumor cells [27], but ctDNA from different cell types have varied characteristics. ctDNA from necrotic tumor cells has various sizes, whereas ctDNA released from apoptotic healthy cells has a uniform size between 185- and 200-bp. Since the major origin of ctDNA in healthy individuals is from apoptotic cells, a preponderance of longer DNA fragments can be considered as an indication for tumors [27]. It has also been evident that tumor patients have increased levels of ctDNA compared to healthy volunteers [28,29]. In a pilot study analyzing ctDNA profiles of lung cancer patients [30], at least two single-nucleotide variations (SNV) in ctDNA were observed in 46 out of 96 (48%) early-stage NSCLC patients, in addition to a single SNV detected in 12 cases. These valuable data indicate that profiling ctDNA can be used to screen early lung cancer.
Liquid biopsy as a means to monitor disease
CTCs to monitor disease recurrence
Wu and colleagues investigated the association between CTC counts and cancer recurrence in untreated lung cancer patients including adenocarcinoma, squamous cell carcinoma, and SCLC [31], where patients with recurrence were found to bear higher levels of CTCs at diagnosis. Reappearance of tumor-specific mutations in the circulation including KRAS and P53 indicates tumor recurrence in lung cancer patients [32]. These data highlight the importance of CTC counts for predicting recurrence. However, studies of larger scales are warranted.
ctDNA predicts treatment response
ctDNA can be used to determine mutations in lung cancer patients. A technique called “cancer personalized profiling by deep sequencing (CAPP-Seq)” has been utilized in lung cancer patients [33], which has an ability to detect various types of somatic mutations. The CAPP-Seq technique also applies to ctDNA. However, as the concentration of ctDNA is low in early-stage lung cancer patients, only approximately 50% of early lung cancer patients harbor ctDNA [6]. Therefore, the value of the CAPP-Seq technique in detecting early lung cancer patients faces challenges. An increased copy number of the MET gene was reported to be determined in lung cancer patients during targeted therapy treatment [34]. It was reported that a microsatellite alteration was detected in the plasma of 71% (5 out 21) patients with small cell lung cancer [35].
According to the mutation patterns, an appropriate targeted therapy can be applied to a certain patient. Amplification of MET genes by determining the copy number in the plasma predicts treatment responses to anti-EGFR therapy in lung cancer patients [36]. The T790M mutation in the membrane receptor EGFR indicates therapeutic resistance to gefitinib and erlotinib in approximately 50% of lung cancer patients [37]. More importantly, the T790M mutation can be also observed in the plasma of recurrent patients [38,39], indicating the feasibility of monitoring treatment resistance by liquid biopsy. The T790M substitution increases the affinity of EGFR for ATP, therefore, inhibits the binding of the inhibitors during targeted therapy [40]. The detection of this mutation has clinical significance, as newer inhibitors (such as the third-generation inhibitor WZ4002) works effectively when the T790M mutation is evident [41]. This sheds a light that it is possible to use liquid biopsy to select a treatment regime in lung cancer patients with certain mutations.
Unbiased approaches can also be applied in liquid biopsy, where genetic mutation markers for treatment resistance can be detected in the blood of cancer patients [42]. It has been reported that the T790M mutation of EGFR was detected in the plasma of a lung cancer patient treated by gefitinib by exome sequencing [43].
ctDNA predicts risks of recurrence
Sozzi and colleagues [32] explored whether the concentration of ctDNA could be used to predict recurrence in patients with NSCLC. Patients with recurrence harbored with higher levels of ctDNA during follow-ups compared to those without regression (34 ng/ml v.s. 345 ng/ml, P < 0.001). More importantly, relapse-free patients showed a trend of reduction of ctDNA levels with time, indicating the dynamic changes of ctDNA has the potential to monitor disease recurrence.
Other markers with predictive values for prognosis of lung cancer patients that can be detected by liquid biopsy
Exosomes are small membrane vesicles originated from cells and are detected in almost all biological fluid, including urine, blood, ascites, and cerebrospinal fluid [44]. The size of exosomes is between 30 and 100 nm. Exosomes are composed of a phospholipid membrane, where proteins involved in membrane transport and fusion, endosomal sorting complex, and tetraspanins are also present, therefore exosomes have an important role in cell-to-cell communication via activating the expression of ligands or transferring signal molecules. Exosomes also affect cancer growth by modulating the local and systemic environment or manipulating the immune system [45]. The profiling of microRNA (miRNA) is significantly different between the primary lung cancer tissues and corresponding normal tissues, which makes miRNA a potential diagnostic marker [46]. It has been reported that exosomes are rich in genetic materials (including miRNA) and protein, and exosomes can transfer genetic information between cells [47]. In a dedicated study performed by Rabinowits and colleagues [48] indicated miRNA derived from circulating exosomes and those collected from lung cancer had a similar profile, indicating the possibility to utilizing circulating exosomes and containing miRNA profiles to detect lung cancer. Moreover, miRNA in exosomes have the capacity to switch the host to a prometastatic phenotype [49].
The secretome was first discovered in a study of proteins secreted by Bacillus subtilis [50], and this concept has been extended to the collection of proteins secreted by a cell, tissue, or organism [51], including the context of cancer [52]. Secreted proteins are involved in many processes related to cancer, such as angiogenesis, differentiation, invasion, and metastasis. Patients with NSCLC occasionally suffer the malignant pleural effusion that has been studied as a source of biomarkers for lung cancer patients [53]. However, the significance of secretome in the pleural effusion of lung cancer patients, and whether a certain protein present in the pleural effusion can be also detected in the circulation, warrants further studies.
Tumor educated platelets [46] have gained interest from researchers in oncology. Confrontation between tumor cells and platelets results in transfer of biomolecule to TEP [54]. Upon activation, platelets undergo specific splicing of pre-mRNAs [55], which leads to a unique mRNA profile that can serve as a means for cancer diagnosis [56]. A study profiling mRNA derived from TEP of cancer patients demonstrated 20 non-protein coding RNAs had altered levels with respect to those from healthy individuals [54]. Moreover, 1453 mRNAs were increased and 793 mRNAs were decreased in TEP of cancer patients comparing to healthy control, which allowed to detect KRAS mutation, EGFR mutation, and MET overexpression. Although roles of TEP and mRNAs derived TEP in lung cancer in terms of diagnosis and prediction have not been fully tested yet, it is worth to warrant a thorough trial in the near future.
Perspective
On May 23rd 2017, the US Food and Drug Administration (FDA) approved pembrolizumab, a PD-1 inhibitor, to treat unresectable or metastatic solid cancers with high microsatellite instability or mismatch repair gene deficiency in adult and pediatric patients, regardless of tumor location [49]. This is the first approved medication for cancer therapy targeting gene mutations rather than tumor sites based on previous encouraging results [57,58]. Although the FDA has not approved a method to detect mismatch repair gene deficiency or microsatellite instability, it is reasonable to speculate that liquid biopsy can be performed to detect mismatch repair gene deficiency in ctDNA or microsatellite instability in CTCs collected from the blood of lung cancer patients to predict treatment responses upon PD-1 blockade. In other cancers like melanoma, results from mouse experiments and human trial indicated that treatment response upon PD-1 blockade relies on the composition of the microbiome in the small intestine and colon [59-62]. Bacterial 16S rRNA is used for bacterial identification and can be detected in the circulation of individuals [63], thus it is possible to use liquid biopsy methods to determine bacterial 16S rRNA to predict the presence of bacterial strains that can predict treatment response upon PD-1 blockade. In the future, investigations are required to test these speculations in lung cancer patients.
Acknowledgements
The National Natural Science foundation of China (Grant No. 81372825), the Hunan Province Science Fund for Distinguished Young Scholars (2018JJ1021), the Key R&D Program of Hunan Province (2017SK2172), the Natural Science Foundation of Hunan Province (2017JJ2003, 2017JJ2004), the Hunan Province Technological Innovation Guiding Plan, Clinical Medical Technology Innovation Guidance Project (2017SK51101), the Health Department project of the Hunan Province (B2017182, B20180378), the Science and Technique Foundation of Chenzhou (JSYF2017021).
Disclosure of conflict of interest
None.
References
- 1.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, Bonaventure A, Valkov M, Johnson CJ, Esteve J, Ogunbiyi OJ, Azevedo E Silva G, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP CONCORD Working Group. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Retrouvey M, Patel Z, Shaves S. US preventive services task force CT lung cancer screening recommendations: community awareness and perceptions. J Am Coll Radiol. 2016;13:R35–37. doi: 10.1016/j.jacr.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Bracht JWP, Mayo-de-Las-Casas C, Berenguer J, Karachaliou N, Rosell R. The present and future of liquid biopsies in non-small cell lung cancer: combining four biosources for diagnosis, prognosis, prediction, and disease monitoring. Curr Oncol Rep. 2018;20:70. doi: 10.1007/s11912-018-0720-z. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Bode AM, Dong Z. Circulating tumor cells: moving biological insights into detection. Theranostics. 2017;7:2606–2619. doi: 10.7150/thno.18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 9.Ilie M, Hofman V, Long-Mira E, Selva E, Vignaud JM, Padovani B, Mouroux J, Marquette CH, Hofman P. “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One. 2014;9:e111597. doi: 10.1371/journal.pone.0111597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soliman SE, Alhanafy AM, Habib MSE, Hagag M, Ibrahem RAL. Serum circulating cell free DNA as potential diagnostic and prognostic biomarker in non small cell lung cancer. Biochem Biophys Rep. 2018;15:45–51. doi: 10.1016/j.bbrep.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wozniak MB, Scelo G, Muller DC, Mukeria A, Zaridze D, Brennan P. Circulating microRNAs as non-invasive biomarkers for early detection of non-small-cell lung cancer. PLoS One. 2015;10:e0125026. doi: 10.1371/journal.pone.0125026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito Y, Tanabe Y, Han J, Fujita T, Tanigaki K, Chen M. Multifunctional porous graphene for high-efficiency steam generation by heat localization. Adv Mater. 2015;27:4302–4307. doi: 10.1002/adma.201501832. [DOI] [PubMed] [Google Scholar]
- 13.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL, Jiang Z, Fang H, Katz RL, Jiang F. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7 doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naito T, Tanaka F, Yoneda K, Takahashi T, Murakami H, Nakamura Y, Tsuya A, Endo M, Kenmotsu H, Kaira K, Shukuya T, Ono A, Akamatsu H, Miura S, Kimura M, Yamamoto N. The prognostic impact of circulating tumor cells in patients with small cell lung cancer. J. Clin. Oncol. 2011;29:10617. doi: 10.1097/JTO.0b013e31823f125d. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Xiao Y, Zhao J, Chen M, Xu Y, Zhong W, Xing J, Wang M. Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology. 2016;21:519–25. doi: 10.1111/resp.12696. [DOI] [PubMed] [Google Scholar]
- 17.Kato T, Yamada K, Nokihara H, Fukui T, Yamamoto N, Sekine I, Kunitoh H, Ohe Y, Koizumi F, Tamura T. Circulating tumor cell analysis in patients with non-small cell lung cancer. J. Clin. Oncol. 2008;26:19026. [Google Scholar]
- 18.Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G, Ranson M, Dive C, Blackhall FH. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 19.Hiltermann TJ, Pore MM, van den Berg A, Timens W, Boezen HM, Liesker JJ, Schouwink JH, Wijnands WJ, Kerner GS, Kruyt FA, Tissing H, Tibbe AG, Terstappen LW, Groen HJ. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann Oncol. 2012;23:2937–2942. doi: 10.1093/annonc/mds138. [DOI] [PubMed] [Google Scholar]
- 20.Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, Hicks RJ, Hampton GM, Amler LC, Pirzkall A, Lackner MR. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res. 2012;18:2391–2401. doi: 10.1158/1078-0432.CCR-11-3148. [DOI] [PubMed] [Google Scholar]
- 21.Bayarri-Lara C, Ortega FG, Cueto Ladron de Guevara A, Puche JL, Ruiz Zafra J, de Miguel-Perez D, Ramos AS, Giraldo-Ospina CF, Navajas Gomez JA, Delgado-Rodriguez M, Lorente JA, Serrano MJ. Circulating tumor cells identify early recurrence in patients with non-small cell lung cancer undergoing radical resection. PLoS One. 2016;11:e0148659. doi: 10.1371/journal.pone.0148659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, Ward T, Blackhall FH, Dive C. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 23.Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, Ward T, Blackhall FH, Dive C. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 24.Mu CY, Huang JA, Chen Y, Chen C, Zhang XG. High expression of PD-L1 in lung cancer may contribute to poor prognosis and tumor cells immune escape through suppressing tumor infiltrating dendritic cells maturation. Med Oncol. 2011;28:682–688. doi: 10.1007/s12032-010-9515-2. [DOI] [PubMed] [Google Scholar]
- 25.Nicolazzo C, Raimondi C, Mancini M, Caponnetto S, Gradilone A, Gandini O, Mastromartino M, Del Bene G, Prete A, Longo F, Cortesi E, Gazzaniga P. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci Rep. 2016;6:31726. doi: 10.1038/srep31726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn MJ, Felip E, Lee JS, Hellmann MD, Hamid O, Goldman JW, Soria JC, Dolled-Filhart M, Rutledge RZ, Zhang J, Lunceford JK, Rangwala R, Lubiniecki GM, Roach C, Emancipator K, Gandhi L KEYNOTE-001 Investigators. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 27.Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, Rossier A, Chen XQ, Anker P. The origin and mechanism of circulating DNA. Ann N Y Acad Sci. 2000;906:161–168. doi: 10.1111/j.1749-6632.2000.tb06608.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhong XY, Ladewig A, Schmid S, Wight E, Hahn S, Holzgreve W. Elevated level of cell-free plasma DNA is associated with breast cancer. Arch Gynecol Obstet. 2007;276:327–331. doi: 10.1007/s00404-007-0345-1. [DOI] [PubMed] [Google Scholar]
- 29.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 30.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, Marafioti T, Kirkizlar E, Watkins TBK, McGranahan N, Ward S, Martinson L, Riley J, Fraioli F, Al Bakir M, Gronroos E, Zambrana F, Endozo R, Bi WL, Fennessy FM, Sponer N, Johnson D, Laycock J, Shafi S, Czyzewska-Khan J, Rowan A, Chambers T, Matthews N, Turajlic S, Hiley C, Lee SM, Forster MD, Ahmad T, Falzon M, Borg E, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Hafez D, Naik A, Ganguly A, Kareht S, Shah R, Joseph L, Marie Quinn A, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Oukrif D, Akarca AU, Hartley JA, Lowe HL, Lock S, Iles N, Bell H, Ngai Y, Elgar G, Szallasi Z, Schwarz RF, Herrero J, Stewart A, Quezada SA, Peggs KS, Van Loo P, Dive C, Lin CJ, Rabinowitz M, Aerts H, Hackshaw A, Shaw JA, Zimmermann BG, consortium TR, consortium P, Swanton C. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C, Hao H, Li L, Zhou X, Guo Z, Zhang L, Zhang X, Zhong W, Guo H, Bremner RM, Lin P. Preliminary investigation of the clinical significance of detecting circulating tumor cells enriched from lung cancer patients. J Thorac Oncol. 2009;4:30–36. doi: 10.1097/JTO.0b013e3181914125. [DOI] [PubMed] [Google Scholar]
- 32.Sozzi G, Conte D, Mariani L, Lo Vullo S, Roz L, Lombardo C, Pierotti MA, Tavecchio L. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001;61:4675–4678. [PubMed] [Google Scholar]
- 33.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW Jr, Alizadeh AA, Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, Lindeman NI, Murphy C, Akhavanfard S, Yeap BY, Xiao Y, Capelletti M, Iafrate AJ, Lee C, Christensen JG, Engelman JA, Janne PA. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen XQ, Stroun M, Magnenat JL, Nicod LP, Kurt AM, Lyautey J, Lederrey C, Anker P. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996;2:1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- 36.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC, Miller V, Ladanyi M, Yang CH, Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, Abdel-Rahman M, Wang X, Levine AD, Rho JK, Choi YJ, Choi CM, Kim SW, Jang SJ, Park YS, Kim WS, Lee DH, Lee JS, Miller VA, Arcila M, Ladanyi M, Moonsamy P, Sawyers C, Boggon TJ, Ma PC, Costa C, Taron M, Rosell R, Halmos B, Bivona TG. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura T, Sueoka-Aragane N, Iwanaga K, Sato A, Komiya K, Kobayashi N, Hayashi S, Hosomi T, Hirai M, Sueoka E, Kimura S. Application of a highly sensitive detection system for epidermal growth factor receptor mutations in plasma DNA. J Thorac Oncol. 2012;7:1369–1381. doi: 10.1097/JTO.0b013e31825f2821. [DOI] [PubMed] [Google Scholar]
- 39.Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F, Kato K. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17:7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 40.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, Engen JR, Wong KK, Eck MJ, Gray NS, Janne PA. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, Marass F, Humphray S, Hadfield J, Bentley D, Chin TM, Brenton JD, Caldas C, Rosenfeld N. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 43.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 45.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 47.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 48.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 49.Rana S, Malinowska K, Zoller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15:281–295. doi: 10.1593/neo.122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. Signal peptide-dependent protein transport in bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev. 2000;64:515–547. doi: 10.1128/mmbr.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volmer MW, Stuhler K, Zapatka M, Schoneck A, Klein-Scory S, Schmiegel W, Meyer HE, Schwarte-Waldhoff I. Differential proteome analysis of conditioned media to detect Smad4 regulated secreted biomarkers in colon cancer. Proteomics. 2005;5:2587–2601. doi: 10.1002/pmic.200401188. [DOI] [PubMed] [Google Scholar]
- 52.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Yu CJ, Wang CL, Wang CI, Chen CD, Dan YM, Wu CC, Wu YC, Lee IN, Tsai YH, Chang YS, Yu JS. Comprehensive proteome analysis of malignant pleural effusion for lung cancer biomarker discovery by using multidimensional protein identification technology. J Proteome Res. 2011;10:4671–4682. doi: 10.1021/pr2004743. [DOI] [PubMed] [Google Scholar]
- 54.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, Schellen P, Verschueren H, Post E, Koster J, Ylstra B, Ameziane N, Dorsman J, Smit EF, Verheul HM, Noske DP, Reijneveld JC, Nilsson RJA, Tannous BA, Wesseling P, Wurdinger T. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28:666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schubert S, Weyrich AS, Rowley JW. A tour through the transcriptional landscape of platelets. Blood. 2014;124:493–502. doi: 10.1182/blood-2014-04-512756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M, Widmark A, Gerritsen WR, Verheul HM, Vandertop WP, Noske DP, Skog J, Wurdinger T. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118:3680–3683. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. 2016;22:813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 58.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragon L, Jacquelot N, Qu B, Ferrere G, Clemenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 62.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, Chang EB, Gajewski TF. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang W, Lederman MM, Hunt P, Sieg SF, Haley K, Rodriguez B, Landay A, Martin J, Sinclair E, Asher AI, Deeks SG, Douek DC, Brenchley JM. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 66.Poudineh M, Aldridge PM, Ahmed S, Green BJ, Kermanshah L, Nguyen V, Tu C, Mohamadi RM, Nam RK, Hansen A, Sridhar SS, Finelli A, Fleshner NE, Joshua AM, Sargent EH, Kelley SO. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat Nanotechnol. 2017;12:274–281. doi: 10.1038/nnano.2016.239. [DOI] [PubMed] [Google Scholar]
- 67.Kwak B, Lee J, Lee D, Lee K, Kwon O, Kang S, Kim Y. Selective isolation of magnetic nanoparticle-mediated heterogeneity subpopulation of circulating tumor cells using magnetic gradient based microfluidic system. Biosens Bioelectron. 2017;88:153–158. doi: 10.1016/j.bios.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 68.Xiong K, Wei W, Jin Y, Wang S, Zhao D, Wang S, Gao X, Qiao C, Yue H, Ma G, Xie HY. Biomimetic immuno-magnetosomes for high-performance enrichment of circulating tumor cells. Adv Mater. 2016;28:7929–7935. doi: 10.1002/adma.201601643. [DOI] [PubMed] [Google Scholar]
- 69.Wen CY, Wu LL, Zhang ZL, Liu YL, Wei SZ, Hu J, Tang M, Sun EZ, Gong YP, Yu J, Pang DW. Quick-response magnetic nanospheres for rapid, efficient capture and sensitive detection of circulating tumor cells. ACS Nano. 2014;8:941–949. doi: 10.1021/nn405744f. [DOI] [PubMed] [Google Scholar]
- 70.Reategui E, Aceto N, Lim EJ, Sullivan JP, Jensen AE, Zeinali M, Martel JM, Aranyosi AJ, Li W, Castleberry S, Bardia A, Sequist LV, Haber DA, Maheswaran S, Hammond PT, Toner M, Stott SL. Tunable nanostructured coating for the capture and selective release of viable circulating tumor cells. Adv Mater. 2015;27:1593–1599. doi: 10.1002/adma.201404677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu J, Wei X, Gan J, Huang L, Shen T, Lou J, Liu B, Zhang JX, Qian K. Multifunctional magnetic particles for combined circulating tumor cells isolation and cellular metabolism detection. Adv Funct Mater. 2016;26:4016–4025. doi: 10.1002/adfm.201504184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang G, Pingle S, Kesari S, Liu YT, Simberg D. Rapid harvesting of highly concentrated circulating tumor cells from bulk human blood with buoyant immuno-microbubbles. Cancer Research. 2016:76. [Google Scholar]
- 73.Kraan J, Sleijfer S, Strijbos MH, Ignatiadis M, Peeters D, Pierga JY, Farace F, Riethdorf S, Fehm T, Zorzino L, Tibbe AG, Maestro M, Gisbert-Criado R, Denton G, de Bono JS, Dive C, Foekens JA, Gratama JW. External quality assurance of circulating tumor cell enumeration using the CellSearch((R)) system: a feasibility study. Cytometry B Clin Cytom. 2011;80:112–118. doi: 10.1002/cyto.b.20573. [DOI] [PubMed] [Google Scholar]
- 74.Lustberg M, Jatana KR, Zborowski M, Chalmers JJ. Emerging technologies for CTC detection based on depletion of normal cells. Recent Results Cancer Res. 2012;195:97–110. doi: 10.1007/978-3-642-28160-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Z, Fusi A, Klopocki E, Schmittel A, Tinhofer I, Nonnenmacher A, Keilholz U. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med. 2011;9:70. doi: 10.1186/1479-5876-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosenberg R, Gertler R, Friederichs J, Fuehrer K, Dahm M, Phelps R, Thorban S, Nekarda H, Siewert JR. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49:150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 77.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, Cote RJ. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koss LG, Melamed MR. Cancer filter deja vu. Science. 2007;318:1864. [PubMed] [Google Scholar]
- 79.Park ES, Jin C, Guo Q, Ang RR, Duffy SP, Matthews K, Azad A, Abdi H, Todenhoefer T, Bazov J, Chi KN, Black PC, Ma H. Continuous flow deformability-based separation of circulating tumor cells using microfluidic ratchets. Small. 2016;12:1909–1919. doi: 10.1002/smll.201503639. [DOI] [PubMed] [Google Scholar]
- 80.Kang YT, Doh I, Byun J, Chang HJ, Cho YH. Label-free rapid viable enrichment of circulating tumor cell by photosensitive polymer-based microfilter device. Theranostics. 2017;7:3179–3191. doi: 10.7150/thno.19686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, Lim WT, Han J, Bhagat AA, Lim CT. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qin X, Park S, Duffy SP, Matthews K, Ang RR, Todenhoefer T, Abdi H, Azad A, Bazov J, Chi KN, Black PC, Ma H. Size and deformability based separation of circulating tumor cells from castrate resistant prostate cancer patients using resettable cell traps. Lab Chip. 2015;15:2278–2286. doi: 10.1039/c5lc00226e. [DOI] [PubMed] [Google Scholar]
- 83.Warkiani ME, Khoo BL, Wu L, Tay AK, Bhagat AA, Han J, Lim CT. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat Protoc. 2016;11:134–148. doi: 10.1038/nprot.2016.003. [DOI] [PubMed] [Google Scholar]
- 84.Hoshiba T, Orui T, Endo C, Sato K, Yoshihiro A, Minagawa Y, Tanaka M. Adhesion-based simple capture and recovery of circulating tumor cells using a blood-compatible and thermo-responsive polymer-coated substrate. Rsc Advances. 2016;6:89103–89112. [Google Scholar]
- 85.Goodall J, Mateo J, Yuan W, Mossop H, Porta N, Miranda S, Perez-Lopez R, Dolling D, Robinson DR, Sandhu S, Fowler G, Ebbs B, Flohr P, Seed G, Nava Rodrigues D, Boysen G, Bertan C, Atkin M, Clarke M, Crespo M, Figueiredo I, Riisnaes R, Sumanasuriya S, Rescigno P, Zafeiriou Z, Sharp A, Tunariu N, Bianchini D, Gillman A, Lord CJ, Hall E, Chinnaiyan AM, Carreira S, de Bono JS. Circulating free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7:1006–1017. doi: 10.1158/2159-8290.CD-17-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C, Hoefler G, Eisner F, Sill H, Samonigg H, Pantel K, Riethdorf S, Bauernhofer T, Geigl JB, Speicher MR. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 87.Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, Ulman MJ, Xega K, Contino G, Alagesan B, Brannigan BW, Milos PM, Ryan DP, Sequist LV, Bardeesy N, Ramaswamy S, Toner M, Maheswaran S, Haber DA. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Catarino R, Ferreira MM, Rodrigues H, Coelho A, Nogal A, Sousa A, Medeiros R. Quantification of free circulating tumor DNA as a diagnostic marker for breast cancer. DNA Cell Biol. 2008;27:415–421. doi: 10.1089/dna.2008.0744. [DOI] [PubMed] [Google Scholar]
- 89.Christensen E, Birkenkamp-Demtröder K, Nordentoft I, Høyer S, van der Keur K, van Kessel K, Zwarthoff E, Agerbæk M, Ørntoft TF, Jensen JB, Dyrskjøt L. Liquid biopsy analysis of FGFR3 and PIK3CA hotspot mutations for disease surveillance in bladder cancer. Eur Urol. 2017;71:961–969. doi: 10.1016/j.eururo.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 90.De Laere B, van Dam PJ, Whitington T, Mayrhofer M, Diaz EH, Van den Eynden G, Vandebroek J, Del-Favero J, Van Laere S, Dirix L, Grönberg H, Lindberg J. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structural variation and splice variant expression patterns. Eur Urol. 2017;72:192–200. doi: 10.1016/j.eururo.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 91.Girotti MR, Gremel G, Lee R, Galvani E, Rothwell D, Viros A, Mandal AK, Lim KH, Saturno G, Furney SJ, Baenke F, Pedersen M, Rogan J, Swan J, Smith M, Fusi A, Oudit D, Dhomen N, Brady G, Lorigan P, Dive C, Marais R. Application of sequencing, liquid biopsies and patient-derived xenografts for personalized medicine in melanoma. Cancer Discov. 2016;6:286–99. doi: 10.1158/2159-8290.CD-15-1336. [DOI] [PubMed] [Google Scholar]


