1. Introduction
Medical research focuses on identifying the causes and deciphering the mechanisms related to a disease, aiming to eventually develop accurate diagnostic tools and effective treatments. With the breakthrough technological advances of the last decades, the “educated” guess that had been previously used for raising scientific hypotheses is rapidly being replaced by the knowledge provided through untargeted high-throughput methods that are able to generate enormous data sets in a short amount of time and in a cost-effective manner. Fortunately, major advances have been also observed in computational mathematics which enables the accurate analysis of the “big” data sets deriving from high-throughput approaches. Here, we summarize the most important “-omics” procedures and describe the current challenges related to their use. Additionally, we describe the novel methods of data-mining and machine learning analysis, and particularly, how they can be used in a hierarchical manner to produce robust results for medicine from “big” data.
2. Omics and their use in medicine
The term “omics” refers to the comprehensive characterization, quantitation and quantification of a large number of molecules, grouped according to fundamental structural or functional biological similarities that they demonstrate. The largest and most important categories so far include:
-
a)
Genomics were the first omics to appear and are the most extensively investigated thus far. Genomics comprise the analysis of a whole genome, aiming to identify genetic variants that are either associated with a specific disease and/or its future prognosis, or are related to the response to a specific treatment [1]. Genome wide association studies (GWAS) are large observational studies with thousands of subjects deriving from multiple populations that usually follow a case-control study design and aim to identify single nucleotide polymorphisms (SNPs) that are more frequently present in the group of cases (i.e. group of subjects with a specific disease) versus the control group (i.e. healthy individuals). The first GWAS was performed in 2005, and included only 96 cases and 50 healthy controls [2]. This study identified two SNPs associated with an increased risk for age-related macular degeneration [2]. Since then, a great number of GWAS have been performed with thousands of subjects in dozens of different diseases [3–5]. GWAS have been very effective at identifying genetic predisposition for certain multifactorial diseases such as metabolic diseases (i.e. coronary heart disease [6], diabetes [7], obesity [8], dyslipidemia [9], hypertension, NAFLD [10]), neurological disorders (i.e. Alzheimer [11], bipolar disorders), certain types of cancer (e.g. breast cancer [12], ovarian cancer [13], prostate cancer [14]), gastroenterological diseases (e.g. Crohn's disease [15]) and autoimmune disorders (e.g. rheumatoid arthritis [16]).
-
b)
Epigenomics analyze the reversal modifications of DNA (e.g. DNA methylation) or DNA-related proteins (histone modification). These modifications affect gene expression and transcription without changing the DNA sequence. Epigenetic modifications are influenced by genetic and environmental factors, can occur at any time over a person's lifetime, can last a lifetime, and can be inherited [17,18]. Epigenome-wide association studies have linked specific epigenetic modifications with cardiovascular disease [19], obesity [20], type 2 diabetes (T2D), cancer [21] and other diseases [22,23]. Methods used to assess epigenetic modifications include histone modification assays such as ChIP-Chip and ChIP-Seq which couple chromatin immunoprecipitation with DNA microarrays or with next generation DNA-sequencing, and DNA methylation assays, which adapt next generation DNA-sequencing procedures.
-
c)
Transcriptomics refers to the quantitation and quantification of all RNA transcripts in a biological sample. This includes analysis both of protein-coding and non-coding transcriptome (long-non coding RNA as well as short RNAs such as microRNAs, small nuclear RNAs, piwi-interacting RNAs etc.). The coding transcriptome is practically an intermediate step in biological processes, linking genome with proteome. Protein-coding transcriptome has been used to identify mechanisms involved in different diseases, as well as combined with genomics and proteomics (Proteogenomics) to discover new genes and their functional relevance. The non-coding transcriptome is not associated with protein formation but can still significantly affect major physiological processes, such as major hormonal pathways [24,25], brown adipose tissue development [26], cell growth, proliferation and differentiation [27,28]. Consequently, specific non-coding RNAs have been associated with several metabolic diseases, neurological disorders and different types of cancer [29].
-
d)
Proteomics refers to the quantitation and quantification of peptides/proteins in biological samples or the investigation of their post-translational modifications and interactions. The post-translational modifications that can be investigated are protein phosphorylation, glycosylation, ubiquitination, nitrosylation and proteolysis. Post-translational modifications can have a major impact on protein function and transport, enzymatic activity and intracellular signaling pathways [30]. Proteomics are performed with mass-spectrometric methods. Protein-protein interactions require affinity purification methods as preparation steps. Proteomics are widely used in different research fields. They are used to identify possible biomarkers of a disease, to detect potential therapeutic targets, and to understand fundamental biological functions.
-
e)
Metabolomics is the investigation of intermediate small molecules and products of metabolism. It commonly refers to molecules <1 kD, including amino acids, fatty acids and carbohydrates. Metabolomics quantify the end products of cellular processes, offering this way a snapshot of the metabolic status of cell(s). The analysis of metabolomics involves first a separation step of the analytes, usually performed with gas or liquid chromatography followed by mass spectrometry. Nuclear magnetic resonance (NMR) spectroscopy can also be used as detection method without prior separation steps. The main advantage of metabolomics is that their findings are downstream processes of proteomics, transcriptomics or genomics, and consequently, they are more strongly related to the final phenotype. Thus, they bridge phenotype with genotype in functional genomics and specifically to identify genetic loci regulating the concentrations of certain metabolites, thus affecting certain biochemical pathways [31]. Additionally, metabolomics profiling is used to identify novel biomarkers in various diseases [32–34] and specifically those that have a major impact on cellular metabolic functions, such as cancer [35], metabolic diseases [25,36,37] and neurological disorders [38]. For example, an area of major interest for metabolomics is breast cancer, where metabolomics profiling has demonstrated great potentials as a biomarker for diagnosing different types of breast cancer, assessing prognosis as well as predicting treatment response [39]. Similarly, numerous metabolomics studies have been performed in different populations with T2D and have identified positive or negative associations of specific metabolite classes (e.g. increase in branched-chain and aromatic amino acids and in lactate and glycolytic intermediates, decrease in tricarbocylic acid cycle intermediates) with the development of the disease [40,41]. Finally, metabolomics are being used in drug development processes, since they are very useful at assessing drug toxicity.
-
f)
Lipidomics is the comprehensive investigation of cellular lipids in biological systems. In fact, the lipidome is part of the metabolome and lipidomics analysis utilizes the same tools as metabolomics. The main difference compared to metabolomics is that lipidomics analyses are able to detect a larger amount of lipids. Therefore, it is particularly important in diseases for which their pathogenesis is closely related to lipid metabolism, such as in obesity [42,43], T2D [44,45], atherosclerosis [46], hypertension [47], cardiovascular disease [48], stroke, metabolic syndrome [25] and non-alcoholic fatty liver disease [49]. In these diseases, lipidomics have been mainly used to identify novel pathways that can be potentially targeted therapeutically. Additionally, lipidomics have been used to detect new biomarkers in breast and prostate cancer as well as in Alzheimer's disease or other neurological disorders [50].
-
g)
Glycomics is the comprehensive investigation of glycan structures i.e. sugars. Glycans derive from complex biosynthetic pathways and can have extreme complicated branched structures. They usually bind to proteins to form the glycoproteins in a process called glycosylation, or conjugate with lipids to create the glycolipids. Glycans can affect the stability and folding of proteins and consequently their function. Glycoproteins are usually located at cell surface and are important for cell to cell recognition, while glycolipids are located often in cell membrane and are important for cell stability. Glycomics analysis is usually performed with mass-spectrometric methods, while lectin and antibody arrays can also provide high-throughput screening of samples containing glycans. Glycomics are less utilized compared to the other omics. Thus far, the particular focus is to assess glycosylation in different cancers (i.e. breast, brain, colon, liver, lung, prostate etc.) [51]. Interestingly, alterations in glycan profile have been far less investigated in metabolic diseases (e.g. obesity, diabetes or non-alcoholic liver disease), despite the fact that most glycoproteins are formed in the liver i.e. an organ which is particularly susceptible to metabolic changes. Glycomics analyses in other liver diseases have led to the development of the GlycoCirrhotest [52], the GlycoFibrotest [53] and the GlycoHCC test [54], which can predict cirrhosis, fibrosis and HCC respectively, based on changes in the glycome profile. In particular, core fucosylated AFP is the success story in glycomics, as it is currently the only FDA-approved test for the detection of HCC [55].
3. From single to multi-omics procedures - advantages, challenges and trends
Selecting a multi-omics approach compared to a single-omic analysis offers some profound advantages but has some serious challenges. A major advantage of the multi-omics analysis is the breadth of the information that it provides. The etiology of the most prevalent diseases (i.e. obesity, T2D, NAFLD, certain types of cancer, Alzheimer disease) is multifactorial. Thus, identification of one specific factor associated with a disease will most probably have limited prognostic or therapeutic value. Additionally, association does not imply causation and associations actually outnumber causations, where many of the reported associations are not reproduced in future studies. The multi-omics analysis allows for the identification of associated factors from different biological processes, i.e. gene expression, protein synthesis and post-translational modifications, cellular metabolic processes, glycosylation, etc., maximizing the available information, and thus, increasing the possibility of identifying the root causes of a disease. A second advantage of multi-omics analysis is the depth of the information it provides. For example, a single change in gene expression may be weakly associated with the pathophysiology of a multifactorial disease. However, when this finding is further supported with alterations in mRNA expression and in protein concentration, the possibility that this gene or protein is an important factor in the pathogenesis of the disease increases. Similarly, individual changes in metabolites, lipids or glycans may have limited translational potential, but when combined, they may reveal important pathways associated with the etiology of a disease.
Despite the obvious advantages of multi-omics, the field has to overcome important challenges. First, the etiology of certain diseases is extremely complex, and related processes have been evolving over long periods of time and are often susceptible to factors that may fluctuate over time and thus be difficult to assess or quantify at any given timepoint. For example, environmental parameters or lifestyle choices may significantly affect the risk for the development or prognosis of a metabolic disease; however, it may still be very difficult to monitor them over time and assess their contribution to a specific outcome at the time of disease manifestation. Additionally, most of the analyses performed have inherent biologic and experimental errors and rely on capturing a snapshot of complex and dynamic biological systems. Consequently, untimely sample collection due to an incorrect experimental design or simply due to randomness can lead to too much “noise” that would not allow us to clearly identify inciting factors or errors (i.e. findings that may not be representative of the condition that is investigated).
The biggest challenge that multi-omics is facing, for which it needs a systematic approach and effective solutions, is how to best and most accurately analyze the already huge, and ever increasing, data sets, while at the same time, minimizing the risk of leading to the wrong scientific conclusions due to false positive results.
There are specific strategies that are being used to reduce the risk of false positive or negative results:
-
a)
Increased sample size: The necessary sample size to have adequate power to detect associations depends on effect size and the heterogeneity of the background noise. If no previous studies are available, the effect size should be estimated a priori. Practically, the investigator must decide not only what it is realistic to expect in terms of effect size but also what should be considered biologically meaningful (beyond statistical significance). In these estimations, financial as well as time limitations may play a significant role. This issue was often observed amongst the early candidate gene studies for several diseases, which were underpowered and led to non-reproducible results. In order to overcome the problem of sample size, several human centric consortia and biobanks have been created to collect either targeted or untargeted anthropometric, demographic and biochemical data as well as biospecimens to be used for further studies. Good examples of such biobanks are the UK biobank [56] or the Million Veterans Program (MVP) [57] that have already collected an until recently unbelievably large volume of data (e.g. the Million Veterans Program (MVP) has been linked with all clinical records of veterans since 1993 through the online medical record or CPRS) as well as biospecimens of >500,000 individuals, which continues to accrue. Another good example is the NAFLD Adult Database and Biobank that consists of >1200 individuals with biopsy-proven NAFLD of different stages coupled with already collected physiologic measures and serum or plasma samples. Such biobanks allow the planning of research studies that would have been previously considered infeasible due to the high costs and time-consuming process related to the performance of a de novo large clinical study.
-
b)
Reduced or integrated heterogeneity of the investigated populations: Most omics studies follow a case-control study design. Cases are usually individuals with a disease and controls are “healthy” individuals, i.e. individuals without the disease. A strategy to reduce heterogeneity is to match both groups as well as possible for several already known, associated factors (e.g. age, BMI, sex for metabolic diseases). In some cases, monozygotic twins are used in order to achieve the highest possible biological proximity. There are two main problems with this approach. First of all, we may not know all the possible associated factors, and thus, we may still have a large variation between groups. Uncontrolled confounding due to unknown or simply unmeasured factors can still introduce confounding. Second and most importantly, in a tightly matched case-control study findings will be tailored for the specific populations selected, and thus, they may have limited applicability to the population at large. Another approach, which is more demanding but much more accurate, is to integrate all or as close to all known factors into our models. Such approaches need advanced mathematical skills and statistical analysis (s. below Artificial Intelligence - Machine Learning, see below). This approach, along with the creation of large studies providing a mountain of data e.g. MVP, allow the creation of nested case-control studies which provide the time-sequence criterion for, although they still cannot fully prove, causality.
-
c)
Reduced heterogeneity related to the methods used for measurements: Omics measurements can demonstrate significant heterogeneity, i.e. laboratory error or variability, depending on the quality of samples, batch effects and instruments that are being used for the measurements. It is, thus, very important that both processing and analysis of samples from cases and controls follows the same procedures. Recent technological advances have allowed for the streamlining of processes and have increased reproducibility of the results. Additionally, technological advances have allowed more specific services to be provided, including the ability to perform omics analysis on single cells or to study more in-depth post-translational protein modifications (i.e. ubiquitinations, phosphorylations etc.). Still, challenges remain with large and/or multicenter studies in which data are expected to have inherent variability of a various degree due to: i) sample collection by various medical staff following different protocols, ii) collection and entering of clinical information by various doctors working in medical facilities that differ from each other, iii) laboratory values that derive from samples that were processed and measured in various clinical chemistry laboratories distributed geographically all over the United States and/or over many decades during which both medical practices and laboratory technology have been evolving. Despite the major fiscal challenges, the authors believe that such studies should repeat the measurements of key variables (e.g. lipid profile for cardiovascular studies or liver tests for NASH studies, etc.) in the context of quality-control efforts, and if large variations are found, in terms of chronology or geographic variability of data entry, certain key variables will have to be reanalyzed for the entirety of the population of a study to minimize variability and enhance certainty for the results.
-
d)
Reduced background noise of data by reducing feature space: Each omic analysis provides measurements/data for dozens and up to thousands of different variables. Thus, it is impossible to avoid false positive results only by increasing sample size, as this will demand a sample size of hundreds of thousands (or more) samples. A classical approach is therefore to try to reduce the feature space by grouping the different variables based on their contribution to the variability observed between the two investigated groups as well as based on their biochemical or functional proximity. In all cases, the use of advanced mathematical models and especially of machine learning techniques (s. below) may significantly improve the accuracy of any findings.
-
e)
Validation of the results by a second or third study: Reproducibility is a major issue not only in omics analysis but in many clinical as well as experimental studies. It is generally recognized that basic research studies, even when published in really high profile journals, face reproducibility issues more frequently than observational human studies and the latter more frequently than randomized, controlled clinical trials. A strategy that has been increasingly adopted over the last few years is to independently validate findings from an omics analysis in a second population with similar characteristics to the first. This approach certainly adds robustness to a study. Another validation strategy which can be combined with the first is to perform an omics analysis in the same population in a later timepoint. This strategy can be very useful in studies where the outcome is not binary (e.g. “presence of the disease: yes, no”) but ordinal or continuous (e.g. stage of the disease) and the subject may change group/stage of the disease over time.
-
f)
Validation of the results in in vitro or in vivo models: Another strategy to add robustness to the findings is to further investigate them using in vitro and/or in vivo models. A good example is genomics studies that have identified novel mutations or genes related to a disease. In many cases, the functional relevance of these findings is further evaluated in cell lines or in knockout rodent models. Similar approaches have been employed in metabolomics or proteomics studies, which indicate a specific pathway related to the outcome of interest (pathogenesis of a disease or response to a treatment). This pathway can be then investigated in animal models of the disease or treatment of interest.
One could envision using more and more advanced analytical approaches (see below) to both minimize error while maximizing efficiency and to incorporate in a study as many of the above omics tools as possible to gain clarity about underlying mechanisms. As technology evolves, one could also envision not only moving to a biological validation of findings by in vitro and in vivo models but also possibly incorporating results of all these experiments into future mathematical models in order to increase the certainty and accuracy of the conclusions. These analytic techniques are evolving and provide increasing power as briefly outlined below.
4. Artificial intelligence - machine learning
Machine learning is a fundamental concept of artificial intelligence that focuses on the progressive improved performance of a computer for a specific task through its ability to “learn” with data. In medicine, machine learning can be used to analyze large data sets, as the ones that derive from multi-omics measurements, and can lead to algorithms with predictive value [45]. The main machine learning categories are supervised and unsupervised learning [58]. In supervised learning, the algorithm is provided with inputs (e.g. omics data) corresponding to specific outputs (e.g. presence of a disease or not), where the information is used to develop a general rule that will link the input to the output. In unsupervised learning, no information is provided, and the algorithm has to train all possible scenarios and find the structure linking the input to the output. Additionally, there are several intermediate categories of machine learning such as the semi-supervised learning, where the algorithm is provided with a limited amount of information, i.e. input data can be much more than the labeled outputs, which is often the case in multi-omics studies. Machine learning tasks typically include: a) dimensionality reduction to reduce the input mass by decreasing the number of random variables under consideration, b) clustering-classification to organize different variables of the input in groups with common characteristics, c) density estimation to assess distribution of input variables in specific space, and d) regression to estimate the relationships among variables and for developing predictive models.
a). Unsupervised and supervised data integration
Multi-omics data integration, in which information from different layers of omics data is combined to discover the coherent biomarkers, is one of the major challenges in “precision medicine”. Considerable work has been done in the field of bioinformatics to develop data integration algorithms (c.f. the review in [59]) among which matrix factorization methods (e.g. sparse canonical correlation analysis [60,61] and partial least squares [62]), Bayesian methods [63], and network-based methods [64] for unsupervised learning, network-based models, and multiple kernel learning methods (e.g. support vector machine [65]) for supervised data integration have been proven to be more effective.
Given the large data set that multi-omics measurements provide, a “hierarchical” approach in which several of the above algorithms as well as novel machine learning tools are combined is necessary for the accurate analysis of the data and for developing predictive models. Suppose that we are interested in developing a diagnostic algorithm, which can distinguish with high sensitivity and specificity people suffering from disease X from healthy individuals. Additionally, we would like the algorithm to be able to classify patients with disease X in 4 groups of increasing disease severity (i.e. X1-X4). Here, we use a case-control study design, where cases are the subjects suffering from disease X in various stages (i.e. X1-X4) and controls are healthy individuals that are matched only for age and sex. In both groups, we collect anthropometric, demographic and biochemical data, and perform untargeted plasma metabolomics, glycomics and lipidomics analysis. The anthropometric, demographic and biochemical variables are in the order of 10, while glycomics measurements are in the order of 100, and metabolomics as well as lipidomics measurements are of the order of 1000. Based on the classic omics statistical analysis and using the “rule of ten” [66], we require screening at least 10,000 subjects with disease X and 10,000 healthy controls. Such sample sizes, however, are rarely available. Different hierarchical approaches may be proposed depending on the type of disease and the number of layers in the available omics data (e.g. metabolomics, glycomics and lipidomics). For example, as the first step, one can apply a dimensionality reduction method such as principle component analysis (PCA) in order to reduce the number of variables under consideration. In PCA, which is a matrix factorization approach, a set of possibly correlated variables is transformed into uncorrelated variables or “principal components”. In many cases, a small number of components is able to cover the vast majority of the variance observed in the study population. In an unsupervised learning setting, other correlation-based analysis such as canonical correlation analysis (CCA) [60,61] and partial least squares (PLS) [62] can alternatively be used. Several variants of these methods with sparse solutions (e.g. sparse CCA and sparse PLS to account for dimensionality reduction) and constraints to identify structures and groups within data sets (e.g. CCA-sparse group) have been used [67] for clustering analysis. In clustering, variables are grouped together based on their similarities. For example, lipid molecules that are measured by the lipidomics analysis are grouped in lipid classes according to their biochemical structure (e.g. sphingolipids, ceramides, glycoglycerolipids, etc.). Similarly, metabolites are divided in large classes, while glycans can also be categorized according to biochemical similarities (e.g. presence of fucose or sialic acids, etc.).
In a supervised learning setting, however, the phenotype labels of samples (e.g. disease or normal) are available and can be used for training the machine learning classification algorithms. Partial least squares-discriminant analysis (PLS-DA) is perhaps the most widely used method applied to metabolomics datasets for multivariate classification and regression analysis [68]. PLS-DA is a technique used to optimize separation between different groups of samples, which is accomplished by linking two data matrices x (i.e. raw data) and y (i.e. groups, class membership, etc.). The method is in fact an extension of PLS, which handles single dependent continuous variable, whereas PLS-DA can handle multiple dependent categorical variables. The main advantage of the PLS-DA algorithm is the availability and handling of highly collinear and noisy data, which are very common outputs from metabolomics experiments. Several caveats, however, have been reported for PLS-DA such as difficulties in the identification of small numbers of variables that are responsible for the separation between two or more groups and its tendency to overfitting [68]. Thus, other alternatives of classification algorithms such as random forests [69] and support vector machines [70] have been recommended and practiced for certain problems as well.
b). Semi-supervised learning (SSL)
As described above, SSL in is an intermediate type between supervised and unsupervised learning [71]. In SSL, the algorithm receives a collection of data points, but only a subset of these data points has associated labels. For example, gene-finding systems can be trained using a semi-supervised approach, in which the input is a collection of annotated genes and an unlabeled whole-genome sequence. The learning procedure begins by constructing an initial gene-finding model on the basis of the labeled subset of the training data alone. Next, the model is used to scan the genome, and tentative labels are assigned throughout the genome. These tentative labels can then be used to improve the learned model, and the procedure iterates until no new genes are found. The semi-supervised approach can in some cases work much better than a fully supervised approach because the model is able to learn from a much larger set of genes — all of the genes in the genome — rather than only the subset of genes that have been identified with high confidence. In biomedicine, SSL although relatively new has been applied to several problems already and has achieved notable results, for example, in the study of protein classification and in functional genomics, among others [72,73]. Due to its capability of learning from both labeled and unlabeled data, SSL is potentially more effective in predicting disease genes.
Semi-supervised learning requires making certain assumptions about the data set [73]. These assumptions of consistency are: (i) nearby points are likely to have the same label and (ii) points on the same structure (typically referred to as a cluster or a manifold) are likely to have the same label [73]. The requirement of the “smoothness” assumption is already fulfilled automatically by employing PCA to identify the proper feature space, and by applying clustering algorithms within each omics category based on molecules biochemical similarities or involvement in common functional pathways.
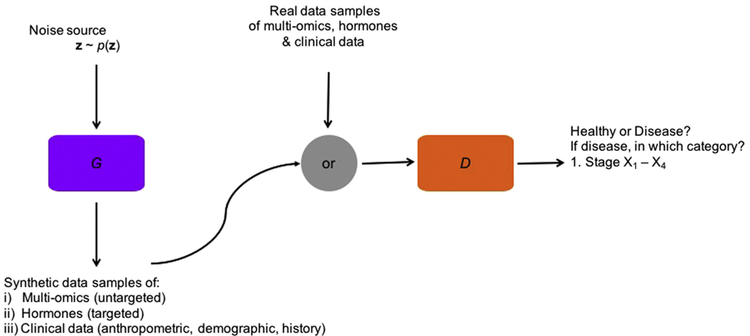
Considering the situation that a large amount of unlabeled data with only a small amount of labeled data are available, SSL is the proper way to train a nonlinear classifier to diagnose different stages in a disease (e.g. X1-X4 in the above example). Furthermore, an extension of generative adversarial networks (GANs) — categorical GANs (CatGANs) that belong to unsupervised machine learning methods can be used to integrate labeled and unlabeled data [74–76]. CatGANs combine both the generative and the discriminative perspective. In particular, the discriminative neural network classifier D predicts the label y for the input x through the conditional distribution p(y|x, D); the adversarial generative neural network G tries to fool the classifier D into accepting bogus input examples, which enforces robustness of the classifier (see Fig.1) hence, both are trained simultaneously and in competition with each other.
Fig. 1.
Visualization of CatGAN with the generator G (in purple) and the discriminative classifier D (in orange) neural networks: The generator creates synthetic data samples of multi-omics, specific hormones and clinical data (anthropometric, demographic or from medical history) from a noise source z. The classifier receives both “fake” and real (disease) data and aims to tell them apart. For a real data sample, the classifier also assigns it to the stage of the disease.
c). Bayesian methods and multi-fidelity data integration
The main advantage of Bayesian methods in data integration is that they can make assumptions not only on different types of data sets with various distributions but also on the correlations among data sets. For example, Bayesian logistic regression may be used for labeled binary (e.g. healthy vs. disease) or categorical (e.g. X1-X4 in the above example) omics data, where uncertainty associated with both the sampling size and the data can be quantified as well. Bayesian logistic regression is typically performed in three steps: First, the likelihood function for the data is written as p(y|x, β), where x and y are the vectors of predictors and binary outcomes, respectively, and β is the vector of unknown regression coefficients. Second, a prior distribution (initial belief) over the known parameters is assumed as p(β). Third, using Bayes theorem, a posterior distribution of the parameters, which is the updated belief about them given evidence, is formed by multiplying the prior distribution by the likelihood function: p(β|x, y) ∝ p(y|x, β) p(β). Note that, one cannot evaluate the closed form posterior but can approximate it by sampling or by employing variational methods. If a Laplace prior is used, sparsity in the model parameters is promoted, which provides a simple and efficient training method. Using Laplacian as prior leads to the posterior parameter distribution that can be accurately approximated as a Gaussian, and, hence, the predictive distribution can be written as the convolution of a sigmoid and Gaussian. If no prior information is known, one can use a non-informative prior, e.g. Cauchy priors.
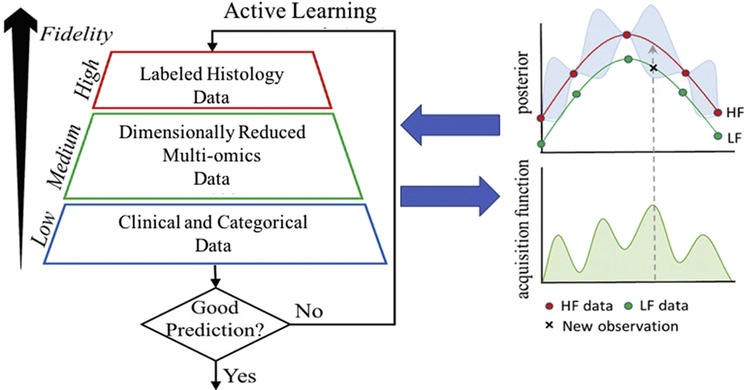
Additionally, autoregressive statistical schemes can be employed to learn the correlations between labeled and unlabeled data (multi-omics and clinical data) in the form of hierarchical multi-fidelity modeling that can be implemented both in terms of neural networks (for classification) as well as Gaussian processes (for regression). In this multi-fidelity approach, one can obtain robust answers by employing a few “gold data” (e.g., histology data for a disease) and a lot of “silver data” (e.g., multi-omics plus clinical data) and fusing them together either with linear or nonlinear autoregressive schemes (see Fig. 2). Nonlinear auto-regressive Gaussian process regression (NARGP) methods have recently been used in learning complex systems and demonstrated very good performance in discovering functional relationships (synergistically) from different types of data [77]. Many existing multi-fidelity algorithms are based on Gaussian process (GP) regression in combination with the linear auto-regressive information (AR1) fusion scheme put forth by Kennedy and O'Hagan [78]. They are effective when low-fidelity models can capture the right trends, and the low- and high-fidelity model outputs exhibit strong linear correlation across the input space [77]. In many cases, however, the low-fidelity may provide some erroneous trends along with the correct trends and in those cases the linear autoregressive schemes pioneered in may fail [78].
Fig. 2.
Multi-fidelity data integration through active leaning: Active learning is combined with the NARGP algorithm to integrate histology, omics and clinical data into a machine-learning predictor. It can also guide us as to what new experiments are needed to enhance predictability, which works by considering the maximum of an acquisition function and obtaining one more point in the parameter space using data at different levels (e.g. low- or high-fidelity – (LF) or (HF)).
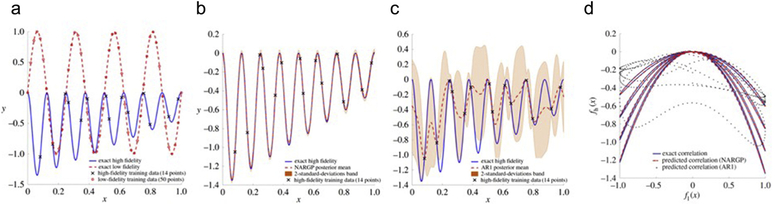
We present a pedagogical example for multi-fidelity information fusion using linear and non-linear auto-regressive models to show the effectiveness of each approach. Let us assume that the low-fidelity model is the function fL(x) = sin(8πx), while the high-fidelity function is . We further assume that we only have access to 50 observations of fL, supplemented by only 14 observations of fH (see Fig. 3). Using this data set, our goal now is to reconstruct the high-fidelity signal as accurately as possible. As shown in Fig. 3(b), AR1 (red curve) fails to discover the real function (blue curve). However, NARGP obtains the right function with very small uncertainty (see Fig. 3(c)). Remarkably, NARGP correctly predicts the true underlying signal even at regions where no high-fidelity data is available and also the low-fidelity model is erroneously providing the opposite trend (e.g. for 0.25 < x < 0.35). The robust NARGP predictions is due to the structure in the prior that enables it to learn the nonlinear and space-dependent cross-correlation between the low- and high-fidelity data (see Fig. 3(d)). This is a key feature in constructing robust multi-fidelity modeling algorithms, as it provides a mechanism to safeguard against wrong trends in the low-fidelity data, while still being able to distill useful information from them. Furthermore, predicting the uncertainty as in this example leads to “active learning” (see Fig. 2), which informs us as to which new experimental or clinical data are required to predict with confidence, i.e. in regions with large uncertainty.
Fig. 3.
Linear (AR1) vs. non-linear (NARGP) Gaussian process regression: (a) Exact low- (red) and high-fidelity (blue) functions along with the observations used for training the multi-fidelity GP models (14 blue points, 50 red points). (b) Exact solution vs. the NARGP posterior mean and uncertainty. (c) AR1 predictions and its uncertainty vs. exact solution. (d) Cross-correlation structure between the exact low- and high-fidelity signals vs. the cross-correlation learnt by the NARGP and AR1 schemes trained on the given multi-fidelity dataset. (Adopted from Perdikaris et al. [77].)
d). Novel machine-learning algorithms
Deep learning, implemented via deep neural networks (DNNs), is a powerful approach to classification, regression and inference problems across disciplines. However, due to the lack of rigorous mathematical foundations of the emerging DNN architectures, their effectiveness is not always guaranteed, and there are currently no established metrics of their performance. New algorithms aim to endow DNNs with uncertainty quantification (UQ) methods for DNNs, i.e. including uncertainty of the DNN as an approximator but also parametric uncertainty due to inherent randomness in the data. Another emerging topic in machine learning is the new concept of meta-learning, i.e. “learning to learn”, which is fundamental in transfer learning, from one situation to another. Meta-learning enables an automated way to optimize the DNN, saving great amounts of human effort and time [79].
We have discussed above both Gaussian processes as well as DNN, with the former being more accurate in general and provide UQ, while the latter can deal with many parameters (dimensions) and can be more easily trained. Neural-net-induced Gaussian process (NNGP) regression inherits both the high expressivity of deep neural networks (DNNs) as well as the uncertainty quantification property of Gaussian processes (GPs). Previous works on NNGP have targeted classification [80], and more recent work has focused on generalizing the NNGP method to function approximation and to solving partial differential equations (PDEs) [81]. Published work on benchmark problems suggest that NNGP combines the best of two worlds, i.e. both high accuracy and easier training, and, thus, has a potential to be used in many emerging applications in the future, especially with multi-fidelity data as discussed above. NNGP is essentially a Gaussian process method for classification or regression but based not on an arbitrary kernel but rather on a kernel produced by data that was fed into a DNN.
5. Conclusions
Omics are high throughput procedures able to provide important information at different levels (gene, mRNA, protein, metabolite level, etc.) in complicated biological systems. The combination of more than one omic analysis (multi-omics approach) can lead to more robust scientific conclusions and can be particularly fruitful at developing diagnostic tools or identifying novel therapeutic targets in multifactorial diseases, such as metabolic diseases, neurological disorders and cancer. Indeed, the combination of the above with clinical data from computerized patient records as well as targeted biochemical and hormonal analyses holds great promise for advancing significantly biomedical research at unprecedented rates, leading to the elucidation of underlying mechanisms as well as discoveries of novel diagnostic and therapeutic tools but is, at the same time, inherently linked to several challenges, such as the computational approach. The use of large, real world, clinical data sets and biobanks that could be used for both traditional and omics analyses, combined with the advanced computational methodology described herein, allow for the performance of phenome-wide association studies- novel and promising tools to assess both potential benefits and adverse effects of therapeutic agents with known pathways and related genes. Such hypothesis generating studies, which would then be validated by other cohort studies and clinical trials, have great potential to catalyze research, in general, and drug discovery and safety efforts, more specifically. The use of advanced and continuously improving machine learning methods is necessary to benefit from analyzing the large data sets produced by multi-omics and other laboratory and clinical data that are been collected and stored at a great cost to our society. Continuous improvements of the methodologies involved and engagement of the best and brightest researchers will be needed to minimize false and unreproducible results and to lead to accurate and effective advances in the biomedical field that would have the potential to provide tangible benefits to our suffering fellow human beings.
Acknowledgments
Funding
The current study was funded by NIH K24DK081913. NP was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) −389891681 (PE 2431/2-1).
Footnotes
Declaration of interest
Authors have nothing to disclose.
Contributor Information
Nikolaos Perakakis, Department of Endocrinology, VA Boston Healthcare System Jamaica Plain, Boston, MA 02130, USA; Division of Endocrinology, Diabetes and Metabolism Beth Israel Deaconess Medical Center, Harvard Medical School Boston, MA 02215, USA.
Alireza Yazdani, Division of Applied Mathematics, Brown University Providence, RI 02906, USA.
George E. Karniadakis, Division of Applied Mathematics, Brown University Providence, RI 02906, USA
Christos Mantzoros, Department of Endocrinology, VA Boston Healthcare System Jamaica Plain, Boston, MA 02130, USA; Division of Endocrinology, Diabetes and Metabolism Beth Israel Deaconess Medical Center, Harvard Medical School Boston, MA 02215, USA.
References
- [1].Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol 2017;18 (1):83 10.1186/s13059-017-1215-1 [Epub 2017/05/10. PubMed PMID: ; PubMed Central PMCID: PMC5418815]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science 2005;308(5720):419–21. 10.1126/science.1110359 [Epub 2005/03/12. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [3].Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447(7145):661–78. 10.1038/nature05911 [Epub 2007/06/08. PubMed PMID: ; PubMed Central PMCID: PMC2719288]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478(7367):103–9. 10.1038/nature10405 [Epub 2011/09/13. PubMed PMID: ; PubMed Central PMCID: PMC3340926]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet 2012;90(1):7–24. 10.1016/j.ajhg.2011.11.029 [Epub 2012/01/17 PubMed PMID: ; PubMed Central PMCID: PMC3257326]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang X, Johnson AD, Hendricks AE, Hwang SJ, Tanriverdi K, Ganesh SK, et al. Genetic associations with expression for genes implicated in GWAS studies for atherosclerotic cardiovascular disease and blood phenotypes. Hum Mol Genet 2014;23(3): 782–95. 10.1093/hmg/ddt461 [Epub 2013/09/24. PubMed PMID: ; PubMed Central PMCID: PMC3900869]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci 2010;1212:59–77. 10.1111/j.1749-6632.2010.05838.x [Epub 2010/11/26. PubMed PMID: ; PubMed Central PMCID: PMC3057517]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fall T, Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Mol Cell Endocrinol 2014;382(1):740–57. 10.1016/j.mce.2012.08.018 [Epub 2012/09/12. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [9].Pirim D, Wang X, Niemsiri V, Radwan ZH, Bunker CH, Hokanson JE, et al. Resequencing of the CETP gene in American whites and African blacks: association of rare and common variants with HDL-cholesterol levels. Metabolism 2016;65 (1):36–47. 10.1016/j.metabol.2015.09.020 [Epub 2015/12/20. PubMed PMID: ; PubMed Central PMCID: PMC4684899]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang X, Yang W, Wang J, Meng Y, Guan Y, Yang J. FAM3 gene family: a promising therapeutical target for NAFLD and type 2 diabetes. Metabolism 2018;81:71–82. 10.1016/j.metabol.2017.12.001 [Epub 2017/12/10. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [11].Chouraki V, Seshadri S. Genetics of Alzheimer's disease. Adv Genet 2014;87:245–94. 10.1016/B978-0-12-800149-3.00005-6 [Epub 2014/10/15. PubMed PMID: 25311924]. [DOI] [PubMed] [Google Scholar]
- [12].Fachal L, Dunning AM. From candidate gene studies to GWAS and post-GWAS analyses in breast cancer. Curr Opin Genet Dev 2015;30:32–41. 10.1016/j.gde.2015.01.004 [Epub 2015/03/03. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [13].Kar SP, Tyrer JP, Li Q, Lawrenson K, Aben KK, Anton-Culver H, et al. Network-based integration of GWAS and gene expression identifies a HOX-centric network associated with serous ovarian cancer risk. Cancer Epidemiol Biomarkers Prev 2015;24 (10):1574–84. 10.1158/1055-9965.EPI-14-1270 [Epub 2015/07/26. PubMed PMID: ; PubMed Central PMCID: PMC4592449]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet 2014;46(10):1103–9. 10.1038/ng.3094 [Epub 2014/09/15. PubMed PMID: ; PubMed Central PMCID: PMC4383163]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Alonso A, Domenech E, Julia A, Panes J, Garcia-Sanchez V, Mateu PN, et al. Identification of risk loci for Crohn's disease phenotypes using a genome-wide association study. Gastroenterology 2015;148(4):794–805. 10.1053/j.gastro.2014.12.030 [Epub 2015/01/06. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [16].Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014;506(7488):376–81. 10.1038/nature12873 [Epub 2014/01/07. PubMed PMID: ; PubMed Central PMCID: PMC3944098]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature 2013;502(7472):489–98. 10.1038/nature12752 [Epub 2013/10/25. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [18].Taudt A, Colome-Tatche M, Johannes F. Genetic sources of population epigenomic variation. Nat Rev Genet 2016;17(6):319–32. 10.1038/nrg.2016.45 [Epub 2016/05/10. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [19].Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One 2010;5(3):e9692 10.1371/journal.pone.0009692 [Epub 2010/03/20. PubMed PMID: ; PubMed Central PMCID: PMC2837739]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Weihrauch-Bluher S, Richter M, Staege MS. Body weight regulation, socioeconomic status and epigenetic alterations. Metabolism 2018. 10.1016/j.metabol.2018.03.006 [Epub 2018/03/13. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [21].Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 2001;10(7):687–92 [Epub 2001/03/21. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [22].Raghuraman S, Donkin I, Versteyhe S, Barres R, Simar D. The emerging role of epigenetics in inflammation and Immunometabolism. Trends Endocrinol Metab 2016;27 (11):782–95. 10.1016/j.tem.2016.06.008 [Epub 2016/10/23. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [23].Trzepizur W, Cortese R, Gozal D. Murine models of sleep apnea: functional implications of altered macrophage polarity and epigenetic modifications in adipose and vascular tissues. Metabolism 2018;84:44–55. 10.1016/j.metabol.2017.11.008 [Epub 2017/11/21. PubMed PMID: ; PubMed Central PMCID: PMC5955762]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol 2015;11(3):151–60. 10.1038/nrendo.2014.229 [Epub 2015/01/07. PubMed PMID: ; PubMed Central PMCID: PMC4376378]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Murri M, Insenser M, Fernandez-Duran E, San-Millan JL, Luque-Ramirez M, Escobar-Morreale HF. Non-targeted profiling of circulating microRNAs in women with polycystic ovary syndrome (PCOS): effects of obesity and sex hormones. Metabolism 2018. 10.1016/j.metabol.2018.01.011 [Epub 2018/02/08. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [26].Alvarez-Dominguez JR, Bai Z, Xu D, Yuan B, Lo KA, Yoon MJ, et al. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab 2015;21(5):764–76. 10.1016/j.cmet.2015.04.003 [Epub 2015/04/30. PubMed PMID: ; PubMed Central PMCID: PMC4429916]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li J, Tian H, Yang J, Gong Z. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol 2016;35(9):459–70. 10.1089/dna.2015.3187 [Epub 2016/05/24. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [28].Xu J, Liu S. Noncoding RNAs in cancer cell plasticity. Adv Exp Med Biol 2016;927: 173–89. 10.1007/978-981-10-1498-7_6 [Epub 2016/07/05. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [29].Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 2014;65(6): 1140–51. 10.1016/j.eururo.2013.12.003 [Epub 2014/01/01. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [30].Pagel O, Loroch S, Sickmann A, Zahedi RP. Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert Rev Proteomics 2015;12(3):235–53. 10.1586/14789450.2015.1042867 [Epub 2015/05/09. PubMed PMID: ; PubMed Central PMCID: PMC4487610]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet 2008;4(11):e1000282 10.1371/journal.pgen.1000282 [Epub 2008/12/02. PubMed PMID: ; PubMed Central PMCID: PMC2581785 products and services in the field of targeted quantitative metabolomics research. The other authors have no competing interests to declare]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Perng W, Rifas-Shiman SL, McCulloch S, Chatzi L, Mantzoros C, Hivert MF, et al. Association of cord blood metabolites with perinatal characteristics, newborn anthropometry, and cord blood hormones in project viva. Metabolism 2017;76:11–22 10.1016/j.metabol.2017.07.001 [Epub 2017/10/11. PubMed PMID: ; PubMed Central PMCID: PMC5675164]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Alvarez JA, Chong EY, Walker DI, Chandler JD, Michalski ES, Grossmann RE, et al. Plasma metabolomics in adults with cystic fibrosis during a pulmonary exacerbation: a pilot randomized study of high-dose vitamin D3 administration. Metabolism 2017;70: 31–41. 10.1016/j.metabol.2017.02.006 [Epub 2017/04/14. PubMed PMID: ; PubMed Central PMCID: PMC5407388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chorell E, Hall UA, Gustavsson C, Berntorp K, Puhkala J, Luoto R, et al. Pregnancy to postpartum transition of serum metabolites in women with gestational diabetes. Metabolism 2017;72:27–36. 10.1016/j.metabol.2016.12.018 [Epub 2017/06/24. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [35].Armitage EG, Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J Pharm Biomed Anal 2014;87:1–11. 10.1016/j.jpba.2013.08.041 [Epub 2013/10/05. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [36].Chen H, Miao H, Feng YL, Zhao YY, Lin RC. Metabolomics in dyslipidemia. Adv Clin Chem 2014;66:101–19 [Epub 2014/10/28. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [37].Zhang AH, Qiu S, Xu HY, Sun H, Wang XJ. Metabolomics in diabetes. Clin Chim Acta 2014;429:106–10. 10.1016/j.cca.2013.11.037 [Epub 2013/12/11. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [38].Jove M, Portero-Otin M, Naudi A, Ferrer I, Pamplona R. Metabolomics of human brain aging and age-related neurodegenerative diseases. J Neuropathol Exp Neurol 2014; 73(7):640–57. 10.1097/NEN.0000000000000091 [Epub 2014/06/12. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [39].McCartney A, Vignoli A, Biganzoli L, Love R, Tenori L, Luchinat C, et al. Metabolomics in breast cancer: a decade in review. Cancer Treat Rev 2018;67:88–96. 10.1016/j.ctrv.2018.04.012 [Epub 2018/05/19. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [40].Gonzalez-Franquesa A, Burkart AM, Isganaitis E, Patti ME. What have metabolomics approaches taught us about type 2 diabetes? Curr Diab Rep 2016;16(8):74 10.1007/s11892-016-0763-1 [Epub 2016/06/21. PubMed PMID: ; PubMed Central PMCID: PMC5441387]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol 2012;8:615 10.1038/msb.2012.43 [Epub 2012/09/27. PubMed PMID: ; PubMed Central PMCID: PMC3472689]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li F, Jiang C, Larsen MC, Bushkofsky J, Krausz KW, Wang T, et al. Lipidomics reveals a link between CYP1B1 and SCD1 in promoting obesity. J Proteome Res 2014;13(5): 2679–87. 10.1021/pr500145n [Epub 2014/04/02. PubMed PMID: ; PubMed Central PMCID: PMC4018097]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bray GA, Redman LM, de Jonge L, Rood J, Sutton EF, Smith SR. Plasma fatty acylcarnitines during 8 weeks of overfeeding: relation to diet energy expenditure and body composition: the PROOF study. Metabolism 2018;83:1–10. 10.1016/j.metabol.2018.01.019 [Epub 2018/01/29. PubMed PMID: ]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Markgraf DF, Al-Hasani H, Lehr S. Lipidomics-reshaping the analysis and perception of type 2 diabetes. Int J Mol Sci 2016;17(11). 10.3390/ijms17111841 [Epub 2016/11/10. PubMed PMID: ; PubMed Central PMCID: PMC5133841]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Suvitaival T, Bondia-Pons I, Yetukuri L, Poho P, Nolan JJ, Hyotylainen T, et al. Lipidome as a predictive tool in progression to type 2 diabetes in Finnish men. Metabolism 2018;78:1–12. 10.1016/j.metabol.2017.08.014 [Epub 2017/09/25. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [46].Ekroos K, Janis M, Tarasov K, Hurme R, Laaksonen R. Lipidomics: a tool for studies of atherosclerosis. Curr Atheroscler Rep 2010;12(4):273–81. 10.1007/s11883-010-0110-y [Epub 2010/04/29. PubMed PMID: ; PubMed Central PMCID: PMC2878593]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kulkarni H, Meikle PJ, Mamtani M, Weir JM, Barlow CK, Jowett JB, et al. Plasma lipidomic profile signature of hypertension in Mexican American families: specific role of diacylglycerols. Hypertension 2013;62(3):621–6. 10.1161/HYPERTENSIONAHA.113.01396 [Epub 2013/06/27. PubMed PMID: ; PubMed Central PMCID: PMC3789127]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U, et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation 2014;129(18):1821–31. 10.1161/CIRCULATIONAHA.113.002500 [Epub 2014/03/14. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [49].Gorden DL, Myers DS, Ivanova PT, Fahy E, Maurya MR, Gupta S, et al. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res 2015;56(3): 722–36. 10.1194/jlr.P056002 [Epub 2015/01/20. PubMed PMID: ; PubMed Central PMCID: PMC4340319]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yang L, Li M, Shan Y, Shen S, Bai Y, Liu H. Recent advances in lipidomics for disease research. J Sep Sci 2016;39(1):38–50. 10.1002/jssc.201500899 [Epub 2015/09/24. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [51].Drake RR. Glycosylation and cancer: moving glycomics to the forefront. Adv Cancer Res 2015;126:1–10. 10.1016/bs.acr.2014.12.002 [Epub 2015/03/03. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [52].Callewaert N, Van Vlierberghe H, Van Hecke A, Laroy W, Delanghe J, Contreras R. Noninvasive diagnosis of liver cirrhosis using DNA sequencer-based total serum protein glycomics. Nat Med 2004;10(4):429–34 [Epub 2004/05/22. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [53].Vanderschaeghe D, Laroy W, Sablon E, Halfon P, Van Hecke A, Delanghe J, et al. GlycoFibroTest is a highly performant liver fibrosis biomarker derived from DNA sequencer-based serum protein glycomics. Mol Cell Proteomics 2009;8(5):986–94. 10.1074/mcp.M800470-MCP200 [Epub 2009/02/03. PubMed PMID: ; PubMed Central PMCID: PMC2689777]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu XE, Desmyter L, Gao CF, Laroy W, Dewaele S, Vanhooren V, et al. N-glycomic changes in hepatocellular carcinoma patients with liver cirrhosis induced by hepatitis B virus. Hepatology 2007;46(5):1426–35. 10.1002/hep.21855 [Epub 2007/08/09. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [55].Mehta A, Herrera H, Block T. Glycosylation and liver cancer. Adv Cancer Res 2015; 126:257–79. 10.1016/bs.acr.2014.11.005 [Epub 2015/03/03. PubMed PMID: ; PubMed Central PMCID: PMC4634841]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12(3):e1001779 10.1371/journal.pmed.1001779[Epub 2015/04/01. PubMed PMID: ; PubMed Central PMCID: PMC4380465]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, et al. Million veteran program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016;70:214–23. 10.1016/j.jclinepi.2015.09.016 [Epub 2015/10/07. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [58].Lecun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- [59].Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front Genet 2017;8:84 10.3389/fgene.2017.00084 [Epub 2017/07/04. PubMed PMID: ; PubMed Central PMCID: PMC5472696]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Le Cao KA, Martin PG, Robert-Granie C, Besse P. Sparse canonical methods for biological data integration: application to a cross-platform study. BMC Bioinf 2009;10:34 10.1186/1471-2105-10-34 [Epub 2009/01/28. PubMed PMID: ; PubMed Central PMCID: PMC2640358]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Witten DM, Tibshirani RJ. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat Appl Genet Mol Biol 2009;8 10.2202/1544-6115.1470 [Article28. Epub 2009/07/04. PubMed PMID: ; PubMed Central PMCID: PMC2861323]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Li W, Zhang S, Liu CC, Zhou XJ. Identifying multi-layer gene regulatory modules from multi-dimensional genomic data. Bioinformatics 2012;28(19):2458–66. 10.1093/bioinformatics/bts476 [Epub 2012/08/07. PubMed PMID: ; PubMed Central PMCID: PMC3463121]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kirk P, Griffin JE, Savage RS, Ghahramani Z, Wild DL. Bayesian correlated clustering to integrate multiple datasets. Bioinformatics 2012;28(24):3290–7. 10.1093/bioinformatics/bts595 [Epub 2012/10/11. PubMed PMID: ; PubMed Central PMCID: PMC3519452]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bonnet E, Calzone L, Michoel T. Integrative multi-omics module network inference with Lemon-Tree. PLoS Comput Biol 2015;11(2):e1003983 10.1371/journal.pcbi.1003983 [Epub 2015/02/14. PubMed PMID: ; PubMed Central PMCID: PMC4332478]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Smirnov P, Safikhani Z, El-Hachem N, Wang D, She A, Olsen C, et al. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics 2016;32 (8):1244–6. 10.1093/bioinformatics/btv723 [Epub 2015/12/15. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [66].Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15(4):361–87. [Epub 1996/02/28. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [67].Lin D, Zhang J, Li J, Calhoun VD, Deng HW, Wang YP. Group sparse canonical correlation analysis for genomic data integration. BMC Bioinf 2013;14:245 10.1186/1471-2105-14-245 [Epub 2013/08/14. PubMed PMID: ; PubMed Central PMCID: PMC3751310]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, et al. A tutorial review: metabolomics and partial least squares-discriminant analysis—a marriage of convenience or a shotgun wedding. Anal Chim Acta 2015;879:10–23. 10.1016/j.aca.2015.02.012 [Epub 2015/05/24. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [69].Menze BH, Kelm BM, Masuch R, Himmelreich U, Bachert P, Petrich W, et al. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinf 2009;10:213 10.1186/1471-2105-10-213 [Epub 2009/07/14. PubMed PMID: ; PubMed Central PMCID: PMC2724423]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mahadevan S, Shah SL, Marrie TJ, Slupsky CM. Analysis of metabolomic data using support vector machines. Anal Chem 2008;80(19):7562–70. 10.1021/ac800954c [Epub 2008/09/05. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [71].Zhu H, Goldberg AB. Introduction to semi-supervised learning. Synthesis Lectures on Artificial Intelligence and Machine Learning; 2009. [Google Scholar]
- [72].Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 2007;4(11):923–5. 10.1038/nmeth1113 [Epub 2007/10/24. PubMed PMID: ]. [DOI] [PubMed] [Google Scholar]
- [73].Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet 2015;16(6):321–32. 10.1038/nrg3920 [Epub 2015/05/08. PubMed PMID: ; PubMed Central PMCID: PMC5204302]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial networks. ArXiv e-prints [Internet] 2014. June 01 Available from: https://ui.adsabs.harvard.edu/#abs/2014arXiv1406.2661G. [Google Scholar]
- [75].Odena A Semi-supervised learning with generative adversarial networks. ArXiv e-prints [Internet] 2016. June 01 Available from: https://ui.adsabs.harvard.edu/#abs/2016arXiv160601583O. [Google Scholar]
- [76].Springenberg JT. Unsupervised and semi-supervised learning with categorical generative adversarial networks. ArXiv e-prints [Internet] 2015. November 01 Available from: https://ui.adsabs.harvard.edu/#abs/2015arXiv151106390S. [Google Scholar]
- [77].Perdikaris P, Raissi M, Damianou A, Lawrence ND, Karniadakis GE. Nonlinear information fusion algorithms for data-efficient multi-fidelity modelling. Proc R Soc A 2017;473(2198). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kennedy M, O'Hagan A. Predicting the output from a complex computer code when fast approximations are available. Biometrika 2000;87(1):1–13. [Google Scholar]
- [79].Finn C, Abbeel P, Levine S. Model-agnostic meta-learning for fast adaptation of deep networks. ArXiv e-prints [Internet] 2017. March 01 Available from: https://ui.adsabs.harvard.edu/#abs/2017arXiv170303400F. [Google Scholar]
- [80].Lee J, Bahri Y, Novak R, Schoenholz SS, Pennington J, Sohl-Dickstein J. Deep neural networks as Gaussian processes. ArXiv e-prints [Internet] 2017. October 01 Available from: https://ui.adsabs.harvard.edu/#abs/2017arXiv171100165L. [Google Scholar]
- [81].Pang G, Yang L, Karniadakis GE. Neural-net-induced Gaussian process regression for function approximation and PDE solution. ArXiv e-prints [Internet] 2018. June 01 Available from: https://ui.adsabs.harvard.edu/#abs/2018arXiv180611187P [Google Scholar]