Principal component analysis (PCA) is often used to describe overall population structure—patterns of relatedness arising from past demographic history—among a set of genomes. Here, Li and Ralph describe how the patterns uncovered by....
Keywords: local PCA, visualization, population structure, genomic landscape
Abstract
Population structure leads to systematic patterns in measures of mean relatedness between individuals in large genomic data sets, which are often discovered and visualized using dimension reduction techniques such as principal component analysis (PCA). Mean relatedness is an average of the relationships across locus-specific genealogical trees, which can be strongly affected on intermediate genomic scales by linked selection and other factors. We show how to use local PCA to describe this intermediate-scale heterogeneity in patterns of relatedness, and apply the method to genomic data from three species, finding in each that the effect of population structure can vary substantially across only a few megabases. In a global human data set, localized heterogeneity is likely explained by polymorphic chromosomal inversions. In a range-wide data set of Medicago truncatula, factors that produce heterogeneity are shared between chromosomes, correlate with local gene density, and may be caused by linked selection, such as background selection or local adaptation. In a data set of primarily African Drosophila melanogaster, large-scale heterogeneity across each chromosome arm is explained by known chromosomal inversions thought to be under recent selection and, after removing samples carrying inversions, remaining heterogeneity is correlated with recombination rate and gene density, again suggesting a role for linked selection. The visualization method provides a flexible new way to discover biological drivers of genetic variation, and its application to data highlights the strong effects that linked selection and chromosomal inversions can have on observed patterns of genetic variation.
WRIGHT (1949) defined “population structure” to encompass “such matters as numbers, composition by age and sex, and state of subdivision,” where “subdivision” refers to restricted migration between subpopulations. The phrase is also commonly used to refer to the genetic patterns that result from this process, as for instance reduced mean relatedness between individuals from distinct populations. However, it is not necessarily clear what aspects of demography should be included in the concept. For instance, Blair (1943) defines population structure to be the sum total of “such factors as size of breeding populations, periodic fluctuation of population size, sex ratio, activity range, and differential survival of progeny” (emphasis added). The definition is similar to Wright’s, but differs in including the effects of natural selection. On closer examination, incorporating differential survival or fecundity makes the concept less clear: should a randomly mating population consisting of two types that exhibit partial postzygotic reproductive isolation from each other be said to show population structure or not? Whatever the definition, it is clear that due to natural selection, the effects of population structure—the realized patterns of genetic relatedness—differ depending on which portion of the genome is being considered. For instance, strongly locally adapted alleles of a gene will be selected against in migrants to different habitats, increasing genetic differentiation between populations near to this gene. Similarly, newly adaptive alleles spread first in local populations. These observations motivate many methods to search for genetic loci under selection, as for example in Huerta-Sánchez et al. (2013), Martin et al. (2016), and Duforet-Frebourg et al. (2016).
These realized patterns of genetic relatedness summarize the shapes of the genealogical trees at each location along the genome. Since these trees vary along the genome, so does relatedness, but averaging over sufficiently many trees we hope to get a stable estimate that does not depend much on the genetic markers chosen. This is not guaranteed; for instance, relatedness on sex chromosomes is expected to differ from the autosomes, and positive or negative selection on particular loci can dramatically distort shapes of nearby genealogies (Charlesworth et al. 1993; Barton 2000; Kim and Stephan 2002). Indeed, many species show chromosome-scale variation in diversity and divergence (e.g., Langley et al. 2012); species phylogenies can differ along the genome due to incomplete lineage sorting, adaptive introgression, and/or local adaptation (e.g., Ellegren et al. 2012; Nadeau et al. 2012; Pease and Hahn 2013; Vernot and Akey 2014; Pool 2015); and theoretical expectations predict that geographic patterns of relatedness should depend on selection (Charlesworth et al. 2003).
Patterns in genome-wide relatedness are often summarized by applying principal component analysis (PCA; Patterson et al. 2006) to the genotype matrix, as inspired by the pioneering work of Menozzi et al. (1978). The results of PCA can be related to the genealogical history of the samples, such as time to most recent common ancestor and migration rate between populations (Novembre and Stephens 2008; McVean 2009), and sometimes produce “maps” of population structure that reflect the samples’ geographic origin distorted by rates of gene flow (Novembre et al. 2008).
Modeling such background kinship between samples is essential to genome-wide association studies (GWAS; Price et al. 2006; Astle and Balding 2009), and so understanding variation in kinship along the genome could lead to more generally powerful methods and may be essential for doing GWAS in species with substantial heterogeneity in realized patterns of mean relatedness along the genome.
Others have applied PCA to windows of the genome. Ma and Amos (2012) used local PCA much as we do to identify putative chromosomal inversions. Bryc et al. (2010) and Brisbin et al. (2012) used PCA to infer tracts of local ancestry in recently admixed populations, but by projecting each genomic window onto the axes of a single, globally defined PCA rather than doing PCA separately on each window.
A note on nomenclature: in this work we describe variation in patterns of relatedness using local PCA, where “local” refers to proximity along the genome. A number of general methods for dimensionality reduction also use a strategy of “local PCA” (e.g., Kambhatla and Leen 1997; Roweis and Saul 2000; Weingessel and Hornik 2000; Manjón et al. 2013), performing PCA not on the entire data set but instead on subsets of observations, providing local pictures that are then stitched back together to give a global picture. At first sight, this differs from our method in that we restrict to subsets of variables instead of subsets of observations. However, if we flip perspectives and think of each genetic variant as an observation, our method shares common threads, although our method does not subsequently use adjacency along the genome, as we aim to identify similar regions that may be distant.
It is common to describe variation along the genome of simple statistics such as FST and to interpret the results in terms of the action of selection (e.g., Turner et al. 2005; Ellegren et al. 2012). However, a given pattern (e.g., valleys of FST) can be caused by more than one biological process (Cruickshank and Hahn 2014; Burri et al. 2015), which in retrospect is unsurprising given that we are using a single statistic to describe a complex process. It is also common to use methods such as PCA to visualize large-scale patterns in mean genome-wide relatedness. In this paper, we show if and how patterns of mean relatedness vary systematically along the genome, in a way particularly suited to large samples from geographically distributed populations. Geographic population structure sets the stage by establishing background patterns of relatedness, our method then describes how this structure is affected by selection and other factors. The method is descriptive: it does not aim to identify outlier loci, but rather to describe larger-scale variation shared by many parts of the genome and give clues about the source of this variation.
Materials and Methods
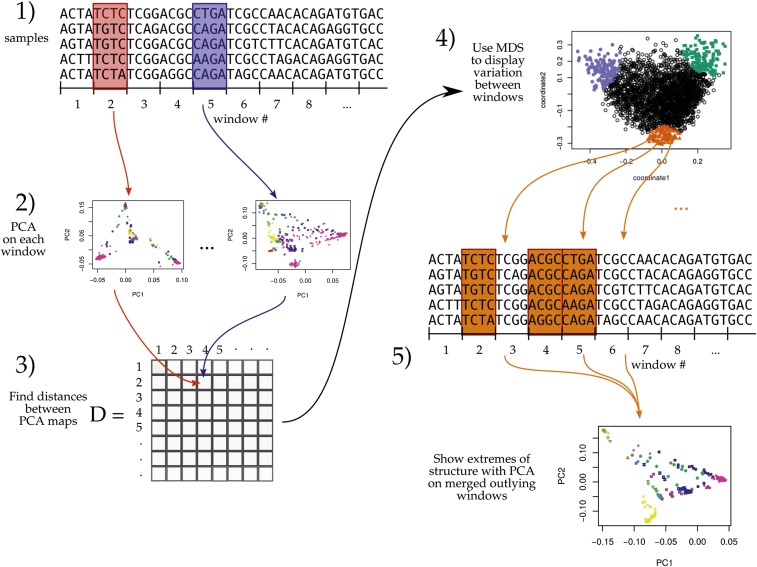
As depicted in Figure 1, the general steps to the method are: (1) divide the genome into windows, (2) summarize the patterns of relatedness in each window, (3) measure dissimilarity in relatedness between each pair of windows, (4) visualize the resulting dissimilarity matrix using multidimensional scaling (MDS), and (5) combine similar windows to more accurately visualize local effects of population structure using PCA.
Figure 1.
An illustration of the method; see Materials and Methods for details. MDS, multidimensional scaling; PC, principal component; PCA, PC analysis.
PCA in genomic windows
To begin, we first recoded sampled genotypes as numeric matrices in the usual manner, by recording the number of nonreference alleles seen at each locus for each sample. We then divided the genome into contiguous segments (windows) and applied PCA as described in McVean (2009) separately to the submatrices that corresponded to each window. The choice of window length entails a tradeoff between signal and noise, since shorter windows allow better resolution along the genome but provide less precise estimates of relatedness. A method for choosing a window length to balance these considerations is given in the Appendix.
Precisely, denote by Z the recoded genotype matrix for a given window (L is the number of SNPs and N is the sample size), and by the mean of nonmissing entries for allele s, so that where the sum is over the ns nonmissing genotypes. We first compute the mean-centered matrix, X, as and preserving missingness (this mean-centering makes the result independent of the choice of reference allele, exactly if there is no missing data, and approximately otherwise). Next, we find the covariance matrix of X, denoted C, as where all sums are over the mij sites, where both sample i and sample j have nonmissing genotypes. The principal components (PCs) are the eigenvectors of C, normalized to have Euclidean length equal to 1, and ordered by magnitude of the eigenvalues.
The top two-to-five PCs are generally good summaries of population structure; for ease of visualization we usually only use the first two (referred to as PC1 and PC2), and check that results hold using more. The above procedure can be performed on any subset of the data; for future reference, denote by PC1j and PC2j the result after applying to all SNPs in the jth window (however, note that our measure of dissimilarity between windows does not depend on PC ordering).
Similarity of patterns of relatedness between windows
We think of the local effects of population structure as being summarized by the relative position of the samples in the space defined by the top PCs. However, we do not compare patterns of relatedness of different genomic regions by directly comparing the PCs, since rotations or reflections of these imply identical patterns of relatedness. Instead, we compare the low-dimensional approximations of the local covariance matrices obtained using the top k PCs, which is invariant under reordering of the PCs, reflections, and rotations, and yet contains all other information about the PCs (for results shown here, we use ) Furthermore, to remove the effect of artifacts such as mutation rate variation, we also rescale each approximate covariance matrix to be of similar size (precisely, so that the underlying data matrix has trace norm equal to 1).
To do this, define the matrix so that the column of is equal to the PC of the window, multiplied by where is the eigenvalue of the genetic covariance matrix. Then, the rescaled, rank k approximate covariance matrix for the ith window is
| (1) |
To measure the similarity of patterns of relatedness for the ith and jth windows, we then use Euclidean distance Dij between the matrices M(i) and M(j), defined by
The goal of comparing PC plots up to rotation and reflection turned out to be equivalent to comparing rank-k approximations to local covariance matrices. This suggests instead directly comparing entire local covariance matrices. However, with thousands of samples and tens of thousands of windows, computing the distance matrix would take months of CPU time while, as defined above, D can be computed in minutes using the following method. Since for square matrices A and B, then due to the orthogonality of eigenvectors and the cyclic invariance of trace, Dij can be computed efficiently as
| (2) |
Visualization of results
We use MDS to visualize relationships between windows as summarized by the dissimilarity matrix D. MDS produces a set of m coordinates for each window that give the arrangement in m-dimensional space that best recapitulates the original distance matrix. For results here, we use to produce one- or two-dimensional visualizations of relationships between windows’ patterns of relatedness.
We then locate variation in patterns of relatedness along the genome by choosing collections of windows that are nearby in MDS coordinates and map their positions along the genome. A visualization of the effects of population structure across the entire collection is formed by extracting the corresponding genomic regions and performing PCA on all aggregated regions.
Testing
We tested the method using two types of simulation. First, to verify expected behavior, we simulated “genomes” as an independent sequence of correlated Gaussian “genotypes,” using a different covariance matrix in the first quarter, middle half, and last quarter of the chromosome. The details of the simulation, also designed to detect sensitivity to PC switching, are given in the Appendix. To verify robustness to missing data, we ran the method after randomly dropping 50% of the genotypes in the first half of the genome; if the method is misled by missing data, then it will distinguish the two halves of the chromosome rather than the segments having different covariance matrices.
To provide a realistic test, we next used forward-time, individual-based simulations, implemented using SLiM v3 (Haller and Messer 2017), which are described in detail in the Appendix. To provide realistic population structure for PCA to identify, each simulation had at least 5000 diploid individuals, living across a continuous square range, with Gaussian dispersal and local density-dependent competition. Each genome was modeled on human chromosome 7, which is -bp long, with an overall recombination rate of 1.6785 crossovers per chromosome per generation. To improve speed, we used tskit (Kelleher et al. 2018) to record tree sequences in SLiM (Haller et al. 2018) and to add neutral mutations afterward, at a rate of 10−9 per bp per generation. Most simulations were neutral, but we also included linked selection of two types. First, we introduced selected mutations into two regions, which extended from one-third to one-half and from five-sixths to the end of the genome respectively. These had selection coefficients from a Gamma distribution with shape 2 and mean 0.005 at a rate of 10−10 per bp, which were either beneficial (with probability 1/30) or deleterious (otherwise). Second, to roughly model a recent expansion followed by local adaptation, we introduced mutations in the same manner as above, except that mutations were no longer unconditionally deleterious or beneficial: each selection coefficient was multiplied by a factor depending on the spatial location of the individual being evaluated, varying linearly from −1 at the left side of the range to +1 at the right edge. In all simulations, genome-wide PCA displayed a map of the population range, as expected.
Data sets
We applied the method to genomic data sets with good geographic sampling: 380 African Drosophila melanogaster from the Drosophila Genome Nexus (Lack et al. 2015), a worldwide data set of humans, 3965 humans from several locations worldwide from the POPRES data set (Nelson et al. 2008), and 263 Medicago truncatula from 24 countries around the Mediterranean basin (a range-wide data set of the partially selfing weedy annual plant from the M. truncatula HapMap Project) (Tang et al. 2014), as summarized in Table 1.
Table 1. Descriptive statistics for each data set used.
| Species | Number of SNPs per window | Mean window length (bp) | Mean number of windows per chromosome | Mean % variance explained by top two principal components |
|---|---|---|---|---|
| D. melanogaster | 1,000 | 9,019 | 2,674 | 0.53 |
| Human | 100 | 636,494 | 203 | 0.55 |
| M. truncatula | 10,000 | 102,580 | 467 | 0.50 |
D. melanogaster:
We used whole-genome sequencing data from the Drosophila Genome Nexus (http://www.johnpool.net/genomes.html; Lack et al. 2015), consisting of the Drosophila Population Genomics Project phases 1–3 (Langley et al. 2012; Pool et al. 2012) and additional African genomes (Lack et al. 2015). After removing 20 genomes with > 8% missing data, we were left with 380 samples from 16 countries across Africa and Europe. Since the Drosophila samples are from inbred lines or haploid embryos, we treat the samples as haploid when recoding: regions with residual heterozygosity were marked as missing in the original data set; we also removed positions with > 20% missing data. Each chromosome arm we investigated (X, 2L, 2R, 3L, and 3R) has 2–3 million SNPs; PCA plots for each arm are shown in Supplemental Material, Figure S1.
Human:
We also used genomic data from the entire POPRES data set (Nelson et al. 2008), which has array-derived genotype information for 447,267 SNPs across the 22 autosomes of 3965 samples in total: 346 African-Americans, 73 Asians, 3187 Europeans, and 359 Indian Asians. Since these data derive from genotyping arrays, the SNP density is much lower than the other data sets, which are each derived from whole-genome sequencing. We excluded the sex chromosomes and the mitochondria. PCA plots for each chromosome, separately, are shown in Figure S2.
M. truncatula:
Finally, we used whole-genome sequencing data from the M. truncatula HapMap Project (Tang et al. 2014), which has 263 samples from 24 countries, primarily distributed around the Mediterranean basin. Each of the eight chromosomes has 3–5 million SNPs; PCA plots for these are shown in Figure S3. We did not use the mitochondria or chloroplasts.
Data availability
The methods described here are implemented in an open-source R package available at https://github.com/petrelharp/local_pca, as well as scripts to perform all analyses from VCF files at various parameter settings.
Data sets are available as follows: human (POPRES) at dbGaP with accession number phs000145.v4.p2, Medicago at the Medicago HapMap http://www.medicagohapmap.org/, and Drosophila at the Drosophila Genome Nexus, http://www.johnpool.net/genomes.html. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7324526.
Results
In all three data sets—a worldwide sample of humans, African D. melanogaster, and a range-wide sample of M. truncatula—PCA plots vary along the genome in a systematic way, showing strong chromosome-scale correlations. This implies that variation is due to meaningful heterogeneity in a biological process, since noise due to randomness in choice of local genealogical trees is not expected to show long-distance correlations. Below, we discuss the results and likely underlying causes.
Validation
Simple non-population-based simulations with Gaussian genotypes showed that the method performs as expected, clearly separating regions of the genome with different underlying covariance matrices without being affected by extreme differences in amount of missing data (Figure S4). This simulation also verifies insensitivity to ordering of top PCs, since it was performed using a covariance matrix with the top two eigenvalues equal, so that the order of empirical eigenvectors (PCs) switches randomly.
Individual-based simulations using SLiM (Haller and Messer 2017) allowed us to test the effects of recombination and mutation rate variation, as well as linked selection. As expected, varying recombination rate stepwise by a factor of 64 did not induce patterns in the MDS visualizations correlated with recombination rate (Figure S5). Since varying mutation rate with a fixed recombination map is equivalent to varying the recombination map and remapping windows, this also indicates that the method is not misled by variation in mutation rate. On the other hand, a recombination map with hotspots [the HapMap human female map for chromosome 7 (International Hap Map Consortium et al. 2007)] induced outliers at long regions of low recombination rate (also as expected).
Simulations with linked selection produced mixed results (Figure S6). The method strongly identified the distinct regions in the simulation with spatially varying linked selection. It also identified the regions (although less unambiguously) with constant selection and stepwise varying recombination rate, but did not clearly identify them with constant recombination rate. The differences in power between these three scenarios are likely explained by varying strength of linked selection; the simulation with spatially varying selection was also the case with strongest positive selection, and recombination rates were overall lower in the simulation with stepwise varying recombination rate than in the simulation with constant rate. These tests are not meant to be a comprehensive survey of linked selection, but only to demonstrate that linked selection can produce signals similar to what we see in real data.
D. melanogaster
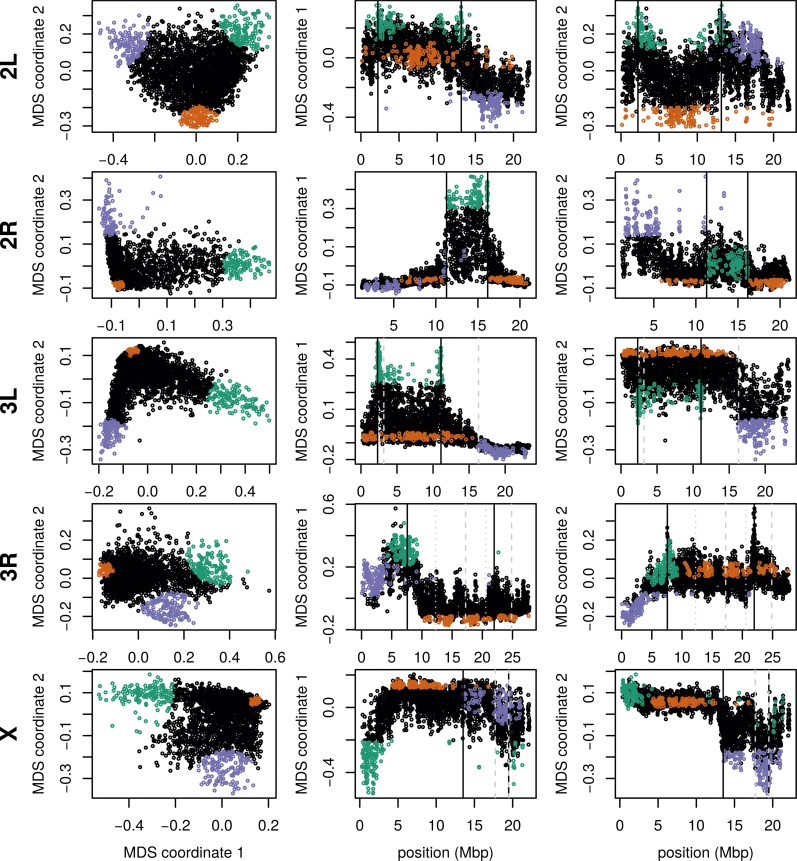
We applied the method to windows of average length 9 kbp across chromosome arms 2L, 2R, 3L, 3R and X separately. The first column of Figure 2 is an MDS visualization of the matrix of dissimilarities between genomic windows: in other words, genomic windows that are closer to each other in the MDS plot show more similar patterns of relatedness. For each chromosome arm, the MDS visualization roughly resembles a triangle, sometimes with additional points. Since the relative position of each window in this plot shows the similarity between windows, this suggests that there are at least three extreme manifestations of population structure typified by windows found in the “corners” of the figure, and that other windows’ patterns of relatedness may be a mixture of those extremes. The next two columns of Figure 2 depict the two MDS coordinates of each window, plotted against the window’s position along the genome, to show how the plot of the first column is laid out along the genome. The patterns did not depend on the number of PCs used (see Figure S7 for the same plot with PCs) and are only weakly correlated with variation in missingness (see Figure S8).
Figure 2.
Variation in patterns of relatedness for windows across D. melanogaster chromosome arms. In all plots, each point represents one window along the genome. The first column shows the MDS visualization of relationships between windows, and the second and third columns show the two MDS coordinates against the midpoint of each window; rows correspond to chromosome arms. Colors are consistent for plots in each row. Vertical lines show the breakpoints of known polymorphic inversions. Solid black lines are for the inversions we used in Figure 3, while dotted gray lines are for other known inversions. MDS, multidimensional scaling.
To help visualize how clustered windows with similar patterns of relatedness are along each chromosome arm, we selected three extreme windows in the MDS plot and the 5% of windows that are closest to it in the MDS coordinates, then highlighted these windows’ positions along the genome, and created PCA plots for the windows, combined. Representative plots are shown for three groups of windows on each chromosome arm in Figure 2 (groups are shown in color) and in Figure S9 (PCA plots). The latter plots are quite different, showing that genomic windows in different regions of the MDS plot indeed show quite different patterns of relatedness.
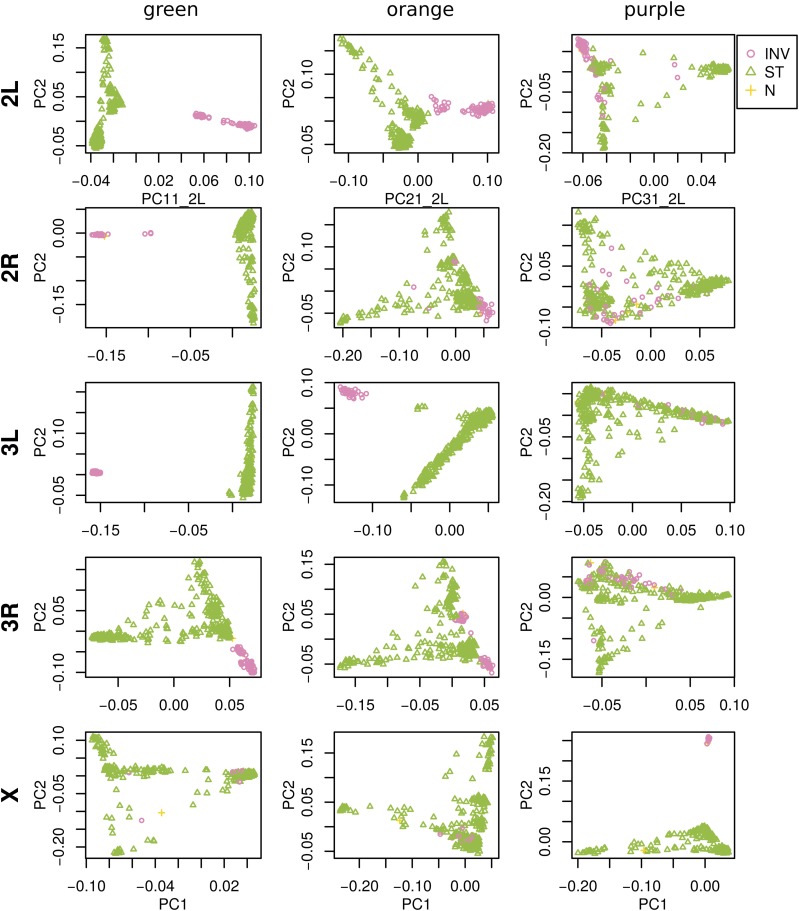
The most striking variation in patterns of relatedness turns out to be explained by several large inversions that are polymorphic in these samples, as discussed in Corbett-Detig and Hartl (2012) and Langley et al. (2012). To depict this, Figure 3 shows the PCA plots in Figure S9 recolored by the orientation of the inversion for each sample. Taking chromosome arm 2L as an example, the two regions of similar, extreme patterns of relatedness shown in green in the first row of Figure 2 lie directly around the breakpoints of the inversion In(2L)t, and the PCA plots in the first rows of Figure 3 show that patterns of relatedness here are mostly determined by inversion orientation. The regions shown in purple on chromosome 2L lie near the centromere and have patterns of relatedness reflective of two axes of variation, seen in Figure 3 and Figure S9, which correspond roughly to latitude within Africa and to degree of “cosmopolitan” admixture, respectively [see Lack et al. (2015) for more about admixture in this sample]. The regions shown in orange on chromosome 2L mostly lie inside the inversion and show patterns of relatedness that are a mixture between the other two, as expected due to recombination within the (long) inversion (Guerrero et al. 2011). Similar results are found in other chromosome arms, albeit complicated by the coexistence of more than one polymorphic inversion; however, each breakpoint visibly affects patterns in the MDS coordinates (see vertical lines in Figure 2).
Figure 3.
PCA plots for the three sets of genomic windows colored in Figure 2, on each chromosome arm of D. melanogaster. In all plots, each point represents a sample. The first column shows the combined PCA plot for windows whose points are colored green in Figure 2; the second is for orange windows; and the third is for purple windows. In each, samples are colored by orientation of the polymorphic inversions In(2L)t, In(2R)NS, In(3L)OK, In(3R)K, and In(1)A, respectively [data from Lack et al. (2015)]. In each, “INV” denotes an inverted genotype, “ST” denotes the standard orientation, and “N” denotes unknown. PC, principal component; PCA, PC analysis.
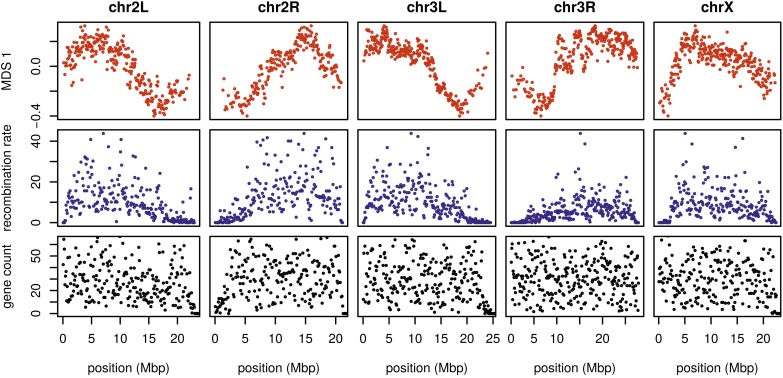
To see how patterns of relatedness vary in the absence of polymorphic inversions, we performed the same analyses after removing, for each chromosome arm, any samples carrying inversions on that arm. In the results, shown in Figure 4 and Figure S10, the striking peaks associated with inversion breakpoints are gone, and previously smaller-scale variation now dominates the MDS visualization. For instance, the majority of the variation along 3L in Figure 2 is on the left end of the arm, dominated by two large peaks around the inversion breakpoints; there is also a relatively small dip on the right end of the arm (near the centromere). In contrast, Figure 4 and Figure S10 show that after removing polymorphic inversions, remaining structure is dominated by the dip near the centromere. Without inversions, variation in patterns of relatedness shown in the MDS plots follows similar patterns to that previously seen in D. melanogaster recombination rate and diversity (Langley et al. 2012; Mackay et al. 2012). Indeed, correlations between the recombination rate in each window and the position on the first MDS coordinate are highly significant (Spearman’s Figure 4 and Figure S11). This is consistent with the hypothesis that variation is due to selection, since the strength of linked selection increases with local gene density, measured in units of recombination distance. The number of genes—measured as the number of transcription start and end sites within each window—was not significantly correlated with MDS coordinate
Figure 4.
The effects of population structure without inversions is correlated to recombination rate in D. melanogaster. The first plot (in red) shows the first MDS coordinate along the genome for windows of 10,000 SNPs, obtained after removing samples with inversions (a plot analogous to Figure 2 is shown in Figure S10). The second plot (in blue) shows local average recombination rates in cM/Mbp, obtained as midpoint estimates for 100-kbp windows from the Drosophila recombination rate calculator (Fiston-Lavier et al. 2010) release 5, using rates from Comeron et al. (2012). The third plot (in black) shows the number of genes’ transcription start and end sites within each 100-kbp window, divided by two. Transcription start and end sites were obtained from the RefGene table from the University of California Santa Cruz browser. The histone gene cluster on chromosome arm 2L is excluded. chr, chromosome; MDS, multidimensional scaling.
Human
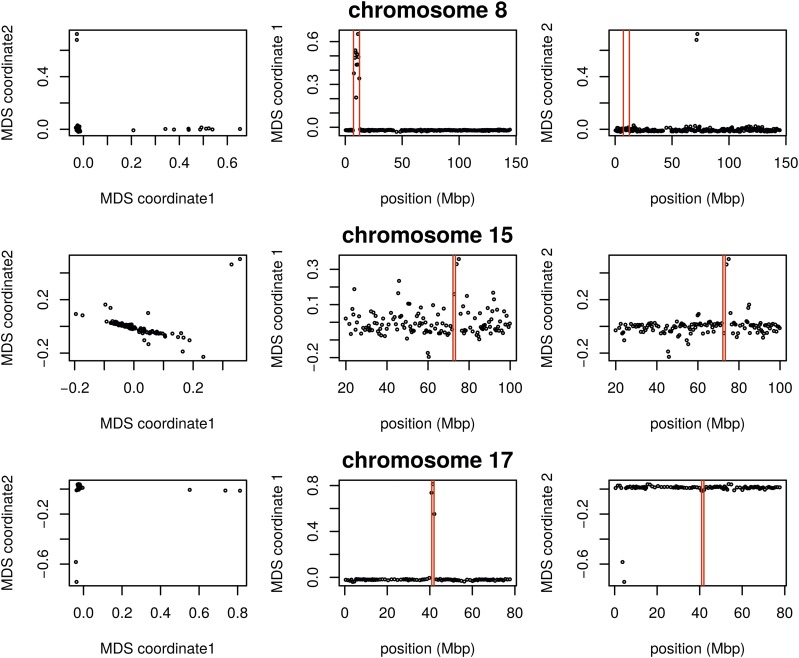
As we did for the Drosophila data, we applied our method separately to all 22 human autosomes. On each, variation in patterns of relatedness was dominated by a small number of windows having similar patterns of relatedness to each other that differed dramatically from the rest of the chromosome. These may primarily be inversions: outlying windows coincide with three of the six large polymorphic inversions described in Antonacci et al. (2009), notably a particularly large, polymorphic inversion on 8p23 (Figure 5). Similar plots for all chromosomes are shown in Figure S12, Figure S13, and Figure S14. PCA plots of many outlying windows show a characteristic trimodal shape (shown for chromosome 8 in Figure S15), presumably distinguishing samples having each of the three diploid genotypes for each inversion orientation (although we do not have data on orientation status). This trimodal shape has been proposed as a method to identify inversions (Ma and Amos 2012), but distinguishing this hypothesis from others, such as regions of low recombination rate, would require additional data.
Figure 5.
Variation in structure between windows on human chromosomes 8, 15, and 17. Each point in each plot represents a window. The first column shows the MDS visualization of relationships between windows; the second and third columns show the two MDS coordinates of each window against its position (midpoint) along the chromosome. Rows, from top to bottom show chromosomes 8, 15, and 17. The vertical red lines show the breakpoints of known inversions from Antonacci et al. (2009). MDS, multidimensional scaling.
We also applied the method on all 22 autosomes together, and found that, remarkably, the inversion on chromosome 8 is still the most striking outlying signal (Figure S16). Further investigation with a denser set of SNPs, allowing a finer genomic resolution, may yield other patterns.
M. truncatula
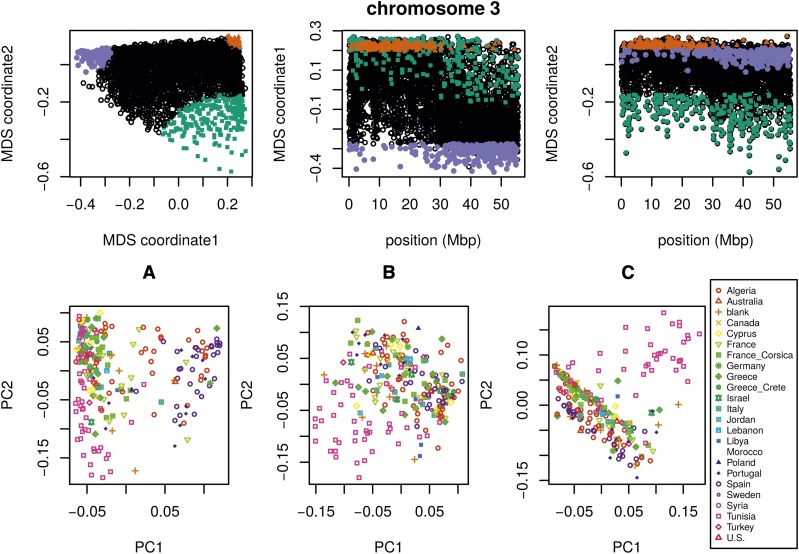
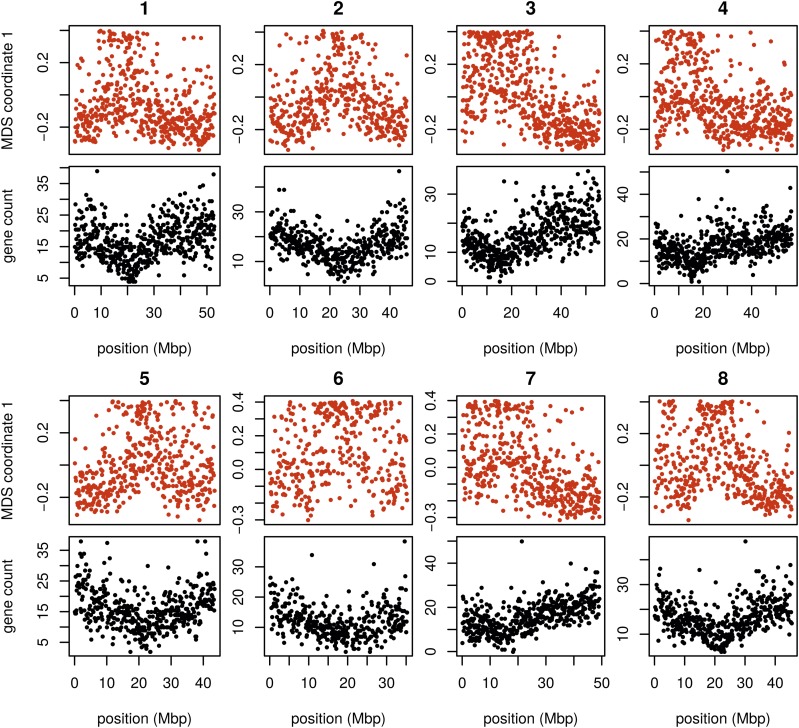
Unlike the other two species, the method applied separately on all eight chromosomes of M. truncatula showed similar patterns of gradual change in patterns of relatedness across each chromosome, with no indications of chromosome-specific patterns. This consistency suggests that the factor affecting the population structure for each chromosome is the same, as might be caused by varying strengths of linked selection. To verify that variation in the effects of population structure is shared across chromosomes, we applied the method to all chromosomes together. Results for chromosome 3 are shown in Figure 6 and other chromosomes are similar: across chromosomes, the high values of the first MDS coordinate coincide with the position of the heterochromatic regions surrounding the centromere, which often have lower gene density and may therefore be less subject to linked selection. To verify that this is a possible explanation, we counted the number of genes found in each window using gene models in Mt4.0 from jcvi.org (Tang et al. 2014), which are shown juxtaposed with the first MDS coordinate of each window in Figure 7 and are significantly correlated, as shown in Figure S17 (values shown are the number of start and end positions of each predicted mRNA transcript, divided by two, assigned to the nearest window.) However, other genomic features, such as distance to centromere, show roughly the same patterns, so we cannot rule out alternative hypotheses. In particular, fine-scale recombination rate estimates are not available in a form mappable to Mt4.0 coordinates [although those in Paape et al. (2012) appear visually similar].
Figure 6.
MDS visualization of patterns of relatedness on M. truncatula chromosome 3, with corresponding PCA plots (upper panels). Each point in the plot represents a window; the structure revealed by the MDS plot is strongly clustered along the chromosome, with windows in the upper-right corner of the MDS plot (colored red) clustered around the centromere, windows in the upper-left corner (purple) furthest from the centromere, and the remaining corner (green) intermediate. Plots for remaining chromosomes are shown in Figure S18. PCA plots for the sets of genomic windows colored (A) green, (B) orange, and (C) purple in the upper panels (lower panels). Each point corresponds to a sample, colored by country of origin. Plots for remaining chromosomes are shown in Figure S19. MDS, multidimensional scaling; PC, principal component; PCA, PC analysis.
Figure 7.
MDS coordinate and gene density for each window in the Medicago genome, for chromosomes 1–8 (numbered above each pair of figures). For each chromosome, the red plot above is first coordinate of MDS against the middle position of each window along each chromosome. The black plot below is gene count for each window against the middle position of each window. MDS, multidimensional scaling.
The results were highly consistent across window sizes, window types (SNPs or bp), and number of PCs, as shown in Table S2.
Discussion
Our investigations have found substantial variation in the patterns of relatedness formed by population structure across the genomes of three diverse species, revealing distinct biological processes driving this variation in each species. More investigation, particularly on more species and data sets, will help to uncover what aspects of species history can explain these differences. With growing appreciation of the heterogeneous effects of selection across the genome, especially the importance of adaptive introgression and hybrid speciation (Fitzpatrick et al. 2010; Staubach et al. 2012; Hufford et al. 2013; Brandvain et al. 2014; Pool 2015), local adaptation (Lenormand 2002; Wang and Bradburd 2014), and inversion polymorphisms (Kirkpatrick 2010; Kirkpatrick and Barrett 2015), local PCA may prove to be a useful exploratory tool to discover important genomic features.
We now discuss possible implications of this variation in the effects of population structure, the impact of various parameter choices in implementing the method, and possible additional applications.
Chromosomal inversions
A major driver of variation in patterns of relatedness in the two data sets we examined seems to be inversions. This may be common, but the example of M. truncatula shows that polymorphic inversions are not ubiquitous. PCA has been proposed as a method for discovering inversions (Ma and Amos 2012); however, the signal left by inversions likely cannot be distinguished from long haplotypes under balancing selection or simply regions of reduced recombination without additional lines of evidence. Inversions show up in our method because across the inverted region, most gene trees share a common split that dates back to the origin of the inversion. However, in many applications, inversions are a nuisance. For instance, SMARTPCA (Patterson et al. 2006) reduces their effect on PCA plots by regressing out the effect of linked SNPs on each other. Removing samples with the less common orientation of each inversion reduced, but did not eliminate, the signal of inversions seen in the D. melanogaster data set, demonstrating that the genomic effects of transiently polymorphic inversions may outlast the inversions themselves.
Genealogical noise?
The field of phylogenetics has long had to deal with the fact that there can be a great number of different local phylogenies along the genome, even between species (Avise et al. 1983; Pamilo and Nei 1988; Hobolth et al. 2007). The within-species patterns we observe might contribute to such incomplete lineage sorting among future descendant species of a given population. The neutral distribution of variation in these patterns has been used to infer demographic history, both between species (Slatkin and Pollack 2006) and within species (Beeravolu et al. 2018). If these distinct phylogenies are merely a result of neutral stochasticity, there is not expected to be a correlation between local phylogeny and other genomic features. However, in some cases, the local assortment of ancestral diversity and subsequent introgression between sister taxa shows clear signs of selection, as for instance in the wild tomato clade (Pease et al. 2016).
The effect of selection
Neutral processes are not expected to produce the chromosome-scale correlations we see in patterns of relatedness in the M. truncatula and D. melanogaster data sets, because correlations in patterns of relatedness induced by neutral processes should extend no further than does linkage disequilibrium (i.e., much less than a chromosome’s length). This suggests that they are produced by linked selection, a hypothesis backed up by correlations with gene density and recombination rate. We have also shown with simulations that linked selection can, in at least some circumstances, produce the sorts of patterns we observe. How might selection cause variation in patterns of relatedness? For instance, background selection (the effect on linked sites of selection against deleterious mutations) (Charlesworth et al. 1993, Charlesworth 2013) can informally be thought of as reducing the number of potential contributors to the gene pool in regions of the genome with many possible deleterious mutations (Hudson and Kaplan 1995). For this reason, if it acts in a spatial context, it is expected to induce samples from nearby locations to cluster together more frequently. Therefore, regions of the genome harboring many targets of local adaptation may show similar patterns, since migrant alleles in these regions will be selected against, and so locally gene trees will more closely reflect spatial proximity. Other forms of selection, such as hard sweeps on new mutations, repeated selection on standing variation, local adaptation, or temporally fluctuating selection, could clearly lead to variation in geographic patterns of relatedness in a similar way.
Another possible contributor is recent admixture between previously separated populations, the effects of which were not uniform across the genome due to selection. For instance, it has been hypothesized that large-scale variation in amount of introgressed Neanderthal DNA along the genome is due to selection against Neanderthal genes, leading to greater introgression in regions of lower gene density (Harris and Nielsen 2016; Juric et al. 2016). African D. melanogaster are thought to have a substantial amount of recently introgressed genome from cosmopolitan sources; if selection regularly favors genes from one origin, this could lead to substantial variation in patterns of relatedness correlated with local gene density.
There has been substantial debate over the relative impacts of different forms of selection (e.g., Charlesworth et al. 1997; Charlesworth 2012; Hedrick 2013; Pease and Hahn 2013; Burri et al. 2015; Corbett-Detig et al. 2015; Harris and Nielsen 2016; Martin et al. 2016; Phung et al. 2016; Stankowski et al. 2018). These have been difficult to disentangle in part because, for the most part, theory makes predictions that are only strictly valid in randomly mating (i.e., unstructured) populations, and it is unclear to what extent the spatial structure observed in most real populations will affect these predictions. Developing a method to distinguish these forms of selection from each other and from the effects of demography is a major challenge to the field. It may be possible to make progress using statistics that make stronger use of spatial information, such as the variation in relatedness that we observe here, similar to the method of Beeravolu et al. (2018).
Parameter choices
There are several choices in the method that may in principle affect the results. As with whole-genome PCA, the choice of samples is important, as variation not strongly represented in the sample will not be discovered. The effects of strongly imbalanced sampling schemes are often corrected by dropping samples in overrepresented groups; but downweighting may be a better option that does not discard data. Next, the choice of window size may be important, although in our applications, results were not sensitive to this. Finally, which collections of genomic regions are compared to each other (steps 3 and 4 in Figure 1), along with the method used to discover common structure, will affect results. We used MDS, applied to either each chromosome separately or to the entire genome; for instance, human inversions are clearly visible as outliers when compared to the rest of their chromosomes, but genome-wide, their signal is obscured by the numerous other signals of comparable strength.
Besides window length, there is also the question of how to choose windows. In these applications, we have used nonoverlapping windows with equal numbers of polymorphic sites. However, we found little change in results when using different window sizes or when measuring windows in physical distance (in base pairs).
Finally, our software allows different choices for how many PCs to use in approximating the structure of each window (k in Equation 1) and how many MDS coordinates to use when describing the distance matrix between windows, but in our exploration, changing these has not produced dramatically different results. However, this choice could in some situations be important: for instance, if the kth and PCs are sufficiently different but have similar eigenvalues, then small amounts of noise could cause these to switch, leading to spuriously inferred differences between windows in which one or the other was included in the top k PCs. This does not seem to be a problem in our applications, as changing the number of PCs did not affect the qualitative results. These choices are all part of more general techniques in dimension reduction and high-dimensional data visualization; we encourage the user to experiment.
Applications
So-called cryptic relatedness between samples has been one of the major sources of confounding in GWAS, and so methods must account for it by modeling population structure or kinship (Astle and Balding 2009; Yang et al. 2014). Modern “mixed model” methods (e.g., Loh et al. 2015) account for this with either a single, genome-wide kinship matrix or one constructed using only sites unlinked to the focal SNP. Since the effects of population structure are not constant along the genome, this could in principle lead to an inflation of false positives in parts of the genome with stronger population structure than the genome-wide average. A method such as ours might be used to estimate local kinship matrices, thus providing a more sensitive correction, although doing so without removing the signal itself could be challenging. Fortunately, in our human data set this does not seem likely to have a strong effect: most variation is due to small, independent regions, possibly primarily inversions, and so may not have a major effect on GWAS. In the other species we examined, particularly D. melanogaster, treating population structure as a single quantity would entail a substantial loss of power and could potentially be misleading.
Acknowledgments
We thank John Pool, Russ Corbett-Detig, Matilde Cordeiro, and Peter Chang for assistance with obtaining data and interpreting results (especially the inversion status of D. melanogaster samples); Jaime Ashander and Jerome Kelleher for providing assistance in performing the simulations; and Yaniv Brandvain, Barbara Engelhardt, Charles Langley, Graham Coop, and Jeremy Berg for helpful comments and for encouraging the project. Work on this project was supported by NSF grant number 1262645 (DBI) to PR. The authors declare no conflicts of interest.
Appendix
Choosing window length
The choice of window length entails a balance between signal and noise. In very short windows, genealogies of the samples will only be represented by a few trees, so variation between windows represents demographic noise rather than meaningful variation in patterns of relatedness. Longer windows generally have more distinct trees (and SNPs), allowing for less noisy estimation of local patterns of relatedness. However, to better resolve meaningful signal, i.e., differences in patterns of relatedness along the genome, we would like reasonably short windows.
Since we summarize patterns of relatedness using relative positions in the principal component maps, we quantify “noise” as the standard error of a sample’s position on PC1 in a particular window, averaged across windows and samples, and “signal” as the standard deviation of the sample’s position on PC1 over all windows, averaged over samples. The definition of eigenvectors does not specify their sign, and so when comparing between windows we choose signs to best match each other: after choosing , for instance, if u is the first eigenvector obtained from the covariance matrix for window j, then we next choose , where the sign is chosen according to which of or is smaller.
After doing this, the mean variance across windows is
where is the position of the individual on in window j, and . We estimate the standard error for each using the block jackknife (Efron, 1982; Busing et al. 1999): we divide the window into 10 equal-sized pieces, and let denote the first principal component of this region found after removing the piece; then the estimate of the squared standard error is . Averaging over samples and windows,
For the main analysis, we defined windows to each consist of the same number of neighboring SNPs, and calculated and for a range of window sizes (i.e., numbers of SNPs). For our main results we chose the smallest window for which was consistently larger than (but checked other sizes); the values for various window sizes across Drosophila chromosomes are shown in Table S1. In the cases we examined, we found nearly identical results after varying window size, and choosing windows to be of the same physical length (in bp) rather than in numbers of SNPs.
Simulations
We implemented two types of simulation: first, simple simulations of Gaussian “genotypes” where the expectation of variation in “population structure” was clear; and next, individual-based simulations with explicit genomes, using SLiM.
Gaussian simulations
We simulated genotypes at each locus independently, drawing each vector of genotypes from a multivariate Gaussian distribution with zero mean and covariance matrix . Sampled individuals came from three populations, and each depends on which populations the individuals i and j are in, as well as the location along the chromosome. There are three population-level mean relatedness matrices along the genome, which apply to the first quarter (), the middle half (), and the last quarter (), respectively:
If indiviudals i and j are in populations and respectively, then the covariance between their genotypes is , using the appropriate S for that segment of the genome. The variance of individual i’s genotype is .
We first created “genotypes” in this way with fifty individuals from each of the three populations; running our method on a genome with 99 windows of 400 loci each produced the first plot in Supplementary Figure S4. These matrices are chosen so that the top two eigenvalues are the same (both 50.1), and so the ordering of the top two PCs is arbitrary. If our method was sensitive to PC ordering, then half the windows in each region that have one ordering would cluster with each other, separate from the other half.
We then marked each genotype in the first half of the chromosome as missing, independently, with probability and ran our method again, producing the second plot of Supplementary Figure S4. If our method was influenced by missing data, we would expect the first half of the chromosome to separate from the second in the MDS plot.
SLiM simulations
Our SLiM simulations were constructed as follows. Individuals are diploid, and genomes have a length of 153,520,244 bp. Recombination was either (a) flat, with a constant rate of ; (b) according to the human female HapMap map for chromosome 7; or (c) constant in each of seven equal-sized regions, beginning at , descending by a factor of four for three steps, and then ascending by a factor of four for three steps, so that the middle seventh has the lowest recombination rate, and the outer two sevenths has a rate 64 times higher. Selected mutations are introduced at a rate of per bp per individual per generation, and have selection coefficients drawn from a Gamma distribution with mean 0.005 and shape 2; each coefficient are either positive or negative with probabilities and respectively. Each simulation was run for 50,000 generations.
Each individual has a spatial position in the two-dimensional square of width . Each time step, each individual chooses the nearest other to mate with, producing a random, Poisson distributed number of offspring with mean . Offspring are assigned random spatial locations displaced from their parent’s by a bivariate Gaussian with mean zero and standard deviation , reflected to stay within the habitat range.
Each individual survives to the next time step with probability equal to their fitness. Fitness values are determined multiplicatively by the effects of each mutation, but are multiplied by an additional factor determined by the local density of individuals. This factor is equal to , where is the carrying capacity per circle of radius σ; is the mean equilibrium population density; and C is the sum of a Gaussian kernel with standard deviation between the focal individual and all other individuals within distance . To avoid edge effects, fitnesses are further multiplied by , where z is the distance to the nearest boundary. This produces populations that fluctuate at equilibrium around 6,000 individuals in total, fairly evenly spread across the square.
In one additional simulation, we modified fitnesses by multiplying the selective effect of each allele in each individual by multiplying it by , where x is the x coordinate of the individual. This makes the effect of each allele opposite on the left and on the right, and neutral in the middle, and leads to a moderate number of balanced polymorphisms.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7324526.
Communicating editor: J. Novembre
Literature Cited
- Antonacci F., Kidd J. M., Marques-Bonet T., Ventura M., Siswara P., et al. , 2009. Characterization of six human disease-associated inversion polymorphisms. Hum. Mol. Genet. 18: 2555–2566. 10.1093/hmg/ddp187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle W., Balding D. J., 2009. Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 24: 451–471. 10.1214/09-STS307 [DOI] [Google Scholar]
- Avise J. C., Shapira J. F., Daniel S. W., Aquadro C. F., Lansman R. A., 1983. Mitochondrial DNA differentiation during the speciation process in Peromyscus. Mol. Biol. Evol. 1: 38–56. [DOI] [PubMed] [Google Scholar]
- Barton N. H., 2000. Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1553–1562. 10.1098/rstb.2000.0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeravolu C. R., Hickerson M. J., Frantz L. A. F., Lohse K., 2018. Able: blockwise site frequency spectra for inferring complex population histories and recombination. Genome Biol. 19: 145 10.1186/s13059-018-1517-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair A. P., 1943. Population structure in toads. Am. Nat. 77: 563–568. 10.1086/281161 [DOI] [Google Scholar]
- Brandvain Y., Kenney A. M., Flagel L., Coop G., Sweigart A. L., 2014. Speciation and introgression between Mimulus nasutus and Mimulus guttatus. PLoS Genet. 10: e1004410 10.1371/journal.pgen.1004410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbin A., Bryc K., Byrnes J., Zakharia F., Omberg L., et al. , 2012. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum. Biol. 84: 343–364. 10.3378/027.084.0401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K., Auton A., Nelson M. R., Oksenberg J. R., Hauser S. L., et al. , 2010. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl. Acad. Sci. USA 107: 786–791. 10.1073/pnas.0909559107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R., Nater A., Kawakami T., Mugal C. F., Olason P. I., et al. , 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25: 1656–1665. 10.1101/gr.196485.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busing F. M., Meijer E., Van Der Leeden R., 1999. Delete-m jackknife for unequal m. Stat. Comput. 9: 3–8. 10.1023/A:1008800423698 [DOI] [Google Scholar]
- Charlesworth B., 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190: 5–22. 10.1534/genetics.111.134288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2013. Background selection 20 years on: the Wilhelmine E. Key 2012 invitational lecture. J. Hered. 104: 161–171. 10.1093/jhered/ess136 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Nordborg M., Charlesworth D., 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70: 155–174. 10.1017/S0016672397002954 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., Barton N. H., 2003. The effects of genetic and geographic structure on neutral variation. Annu. Rev. Ecol. Evol. Syst. 34: 99–125. 10.1146/annurev.ecolsys.34.011802.132359 [DOI] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Hartl D. L., 2012. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8: e1003056 (erratum: PLoS Genet. 9) 10.1371/journal.pgen.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Hartl D. L., Sackton T. B., 2015. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 13: e1002112 10.1371/journal.pbio.1002112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank T. E., Hahn M. W., 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 23: 3133–3157. 10.1111/mec.12796 [DOI] [PubMed] [Google Scholar]
- Duforet-Frebourg N., Luu K., Laval G., Bazin E., Blum M. G., 2016. Detecting genomic signatures of natural selection with principal component analysis: application to the 1000 genomes data. Mol. Biol. Evol. 33: 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B., 1982. The Jackknife, the Bootstrap and Other Resampling Plans. Society for Industrial and Applied Mathematics, Philadelphia: 10.1137/1.9781611970319 [DOI] [Google Scholar]
- Ellegren H., Smeds L., Burri R., Olason P. I., Backström N., et al. , 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491: 756–760. 10.1038/nature11584 [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier A.-S., Singh N. D., Lipatov M., Petrov D. A., 2010. Drosophila melanogaster recombination rate calculator. Gene 463: 18–20. 10.1016/j.gene.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick B. M., Johnson J. R., Kump D. K., Smith J. J., Voss S. R., et al. , 2010. Rapid spread of invasive genes into a threatened native species. Proc. Natl. Acad. Sci. USA 107: 3606–3610. 10.1073/pnas.0911802107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero R. F., Rousset F., Kirkpatrick M., 2011. Coalescent patterns for chromosomal inversions in divergent populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 430–438. 10.1098/rstb.2011.0246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller B. C., Messer P. W., 2017. SLiM 2: flexible, interactive forward genetic simulations. Mol. Biol. Evol. 34: 230–240. 10.1093/molbev/msw211 [DOI] [PubMed] [Google Scholar]
- Haller B. C., Galloway J., Kelleher J., Messer P. W., Ralph P. L., 2018. Tree-sequence recording in SLiM opens new horizons for forward-time simulation of whole genomes. bioRxiv. Available at: 10.1101/407783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K., Nielsen R., 2016. The genetic cost of Neanderthal introgression. Genetics 203: 881–891. 10.1534/genetics.116.186890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22: 4606–4618. 10.1111/mec.12415 [DOI] [PubMed] [Google Scholar]
- Hobolth A., Christensen O. F., Mailund T., Schierup M. H., 2007. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3: e7 10.1371/journal.pgen.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1995. Deleterious background selection with recombination. Genetics 141: 1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sánchez E., DeGiorgio M., Pagani L., Tarekegn A., Ekong R., et al. , 2013. Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol. Biol. Evol. 30: 1877–1888. 10.1093/molbev/mst089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford M. B., Lubinksy P., Pyhäjärvi T., Devengenzo M. T., Ellstrand N. C., et al. , 2013. The genomic signature of crop-wild introgression in maize. PLoS Genet. 9: e1003477 10.1371/journal.pgen.1003477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. Frazer K. A., Ballinger D. G., Cox D. R., Hinds D. A., et al. , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. 10.1038/nature06258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric I., Aeschbacher S., Coop G., 2016. The strength of selection against Neanderthal introgression. PLoS Genet. 12: e1006340 10.1371/journal.pgen.100634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambhatla N., Leen T. K., 1997. Dimension reduction by local principal component analysis. Neural Comput. 9: 1493–1516. 10.1162/neco.1997.9.7.1493 [DOI] [Google Scholar]
- Kelleher J., Thornton K. R., Ashander J., Ralph P. L., 2018. Efficient pedigree recording for fast population genetics simulation. PLoS Comput. Biol. 14: e1006581 10.1371/journal.pcbi.1006581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., 2010. How and why chromosome inversions evolve. PLoS Biol. 8: e1000501 10.1371/journal.pbio.1000501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Barrett B., 2015. Chromosome inversions, adaptive cassettes and the evolution of species’ ranges. Mol. Ecol. 24: 2046–2055. 10.1111/mec.13074 [DOI] [PubMed] [Google Scholar]
- Lack J. B., Cardeno C. M., Crepeau M. W., Taylor W., Corbett-Detig R. B., et al. , 2015. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199: 1229–1241. 10.1534/genetics.115.174664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C., Schrider D. R., et al. , 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. 10.1534/genetics.112.142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T., 2002. Gene flow and the limits to natural selection. Trends Ecol. Evol. 17: 183–189. 10.1016/S0169-5347(02)02497-7 [DOI] [Google Scholar]
- Loh P. R., Tucker G., Bulik-Sullivan B. K., Vilhjálmsson B. J., Finucane H. K., et al. , 2015. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47: 284–290. 10.1038/ng.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Amos C. I., 2012. Investigation of inversion polymorphisms in the human genome using principal components analysis. PLoS One 7: e40224 10.1371/journal.pone.0040224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjón J. V., Coupé P., Concha L., Buades A., Collins D. L., et al. , 2013. Diffusion weighted image denoising using overcomplete local PCA. PLoS One 8: e73021 10.1371/journal.pone.0073021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. H., Möst M., Palmer W. J., Salazar C., McMillan W. O., et al. , 2016. Natural selection and genetic diversity in the butterfly Heliconius melpomene. Genetics 203: 525–541. 10.1534/genetics.115.183285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G., 2009. A genealogical interpretation of principal components analysis. PLoS Genet. 5: e1000686 10.1371/journal.pgen.1000686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menozzi P., Piazza A., Cavalli-Sforza L., 1978. Synthetic maps of human gene frequencies in Europeans. Science 201: 786–792. 10.1126/science.356262 [DOI] [PubMed] [Google Scholar]
- Nadeau N. J., Whibley A., Jones R. T., Davey J. W., Dasmahapatra K. K., et al. , 2012. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 343–353. 10.1098/rstb.2011.0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. R., Bryc K., King K. S., Indap A., Boyko A. R., et al. , 2008. The population reference sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. 83: 347–358. 10.1016/j.ajhg.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Stephens M., 2008. Interpreting principal component analyses of spatial population genetic variation. Nat. Genet. 40: 646–649. 10.1038/ng.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A. R., et al. , 2008. Genes mirror geography within Europe. Nature 456: 98–101 (erratum: Nature 456: 274) 10.1038/nature07331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paape T., Zhou P., Branca A., Briskine R., Young N., et al. , 2012. Fine-scale population recombination rates, hotspots, and correlates of recombination in the Medicago truncatula genome. Genome Biol. Evol. 4: 726–737. 10.1093/gbe/evs046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo P., Nei M., 1988. Relationships between gene trees and species trees. Mol. Biol. Evol. 5: 568–583. [DOI] [PubMed] [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease J. B., Hahn M. W., 2013. More accurate phylogenies inferred from low-recombination regions in the presence of incomplete lineage sorting. Evolution 67: 2376–2384. 10.1111/evo.12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease J. B., Haak D. C., Hahn M. W., Moyle L. C., 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14: e1002379 10.1371/journal.pbio.1002379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung T. N., Huber C. D., Lohmueller K. E., 2016. Determining the effect of natural selection on linked neutral divergence across species. PLoS Genet. 12: e1006199 10.1371/journal.pgen.1006199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., 2015. The mosaic ancestry of the Drosophila genetic reference panel and the D. melanogaster reference genome reveals a network of epistatic fitness interactions. Mol. Biol. Evol. 32: 3236–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080 10.1371/journal.pgen.1003080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Roweis S. T., Saul L. K., 2000. Nonlinear dimensionality reduction by locally linear embedding. Science 290: 2323–2326. 10.1126/science.290.5500.2323 [DOI] [PubMed] [Google Scholar]
- Slatkin M., Pollack J. L., 2006. The concordance of gene trees and species trees at two linked loci. Genetics 172: 1979–1984. 10.1534/genetics.105.049593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankowski S., Chase M. A., Fuiten A. M., Ralph P. L., Streisfeld M. A., 2018. The tempo of linked selection: rapid emergence of a heterogeneous genomic landscape during a radiation of monkeyflowers. bioRxiv. Available at: 10.1101/342352. [DOI] [Google Scholar]
- Staubach F., Lorenc A., Messer P. W., Tang K., Petrov D. A., et al. , 2012. Genome patterns of selection and introgression of haplotypes in natural populations of the house mouse (Mus musculus). PLoS Genet. 8: e1002891 10.1371/journal.pgen.1002891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Krishnakumar V., Bidwell S., Rosen B., Chan A., et al. , 2014. An improved genome release (version mt4.0) for the model legume Medicago truncatula. BMC Genomics 15: 312 10.1186/1471-2164-15-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Hahn M. W., Nuzhdin S. V., 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3: e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B., Akey J. M., 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343: 1017–1021. 10.1126/science.1245938 [DOI] [PubMed] [Google Scholar]
- Wang I. J., Bradburd G. S., 2014. Isolation by environment. Mol. Ecol. 23: 5649–5662. 10.1111/mec.12938 [DOI] [PubMed] [Google Scholar]
- Weingessel A., Hornik K., 2000. Local PCA algorithms. Neural networks. IEEE Transactions on 11: 1242–1250. [DOI] [PubMed] [Google Scholar]
- Wright S., 1949. The genetical structure of populations. Ann. Eugen. 15: 323–354. 10.1111/j.1469-1809.1949.tb02451.x [DOI] [PubMed] [Google Scholar]
- Yang J., Zaitlen N. A., Goddard M. E., Visscher P. M., Price A. L., 2014. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 46: 100–106. 10.1038/ng.2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The methods described here are implemented in an open-source R package available at https://github.com/petrelharp/local_pca, as well as scripts to perform all analyses from VCF files at various parameter settings.
Data sets are available as follows: human (POPRES) at dbGaP with accession number phs000145.v4.p2, Medicago at the Medicago HapMap http://www.medicagohapmap.org/, and Drosophila at the Drosophila Genome Nexus, http://www.johnpool.net/genomes.html. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7324526.