Abstract
The body’s external surfaces and the insides of biological tubes, like the vascular system, are lined by a lipid-, glycoprotein-, and glycosaminoglycan-rich apical extracellular matrix (aECM). aECMs are the body’s first line of defense against infectious agents and promote tissue integrity and morphogenesis, but are poorly described relative to basement membranes and stromal ECMs. While some aECM components, such as zona pellucida (ZP) domain proteins, have been identified, little is known regarding the overall composition of the aECM or the mechanisms by which different aECM components work together to shape epithelial tissues. In Caenorhabditis elegans, external epithelia develop in the context of an ill-defined ZP-containing aECM that precedes secretion of the collagenous cuticle. C. elegans has 43 genes that encode at least 65 unique ZP proteins, and we show that some of these comprise distinct precuticle aECMs in the embryo. Previously, the nidogen- and EGF-domain protein DEX-1 was shown to anchor dendrites to the C. elegans nose tip in concert with the ZP protein DYF-7. Here, we identified a new, strong loss-of-function allele of dex-1, cs201. dex-1 mutants die as L1 larvae and have a variety of tissue distortion phenotypes, including excretory defects, pharyngeal ingression, alae defects, and a short and fat body shape, that strongly resemble those of genes encoding ZP proteins. DEX-1 localizes to ZP-containing aECMs in the tissues that show defects in dex-1 mutants. Our studies suggest that DEX-1 is a component of multiple distinct embryonic aECMs that shape developing epithelia, and a potential partner of multiple ZP proteins.
Keywords: C. elegans, extracellular matrix, zona pellucida, cuticle, excretory system
APICAL extracellular matrices (aECMs) line exposed epithelial surfaces, such as the epidermis and the insides of tubes. For example, the stratum corneum lines the outside of the skin (Feingold 2007), a glycocalyx lines the inside of blood vessels (Reitsma et al. 2007), and surfactant coats the inside of lung alveoli (Pérez-Gil 2008). aECMs serve as primary barriers to infectious agents and have important roles in tissue shaping and integrity (Plaza et al. 2010; Johansson et al. 2013; Tarbell and Cancel 2016). However, aECMs are poorly characterized relative to stromal ECMs or basement membranes, in part due to their fragility under fixation conditions (Chappell et al. 2009; Luschnig and Uv 2014). It is clear that aECMs are lipid-, glycoprotein-, and glycosaminoglycan-rich structures that often form distinct layers (Chappell et al. 2009; Johansson et al. 2013; Gill et al. 2016). Despite the importance of aECMs, individual aECM components’ functions, and their relationships with other components, are often poorly described.
Zona pellucida (ZP) proteins can be found in high abundance in many apical matrices (Plaza et al. 2010). For example, the founding members of the family, ZP1, ZP2, and ZP3, are found in the mammalian egg-coat, the ZP, where they both enable fertilization and serve as a barrier to polyspermy (Gupta 2018). Other ZP proteins are found in the lumen of the vascular system [endoglin and betaglycan (ten Dijke et al. 2008)], the kidney tubules [uromodulin and oit3 (Zaucke et al. 2010; Yan et al. 2012)], or in the tectorial membrane of the inner ear [tectorins (Legan et al. 1997)]. Loss or dysfunction of these ZP proteins can cause hereditary hemorrhagic telangiectasia (HHT), chronic kidney disease, or deafness, respectively (Rampoldi et al. 2011; Richardson et al. 2011; Saito et al. 2017). In invertebrates such as Caenorhabditis elegans and Drosophila, various ZP proteins are part of the cuticle or exoskeleton (Sebastiano et al. 1991; Muriel et al. 2003; Sapio et al. 2005; Fernandes et al. 2010). While some ZP proteins are involved in signaling via protein–protein interactions, others polymerize to form matrix fibrils with presumed structural roles (Jovine et al. 2002; Bokhove et al. 2016; Saito et al. 2017). Better understanding of how ZP-containing matrices shape tissues will require identification of other components of these matrices.
The nematode Caenorhabditis elegans contains multiple different types of aECMs, and is a tractable model system for studying aECM composition, assembly, and function. Most external surfaces of larvae and adults are lined by a cuticle composed primarily of collagens (Page and Johnstone 2007), with different ZP proteins termed cuticlins (CUT proteins) contributing to stage-specific lateral ridges or alae (Sebastiano et al. 1991; Muriel et al. 2003; Sapio et al. 2005). In contrast, the pharynx or foregut is lined by a different type of cuticle containing the polysaccharide chitin (Zhang et al. 2005), and internal epithelia such as the gut are not lined by cuticle, although their alternative aECMs have not yet been described. Finally, in the embryo, cuticles are not secreted until the completion of epidermal elongation and morphogenesis, and epithelial tissue development occurs in the context of a still ill-defined precuticular or “sheath” matrix (Priess and Hirsh 1986). Multiple ZP proteins, including FBN-1, NOAH-1, NOAH-2, and LET-653, are components of this early precuticular matrix, and play important roles in embryonic tissue shaping (Kelley et al. 2015; Gill et al. 2016; Vuong-Brender et al. 2017). Other apparent components of this early matrix include members of the extracellular leucine-rich repeat only (eLRRon) (Mancuso et al. 2012) and lipocalin (Forman-Rubinsky et al. 2017) families, which also play important roles in the shaping or protection of mammalian tissues (Paragas et al. 2012; Chen and Birk 2013), suggesting similarities between the C. elegans precuticular matrix and ECMs of other organisms. Such similarities make this early matrix of particular interest.
C. elegans has several advantages for studying aECM. First, aECM components can be easily visualized in live animals using fluorescent fusion proteins, avoiding typical aECM destruction by fixation. Second, unbiased genetic screening approaches can be used to identify new matrix components. Foundational studies on ZP components of the aECM suggest some of the phenotypes that we would expect to see in new precuticle matrix mutants. For example, FBN-1 anchors the pharynx to the nose tip as the embryo elongates. In fbn-1 mutants, the pharynx recedes into the animal’s interior (Pharynx Ingressed, Pin), such that the animal cannot feed (Kelley et al. 2015). NOAH-1 and NOAH-2 maintain epidermal integrity during embryonic elongation; noah-1 and noah-2 mutants experience epidermal rupture (Vuong-Brender et al. 2017). Finally, LET-653 maintains the tiny excretory duct tube during its elongation. The unicellular duct tube is the middle tube of the C. elegans excretory system, which is required for osmoregulation (Nelson et al. 1983; Nelson and Riddle 1984). During embryogenesis, the duct tube elongates from a simple toroid to a more complex shape with a looped lumen (Sundaram and Buechner 2016). In let-653 mutants, the duct lumen fragments and develops dilations (Gill et al. 2016). LET-653 is also required for proper morphogenesis of cuticular alae in L1 larvae and adults (Gill et al. 2016; Forman-Rubinsky et al. 2017). Here, we determined that these different ZP mutant phenotypes can be explained in part by nonuniform tissue distributions of different embryonic ZP proteins, thereby defining a diverse set of precuticular aECMs.
We sought new mutants with ZP-like phenotypes and identified a new allele of the previously described gene dex-1. DEX-1 is a proposed partner of the ZP protein DYF-7 in a dendritic cap matrix that anchors sensory dendrites and glia to the nose tip (Heiman and Shaham 2009; Low et al. 2018). In both dex-1 and dyf-7 mutants, dendrites retract into the body cavity under tension from elongation forces in the embryo. dex-1 encodes a nidogen- and EGF-domain transmembrane protein related to the conserved basement membrane protein nidogen and to other mammalian matrix proteins including alpha-tectorin, MUC4, and SNED1. Here, we show that DEX-1 localizes to diverse ZP-containing aECMs in the embryo and to L1 cuticle alae. The originally described dex-1 allele leaves the nidogen and EGF domains intact and is hypomorphic. By characterizing a new, stronger allele that disrupts those domains, we show that DEX-1 is important for a variety of aECM-dependent tissue shaping processes in the embryo. DEX-1 is not normally expressed beyond the L1 larval stage, but in the accompanying manuscript (Flatt et al., 2019), we show that starvation signaling upregulates DEX-1 to remodel cuticle structure and generate alae in dauer larvae. Together, our studies suggest that DEX-1 is a component of diverse aECMs and shapes tissues in concert with many different C. elegans ZP proteins.
Materials and Methods
Worm strains, alleles, and transgenes
All strains were derived from Bristol N2 and were grown at 20° under standard conditions (Brenner 1974). dex-1 mutants were obtained from mothers rescued with a dex-1(+) transgene or homozygous for dex-1. Mutants used included: dex-1(ns42) I (Heiman and Shaham 2009), dex-1(cs201) I (this study), fbn-1(ns283) III (Kelley et al. 2015), dyf-7(ns117) X and dyf-7(ns119) X (Heiman and Shaham 2009). Transgenes used included: aaaEx78 (mini-FBN-1::mCherry) (Kelley et al. 2015), cbcEx1 (dex-1pro,long::GFP,unc-122pro::GFP) (Flatt et al., 2019), hmnEx144 (DYF-7::SfGFP) (Low et al. 2018), csEx402 (dex-1pro::DEX-1, unc-119pro::GFP) (this study), csEx503 (dex-1pro::DEX-1(ecto)::SfGFP, lin-48pro::mRFP) (this study), csEx808 (dex-1pro::ssSfGFP::DEX-1, lin-48pro::mRFP) (this study), csIs61 (RDY-2::GFP, lin-48pro::mRFP) I (Gill et al. 2016), csIs66 (let-653pro::ssSfGFP::LET-653(ZP), let-653pro:PH-mCherry, lin-48p::mRFP) X, jcIs1 (AJM-1::GFP, rol-6d) IV (Köppen et al. 2001), nsEx1339 (pMH118 = pJIM20dex-1pro:his-24-mCherry, pRF4) (Heiman and Shaham 2009), nsEx1404(pMH136 = pJIM20dex-1prolong:his-24-mCherry, pRF4) (Heiman and Shaham 2009), qnEx59 (dct-5pro:: mCherry, unc-119+) (Abdus-Saboor et al. 2011), and zwEx113 (inx-13pro::GFP, lin-15+) (Altun et al. 2009). CRISPR insertions used included: noah-1[mc68 (noah-1::mCherry)] I and noah-2[mc93 (noah-2::gfp)] IV (Vuong-Brender et al. 2017). See Supplemental Material, Table S1 for a complete list of strains used in this study.
Bioinformatic analyses of C. elegans ZP proteins
To survey the full complement of ZP proteins encoded by the C. elegans genome, we used Uniprot (The UniProt Consortium 2018) to identify all predicted C. elegans ZP proteins (Figure S1 and Table S2), and then aligned the ZP domain sequences using M-Coffee, a multiple sequence alignment package (Wallace et al. 2006; Moretti et al. 2007) (Figure S2). We also cross-referenced these predictions with WormBase to identify all predicted splice isoforms (Lee et al. 2018); in most cases, alternative splicing affected domains outside of the ZP domain, but in six cases the ZP domain was affected (Figure S1 and S2). This analysis showed that C. elegans has 43 genes encoding 65 unique ZP proteins (Figure S1), many more than humans (Bokhove and Jovine 2018) or Drosophila (Fernandes et al. 2010). In literature searches, we found that only 11 out of 43 C. elegans ZP genes have been previously studied, as summarized in Table S3 and the Introduction.
Most ZP domains consist of two independently folding subdomains, ZPn and ZPc, each of which contains at least four conserved cysteines (#1–8) involved in disulfide bond formation (Bokhove and Jovine 2018) (Figure S1). Most human ZPc domains contain two additional cysteines (a, b) that may confer unique folding conformations (Figure S2) (Lin et al. 2011; Bokhove and Jovine 2018). Likewise, we observed additional cysteines in many C. elegans ZPc domains (Figure S1 and S2). We defined three classes of C. elegans ZP proteins depending on whether the ZPc domain has four, six, or eight cysteines (Figure S1), and we defined subfamilies within the large class III depending on which specific conserved cysteines are present (Figure S1 and S2). We also found five C. elegans ZP proteins contain a ZPc domain but lack a ZPn domain (Figure S1 and S2).
Staging and microscopy
Embryos were selected at the 1.5-fold stage and then incubated for the number of hours indicated before mounting for imaging. Larvae were staged by hours after hatch, or by size and gonad morphology. Fluorescent, differential interference contrast (DIC), and Dodt (an imaging technique that simulates DIC; Dodt and Zieglgänsberger 1990) images were captured on a compound Zeiss Axioskop fitted with a Leica DFC360 FX camera, or with a Leica TCS SP8 confocal microscope. Images were processed and merged using ImageJ. Duct-canal secretory junction dimensions and larval width were measured using the straight-line tool in ImageJ, while pore autocellular junction and larval length measurements were performed using the freehand line tool in ImageJ. For each specimen, measurements were made in triplicate and then averaged. To measure fluorescence intensity, a box was drawn around the entire embryo. The image was cropped to the dimensions of the box and 10 consecutive slices from the embryo’s midsection (where the duct and pore are located) were chosen. Total fluorescence intensity was measured within the cropped images in each chosen slice. Fluorescence intensity measurements were averaged across all values from an individual embryo.
dex-1 mutant isolation and allele identification
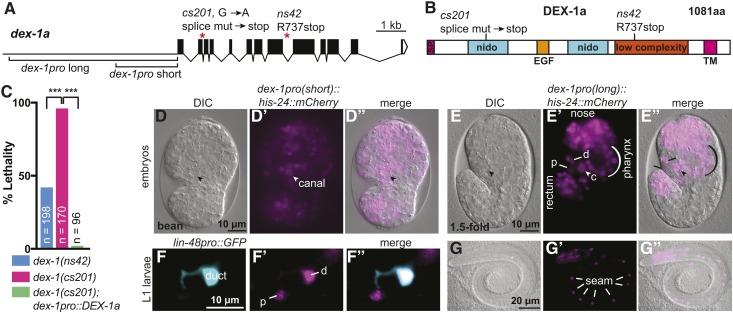
dex-1(cs201) was isolated based on its rod-like lethal phenotype after standard ethylmethanesulfonate (EMS) mutagenesis (Brenner 1974) of strain UP2214 (unc-119; jcIs1; qnEx59), as previously described (Gill et al. 2016). The mutant was outcrossed three times, and the linked and unlinked markers were removed (and then reintroduced as appropriate) prior to analysis. The dex-1 lesion was detected after whole-genome sequencing followed by bioinformatics analyses with Cloudmap (Minevich et al. 2012), and was determined to be causal based on transgenic rescue experiments (Figure 2) and analysis of the allele ns42. cs201 changes the splice donor at intron 4 from G to A.
Figure 2.
dex-1 is an essential gene and broadly expressed in the embryo. (A) dex-1 promoter regions, gene structure and mutant alleles. (B) DEX-1a protein schematic and locations of mutant allele lesions. SP: signal peptide; Nido: nidogen domains; EGF: EGF-like domain. The low complexity region was previously proposed to have homology to the sperm protein zonadhesin (Heiman and Shaham 2009). We were not able to detect that similarity. TM: transmembrane domain. (C) Both ns42 and cs201 mutants show larval lethality, with a higher percentage in cs201 mutants. This lethality was efficiently rescued by a transgene encoding DEX-1a driven by the short dex-1 promoter (see A). *** P < 0.0001. Fisher’s exact test. (D and E) dex-1 transcriptional reporters were widely expressed in embryonic epithelia, including the epidermis, pharynx, nose tip, rectum, duct (d), pore (p), and canal nucleus (arrowhead, c). DIC: differential interference contrast. (D–D”) Bean stage embryo, ventral view. (E–E”) 1.5-fold stage embryo, lateral view. (F–G) dex-1 transcriptional reporter expression persisted in L1 larvae. (F–F”) Expression in the duct (labeled with lin-48pro::GFP) (d) and pore (p). (G–G”) Expression in seam epidermal cells (lines). Reporters contain either the short (D and F) or long (E and G) version of the dex-1 promoter, as shown in (A); both gave similar expression patterns.
RT-PCR
For RT-PCR analysis, RNA was extracted from mixed populations of dex-1(cs201) and N2 using the RNeasy Mini Kit (Qiagen). Genomic DNA was then removed from RNA samples using the TURBO DNA-Free Kit (ThermoFisher Scientific) and reverse transcribed using First Strand DNA Synthesis (New England Biolabs). A fragment from dex-1 exon 4–exon 5 was PCR amplified from dex-1(cs201) and WT cDNA using primers oJC342 (GTCAGCACTTATTATTACTTGGG) and oJC344 (CTGTATATGAAGTTGGCAAATGTTCCG), and then sequenced with primer oJC342. The predominant band obtained from dex-1(cs201) cDNA contained a small portion of intron 4 and then spliced to exon 5 (Figure S3). Larger molecular weight products, including a completely unspliced product, were observed with lower abundance and frequency. All products encoded a stop codon near the failed splice site. We saw no evidence for normal splicing in cs201 mutants.
dex-1 reporters
dex-1 rescue plasmid pMH7 (dex-1pro::DEX-1), 5.7kb dex-1pro::GFP transcriptional reporter plasmid pMH111, SuperfolderGFP source plasmid pCW11, and dex-1 transcriptional reporter strains OS2336 and OS2438 all were obtained from Max Heiman (Harvard University). The dex-1 rescue transgene csEx402 was generated by coinjecting 30 ng/μl pMH7 with 30 ng/μl pIM175 (unc-119pro::GFP). The 5.7 kb dex-1pro::GFP reporter cbcEx1 was generated by coinjecting 20 ng/μl pMH111 with 80 ng/μl of unc-122pro::GFP as the coinjection marker (K.M.F. et al., unpublished results). To build DEX-1(ecto)::SfGFP (pJC15), a 2.1-kb fragment containing the dex-1 promoter and upstream regulatory region was digested from pMH7 with restriction enzymes XbaI and AgeI and cloned into pPD49.26. DEX-1(ecto) was PCR amplified from pMH11 with primers oJC19 (GGGAAAGCTAGCAAAATGCGCAATAGGCATGCCCTTC) and oJC32 (GAAAAGTTCTTCTCCTTTGCTTTGAGTTGTACTCTGGGCACG) and PCR sewn with SfGFP using primers oJC32 (CGTGCCCAGAGTACAACTCAAAGCAAAGGAGAAGAACTTTTC) and oJC18 (GGGAGATCTGATCCTTTGTAGAGCTCATCCATGCC), then inserted into pPD49.26 behind the dex-1 promoter. To build ssSfGFP::DEX-1 (pJC24), SfGFP was PCR amplified from pCW11 using primers oJC6 (GGGAGATCTAAGCAAAGGAGAAGAACTTTTCACTGG) and oJC18 (GGGAGATCTGATCCTTTGTAGAGCTCATCCATGCC) and inserted within pMH7 at an existing BglII restriction site. Plasmids were coinjected at concentration of 30 ng/μl with marker pHS4 (lin-48pro::mRFP) at concentration 50 ng/μl.
Data availability statement
All strains and plasmids are available upon request. Supplemental data files are available at https://figshare.com/. Table S1 contains complete genotypes of all strains used. Table S2 contains GenBank and Uniprot accession numbers for all C. elegans ZP protein sequences. Table S3 contains summary information for 11 previously studied ZP proteins. Figure S1 contains a diagram of C. elegans ZP proteins sorted by ZPc cysteine number. Figure S2 contains multiple sequence alignments for all C. elegans ZP proteins. Figure S3 contains RT-PCR results for dex-1(cs201). Figure S4 contains images of dyf-7 mutants. Figure S5 contains images of dex-1 mutant excretory canals. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7312148.
Results
Precuticle ZP composition varies between tissues
The epidermal precuticle ensheaths the developing embryo beginning between the bean and 1.5-fold stages (Priess and Hirsh 1986), and later serves as a template for building the cuticle (Costa et al. 1997; Forman-Rubinsky et al. 2017; Katz et al. 2018). A morphologically similar aECM lines developing tubes, such as the excretory duct and pore, that connect to the epidermis (Mancuso et al. 2012), but possible molecular distinctions among these precuticle matrices have not been described.
We are particularly interested in the composition of the excretory duct and pore precuticle, which we previously showed contains the ZP protein LET-653 (Gill et al. 2016). Since ZP proteins often function together, we wondered if other family members might also be present in this matrix. Bioinformatic analyses using Uniprot and Wormbase revealed that C. elegans contains 43 genes that encode at least 65 different ZP proteins (Materials and Methods, Figure S1 and S2). These ZP proteins fall into three classes based on the number and spacing of cysteines (Figure S1). Of the 11 ZP proteins that have been studied, three others besides LET-653 have been previously found in the C. elegans precuticle (Kelley et al. 2015; Vuong-Brender et al. 2017) (Table S3). Available RNA-seq data suggest that the corresponding genes are transcribed in the excretory duct and pore (Burdick et al. 2016; Cao et al. 2017). To determine if any of these other known precuticle ZP proteins contribute to the excretory duct and pore aECM, we examined existing mutants and translational reporters.
FBN-1 contains a C-terminal ZP domain attached to a protein with overall similarity to human fibrillin (which is not a ZP protein) (Kelley et al. 2015); 62% (n = 292) of fbn-1(ns283) strong hypomorphic mutants arrested as embryos or larvae, but they did not exhibit “rod-like lethality” or any visible dilations indicative of excretory system defects. Although a mini-FBN-1::mCherry fusion containing the FBN-1 signal peptide, ZP domain, and C-terminal transmembrane domain lined the embryonic epidermis, buccal cavity, and rectum, we did not observe any accumulation in the excretory duct or pore (Figure 1B). However, mini-FBN-1::mCherry did appear in the excretory duct and pore lumen during later larval molt cycles (Figure 1C), suggesting potential stage-specific partners.
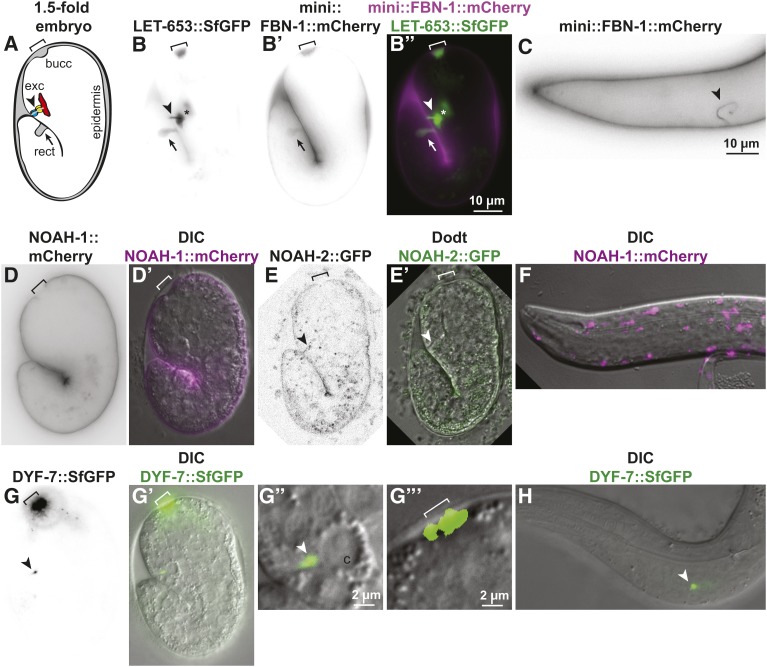
Figure 1.
ZP proteins localize to aECMs in distinct tissues. ZP fusion proteins, expressed under their endogenous promoters, showed different patterns in elongating 1.5- to 2-fold embryos (A, B, D, E, and G) and L1 larvae (C, F, and H). All tags are inserted N-terminally to the predicted ZP cleavage site, except in the case of LET-653, where the tag is C-terminal but the cleaved product remains associated with the matrix (Gill et al. 2016). Arrowhead: excretory system; bracket: buccal cavity and nose tip; arrow: rectum. (A) Cartoon of 1.5-fold embryo, indicating major regions of aECM accumulation. Cytoplasm: white; aECM: gray; apical membranes: black. (B) LET-653::SfGFP (Gill et al. 2016) strongly lined the duct/pore lumen (Gill et al. 2016), and was also visible in or near the buccal cavity, rectum, and epidermis. Asterisk indicates neuronal coinjection marker ttx3-pro::GFP present in the FBN-1 transgene array. (B’) mini-FBN-1::mCherry (Kelley et al. 2015) strongly lined the buccal cavity, rectum, and epidermis, but was not detected in the duct/pore lumen at the 1.5- to 2-fold (n = 25) or 3-fold (n = 25) stages. (B”) Merged images illustrate distinct localization. (C) mini-FBN-1::mCherry did line the excretory duct and pore lumen in larvae. (D and D’) NOAH-1::mCherry (Vuong-Brender et al. 2017) was restricted to the region closely apposed to the epidermis and excluded from the buccal cavity, rectum, and duct/pore lumen (n = 20). (E and E’) NOAH-2::GFP (Vuong-Brender et al. 2017) was faintly visible lining the epidermis and potentially seen along the duct/pore lumen, but was never visualized at the nose tip (n = 11). F) NOAH-1::mCherry appeared intracellular in L1 larvae (n = 10). (G and G’) DYF-7::SfGFP (Low et al. 2018) localized specifically to the nose tip and a region near the duct canal junction (n = 19). (G” and G”’) show magnifications of the excretory canal (c) and nose regions, respectively. (H) DYF-7::SfGFP remained associated with the canal region in L1 larvae (n = 15).
NOAH-1 and NOAH-2 are related to Drosophila sensory matrix protein NOMP-A (Chung et al. 2001). Like LET-653, they each contain plasminogen-like (PAN) domains in addition to ZP domains. noah-1 and noah-2 mutants arrest as embryos prior to excretory tube morphogenesis (Vuong-Brender et al. 2017), precluding their analysis. Like mini-FBN-1::mCherry, CRISPR-generated NOAH-1::mCherry and NOAH-2::GFP were both visible lining the epidermis (Vuong-Brender et al. 2017). However, NOAH-1 was absent from the rectum, buccal cavity, and excretory duct and pore lumen (Figure 1D). NOAH-2 also could not be reliably detected in these areas, but it was extremely faint overall and therefore more difficult to evaluate (Figure 1E). In early L1 larvae, NOAH-1 appeared primarily intracellular (Figure 1F), as previously described (Vuong-Brender et al. 2017).
We also examined mutants and reporters for the dendritic cap matrix ZP protein DYF-7, whose expression has been previously reported in the excretory canal cell (Heiman and Shaham 2009). Some dyf-7(ns119) null mutants arrested as L1s or disappeared from the plates (22%, n = 125), and many of these exhibited an ingressed or unattached pharynx (Pin or Pun) phenotype similar to that described for fbn-1 mutants (Kelley et al. 2015) (Figure S4), but the excretory duct and pore appeared normal in all animals examined (n = 35) (Figure S4). In 1.5-fold embryos and L1 larvae, a discrete spot of DYF-7::SfGFP was observed near the excretory canal cell, but it did not extend through the duct and pore lumens (Figure 1, G and H). The vast majority of DYF-7::SfGFP accumulated near the nose tip, as previously described (Heiman and Shaham 2009; Low et al. 2018), and we did not detect any signal in the remainder of the epidermis (Figure 1, G and H).
While we cannot exclude very low levels and/or redundant functions for some of these ZP proteins, we conclude that FBN-1, NOAH-1/2, and DYF-7 are unlikely to be key partners of LET-653 in shaping the excretory duct and pore. Rather, ZP composition varies among different regions of the embryonic precuticle, which may more accurately be described as a collection of distinct matrices. The different localization patterns of these various ZP proteins could explain in part the rather different phenotypes that result from mutations in these genes.
dex-1 strong loss-of-function mutants are larval lethal
To identify matrix proteins present in the excretory duct and pore, we analyzed additional hits from the screen for excretory lethality that isolated let-653 mutants (Gill et al. 2016). One mutant, cs201, corresponded to a new allele of dex-1 (Figure 2, A–C). Most cs201 mutants arrested as L1 larvae (Figure 2C), although a small proportion survived to adulthood; these escapers yielded progeny with the same L1 arrest phenotype, suggesting no significant impact of maternal genotype on the dex-1 phenotype. cs201 mapped to the left arm of chromosome III based on balancer mapping with hT2 and qC1. Whole genome sequencing revealed a G-to-A change at the splice donor of dex-1 intron 4 (Figure 2A). Via RT-PCR, we confirmed that this mutation perturbed splicing and led to inclusion of a premature stop that truncates the DEX-1 protein (if any is made) within the first nidogen domain (Figure 2B and Figure S3). We were able to rescue dex-1(cs201) lethality with a transgene expressing the dex-1a cDNA under control of its endogenous 2.1-kb 5′ regulatory region (dex-1pro short) (Figure 2, A and C). We conclude that cs201 is a severe loss-of-function and likely null allele of dex-1.
dex-1 encodes a predicted ECM protein with two nidogen domains, an EGF-like domain, and a low complexity region, followed by a transmembrane domain and a short cytoplasmic tail (Figure 2B, and see Heiman and Shaham 2009). Nidogen (nido) domains were first identified in the basement membrane protein nidogen and are thought to be involved in protein–protein interactions, although their precise role is unclear (Ho et al. 2008). Calcium-binding EGF domains are found in many matrix proteins and can act in signaling or bind calcium ions to stiffen the region between two important domains (Appella et al. 1988; Davis 1990; Selander-Sunnerhagen et al. 1992). The previously described dex-1(ns42) allele has potential to encode a truncated protein that retains the nido and EGF domains (Figure 2B) (Heiman and Shaham 2009), but dex-1(cs201) eliminates these domains (Figure 2B). Some dex-1(ns42) mutants do die as L1 larvae, but dex-1(cs201) lethality is more penetrant, consistent with cs201 being a more severe allele (Figure 2C). Although DEX-1 was previously proposed to be a sensory matrix partner of the ZP protein DYF-7 (Heiman and Shaham 2009), our analysis showed that the dex-1 null phenotype is much more severe than that of dyf-7. We therefore hypothesized that DEX-1 may be a matrix component with some DYF-7-independent, non-neuronal functions.
dex-1 is broadly expressed in epithelial cells of embryos
To determine when and where dex-1 is expressed, we used transcriptional reporters (kindly provided by Max Heiman) containing promoter regions upstream of the dex-1 start site fused to an mCherry-tagged histone. We examined two promoters: one 2.1-kb fragment that was previously used in the rescuing dex-1 transgene, and a second containing the entire 5.7-kb region between dex-1 and the nearest upstream gene (Figure 2A). Both transcriptional reporters were broadly expressed in embryos and L1 larvae, including in cells relevant to the phenotypes discussed below (Figure 2, D–G). Both reporters showed expression in the canal, duct, and pore of the excretory system and the seam cells of the epidermis, which sit under the alae and are presumed to secrete factors required for alae development (Figure 2, D and E) (Sapio et al. 2005). The reporters were also active in a number of cells in the pharynx and near the nose tip, where dendrites are anchored, as previously described (Heiman and Shaham 2009). Although both reporters were active in L1 larvae, (Figure 2, F and G) they were progressively fainter in subsequent larval stages and were not observed in adults. Notably, a 5.7 kb dex-1pro::GFP reporter was not detected in L1 larvae (n = 10), suggesting that persistent expression of the histone::mCherry reporter in L1s may be due to protein perdurance (see also Flatt et al., 2019). This is consistent with modENCODE RNA seq data (Celniker et al. 2009), which shows robust dex-1 expression in embryos but little or no dex-1 expression at larval stages. We conclude that dex-1 is expressed in epithelial tissues that build external aECMs during embryogenesis.
dex-1 mutants have excretory defects with some similarities to let-653 mutants
dex-1(cs201) was isolated in a genetic screen for mutants with excretory system defects. To determine the extent and nature of excretory defects in dex-1 mutants, we first examined L1 larvae whose excretory systems were labeled with the external epithelial marker dct-5pro::mCherry and junction marker AJM-1::GFP. 22% (n = 47/212) of L1 larvae had excretory defects, with large dilations in the duct and canal tubes. In the larvae with dilations, 95% (n = 45/47) of the pore tubes lacked an autocellular junction (Figure 3, B, C, E, and F). The tube dilations were luminal, as visualized with the apical membrane marker RDY-2::GFP (Figure 3, D and G). This phenotype, though less penetrant, strongly resembled that of let-653 mutants (Gill et al. 2016). Reduced penetrance of excretory defects in dex-1 compared with let-653 mutants indicates that, while the two proteins may act in concert, LET-653 has additional DEX-1-independent functions in the excretory system.
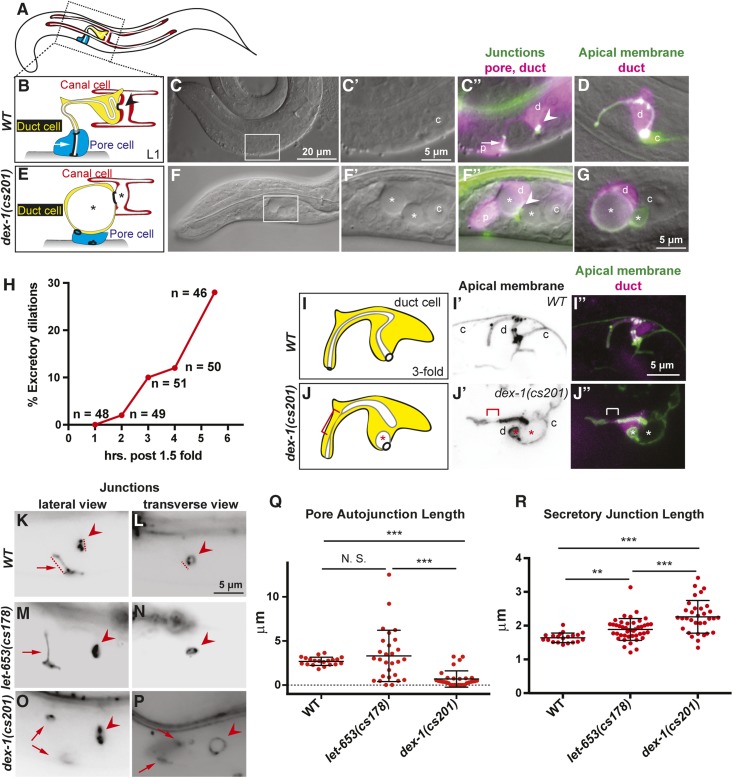
Figure 3.
dex-1 mutants have excretory duct, pore, and canal tube morphological defects. (A) A cartoon of an L1 larva with the canal, duct, and pore indicated. (B) A cartoon of the WT L1 excretory system. The canal (red) is a seamless, unicellular tube that extends four lumenized arms along the length of the animal. Its lumen empties into the duct (yellow), another seamless, unicellular tube, which has a looped lumen. The duct connects to the pore (blue), a seamed, unicellular tube whose autocellular junction runs parallel to its lumen. The pore lumen opens to the outside environment. Arrow indicates pore autocellular junction, and arrowhead indicates ring-like duct-canal secretory junction. (C) WT L1 larva. Box shows region magnified in subsequent panels. (C’ and C”) Inset of C showing the excretory system. c; canal nucleus. (C”) The duct (d) and pore (p) were labeled by the epithelial marker dct-5pro::mCherry. The cell junctions were labeled by AJM-1::GFP. Pore autocellular junction; arrow. Duct-canal secretory junction; arrowhead. (D) WT L1 labeled by duct marker lin-48pro::mRFP and apical membrane marker RDY-2::GFP; the latter also marks the canal lumen (c). (E) Cartoon of dex-1 mutant phenotype. The canal and duct lumens contained large dilations (*). The pore autocellular junction is missing, but its ring junctions remain and the pore has lost its characteristic shape. (F–G) dex-1(cs201) mutant L1 larvae. (F’ and F”) Inset of F showing the excretory system with markers as above. (G) dex-1(cs201) mutant with large dilation visible within the duct lumen, visualized by markers as above. (H) Timeline of excretory dilation appearance. dex-1(cs201) embryos were evaluated for presence of duct/canal luminal dilations using the apical membrane marker RDY-2::GFP. Embryos were picked at 1.5-fold stage and maintained at 20°C for the time indicated. Dilations recorded as present were at least double the normal lumen diameter. Dilations of this size were never observed in WT (Pu et al. 2017). (I–J) Cartoons of WT and dex-1(cs201) duct cells at the 1.5 + 3 hr stage when luminal dilations first appeared. Cytoplasm: yellow; Apical membrane: gray; Lumen: white. (I’–J”) Maximum Z projections of 10–15 slices through the middle of the duct cell. Apical membranes marked by RDY-2::GFP and duct by lin-48pro::mRFP. The WT lumen appeared even in diameter. The dex-1 lumen was discontinuous and has several dilations (*). There was also a region with very thin lumen (bracket). (K–P) Lateral and transverse views of excretory system junctions labeled with AJM-1::GFP. Arrows indicate pore autocellular junction, and arrowheads indicate ring-like duct-canal secretory junction. See cartoons (B and E) for reference. (K–L) WT. Dashed lines indicate axes along which measurements were taken. (M–N) let-653(cs178). M shows an example of a mutant with an elongated pore autocellular junction. (O and P). dex-1(cs201) mutant L1 larvae without a pore autocellular junction, but with remaining pore-duct and pore-epidermis ring junctions (arrows) and an expanded secretory junction. Note some bleedthrough of dct-5pro::mCherry signal in (P). (Q and R) Quantification of pore autocellular junction and secretory junction length in newly hatched L1 larvae. All let-653(cs178) mutants had excretory dilations; only those dex-1(cs201) mutants with dilations were measured. Absent pore autocellular junctions were measured as being of length 0 and were included in statistical analysis. *** P < 0.0001, ** P < 0.001, N. S. = not significant. Mann-Whitney U-test. let-653 mutant pore autocellular junctions showed greater variation in length, but the mean length was not statistically different from WT.
To determine when excretory defects first appeared, we examined embryos at different developmental time points. DEX-1 luminal dilations began to appear during embryonic elongation (Figure 3H). By 3 hr after 1.5-fold, when embryos have reached threefold elongation, 7/51 embryos had luminal dilations of varying size in the duct and canal. In four embryos, the duct lumen dilation seemed to encompass the entire region of visible lumen, similar to what we had observed in L1 larvae. However, in one embryo, we could observe irregular thick and thin regions of the duct lumen downstream of the dilation (Figure 3, I and J). We posit that this may be a transient state at the start of lumen distortion and that dilations, once appeared, rapidly and severely distorted the lumen. This phenotype is similar to that seen in let-653 mutants, where the duct and canal lumens develop dilations concurrent with downstream luminal discontinuities (Gill et al. 2016), suggesting that dilations are caused by tube content accumulation upstream of the block. We conclude that, like let-653, dex-1 promotes normal duct and pore lumen diameter during tube morphogenesis.
Unlike let-653, dex-1 was expressed in the canal cell (Heiman and Shaham 2009; Gill et al. 2016; Figure 2, D and E). To test if the canal had duct-independent phenotypes in dex-1 mutants, we analyzed newly hatched L1s that did not have duct luminal dilations using the cytoplasmic marker inx-13pro::GFP or the apical membrane marker RDY-2::GFP (Figure S5). While the canal lumen took a tortuous path (potentially due to the shortened and widened body length—see below), canal outgrowth remained proportional to the body and there were no additional distortions in diameter (Figure S5). In contrast, we observed that animals with duct luminal defects had canal cells with large dilations and very short, severely distorted arms. In summary, we found no evidence that DEX-1 influences canal lumen diameter independently of its effect on the duct and pore lumen.
dex-1 and let-653 mutants have different excretory junction morphologies
We noticed that, despite the similarity in their duct lumen phenotypes, dex-1 and let-653 had different excretory junction morphologies. In WT, an autocellular junction holds the pore in the shape of a tube, and ring-shaped junctions link the different unicellular tubes together (Figure 3B) (Sundaram and Buechner 2016). Both types of junctions were altered in the mutants.
We used the epithelial junction marker AJM-1::GFP to measure the length of the junctions in newly hatched let-653 and dex-1 mutant L1 larvae. dex-1 mutants with excretory dilations rarely had any pore autocellular junction, but the ring junction linking the duct and pore sometimes remained (Figure 3, E, K, and O), suggesting that the pore autocellular junction had never stably formed or had ruptured. The few remaining pore autocellular junctions were of normal length (Figure 3Q). In contrast, most newly hatched let-653 mutants with excretory dilations still had the pore autocellular junction, but this junction was often elongated, occasionally to as much as four times the normal length (Figure 3, K, M, and Q).
dex-1 mutants often had severe distortions in the secretory junction, the complex ring junction that links the duct, canal, and gland cells. In dex-1 mutants with lumen dilations, secretory junctions were widened to up to double the diameter of those seen in wild type (Figure 3, L, O, P, and R). Similar distortions were observed in dex-1(ns42); dyf-7(ns117) mutants (Figure S4), which were previously described to exhibit a synthetic excretory dilation phenotype (Heiman and Shaham 2009). We never saw loss of this junction. In contrast, we saw only slightly widened secretory junctions in let-653 mutants, despite equally severe lumen dilations (Figure 3, L, N, and R).
We propose that luminal dilations impose stress on junctions, and that the different junction phenotypes seen in dex-1 and let-653 mutants reflect these genes’ distinct roles in resisting that stress. As dex-1 loss of function led to more severe defects in both the pore autocellular junction and the secretory junction, DEX-1 may act with additional matrix proteins, including DYF-7, and therefore have a bigger role than LET-653 in resisting circumferential expansive forces.
dex-1 mutants have a Pin (pharynx ingressed) phenotype resembling that of fbn-1 mutants
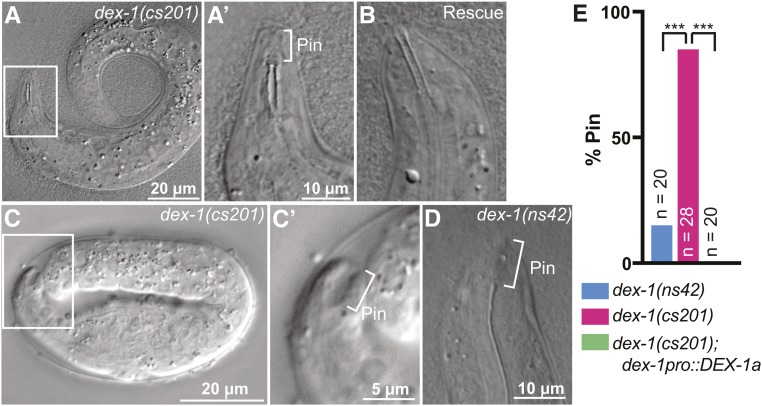
As less than a quarter of dex-1(cs201) mutants had obvious excretory defects, we inferred that the majority arrested for some other reason(s). To determine the cause of dex-1 mutants’ high larval lethality rate, we examined newly hatched larvae via DIC microscopy. A high proportion of newly hatched dex-1(cs201) mutants had ingressed pharynxes (Pin phenotype), which occluded the pharyngeal opening to prevent feeding (Figure 4, A and E). The dex-1pro::DEX-1 transgene efficiently rescued this Pin phenotype (Figure 4, B and E). We could detect a Pin phenotype starting at the twofold stage of embryonic elongation, as has been reported for fbn-1 mutants, which fail to maintain the pharyngeal-epidermal attachment under tension from embryonic elongation (Kelley et al. 2015) (Figure 4C). Some dex-1(ns42) mutants also had a Pin defect (Figure 4, D and E). We conclude that, like fbn-1 and dyf-7, dex-1 helps anchor the pharynx to the nose tip.
Figure 4.
dex-1 mutants are Pin (pharynx ingressed). (A) dex-1(cs201) L1 larva. Box indicates region that will be magnified in subsequent panel. (A’) The pharynx opening was posterior to the nose tip (bracket) and presumably did not open to the outside environment. (B) dex-1 cDNA efficiently rescued the pharyngeal phenotype. (C) twofold dex-1 mutant embryo. (C’) The pharynx was ingressed (bracket) but attached to the nose tip epithelium, leaving a large “keyhole” indicative of a true Pin phenotype (Kelley et al. 2015). (D) The Pin phenotype was also observed in dex-1(ns42) mutants. (E) Quantification of frequency of Pin phenotype in newly hatched L1 larvae. *** P < 0.0001, Fisher’s exact test.
dex-1 mutants have body shape and alae defects resembling cut mutants
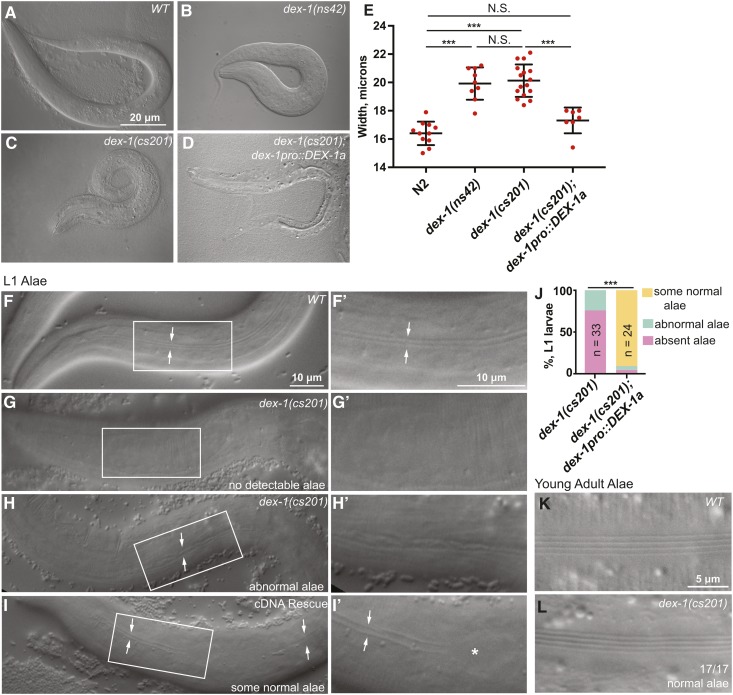
We also noticed that dex-1 mutant L1 larvae were short and fat (Dpy). C. elegans embryos elongate from a round ball of cells into a long, narrow cylinder. Failure to create a proper aECM can result in failure to fully elongate or to maintain body elongation (Labouesse 2012; Vuong-Brender et al. 2017). To measure the extent of the dex-1 Dpy phenotype, we used ImageJ to measure the length and width of newly hatched L1 larvae. Both ns42 and cs201 mutants are Dpy, and this phenotype was rescued by the dex-1pro::DEX-1 transgene (Figure 5, A–E and Figure S5). We conclude that, like multiple ZP proteins (Table 1), DEX-1 promotes normal body dimensions.
Figure 5.
dex-1 mutants have L1 cuticle defects. (A–D) dex-1 mutants were visibly short and fat (Dpy). DIC images of newly hatched L1 larvae. (E) Quantification of hatchling width. Measurements were performed by drawing a line across the animal’s width at the region of the pharynx terminal bulb and recording line length in ImageJ. All measurements were taken three times and then averaged to find a final, recorded value. *** P < 0.0001, Mann-Whitney U-test. (F–I) DIC images of alae in newly hatched L1 larvae. (F and F’) WT L1 larvae had distinct alae, visible as two parallel ridges (arrows). Box indicates region magnified in subsequent panel. (G–H’) dex-1(cs201) L1 alae were deformed or absent. (I and I’) dex-1pro::DEX-1a transgene used to rescue lethality (see Figure 2D) rescued alae variably, with regions where alae were normal (arrows) and regions where they were missing (*). Most L1 larvae had at least some regions with normal alae. (J) Quantification of alae phenotypes. *** P < 0.0001, Freeman-Halton Fisher’s exact test. (K and L) DIC images of newly molted adults. 17/17 dex-1(cs201) mutants had normal adult alae.
Table 1. Summary of shared dex-1 and ZP mutant phenotypes.
| dex-1 mutant phenotype | ZP mutants with this phenotype | References |
|---|---|---|
| Dendrite extension defect (Dex) | dyf-7 | Heiman and Shaham 2009 |
| Excretory tube defects (Exc) | let-653, | Gill et al. 2016; |
| dyf-7 (synthetic with dex-1) | Heiman and Shaham 2009; this study | |
| Pharynx ingressed (Pin) | fbn-1, | Kelley et al. 2015; |
| dyf-7 | This study | |
| L1 Dpy/alae defects | cut-3, cut-5 | Sapio et al. 2005; |
| This study | ||
| Dauer Dpy/alae defects | cut-6, | Muriel et al. 2003; |
| cut-1, cut-5 | Sapio et al. 2005; | |
| Flatt et al., 2019 |
All known ZP proteins that promote proper body shape (CUT-1, CUT-3, CUT-5, and CUT-6) also influence the development of alae, ridged cuticular structures that line L1 larvae, dauers, and adults (Table S3) (Sebastiano et al. 1991; Muriel et al. 2003; Sapio et al. 2005). To test if dex-1 mutants had alae defects, we performed DIC microscopy of newly hatched L1 larvae. Most dex-1 mutants lacked L1 alae, and, when alae were present, they appeared flattened or misshapen (Figure 5, F–J). Alae defects were partially rescued by the dex-1pro::DEX-1 transgene that had rescued lethality, but rescue was quite variable along the length of the animal (Figure 5, I and J). In contrast to the high penetrance of defects in L1 alae, dex-1 mutants that survived to adulthood developed normal adult alae (Figure 5, K and L). We conclude that dex-1 is required to build normal L1 alae but is dispensable for building the adult cuticle, consistent with dex-1 being primarily expressed in embryos (Figure 2). In the accompanying paper, we show that dex-1 is also required to build alae in the dauer larva, and that it genetically interacts with the cuticlin cut-5 (Flatt et al., 2019).
In summary, dex-1 mutants have a variety of phenotypes (Exc, Pin, Dpy, alae defects) that resemble those of ZP mutants (Table 1), suggesting that DEX-1 could be a component of the embryonic precuticle.
DEX-1 localizes to apical surfaces of embryos and larvae
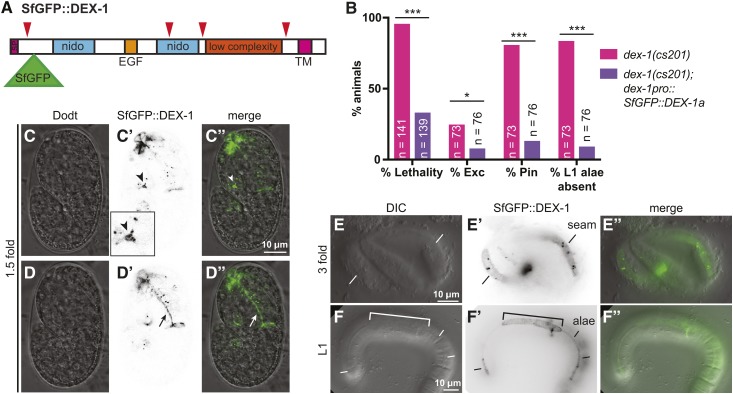
To determine where DEX-1 localizes, we generated a tagged version of full-length DEX-1 isoform a. Previous attempts to visualize DEX-1 have relied on constructs that tagged DEX-1 at its cytoplasmic C-terminus (Heiman and Shaham 2009). However, like other matrix proteins, DEX-1 may be cleaved to release its functional ectodomain. Indeed, ProP (Duckert et al. 2004) predicted several potential sites of cleavage by a furin-type proprotein convertase (Figure 6A), all of which would release an extracellular peptide into the ECM. We therefore propose that localization of an N-terminally tagged DEX-1 should better reflect the places where DEX-1 is likely to function. We inserted Superfolder GFP (SfGFP), a GFP variant that is stable in oxidative, extracellular environments (Pédelacq et al. 2006; Aronson et al. 2011), into DEX-1 after the signal peptide to generate SfGFP::DEX-1 (Figure 6A).
Figure 6.
DEX-1 localizes to the aECM. (A) Model of SfGFP::DEX-1a protein showing position of SfGFP insertion after the signal peptide. Red arrowheads indicate potential cleavage sites predicted by ProP (Duckert et al. 2004). SfGFP was inserted downstream of the first predicted cleavage site in order to tag the portion of DEX-1 containing the nido and EGF domains. (B) SfGFP::DEX-1a rescued dex-1(cs201) lethality, excretory, Pin, and alae defects. *** P < 0.0001, * P < 0.01. Fisher’s exact test. (C–D”) In WT 1.5-fold embryos, SfGFP::DEX-1a localized to apical surfaces. Single Z-slices from a confocal Z-stack. (C–C”) DEX-1 was present in the duct/pore lumen (arrowhead) and particularly strong near the duct-canal junction (arrowhead). This region is enlarged in the inset. (D–D”) DEX-1 lines the pharyngeal lumen (arrow). (E–E”) threefold embryo. DEX-1 lines a strip of seam epithelium. (F–F”) In L1 larvae, DEX-1 lines alae. Bracket indicates a region of missing or abnormal alae, suggesting dominant negative effects related to inappropriately high expression level. All images shown are of WT animals expressing SfGFP::DEX-1a.
SfGFP::DEX-1 rescued dex-1(cs201) lethality, Pin, excretory, and L1 alae defects (Figure 6B). This fusion protein was first observed around the time of ventral enclosure. In 1.5-fold embryos, it accumulated in the duct and pore lumen, along the nose tip, and along the apical surface of the developing pharynx (Figure 6, C and D). SfGFP::DEX-1 was not observed along the dorsal or ventral epidermis, but later, in threefold embryos, it lined the apical surface of the seam cells, where alae would later appear (Figure 6E). After hatch, SfGFP::DEX-1 was rarely seen in the duct and pore lumen or the nose tip, but prominently lined alae of L1 larvae. However, high levels of SfGFP::DEX-1 sometimes caused flattened or absent alae in a WT background, suggesting a dominant negative effect (Figure 6F; Flatt et al., 2019). Correspondingly, this construct showed variable rescue of dex-1 mutant alae defects, with regions of good rescue corresponding to modest levels of SfGFP::DEX-1, suggesting that expression level and stoichiometry are very important. We conclude that SfGFP::DEX-1 is present along apical surfaces during embryonic and L1 stages, consistent with DEX-1 being part of the precuticular aECM of many external epithelia, as well as part of the special cuticular aECM of alae.
DEX-1 can act via its ecto domain in some, but not all, tissues
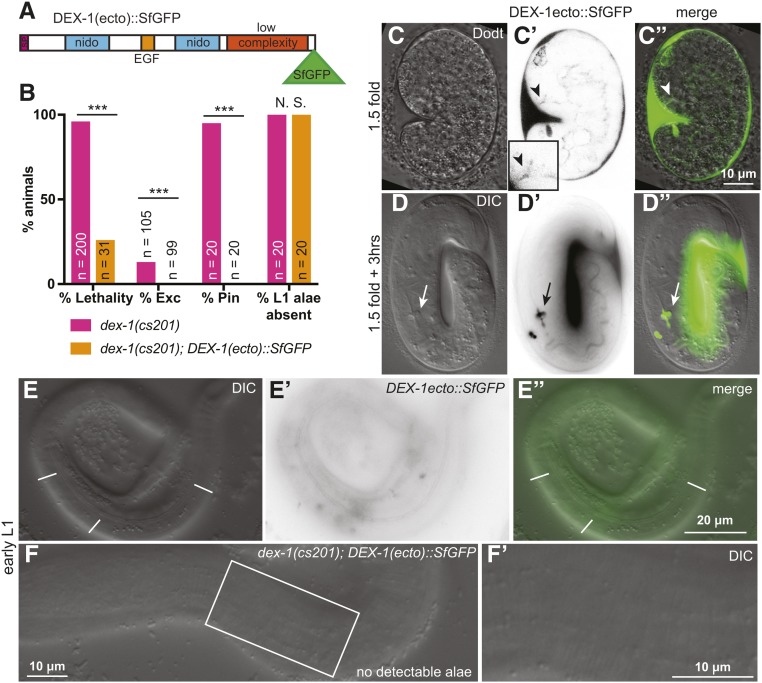
Previously, a version of DEX-1 that lacked its transmembrane and cytoplasmic domains, mimicking the predicted DEX-1 cleavage product, was shown to retain some function in anchoring dendrites at the nose tip (Heiman and Shaham 2009). To test whether the ecto domain could localize properly and function for its other roles, we generated a construct containing the ectodomain tagged with SfGFP [DEX-1(ecto)::SfGFP] (Figure 7A). DEX-1(ecto)::SfGFP rescued dex-1(cs201) lethality, Pin, and excretory phenotypes, but failed to rescue the L1 alae defects (Figure 7, B and F). Accordingly, in embryos, DEX-1(ecto)::SfGFP localized strongly to the nose tip, pharyngeal lumen, and developing duct and pore lumen, but failed to attach to the seam epidermis and rather appeared to fill the space between the embryo and the eggshell (Figure 7, C and D). Unlike full-length DEX-1, DEX-1(ecto)::SfGFP accumulated strongly in the gut and at the developing pharyngeal grinder, a chitinous “tooth” that breaks down bacteria (Zhang et al. 2005; George-Raizen et al. 2014) (Figure 7D). After hatch, DEX-1(ecto)::SfGFP was diffusely associated with the cuticle but was not localized to the alae, unlike full-length DEX-1 (Figure 7E). We conclude that the DEX-1 ecto domain is sufficient for function in some tissues, but that DEX-1 requires its transmembrane domain, short cytoplasmic tail, or possibly a region near the SfGFP tag, to anchor along developing L1 alae.
Figure 7.
DEX-1(ecto) retains some, but not all, functions. (A) Model of truncated DEX-1 protein showing position of SfGFP insertion after the low complexity domain, at the position of one predicted cleavage site. The truncation matches that generated in Heiman and Shaham (2009). (B) DEX-1(ecto)::SfGFP rescued dex-1(cs201) lethality, Pin, and Exc phenotypes, but did not rescue dex-1(cs201) alae defects. *** P < 0.0001, Fisher’s Exact Test. (C–D”) In 1.5-fold embryos, DEX-1(ecto)::SfGFP localized to apical surfaces. Single Z-slices from a confocal Z-stack. (C–C”) DEX-1 was present in the duct/pore lumen (arrowhead). (D–D”) threefold embryo. DEX-1(ecto)::SfGFP accumulated between the embryo and the eggshell and within the gut lumen but was not visible at the epidermal surface. (E–E”) In L1 larvae, DEX-1(ecto)::SfGFP showed diffuse signal over the entire the cuticle region but did not specifically label alae above background. (F and F’) DEX-1(ecto)::SfGFP failed to rescue dex-1(cs201) alae defects.
dex-1 is not required for LET-653(ZP) secretion or accumulation
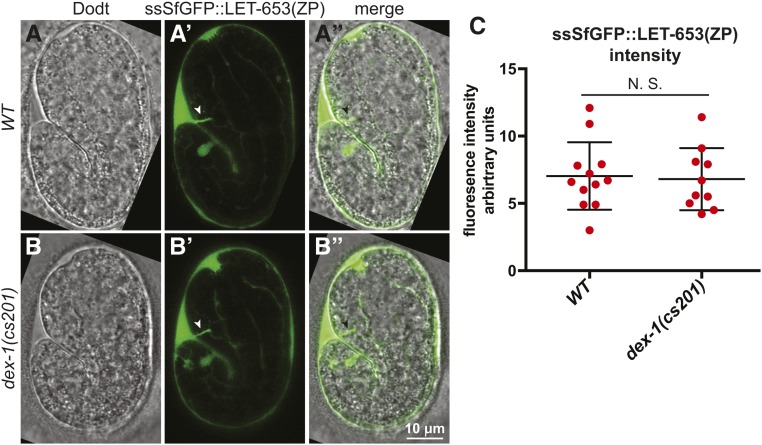
As DEX-1 localizes to the same aECMs as many ZP proteins, we hypothesized that DEX-1 could promote ZP protein aECM integration. To test whether DEX-1 impacted ZP protein secretion or accumulation, we expressed a functional, tagged LET-653 ZP domain (ssSfGFP::LET-653(ZP)) (Gill et al. 2016) in dex-1 mutants. In 1.5-fold embryos, ssSfGFP::LET-653(ZP) localized normally to the duct/pore lumen, the nose tip, and the extra-embryonic space (Figure 8, A and B). To confirm that LET-653(ZP) accumulated to normal levels, we measured the total fluorescence intensity of ssSfGFP::LET-653(ZP) across the entire embryo in a set of 10 confocal Z-slices through the middle of the embryo. We observed no difference in fluorescence intensity between WT and dex-1 mutants (Figure 8C). To test whether dex-1 promotes LET-653(ZP) clearance, we monitored embryos after 1.5-fold stage. In both WT and dex-1(cs201) embryos, LET-653(ZP) was efficiently cleared from the duct/pore lumen by 5 hr after 1.5-fold stage (WT: n = 50; dex-1: n = 20). We conclude that dex-1 does not influence LET-653 secretion, localization, or clearance.
Figure 8.
dex-1 is not required for LET-653 localization or accumulation. (A–B”) Single confocal slices of WT and dex-1(cs201) mutant 1.5-fold embryos expressing ssSfGFP::LET-653(ZP) (Gill et al. 2016), which localized to the duct/pore lumen, nose tip, and extra-embryonic space. (C) Quantification of total ssSfGFP::LET-653(ZP) fluorescence taken from 10 consecutive Z-slices through the embryo center. N.S., no significant difference, Mann Whitney U-test.
Discussion
aECMs shape both planar and tubular epithelia during development. Mutation or environmental disruption of aECMs is associated with a variety of human diseases (Rampoldi et al. 2011; Richardson et al. 2011; Johansson et al. 2013; Saito et al. 2017). Despite their importance, relatively few aECM components have been described. One class of key aECM proteins are ZP domain proteins. This study, along with an accompanying study (K.M.F. et al., unpublished results), shows that the nidogen- and EGF-domain protein DEX-1 is an essential aECM component that acts in concert with many different ZP proteins to shape epithelial tissues in C. elegans.
Modular design of ZP-containing aECMs in C. elegans
Our survey of C. elegans ZP proteins revealed a large number and sequence diversity of ZP proteins. We identified 43 genes encoding 65 ZP proteins, more than double the number found in humans (Bokhove and Jovine 2018). This large number of ZP proteins is not typical of Nematoda or Ecdysozoa, as some nematodes and many insects, including Drosophila, contain similar numbers of ZP proteins as humans (Fernandes et al. 2010; UniProt Consortium T 2018). So far, only 11 C. elegans ZP proteins have been studied, all of which are required for epithelial or matrix shaping (Table S3). However, many genes encoding ZP proteins show the cyclic expression characteristic of cuticle and precuticle genes (Hendriks et al. 2014), and may encode additional cuticle or precuticle components.
Different sets of ZP proteins are found in different C. elegans aECMs. Previous work suggested that the morphologically distinct alae of L1 larvae, dauer larvae, and adults contain partially overlapping but distinct sets of CUT proteins (Sebastiano et al. 1991; Muriel et al. 2003; Sapio et al. 2005) (Table S3). Similarly, we showed that different subsets of ZP proteins are found within different regions of the embryonic precuticle, which is in fact a collection of distinct matrices. For example, LET-653, FBN-1, NOAH-1, and NOAH-2 are all found in or near the epidermal sheath, but, of these, only LET-653 is also visible in the excretory duct and pore lumen. LET-653, FBN-1, and DYF-7, but not NOAH-1 or NOAH-2, are abundant at the nose tip. None of these four proteins are in the precuticle of the pharyngeal lumen, which later develops a chitinous cuticle that is very different from the collagenous cuticle of the epidermis (Zhang et al. 2005). The different localization patterns of these various ZP proteins could, in part, explain the rather different phenotypes that result from mutations in the corresponding genes.
Different ZP components likely confer different physical properties to aECMs, which are specialized for the types of stresses each tissue experiences. For example, embryonic elongation puts the pharyngeal-epidermal connection, the glial-epidermal connection, and the epidermis under tension that ZP proteins (FBN-1, DYF-7, and NOAH-1 and -2, respectively) resist (Heiman and Shaham 2009; Kelley et al. 2015; Vuong-Brender et al. 2017; Low et al. 2018). The duct/pore lumen is likely under stress from fluid flow as well as from morphogenetic forces associated with duct lumen elongation, which may exert both circumferential and longitudinal pressures that must be countered by LET-653 to maintain uniform lumen diameter (Gill et al. 2016). Meanwhile, CUT proteins are thought to mediate apical constriction to narrow the worm’s body and to create the buckled cuticle that is alae (Sapio et al. 2005). C. elegans may have so many ZP proteins because it needs different types of aECM in different body regions and at different developmental stages.
DEX-1 is a component of multiple aECMs and shapes epithelia
Heiman and Shaham (2009) previously characterized dex-1 for its dendrite-anchoring function. The present study, along with the accompanying study (Flatt et al., 2019), demonstrates that dex-1 is also required to shape many epithelial tissues, including the pharynx, excretory system, and epidermis. We showed that DEX-1 localizes to diverse ZP-containing precuticle matrices along apical surfaces of the tissues whose shaping requires dex-1. In the accompanying manuscript (Flatt et al., 2019), we showed that DEX-1 also localizes to apical surfaces of dauer seam cells and acts cell-autonomously to shape dauer alae. Although we did not specifically test cell autonomy in other stages or tissues, the data from (Flatt et al., 2019) and from (Heiman and Shaham 2009) suggest that DEX-1 incorporates into aECMs near its site of synthesis to shape tissues locally.
Consistent with the modular nature of C. elegans aECMs, DEX-1 is a broad but not ubiquitous aECM component. For example, DEX-1 is not present in the epidermal sheath, and dex-1 expression is confined to embryonic and dauer stages (this study and Flatt et al., 2019). Together with the incomplete penetrance of most dex-1 mutant phenotypes, these data indicate that aECMs can sometimes function without DEX-1. Other proteins may compensate for the absence of dex-1 at later stages. Alternatively, the rapid and extensive tissue remodeling that occurs during embryonic and dauer stages may create a special requirement for DEX-1.
DEX-1 domains and mammalian relatives
DEX-1 contains nido, EGF, and low complexity domains, all of which are potential interaction domains found in other matrix proteins. The low complexity region of DEX-1 has been called zonadhesin-like and is proposed to mediate ZP interactions (Heiman and Shaham 2009) by analogy to mammalian zonadhesin—a sperm protein that binds to the oocyte’s ZP-rich ZP (Tardif and Cormier 2011). However, any similarity of DEX-1 to zonadhesin is quite subtle, such that we could not detect it, and our data comparing dex-1(cs201) to dex-1(ns42) suggest that the nido and/or EGF domains may be more critical determinants of DEX-1 function.
Mammalian proteins with nido and EGF domains include the basement membrane protein nidogen (Ho et al. 2008), the inner ear tectorial membrane component alpha-tectorin (Legan et al. 1997), the putative metastasis factor SNED1 (Naba et al. 2014), and the large mucin MUC4 (Senapati et al. 2011). Each of these proteins also contains additional domains, however, and the functions of their nido domains remain unclear. Here, and in the accompanying manuscript (Flatt et al., 2019), we showed that a secreted form of DEX-1 can bind to diverse aECMs in C. elegans; this should be an excellent system in which to test the requirements for nido domains and ultimately identify specific binding partners and functions of this mysterious domain.
One surprising result from our current analysis is that the secreted form of DEX-1 could not properly localize to or shape L1 alae, whereas it retained all other tested functions, including its function in dauer alae (Flatt et al., 2019). These data suggest that DEX-1 has unique partners or modes of regulation in different tissues, consistent with the concept of modular aECMs.
What is the relationship between DEX-1 and ZP proteins?
dex-1 mutants share a variety of tissue distortion phenotypes with previously described ZP mutants. Heiman and Shaham (2009) initially reported that dex-1 promotes dendrite anchoring to the nose tip alongside the ZP protein DYF-7. We showed that dex-1 mutant excretory tube distortions strongly resemble those of let-653 mutants, and pharyngeal tube distortions resemble those of fbn-1 and dyf-7 mutants. Furthermore, dex-1 mutants also have the short and fat body shape and missing alae of many cut mutants (this study and Flatt et al., 2019). Consistent with these pleiotropic requirements, DEX-1 and these ZP proteins localize to many of the same apical surfaces. Together, these similarities in both phenotype and localization suggest that dex-1 functions in concert with many different ZP proteins.
We cannot yet distinguish between models where DEX-1 functions as a direct partner of ZP proteins or simply as another component of complex ZP-containing aECMs. DEX-1 domain structure suggests that it might be a direct ZP binding partner, as first proposed by Heiman and Shaham (2009); however, direct interactions have not yet been demonstrated, and using LET-653(ZP) as a test case, we did not find any evidence that DEX-1 promotes ZP protein secretion, localization, or clearance. Furthermore, aECMs are complex and contain multiple discreet layers (Chappell et al. 2009; Johansson et al. 2013; Gill et al. 2016), and it has not yet been possible to distinguish whether DEX-1 and ZP proteins are in the same layer(s). It is also important to note that dex-1 phenotypes were often less penetrant than those of ZP mutants, indicating that ZP proteins must have at least some DEX-1-independent functions.
Our study also shows that, despite substantial overlap, not all ZP-containing matrices also include DEX-1, and not all DEX-1-containing matrices necessarily include ZP proteins. For example, DEX-1 was observed in the precuticle aECM along the pharyngeal lumen, where no ZP proteins have (thus far) been reported. Therefore, DEX-1 could contribute to distinct aECMs through interactions with different types of matrix factors.
Concluding statement
This study (and that of Flatt et al., 2019) now add DEX-1 to the growing list of epithelial-shaping aECM components in C. elegans, along with various collagens (Page and Johnstone 2007), ZP proteins (Table S3), eLRRon proteins (Mancuso et al. 2012), lipocalins (Forman-Rubinsky et al. 2017), and unknown chondroitin proteoglycans (Hwang et al. 2003). Further studies of these factors should identify which are direct vs. indirect partners and elucidate the mechanisms by which different matrix factors assemble into a complex and layered aECM with the appropriate physical properties to shape cells and tissues.
Acknowledgments
We are grateful to Max Heiman, Michel Labouesse, and Alison Frand for providing strains and reagents, and to Max Heiman and Alison Frand for generously sharing unpublished data and advice. We thank Andrea Stout and the UPenn CDB Microscopy Core for training and assistance with confocal microscopy. Some strains were obtained from the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the NIH (grant R01GM058540 to M.V.S., R01GM111566 to N.E.S., T32GM008216 to J.D.C.).
Note added in proof: See Flatt et al. 2019 (pp. 169–183) in this issue for a related work.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7312148.
Communicating editor: D. Greenstein
Literature Cited
- Abdus-Saboor I., Mancuso V. P., Murray J. I., Palozola K., Norris C., et al. , 2011. Notch and Ras promote sequential steps of excretory tube development in C. elegans. Development 138: 3545–3555. 10.1242/dev.068148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun Z. F., Chen B., Wang Z. W., Hall D. H., 2009. High resolution map of Caenorhabditis elegans gap junction proteins. Dev. Dyn. 238: 1936–1950. 10.1002/dvdy.22025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appella E., Weber I. T., Blasi F., 1988. Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett. 231: 1–4. 10.1016/0014-5793(88)80690-2 [DOI] [PubMed] [Google Scholar]
- Aronson D. E., Costantini L. M., Snapp E. L., 2011. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic 12: 543–548. 10.1111/j.1600-0854.2011.01168.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhove M., Jovine L., 2018. Structure of zona pellucida module proteins. Curr. Top. Dev. Biol. 130: 413–442. 10.1016/bs.ctdb.2018.02.007 [DOI] [PubMed] [Google Scholar]
- Bokhove M., Nishimura K., Brunati M., Han L., de Sanctis D., et al. , 2016. A structured interdomain linker directs self-polymerization of human uromodulin. Proc. Natl. Acad. Sci. USA 113: 1552–1557. 10.1073/pnas.1519803113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick J., Walton T., Preston E., Zacharias A., Raj A., et al. , 2016. Overlapping cell population expression profiling and regulatory inference in C. elegans. BMC Genomics 17: 159 10.1186/s12864-016-2482-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Packer J. S., Ramani V., Cusanovich D. A., Huynh C., et al. , 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357: 661–667. 10.1126/science.aam8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Dillon L. A., Gerstein M. B., Gunsalus K. C., Henikoff S., et al. , 2009. Unlocking the secrets of the genome. Nature 459: 927–930. 10.1038/459927a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell D., Jacob M., Paul O., Rehm M., Welsch U., et al. , 2009. The glycocalyx of the human umbilical vein endothelial cell: an impressive structure ex vivo but not in culture. Circ. Res. 104: 1313–1317. 10.1161/CIRCRESAHA.108.187831 [DOI] [PubMed] [Google Scholar]
- Chen S., Birk D. E., 2013. The regulatory roles of small leucine-rich proteoglycans in extracellular matrix assembly. FEBS J. 280: 2120–2137. 10.1111/febs.12136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y. D., Zhu J., Han Y., Kernan M. J., 2001. nompA encodes a PNS-specific, ZP domain protein required to connect mechanosensory dendrites to sensory structures. Neuron 29: 415–428. 10.1016/S0896-6273(01)00215-X [DOI] [PubMed] [Google Scholar]
- Costa M., Draper B. W., Priess J. R., 1997. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev. Biol. 184: 373–384. 10.1006/dbio.1997.8530 [DOI] [PubMed] [Google Scholar]
- Davis C. G., 1990. The many faces of epidermal growth factor repeats. New Biol. 2: 410–419. [PubMed] [Google Scholar]
- Dodt H. U., Zieglgänsberger W., 1990. Visualizing unstained neurons in living brain slices by infrared DIC-videomicroscopy. Brain Res. 537: 333–336. 10.1016/0006-8993(90)90380-T [DOI] [PubMed] [Google Scholar]
- Duckert P., Brunak S., Blom N., 2004. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 17: 107–112. 10.1093/protein/gzh013 [DOI] [PubMed] [Google Scholar]
- Feingold K. R., 2007. Thematic review series: skin lipids. The role of epidermal lipids in cutaneous permeability barrier homeostasis. J. Lipid Res. 48: 2531–2546. 10.1194/jlr.R700013-JLR200 [DOI] [PubMed] [Google Scholar]
- Fernandes I., Chanut-Delalande H., Ferrer P., Latapie Y., Waltzer L., et al. , 2010. Zona pellucida domain proteins remodel the apical compartment for localized cell shape changes. Dev. Cell 18: 64–76. 10.1016/j.devcel.2009.11.009 [DOI] [PubMed] [Google Scholar]
- Flatt K. M., Cohen J. D., Sundaram M. V., Schroeder N. E., 2018. Epidermal remodeling in Caenorhabditis elegans dauers requires the nidogen domain protein DEX-1. Genetics 211: 169–183. 10.1534/genetics.118.301557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman-Rubinsky R., Cohen J. D., Sundaram M. V., 2017. Lipocalins are required for apical extracellular matrix organization and remodeling in Caenorhabditis elegans. Genetics 207: 625–642. 10.1534/genetics.117.300207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George-Raizen J. B., Shockley K. R., Trojanowski N. F., Lamb A. L., Raizen D. M., 2014. Dynamically-expressed prion-like proteins form a cuticle in the pharynx of Caenorhabditis elegans. Biol. Open 3: 1139–1149. 10.1242/bio.20147500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill H. K., Cohen J. D., Ayala-Figueroa J., Forman-Rubinsky R., Poggioli C., et al. , 2016. Integrity of narrow epithelial tubes in the C. elegans excretory system requires a transient luminal matrix. PLoS Genet. 12: e1006205 10.1371/journal.pgen.1006205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K., 2018. The human egg’s zona pellucida. Curr. Top. Dev. Biol. 130: 379–411. 10.1016/bs.ctdb.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Heiman M. G., Shaham S., 2009. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell 137: 344–355. 10.1016/j.cell.2009.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks G. J., Gaidatzis D., Aeschimann F., Grosshans H., 2014. Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell 53: 380–392. 10.1016/j.molcel.2013.12.013 [DOI] [PubMed] [Google Scholar]
- Ho M. S., Bose K., Mokkapati S., Nischt R., Smyth N., 2008. Nidogens-extracellular matrix linker molecules. Microsc. Res. Tech. 71: 387–395. 10.1002/jemt.20567 [DOI] [PubMed] [Google Scholar]
- Hwang H. Y., Olson S. K., Esko J. D., Horvitz H. R., 2003. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature 423: 439–443. 10.1038/nature01634 [DOI] [PubMed] [Google Scholar]
- Johansson M. E., Sjovall H., Hansson G. C., 2013. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 10: 352–361. 10.1038/nrgastro.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovine L., Qi H., Williams Z., Litscher E., Wassarman P. M., 2002. The ZP domain is a conserved module for polymerization of extracellular proteins. Nat. Cell Biol. 4: 457–461. 10.1038/ncb802 [DOI] [PubMed] [Google Scholar]
- Katz S. S., Maybrun C., Frand A. R., 2018. Non-canonical apical constriction shapes emergent matrices in C. elegans. bioRxiv. 10.1101/189951 [DOI] [Google Scholar]
- Kelley M., Yochem J., Krieg M., Calixto A., Heiman M. G., et al. , 2015. FBN-1, a fibrillin-related protein, is required for resistance of the epidermis to mechanical deformation during C. elegans embryogenesis. eLife 4 10.7554/eLife.06565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köppen M., Simske J. S., Sims P. A., Firestein B. L., Hall D. H., et al. , 2001. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat. Cell Biol. 3: 983–991. 10.1038/ncb1101-983 [DOI] [PubMed] [Google Scholar]
- Labouesse M., 2012. Role of the extracellular matrix in epithelial morphogenesis: a view from C. elegans. Organogenesis 8: 65–70. 10.4161/org.20261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Y. N., Howe K. L., Harris T. W., Arnaboldi V., Cain S., et al. , 2018. WormBase 2017: molting into a new stage. Nucleic Acids Res. 46: D869–D874. 10.1093/nar/gkx998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legan P. K., Rau A., Keen J. N., Richardson G. P., 1997. The mouse tectorins. Modular matrix proteins of the inner ear homologous to components of the sperm-egg adhesion system. J. Biol. Chem. 272: 8791–8801. 10.1074/jbc.272.13.8791 [DOI] [PubMed] [Google Scholar]
- Lin S. J., Hu Y., Zhu J., Woodruff T. K., Jardetzky T. S., 2011. Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-beta binding. Proc. Natl. Acad. Sci. USA 108: 5232–5236. 10.1073/pnas.1010689108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low I. I. C., Williams C. R., Chong M. K., McLachlan I. G., Wierbowski B. M., et al. , 2018. Morphogenesis of neurons and glia within an epithelium. bioRxiv. 10.1101/393850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig S., Uv A., 2014. Luminal matrices: an inside view on organ morphogenesis. Exp. Cell Res. 321: 64–70. 10.1016/j.yexcr.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Mancuso V. P., Parry J. M., Storer L., Poggioli C., Nguyen K. C., et al. , 2012. Extracellular leucine-rich repeat proteins are required to organize the apical extracellular matrix and maintain epithelial junction integrity in C. elegans. Development 139: 979–990. 10.1242/dev.075135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., 2012. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192: 1249–1269. 10.1534/genetics.112.144204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti S., Armougom F., Wallace I. M., Higgins D. G., Jongeneel C. V., et al. , 2007. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 35: W645–W648. 10.1093/nar/gkm333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel J. M., Brannan M., Taylor K., Johnstone I. L., Lithgow G. J., et al. , 2003. M142.2 (cut-6), a novel Caenorhabditis elegans matrix gene important for dauer body shape. Dev. Biol. 260: 339–351. 10.1016/S0012-1606(03)00237-9 [DOI] [PubMed] [Google Scholar]
- Naba A., Clauser K. R., Lamar J. M., Carr S. A., Hynes R. O., 2014. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. eLife 3: e01308 10.7554/eLife.01308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson F. K., Riddle D. L., 1984. Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J. Exp. Zool. 231: 45–56. 10.1002/jez.1402310107 [DOI] [PubMed] [Google Scholar]
- Nelson F. K., Albert P. S., Riddle D. L., 1983. Fine structure of the Caenorhabditis elegans secretory-excretory system. J. Ultrastruct. Res. 82: 156–171. 10.1016/S0022-5320(83)90050-3 [DOI] [PubMed] [Google Scholar]
- Page A. P., Johnstone I. L., 2007. The cuticle. WormBook 19: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paragas N., Qiu A., Hollmen M., Nickolas T. L., Devarajan P., et al. , 2012. NGAL-Siderocalin in kidney disease. Biochim. Biophys. Acta 1823: 1451–1458. 10.1016/j.bbamcr.2012.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24: 79–88 (erratum: Nat. Biotechnol. 24: 1170) 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- Pérez-Gil J., 2008. Structure of pulmonary surfactant membranes and films: the role of proteins and lipid-protein interactions. Biochim. Biophys. Acta 1778: 1676–1695. 10.1016/j.bbamem.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Plaza S., Chanut-Delalande H., Fernandes I., Wassarman P. M., Payre F., 2010. From A to Z: apical structures and zona pellucida-domain proteins. Trends Cell Biol. 20: 524–532. 10.1016/j.tcb.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Priess J. R., Hirsh D. I., 1986. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev. Biol. 117: 156–173. 10.1016/0012-1606(86)90358-1 [DOI] [PubMed] [Google Scholar]
- Pu P., Stone C. E., Burdick J. T., Murray J. I., Sundaram M. V., 2017. The lipocalin LPR-1 cooperates with LIN-3/EGF signaling to maintain narrow tube integrity in Caenorhabditis elegans. Genetics 205: 1247–1260. 10.1534/genetics.116.195156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampoldi L., Scolari F., Amoroso A., Ghiggeri G., Devuyst O., 2011. The rediscovery of uromodulin (Tamm-Horsfall protein): from tubulointerstitial nephropathy to chronic kidney disease. Kidney Int. 80: 338–347. 10.1038/ki.2011.134 [DOI] [PubMed] [Google Scholar]
- Reitsma S., Slaaf D. W., Vink H., van Zandvoort M. A., oude Egbrink M. G., 2007. The endothelial glycocalyx: composition, functions, and visualization. Pflugers Arch. 454: 345–359. 10.1007/s00424-007-0212-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson G. P., de Monvel J. B., Petit C., 2011. How the genetics of deafness illuminates auditory physiology. Annu. Rev. Physiol. 73: 311–334. 10.1146/annurev-physiol-012110-142228 [DOI] [PubMed] [Google Scholar]
- Saito T., Bokhove M., Croci R., Zamora-Caballero S., Han L., et al. , 2017. Structural basis of the human endoglin-BMP9 interaction: insights into BMP signaling and HHT1. Cell Reports 19: 1917–1928. 10.1016/j.celrep.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapio M. R., Hilliard M. A., Cermola M., Favre R., Bazzicalupo P., 2005. The Zona Pellucida domain containing proteins, CUT-1, CUT-3 and CUT-5, play essential roles in the development of the larval alae in Caenorhabditis elegans. Dev. Biol. 282: 231–245. 10.1016/j.ydbio.2005.03.011 [DOI] [PubMed] [Google Scholar]
- Sebastiano M., Lassandro F., Bazzicalupo P., 1991. cut-1 a Caenorhabditis elegans gene coding for a dauer-specific noncollagenous component of the cuticle. Dev. Biol. 146: 519–530. 10.1016/0012-1606(91)90253-Y [DOI] [PubMed] [Google Scholar]
- Selander-Sunnerhagen M., Ullner M., Persson E., Teleman O., Stenflo J., et al. , 1992. How an epidermal growth factor (EGF)-like domain binds calcium. High resolution NMR structure of the calcium form of the NH2-terminal EGF-like domain in coagulation factor X. J. Biol. Chem. 267: 19642–19649. [DOI] [PubMed] [Google Scholar]
- Senapati S., Chaturvedi P., Chaney W. G., Chakraborty S., Gnanapragassam V. S., et al. , 2011. Novel INTeraction of MUC4 and galectin: potential pathobiological implications for metastasis in lethal pancreatic cancer. Clin. Cancer Res. 17: 267–274. 10.1158/1078-0432.CCR-10-1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M. V., Buechner M., 2016. The Caenorhabditis elegans excretory system: a model for tubulogenesis, cell fate specification, and plasticity. Genetics 203: 35–63. 10.1534/genetics.116.189357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbell J. M., Cancel L. M., 2016. The glycocalyx and its significance in human medicine. J. Intern. Med. 280: 97–113. 10.1111/joim.12465 [DOI] [PubMed] [Google Scholar]
- Tardif S., Cormier N., 2011. Role of zonadhesin during sperm-egg interaction: a species-specific acrosomal molecule with multiple functions. Mol. Hum. Reprod. 17: 661–668. 10.1093/molehr/gar039 [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Goumans M. J., Pardali E., 2008. Endoglin in angiogenesis and vascular diseases. Angiogenesis 11: 79–89. 10.1007/s10456-008-9101-9 [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium , 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46: 2699 10.1093/nar/gky092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong-Brender T. T. K., Suman S. K., Labouesse M., 2017. The apical ECM preserves embryonic integrity and distributes mechanical stress during morphogenesis. Development 144: 4336–4349. 10.1242/dev.150383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace I. M., O’Sullivan O., Higgins D. G., Notredame C., 2006. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 34: 1692–1699. 10.1093/nar/gkl091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Zhang Z. Z., Huang L. Y., Shen H. L., Han Z. G., 2012. OIT3 deficiency impairs uric acid reabsorption in renal tubule. FEBS Lett. 586: 760–765. 10.1016/j.febslet.2012.01.038 [DOI] [PubMed] [Google Scholar]
- Zaucke F., Boehnlein J. M., Steffens S., Polishchuk R. S., Rampoldi L., et al. , 2010. Uromodulin is expressed in renal primary cilia and UMOD mutations result in decreased ciliary uromodulin expression. Hum. Mol. Genet. 19: 1985–1997. 10.1093/hmg/ddq077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Foster J. M., Nelson L. S., Ma D., Carlow C. K., 2005. The chitin synthase genes chs-1 and chs-2 are essential for C. elegans development and responsible for chitin deposition in the eggshell and pharynx, respectively. Dev. Biol. 285: 330–339. 10.1016/j.ydbio.2005.06.037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and plasmids are available upon request. Supplemental data files are available at https://figshare.com/. Table S1 contains complete genotypes of all strains used. Table S2 contains GenBank and Uniprot accession numbers for all C. elegans ZP protein sequences. Table S3 contains summary information for 11 previously studied ZP proteins. Figure S1 contains a diagram of C. elegans ZP proteins sorted by ZPc cysteine number. Figure S2 contains multiple sequence alignments for all C. elegans ZP proteins. Figure S3 contains RT-PCR results for dex-1(cs201). Figure S4 contains images of dyf-7 mutants. Figure S5 contains images of dex-1 mutant excretory canals. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7312148.