Abstract
Background
Bovine respiratory disease complex (BRDC) is one of the most important sources of loss within the beef cattle industry in the USA. Steps have been taken to reduce the incidence of BRDC through vaccination. Despite the effectiveness of vaccines, large proportions of cattle still experience morbidity and mortality. Identification of genomic regions that are associated with variation in response to vaccination would allow for the selection of individuals genetically predisposed to respond to vaccination based on specific markers, while heritability and accuracy estimates would help facilitate genomic selection. This in turn may lead to selection for beef cattle herds that may have lower incidence rate of BRDC after vaccination. This study utilizes an Angus herd of more than 2000 head of cattle to identify these regions of association.
Results
Genome wide association studies were performed for viral neutralization antibody level and response to vaccination traits against four different viruses associated with BRDC: bovine viral diarrhea virus 1 and 2 (BVDV1 and BVDV2), bovine respiratory syncytial virus (BRSV), and bovine herpesvirus (BHV1). A total of six 1-Mb windows were associated with greater than 1% of the genetic variance for the analyzed vaccination response traits. Heritabilities ranged from 0.08 to 0.21 and prediction accuracy ranged from 0.01 to 0.33 across 7 different vaccination traits.
Conclusions
Although six 1-Mb windows were identified as associated with 1% or greater genetic variance for viral neutralization antibody level and response to vaccination traits, few genes around these windows could readily be considered candidates. This indicates the need for further functional genomic annotation, as these regions appear to be gene deserts. Traits ranged from lowly to moderately heritable, which indicated the potential for selection of individuals that are genetically pre-disposed to respond to vaccination. The relatively low amount of genetic variance accounted for by any 1-Mb window indicated that viral neutralization antibody level and response to vaccination traits are polygenic in nature. Selection for these traits is possible, but likely to be slow due to the low heritabilities and absence of markers with high genetic variation associated with them.
Keywords: Accuracy, Beef cattle, Bovine respiratory disease complex, Genome-wide association study, Heritability, Immune response, Vaccination
Background
Bovine respiratory disease complex (BRDC) is one of the most costly and pervasive disease conditions facing beef cattle producers. With more than $750 million dollars in losses each year due to morbidity, mortality, and performance loss, vaccines have been one method used in an attempt to reduce overall incidence rates and by extension economic losses [1–4]. Despite widespread adoption of vaccination protocols in the beef cattle industry, BRDC remains extremely prevalent due to varying levels of vaccine efficacy [5, 6]. Research into response to vaccination has identified multiple environmental and management variables which impact an individual calf’s ability to mount an antibody response [7–9]. Factors such as maternally derived antibody levels from colostrum, calf age, seasonality, and weaning status all impact a calf’s individual immune system, and result in variability in its response to vaccination [10–12].
In order to improve efficacy of vaccination for BRDC, it is desired that all calves exhibit an adequate immune response after vaccination. If this is the case, calves would likely be better protected against future exposure to BRDC viruses after a multi-shot vaccination procedure and thereby result in less economic loss due to BRDC. One way to improve beef cattle response to vaccination may be through genomic selection. Previously, we reported on variables that influenced response to vaccination against bovine viral diarrhea virus 1 (BVDV1), bovine viral diarrhea virus 2 (BVDV2), bovine respiratory syncytial virus (BRSV), and bovine herpesvirus (BHV1) [13–15]. A genome-wide association study was performed on five different responses to vaccination/viral antibody titer traits across all four viruses (initial titer, final titer, initial vaccination response [IVR], booster vaccination response [BVR], overall vaccination response [OVR]) with an additional two traits analyzed only on BVDV1 and BVDV2 (maternal decay and maternal antibody titer) due to availability of data (Fig. 1). Through heritability and accuracy estimates for response to vaccination and viral antibody titer level traits, producers would be afforded the opportunity to select for cattle with higher immune response to vaccination. As such, the goal of this study was to identify regions of the genome that were associated with response to vaccination and viral antibody level and evaluate the potential for genomic selection for calves with improved immune response to vaccination as has been done in cattle [16] and other species [17].
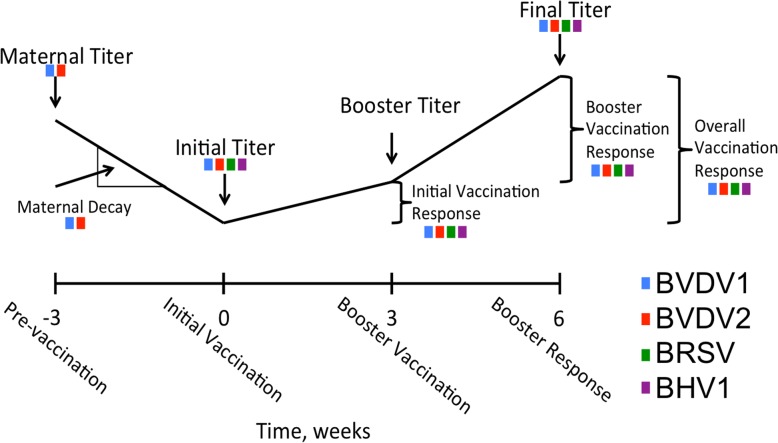
Fig. 1.
Study sample collection timeline. Colored boxes correspond to data availability of that trait to one of the four viruses. Bovine Viral Diarrhea Virus 1 (BVDV1) in blue, Bovine Viral Diarrhea Virus 2 (BVDV2) in red, Bovine Respiratory Syncytial Virus (BRSV) in green, Bovine Herpesvirus (BHV1) in purple. For each virus 3 response variables were calculated: 1) initial vaccination response = booster titer – initial titer; 2) booster vaccination response = final titer – booster titer; 3) overall vaccination response = final titer – initial titer. For BVDV1 and BVDV2 rate of maternal antibody decay was calculated: (initial titer – maternal titer) / number of days between initial titer and maternal titer measurements)
Results
Estimates of genetic and residual variance, and heritability
Estimates of heritabilities and variances were obtained for each viral antibody titer levels and response to vaccination traits (Table 1). Heritability (h2) estimates, which were the proportion of additive genetic variance out of the total phenotypic variance, ranged from 0.08 to 0.22. The greatest spread of heritabilities was found within BVDV2 with a minimum estimated heritability of 0.08 and a maximum of 0.22. In contrast, BRSV had the smallest spread with a minimum heritability of 0.12 and a maximum of 0.18. Maternal decay and maternal antibody titer traits were only available for BVDV1 and BVDV2. Total genetic and residual posterior variances were relatively similar across all viral neutralization antibody level and response to vaccination traits. Maternal decay of BVDV2 titer level had the lowest posterior genetic variance (0.005) and residual variance (0.02). In contrast, no single antibody titer level or response to vaccination trait had both the highest genetic and residual posterior variance.
Table 1.
Posterior estimates of genetic (σg2) and residual (σe2) variance, and heritability (h2)
| Virus | Traita | σg2 | σe2 | h2 |
|---|---|---|---|---|
| BVDV1 | Maternal Decayb | 0.01 | 0.03 | 0.22 |
| Maternal Antibody Titerb | 0.28 | 1.58 | 0.15 | |
| Initial Titer | 0.41 | 1.58 | 0.21 | |
| Initial Vaccination Response | 0.17 | 0.73 | 0.18 | |
| Booster Vaccination Response | 0.33 | 1.42 | 0.19 | |
| Overall Vaccination Response | 0.29 | 1.48 | 0.16 | |
| Final Titer | 0.33 | 1.65 | 0.17 | |
| BVDV2 | Maternal Decayb | 0.005 | 0.02 | 0.21 |
| Maternal Antibody Titerb | 0.23 | 2.03 | 0.10 | |
| Initial Titer | 0.17 | 1.94 | 0.08 | |
| Initial Vaccination Response | 0.11 | 1.16 | 0.09 | |
| Booster Vaccination Response | 0.21 | 2.25 | 0.08 | |
| Overall Vaccination Response | 0.18 | 2.02 | 0.08 | |
| Final Titer | 0.20 | 2.01 | 0.09 | |
| BRSV | Initial Titer | 0.18 | 1.38 | 0.12 |
| Initial Vaccination Response | 0.07 | 0.35 | 0.16 | |
| Booster Vaccination Response | 0.09 | 0.42 | 0.18 | |
| Overall Vaccination Response | 0.09 | 0.47 | 0.16 | |
| Final Titer | 0.08 | 0.49 | 0.14 | |
| BHV1 | Initial Titer | 0.14 | 1.24 | 0.10 |
| Initial Vaccination Response | 0.22 | 1.52 | 0.13 | |
| Booster Vaccination Response | 0.29 | 2.20 | 0.18 | |
| Overall Vaccination Response | 0.30 | 2.28 | 0.12 | |
| Final Titer | 0.27 | 2.36 | 0.10 |
aAll trait titers measured in log2 transformations from serum neutralization dilutions. Maternal Decay measured as log2 titer change per day
bMaternal Decay and Maternal antibody titer data only available for BVDV1 and BVDV2 (Bayes C analysis)
Whole genome association
The amount of genetic variance that every 1-Mb window across the bovine genome could account for was estimated based on the model for a viral neutralization antibody level or response to vaccination trait. Furthermore, the posterior probability of inclusion for each SNP and 1-Mb window was calculated as well. There were no 1-Mb regions across all viral neutralization antibody level and response to vaccination traits for all four viruses that exceeded a 0.9 posterior probability of inclusion. Six 1-Mb windows across the genome across traits could account for greater than 1% of the trait genetic variance (Table 2). The percent variance accounted by these windows ranged from 1.01 to 1.93%. These six 1-Mb windows were associated with four antibody titer levels or response to vaccination traits: two windows for BVDV1 initial titer (Fig. 2), two for BVDV1 overall vaccination response (Fig. 3), one for BVDV2 booster vaccination response (Fig. 4), and the final 1-Mb window for BRSV overall vaccination response (Fig. 5).
Table 2.
1-Mb genomic windows that accounted for greater than 1% genetic variance for response to vaccination traits
| Virus | Traita | Chr_Mbb | Starting SNP | Ending SNP | # of SNP | Genetic Variance (%) | PPIc |
|---|---|---|---|---|---|---|---|
| BVDV1 | Initial Vaccination Titer | 2_24 | rs137131604 | rs109222292 | 220 | 1.64 | 0.57 |
| 18_9 | rs43715906 | rs29024678 | 400 | 1.01 | 0.65 | ||
| Overall Vaccination Response | 4_12 | rs133210106 | rs109951163 | 172 | 1.93 | 0.61 | |
| 29_32 | rs43727482 | rs137609870 | 253 | 1.15 | 0.51 | ||
| BVDV2 | Booster Vaccination Response | 27_16 | rs134611614 | rs43207573 | 300 | 1.03 | 0.54 |
| BRSV | Overall Vaccination Response | 1_144 | rs109534947 | rs42965155 | 296 | 1.03 | 0.49 |
aAll trait titers measured in log2 transformations from serum neutralization dilutions
b1-Mb window defined by chromosome and mega base position. UMD3.1 build of Bos taurus genome
cPosterior probability of inclusion (inclusion rate in BayesB analysis)
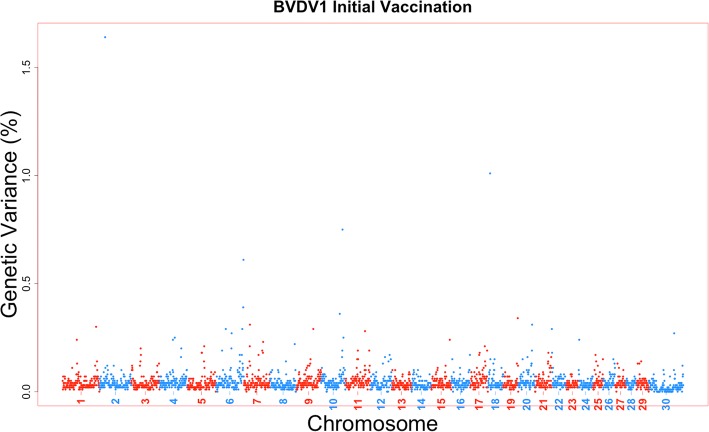
Fig. 2.
Manhattan plot for 1-Mb windows for Bovine viral diarrhea virus type 1 initial titer. A Manhattan plot showing every 1-Mb window by %variance accounted for by that window across the entire Bos taurus genome. Two singular windows exceeded the 1% genetic variance threshold: chromosome 2, Mb 24 with 1.64% genetic variance; chromosome 18, Mb 9 with 1.01% genetic variance
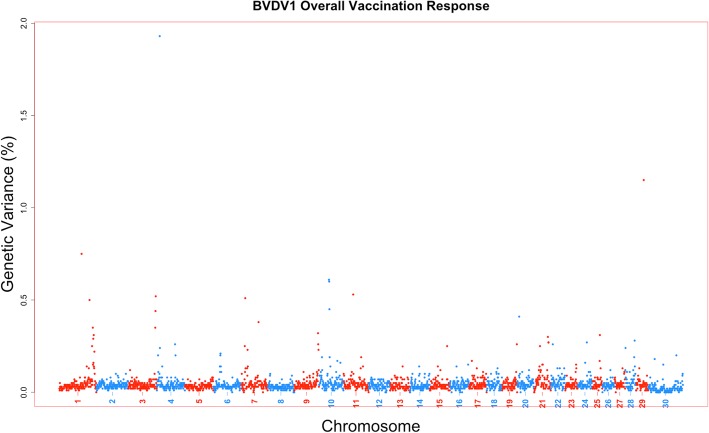
Fig. 3.
Manhattan plot for 1-Mb windows for Bovine viral diarrhea virus type 1 overall vaccination response. A Manhattan plot showing every 1-Mb window by %variance accounted for by that window across the entire Bos taurus genome. Two singular windows exceeded the 1% genetic variance threshold: chromosome 4, Mb 12 with 1.93% genetic variance; chromosome 29, Mb 32 with 1.15% genetic variance
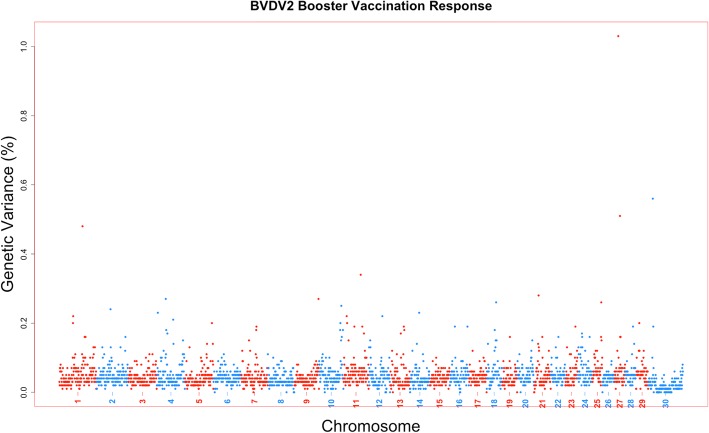
Fig. 4.
Manhattan plot for 1-Mb windows for Bovine viral diarrhea virus type 2 booster vaccination response. A Manhattan plot showing every 1-Mb window by %variance accounted for by that window across the entire Bos taurus genome. One window exceeded the 1% genetic variance threshold: chromosome 27, Mb 16 with 1.03% genetic variance
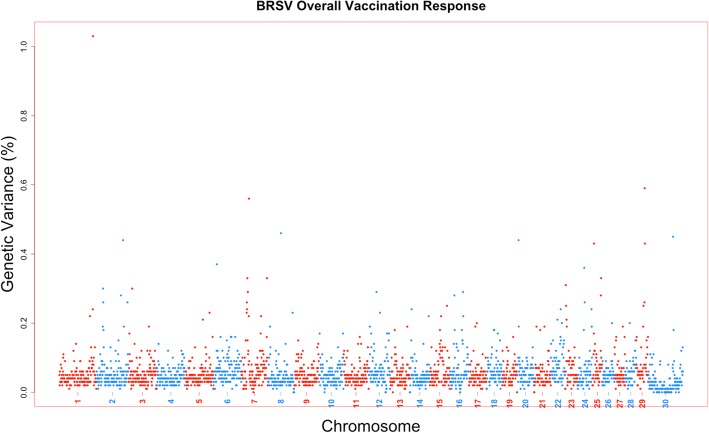
Fig. 5.
Manhattan plot for 1-Mb windows for Bovine respiratory syncytial virus overall vaccination response. A Manhattan plot showing every 1-Mb window by %variance accounted for by that window across the entire Bos taurus genome. One window exceeded the 1% genetic variance threshold: chromosome 1, Mb 144 with 1.03% genetic variance
All genes within these windows were evaluated as potential candidate genes. Unfortunately, none of the genes within these windows were annotated with immune function Gene Ontology terms. However, higher order gene families names, e.g. TRIML, may have some function related to immune system response [18]. Additionally, multiple transcription factors were identified such as EST1, which may have some functional relationship with immune response.
Traits were also analyzed categorically, with phenotypes being set as their integer value. This marginally improved the heritability of the response to vaccination and viral neutralization antibody levels (Table 3). However, in these analyses no windows accounted for greater than 1% of the estimated genetic variation.
Table 3.
Categorical analysis posterior estimates of genetic (σg2) and residual (σe2) variance, and heritability (h2)
| Virus | Traita | σg2 | σe2 | h2 |
|---|---|---|---|---|
| BVDV1 | Maternal Antibody Titerb | 0.33 | 1.00 | 0.25 |
| Initial Vaccination | 0.22 | 1.00 | 0.18 | |
| Initial Vaccination Response | 0.22 | 1.00 | 0.18 | |
| Booster Vaccination Response | 0.15 | 1.00 | 0.13 | |
| Overall Vaccination Response | 0.16 | 1.00 | 0.14 | |
| Final Titer | 0.52 | 1.00 | 0.34 | |
| BVDV2 | Maternal Antibody Titerb | 0.24 | 1.00 | 0.19 |
| Initial Vaccination | 0.15 | 1.00 | 0.13 | |
| Initial Vaccination Response | 0.11 | 1.00 | 0.10 | |
| Booster Vaccination Response | 0.11 | 1.00 | 0.10 | |
| Overall Vaccination Response | 0.11 | 1.00 | 0.10 | |
| Final Titer | 0.27 | 1.00 | 0.21 | |
| BRSV | Initial Vaccination | 0.61 | 1.00 | 0.38 |
| Initial Vaccination Response | 0.17 | 1.00 | 0.14 | |
| Booster Vaccination Response | 0.51 | 1.00 | 0.34 | |
| Overall Vaccination Response | 0.29 | 1.00 | 0.23 | |
| Final Titer | 0.47 | 1.00 | 0.32 | |
| BHV1 | Initial Vaccination | 0.61 | 1.00 | 0.38 |
| Initial Vaccination Response | 0.14 | 1.00 | 0.12 | |
| Booster Vaccination Response | 0.14 | 1.00 | 0.13 | |
| Overall Vaccination Response | 0.15 | 1.00 | 0.13 | |
| Final Titer | 0.51 | 1.00 | 0.34 |
aAll trait titers measured in log2 transformations from serum neutralization dilutions
bMaternal antibody titer data only available for BVDV1 and BVDV2
Correlations and accuracies
Individuals were separated into five groups that exhibited high genetic diversity between groups while maintaining genetic similarity within groups. The number of individuals in each of the five groups can be seen in Table 4. Different numbers of individuals were present in the different clusters between different viral antibody titer levels and responses to vaccination traits was due to different numbers of calves with phenotype data and the presence or absence of various data time points. BVDV2 was the initial viral antigen studied at the inception of this study. As such, the individuals that could be analyzed for the other 3 viral antigens came from a smaller pool and are therefore more variable in the total number utilized. The pooled correlation between direct genetic value estimates and the true phenotype ranged from − 0.10 for BHV1 overall vaccination response to 0.09 for BVDV1 final titer and overall vaccination response. Prediction accuracies ranged from an absolute value of 0.002 to 0.31 (Table 5). Maternal decay traits for BVDV1 and BVDV2 were unable to be estimated due to a failure to converge, and so could not be calculated for correlation or accuracy.
Table 4.
Number of individuals for each K Means group within response to vaccination trait
| K Means Groups | Total | ||||||
|---|---|---|---|---|---|---|---|
| Virus | Traita | 1 | 2 | 3 | 4 | 5 | |
| BVDV1 | Maternal Decayb | 131 | 89 | 126 | 90 | 133 | 569 |
| Maternal Antibody Titerb | 130 | 88 | 125 | 90 | 133 | 566 | |
| Initial Vaccination | 314 | 289 | 306 | 290 | 319 | 1518 | |
| Initial Vaccination Response | 244 | 236 | 253 | 234 | 273 | 1240 | |
| Booster Vaccination Response | 211 | 214 | 213 | 208 | 245 | 1091 | |
| Overall Vaccination Response | 198 | 187 | 210 | 206 | 243 | 1044 | |
| Final Titer | 234 | 236 | 238 | 240 | 281 | 1229 | |
| BVDV2 | Maternal Decayb | 198 | 194 | 150 | 161 | 192 | 895 |
| Maternal Antibody Titerb | 232 | 210 | 185 | 195 | 227 | 1049 | |
| Initial Vaccination | 408 | 353 | 291 | 354 | 332 | 1738 | |
| Initial Vaccination Response | 426 | 353 | 314 | 371 | 351 | 1815 | |
| Booster Vaccination Response | 425 | 350 | 313 | 371 | 347 | 1806 | |
| Overall Vaccination Response | 424 | 350 | 312 | 372 | 347 | 1805 | |
| Final Titer | 425 | 350 | 312 | 372 | 347 | 1805 | |
| BRSV | Initial Vaccination | 312 | 285 | 312 | 281 | 318 | 1508 |
| Initial Vaccination Response | 268 | 217 | 248 | 230 | 246 | 1209 | |
| Booster Vaccination Response | 250 | 200 | 230 | 214 | 222 | 1116 | |
| Overall Vaccination Response | 286 | 256 | 273 | 252 | 270 | 1337 | |
| Final Titer | 287 | 263 | 276 | 261 | 280 | 1367 | |
| BHV1 | Initial Vaccination | 312 | 283 | 301 | 280 | 315 | 1491 |
| Initial Vaccination Response | 312 | 285 | 302 | 281 | 318 | 1498 | |
| Booster Vaccination Response | 254 | 230 | 243 | 218 | 234 | 1179 | |
| Overall Vaccination Response | 256 | 236 | 253 | 226 | 245 | 1216 | |
| Final Titer | 257 | 244 | 256 | 234 | 254 | 1245 | |
aAll trait titers measured in log2 transformations from serum neutralization dilutions. Maternal Decay measured as log2 titer change per day
bMaternal Decay and Maternal antibody titer data only available for BVDV1 and BVDV2
Table 5.
Pooled correlation between phenotype and predicted direct genetic value, and accuracy of prediction
| Virus | Traita | Correlation | Accuracyb |
|---|---|---|---|
| BVDV1 | Maternal Antibody Titerc | 0.02 | 0.04 |
| Initial Vaccination | 0.06 | 0.12 | |
| Initial Vaccination Response | 0.02 | 0.05 | |
| Booster Vaccination Response | 0.08 | 0.19 | |
| Overall Vaccination Response | 0.09 | 0.22 | |
| Final Titer | 0.09 | 0.22 | |
| BVDV2 | Maternal Antibody Titerc | − 0.05 | 0.15 |
| Initial Vaccination | −0.02 | 0.08 | |
| Initial Vaccination Response | 0.04 | 0.15 | |
| Booster Vaccination Response | −0.01 | 0.03 | |
| Overall Vaccination Response | 0.00 | 0.00 | |
| Final Titer | −0.03 | 0.11 | |
| BRSV | Initial Vaccination | 0.07 | 0.22 |
| Initial Vaccination Response | 0.00 | 0.01 | |
| Booster Vaccination Response | −0.05 | 0.11 | |
| Overall Vaccination Response | 0.00 | 0.01 | |
| Final Titer | 0.01 | 0.04 | |
| BHV1 | Initial Vaccination | 0.02 | 0.05 |
| Initial Vaccination Response | 0.05 | 0.13 | |
| Booster Vaccination Response | 0.01 | 0.03 | |
| Overall Vaccination Response | −0.10 | 0.31 | |
| Final Titer | − 0.06 | 0.19 |
aAll trait titers measured in log2 transformations from serum neutralization dilutions
bAccuracy calculated as correlation between direct genetic value and phenotype divided by square root of heritability
cMaternal antibody titer data only available for BVDV1 and BVDV2
Discussion
In this study, we set out to identify regions of the genome that were associated with viral neutralization antibody level and response to vaccination traits for four viruses associated with BRDC. To do so, a genome wide association study was performed to characterize regions based on the proportion of genetic variance they could account for within a trait. Heritability estimates for viral neutralization antibody level and response to vaccination traits were lowly to moderately heritable. Ranging from 0.08 to 0.22, we determined that the heritability of viral neutralization antibody level and response to vaccination traits varied the most in BVDV1 and BVDV2, and the least in BRSV. However, when only traits measured on all four viruses are compared, BVDV1 and BVDV2 have a much more concentrated range in heritabilities than the other two. Additionally, as described in Kramer et al. 2017, BRSV was the singular virus out of the four studied that did not contain individual calves that failed to seroconvert, potentially due to the ubiquitous nature of BRSV [16, 19]. This represented a smaller range in the titer sample data and may impact its ability to be compared to the other three viruses. Heritability estimates for immune related traits in other studies range from lowly (0.11) to highly heritable (0.47), dependent on the immune trait evaluated, whether it is due to a infectious challenge or vaccination, and species [20–24]. The heritability estimates based on estimated direct genetic variation are on the lower end of this previously found range. This is likely due to response to vaccination as a complex trait with many non-genetic influences as noted in Kramer et al. 2017, and will make selection for these antibody titer level and response to vaccination traits slower than if they were more heritable.
Low correlations were calculated between direct genetic value and the true phenotype, with a range of − 0.10 to 0.09. Accuracies also ranged from an absolute value of 0.002 to 0.31. These accuracies are a direct result of low heritabilities as accuracy was calculated as correlation divided by the square root of heritability. While the correlation between true and predicted phenotype is low, and accuracies are on average 0.11, some traits such as BHV1 overall vaccination response and BVDV1 final titer may have a level of accuracy such that they could be utilized in a selection index. These determined correlations and accuracies underscore the point that responses to vaccination traits remain complicated even after fitting management practices and other environmental and physiological factors in a model. With a greater number of individuals, it may be possible to improve the accuracy closer to its maximum, although it will be capped by the heritability estimates of response to vaccination traits (0.28–0.45 range for maximums). If accuracies of genomic predictions can be increased towards these maximums, then the rate of genetic progress for response to vaccination and viral neutralization antibody level traits will increase.
These traits appear to be highly polygenic, as no singular region was able to account for a large proportion of genetic variation and only six regions were estimated to account for more than 1% of that variation (Figs. 2, 3, 4, 5). The polygenic nature of viral neutralization antibody level, response to vaccination, and immune related traits is something that has been previously reported in numerous studies such as those focused on porcine [17, 25], chicken [26, 27], cattle [28–31], and mice [32]. In the six windows that accounted for more than 1% of the genetic variance in this study, no candidate genes were identified as none of the genes in those regions were annotated with Gene Ontology terms associated with immune response functions. The TRIM gene family is present, however, and may have some further functional relevance that is not currently described in published literature [18].
Conclusions
This study aimed to examine response to vaccination against viruses associated with BRDC in Angus calves using genome wide association studies. Vaccination is one of the most widely used tools by producers to reduce and/or prevent BRDC incidence in herds. Efficacy remains lower than desired; therefore selection of cattle for response to vaccination could increase the effectiveness of vaccine use. This would potentially reduce losses associated with BRDC and improve animal welfare. Using serum samples across a 6–9 week period we were able to identify genomic regions associated with multiple response to vaccination traits, and determine the predictive accuracy of response to vaccination. Windows of size 1-Mb were found to account for up to 1.93% of genetic variation, indicating the polygenic nature of response to vaccination. With low to moderate heritabilities across response to vaccination traits, there appears to be room for selection of animals with improved vaccination response. Genes within the associated regions had little functional annotation associated with them, which may be resolved in future builds of the Bos taurus genome along with the help of the Functional Annotation of Animal Genomes (FAANG) initiative. All of this together provides a solid foundation for genetic variation within response to vaccination in Angus calves, and a basis for selection procedures.
Methods
Population
Calves (n = 2518) from a purebred American Angus herd located at Iowa State University Ames, IA, were used in this study. Not all individuals were utilized for all response to vaccination traits due to limited availability of recorded data at specific time points and non-genotyped calves. Therefore, the number of calves analyzed for each response to vaccination trait was less than the total number of genotyped calves. Calves were born in either a spring season or fall season classification, and across multiple years (2006–2012, 2014).
Phenotypic data
Phenotypic data was collected as described in previous work [13–15]. Briefly; individuals were vaccinated with a modified live vaccine (Bovi-Shield Gold 5, Zoetis, Inc. Parsippany NJ). Serum samples were collected from the calves at multiple time points and then a viral neutralization assay was performed for each sample to quantify the level of antibodies present against one of four viruses (BVDV1, BVDV2, BRSV, BHV1). Dilutions were performed to identify the greatest dilution where neutralizing antibodies could still be detected, and a log2 titer was recorded. This was based on the Spearman-Kärber method for initial calculation [33]. Response traits were calculated as the change in titer values between serum sample collection time points (initial, booster, and overall vaccination response), or as the calculated titer value for a given time (maternal, initial, and final antibody titer).
Genotypic data
The BovineSNP50 BeadChip (Illumina, San Diego, CA) and BovineHD BeadChip (Illumina, San Diego, CA) were used by Neogen GeneSeek Operations (Lincoln, NE) to perform SNP genotyping. Individuals were imputed by using the SNPipeline package (Hailin Su, https://github.com/cbkmephisto/SNPipeline) and FImpute [34] from the BovineSNP50 BeadChip to the BovineHD BeadChip using 820 Angus individuals genotyped natively for the Bovine HD BeadChip, both within and outside the ISU herd. Accuracy of imputation was about 97%, and was tested by randomly removing 5000 markers from the 50 k genotype, and then comparing imputed genotypes of these 5000 markers to the original genotypes. 574,662 single nucleotide polymorphisms (SNPs) remained after editing for a minor allele frequency (MAF) of 0.05. All SNPs were assigned a UMD3.1 bovine genome build position [35].
Statistical model
All 574,662 SNPs were used alongside phenotypic titer measurements or differences to determine an estimate of each SNP effect for a respective trait. Single SNP effect estimates were obtained using BayesB approach through the program GenSel [36]. This method assumes a model in which a portion of SNP markers have zero effect on a trait, denoted as a π, and markers not assigned zero effect are used in prediction by estimating individual SNP effects. Models were generally fit as follows:
where y was a vector of response to vaccination trait observations; b is a set of vectors of fixed effects; u is a vector of SNPS as random effects sampled through BayesB; and e was the remaining vector of residuals corresponding to each phenotypic record [37]. Fixed effects and Covariates are explained in detail within, but included the fixed effects of: Year-Season classification, Sex, Dam Age, Weaned Status, Pink Eye Status; covariates of Calf Age within Year-Season classification, Titer score, Titer-by-Titer score, and Average Daily Gain [13]. A π value of 0.999 was used for BayesB [38] analysis of each trait (approximately 575 SNP markers with a non-zero effect), while genetic and residual variances for each trait were estimated using BayesC (initial variances set as half the total phenotypic variance) before being used in BayesB [37, 39]. A total of 50,000 iterations were used with Monte Carlo Markov Chain, with the first 5000 thrown out as a burn in, to obtain posterior means of SNP marker effects and posterior probabilities of inclusion.
BayesB analysis outputs included SNP marker effect estimates, 1-Mb window effect estimates, and a direct genetic value prediction for every individual. Each SNP marker and 1-Mb window was estimated a % genetic variance that it accounted for, and a posterior probability of inclusion to indicate the frequency in which it was included in the BayesB analysis.
Response to vaccination and viral titer level traits were also analyzed as categorical traits in addition to the above analysis, and models for these traits as categorical were fit and run as above. Phenotypes were set as the truncated integer value for a recorded phenotypic measurement (1.0–1.99 = 1, etc.…) rather than rounding to integer values. All other fixed effects, covariates, and the matrix of random marker effects remained the same.
Heritability
Heritability estimates for each trait were obtained through BayesC with a π value of 0. This was chosen due to BayesB shrinking small effects to allow for the detection of relatively larger effects, and therefore biasing heritability estimates [37, 39]. Heritability was calculated as the posterior genetic variance over the total estimated posterior phenotypic variance.
Accuracy of prediction
Prediction accuracies were estimated through cross validation for the direct genetic values that were predicted for each individual in BayesC. Genotyped individuals were initially classified into 9 unequal groups through K-means clustering [40] after creating a genomic relationship matrix [41]. Using this method, each group was made to be as genetically similar as possible within group, while remaining genetically distinct to the other 8 groups. Through this, groups can be used as predictors of other groups due to genetic similarity. The 9 initial groups were clustered down to 5 total groups by merging numerically smaller groups together so that groups were more consistently sized. BayesC with a π value of 0 was used to perform training and validation for these groups as follows: 1 group was excluded from the BayesC π = 0 analysis, and direct genetic values were calculated on the remaining 4 groups of animals. Those single SNP marker estimates were then used validate the final 5th group. Each group was rotated so that every individual was validated once and used as training set four times. Accuracy was calculated by taking the correlation between predicted direct genetic value and phenotypes for all individuals in a given trait divided by the square root of heritability for that trait.
Candidate gene identification
The 1-Mb genomic regions accounting for more than 1% OF genetic variation for response to vaccination traits were identified for candidate gene investigation. A region extending half a mega base on either side of the identified 1-Mb regions, for a total of 2 Mb in length, was analyzed by looking at the biological function of every gene currently annotated in the UMD 3.1 Bos taurus assembly of the genome.
Acknowledgements
The authors thank Mahdi Saatchi and Hailin Su (https://github.com/cbkmephisto/SNPipeline) for their assistance in imputation of genotypes, and Mahdi Saatchi for providing K-means clustering code. James Koltes coordinated, oversaw, and participated in sample collection for this project and coordinated experimental procedures with farm staff.
Funding
This research was supported by NIFA Award No-2012-38420-19286 and No-2017-67011–26043, and using HPC equipment purchased through NSF MRI grant numbers CNS 1229081 and 1205413. All three funding sources were used in the Analysis and Interpretation of data, while No-2017-67011–26043 was used for the writing of this manuscript and was involved in the design of the study.
Availability of data and materials
Phenotype and genotype data is available upon reasonable request.
Abbreviations
- BHV1
Bovine Herpesvirus
- BRSV
Bovine Respiratory Syncytial Virus
- BVDV1
Bovine Viral Diarrhea Virus Type 1
- BVDV2
Bovine Viral Diarrhea Virus Type 2
- BVR
Booster Vaccination Response
- GWAS
Genome-wide Association Study
- IVR
Initial Vaccination Response
- MAF
Minor Allele Frequency
- OVR
Overall Vaccination Response
- SNP
Single Nucleotide Polymorphism
Author’s contributions
JMR and RGT conceived and coordinated the study. LM performed data and statistical analysis. MSM, ED, RGT, and CC were responsible for sample testing and collection. AW, CC, and JMR were instrumental in obtaining funds. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Iowa State University Animal Care and Use Committee (IACUC) approved all animal work before the initiation of this study.
Consent for publication
Not Applicable.
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
L. M. Kramer, Email: lmkramer@iastate.edu
M. S. Mayes, Email: mmayes@iastate.edu
E. D. Downey, Email: eddowney87@gmail.com
R. G. Tait, Jr, Email: rtait@neogen.com.
A. Woolums, Email: Amelia.woolums@msstate.edu
C. Chase, Email: Christopher.Chase@sdstate.edu
J. M. Reecy, Email: jreecy@iastate.edu
References
- 1.Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL, Koohmaraie M, Dikeman ME: Bovine respiratory disease in feedlot cattle: phenotypic, environmental, and genetic correlations with growth, carcass, and longissimus muscle palatability traits. J Anim Sci 2007, 85(8):1885-1892. [DOI] [PubMed]
- 2.Irsik M: BRD and fed cattle performance. Florida Cattlemen’s Association Proceedings 2010.
- 3.Van Eenennaam AL: Integrated Program for Reducing Bovine Respiratory Disease Complex in Beef and Dairy Cattle Coordinated Agricultural Project (BRD CAP). In: American Society of Bovine Practitioners meeting: Sept. 20-22 2012; Montreal, Canada; 2012. https://ucanr.edu/sites/UCCE_LR/files/152458.pdf.
- 4.Schneider MJ, Tait RG, Jr, Busby WD, Reecy JM. An evaluation of bovine respiratory disease complex in feedlot cattle: Impact on performance and carcass traits using treatment records and lung lesion scores. J Anim Sci. 2009;87(5):1821–1827. doi: 10.2527/jas.2008-1283. [DOI] [PubMed] [Google Scholar]
- 5.Theurer ME, Larson RL, White BJ. Systematic review and meta-analysis of the effectiveness of commercially available vaccines against bovine herpesvirus, bovine viral diarrhea virus, bovine respiratory syncytial virus, and parainfluenza type 3 virus for mitigation of bovine respiratory disease complex in cattle. J Am Vet Med Assoc. 2015;246(1):126–142. doi: 10.2460/javma.246.1.126. [DOI] [PubMed] [Google Scholar]
- 6.Vaccination of Cattle and Calves on U.S. Beef Cow-calf Operations. In. Edited by USDA. Veterinary Services: APHIS; 2009. https://www.aphis.usda.gov/animal_health/nahms/beefcowcalf/downloads/beef0708/Beef0708_is_GenVacc.pdf.
- 7.Di Giacomo S, Brito BP, Perez AM, Bucafusco D, Pega J, Rodriguez L, Borca MV, Perez-Filgueira M. Heterogeneity in the Antibody Response to Foot-and-Mouth Disease Primo-vaccinated Calves. Transbound Emerg Dis. 2015;62(3):280–287. doi: 10.1111/tbed.12130. [DOI] [PubMed] [Google Scholar]
- 8.Hulbert LE, Moisa SJ. Stress, immunity, and the management of calves. J Dairy Sci. 2016;99(4):3199–3216. doi: 10.3168/jds.2015-10198. [DOI] [PubMed] [Google Scholar]
- 9.Langel SN, Wark WA, Garst SN, James RE, McGilliard ML, Petersson-Wolfe CS, Kanevsky-Mullarky I. Effect of feeding whole compared with cell-free colostrum on calf immune status: Vaccination response. J Dairy Sci. 2016;99(5):3979–3994. doi: 10.3168/jds.2015-9892. [DOI] [PubMed] [Google Scholar]
- 10.Niewiesk S. Maternal antibodies: clinical significance, mechanism of interference with immune responses, and possible vaccination strategies. Front Immunol. 2014;5:446. doi: 10.3389/fimmu.2014.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roth JA: Evaluation of cell-mediated immunity. In: Large Animal Proceedings of the North American Veterinary Conference: 2008; Orlando, FL; 2008: 49.
- 12.Lippolis KD, Cooke RF, Schubach KM, Brandao AP, da Silva LG, Marques RS, Bohnert DW. Altering the time of vaccination against respiratory pathogens to enhance antibody response and performance of feeder cattle. J Anim Sci. 2016;94(9):3987–3995. doi: 10.2527/jas.2016-0673. [DOI] [PubMed] [Google Scholar]
- 13.Kramer LM, Mayes MS, Fritz-Waters E, Williams JL, Downey ED, Tait RG, Woolums A, Chase C, Reecy JM. Evaluation of responses to vaccination of Angus cattle for four viruses that contribute to bovine respiratory disease complex. J Anim Sci. 2017;95(11):4820–4834. doi: 10.2527/jas2017.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downey ED, Tait RG, Jr, Mayes MS, Park CA, Ridpath JF, Garrick DJ, Reecy JM. An evaluation of circulating bovine viral diarrhea virus type 2 maternal antibody level and response to vaccination in Angus calves. J Anim Sci. 2013;91(9):4440–4450. doi: 10.2527/jas.2012-5890. [DOI] [PubMed] [Google Scholar]
- 15.Tait RG, Jr, Downey ED, Mayes MS, Park CA, Ridpath JF, Garrick DJ, Reecy JM. Evaluation of response to bovine viral diarrhea virus type 2 vaccination and timing of weaning on yearling ultrasound body composition, performance, and carcass quality traits in Angus calves. J Anim Sci. 2013;91(11):5466–5476. doi: 10.2527/jas.2012-5891. [DOI] [PubMed] [Google Scholar]
- 16.Leach RJ, O’Neill RG, Fitzpatrick JL, Williams JL, Glass EJ: Quantitative trait loci associated with the immune response to a bovine respiratory syncytial virus vaccine. PloS one 2012, 7(3):e33526. [DOI] [PMC free article] [PubMed]
- 17.Zanella R, Gava D, Peixoto Jde O, Schaefer R, Ciacci-Zanella JR, Biondo N, da Silva MV, Cantao ME, Ledur MC. Unravelling the genetic components involved in the immune response of pigs vaccinated against influenza virus. Virus Res. 2015;210:327–336. doi: 10.1016/j.virusres.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8(11):849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valarcher JF, Taylor G. Bovine respiratory syncytial virus infection. Vet Res. 2007;38(2):153–180. doi: 10.1051/vetres:2006053. [DOI] [PubMed] [Google Scholar]
- 20.Serao NV, Kemp RA, Mote BE, Willson P, Harding JC, Bishop SC, Plastow GS, Dekkers JC. Genetic and genomic basis of antibody response to porcine reproductive and respiratory syndrome (PRRS) in gilts and sows. Genet Sel Evol. 2016;48(1):51. doi: 10.1186/s12711-016-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manjeet PBL, Sharma R, Dhaka SS, Magotra A, Dev K. Evaluation of genetic and non-genetic factors on foot and mouth disease (FMD) virus vaccine-elicited immune response in Hardhenu (Bos taurus x Bos indicus) cattle. Trop Anim Health Prod. 2017;49(8):1689–1695. doi: 10.1007/s11250-017-1379-4. [DOI] [PubMed] [Google Scholar]
- 22.Bovenhuis H, Bralten H, Nieuwland MG, Parmentier HK. Genetic parameters for antibody response of chickens to sheep red blood cells based on a selection experiment. Poult Sci. 2002;81(3):309–315. doi: 10.1093/ps/81.3.309. [DOI] [PubMed] [Google Scholar]
- 23.Dunkelberger JR, Serao NV, Niederwerder MC, Kerrigan MA, Lunney JK, Rowland RR, Dekkers JC. Effect of a major quantitative trait locus for porcine reproductive and respiratory syndrome (PRRS) resistance on response to coinfection with PRRS virus and porcine circovirus type 2b (PCV2b) in commercial pigs, with or without prior vaccination for PRRS. J Anim Sci. 2017;95(2):584–598. doi: 10.2527/jas.2016.1071. [DOI] [PubMed] [Google Scholar]
- 24.Thompson-Crispi KA, Sewalem A, Miglior F, Mallard BA. Genetic parameters of adaptive immune response traits in Canadian Holsteins. J Dairy Sci. 2012;95(1):401–409. doi: 10.3168/jds.2011-4452. [DOI] [PubMed] [Google Scholar]
- 25.Dunkelberger JR, Serao NVL, Weng Z, Waide EH, Niederwerder MC, Kerrigan MA, Lunney JK, Rowland RRR, Dekkers JCM. Genomic regions associated with host response to porcine reproductive and respiratory syndrome vaccination and co-infection in nursery pigs. BMC Genomics. 2017;18(1):865. doi: 10.1186/s12864-017-4182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamzic E, Buitenhuis B, Herault F, Hawken R, Abrahamsen MS, Servin B, Elsen JM, Pinard-van der Laan MH, Bed’Hom B. Genome-wide association study and biological pathway analysis of the Eimeria maxima response in broilers. Genet Sel Evol. 2015;47:91. doi: 10.1186/s12711-015-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Psifidi A, Banos G, Matika O, Desta TT, Bettridge J, Hume DA, Dessie T, Christley R, Wigley P, Hanotte O, et al. Genome-wide association studies of immune, disease and production traits in indigenous chicken ecotypes. Genet Sel Evol. 2016;48(1):74. doi: 10.1186/s12711-016-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sallam AM, Zare Y, Alpay F, Shook GE, Collins MT, Alsheikh S, Sharaby M, Kirkpatrick BW. An across-breed genome wide association analysis of susceptibility to paratuberculosis in dairy cattle. J Dairy Res. 2017;84(1):61–67. doi: 10.1017/S0022029916000807. [DOI] [PubMed] [Google Scholar]
- 29.Thompson-Crispi KA, Sargolzaei M, Ventura R, Abo-Ismail M, Miglior F, Schenkel F, Mallard BA. A genome-wide association study of immune response traits in Canadian Holstein cattle. BMC Genomics. 2014;15:559. doi: 10.1186/1471-2164-15-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson-Crispi K, Atalla H, Miglior F, Mallard BA. Bovine mastitis: frontiers in immunogenetics. Front Immunol. 2014;5:493. doi: 10.3389/fimmu.2014.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glass EJ, Baxter R, Leach RJ, Jann OC. Genes controlling vaccine responses and disease resistance to respiratory viral pathogens in cattle. Vet Immunol Immunopathol. 2012;148(1-2):90–99. doi: 10.1016/j.vetimm.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosley YC, Radder JE, Berndt A, HogenEsch H. Genome-Wide Association Mapping of the Antibody Response to Diphtheria, Tetanus and Acellular Pertussis Vaccine in Mice. J Infect Dis. 2017;215(3):466–474. doi: 10.1093/infdis/jiw587. [DOI] [PubMed] [Google Scholar]
- 33.Finney DJ. Statistical method in biological assay. 3. England: Oxford University Press, High Wycombe; 1978. [Google Scholar]
- 34.Sargolzaei M, Chesnais JP, Schenkel FS. A new approach for efficient genotype imputation using information from relatives. BMC Genomics. 2014;15:478. doi: 10.1186/1471-2164-15-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS et al: A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol 2009, 10(4):R42. [DOI] [PMC free article] [PubMed]
- 36.Fernando RL GD: GenSel - User manual for portfolio of Genomic Selection related Analyses, Version 3.0. ISU 2009.
- 37.Habier D, Fernando RL, Kizilkaya K, Garrick DJ. Extension of the bayesian alphabet for genomic selection. BMC Bioinformatics. 2011;12:186. doi: 10.1186/1471-2105-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meuwissen TH, Odegard J, Andersen-Ranberg I, Grindflek E. On the distance of genetic relationships and the accuracy of genomic prediction in pig breeding. Genet Sel Evol. 2014;46:49. doi: 10.1186/1297-9686-46-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kizilkaya K, Fernando RL, Garrick DJ. Genomic prediction of simulated multibreed and purebred performance using observed fifty thousand single nucleotide polymorphism genotypes. JAnim Sci. 2010;88(2):544–551. doi: 10.2527/jas.2009-2064. [DOI] [PubMed] [Google Scholar]
- 40.Hartigan JAWM, Algorithm AS. 136: A k-means clustering algorithm. Applied Statistics. 1979;28:100–108. doi: 10.2307/2346830. [DOI] [Google Scholar]
- 41.VanRaden PMVTC, Wiggans GR, Sonstegard TS, Schnabel RD, Taylos JF, Scenkel FS. Invited review: reliability of genomic predictions for North American Holstein bulls. J Dairy Sci. 2009;92(1):16–24. doi: 10.3168/jds.2008-1514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Phenotype and genotype data is available upon reasonable request.