Abstract
In filamentous fungi, homeobox proteins are conserved transcriptional regulators described to control conidiogenesis and fruiting body formation. Eight homeobox (hbx) genes are found in the genome of the aflatoxin-producing ascomycete, Aspergillus flavus. While loss-of-function of seven of the eight genes had little to no effect on fungal growth and development, disruption of hbx1, resulted in aconidial colonies and lack of sclerotial production. Furthermore, the hbx1 mutant was unable to produce aflatoxins B1 and B2, cyclopiazonic acid and aflatrem. In the present study, hbx1 transcriptome analysis revealed that hbx1 has a broad effect on A. flavus gene expression, and the effect of hbx1 increases overtime, impacting more than five thousand protein-coding genes. Among the affected genes, those in the category of secondary metabolism (SM), followed by that of cellular transport, were the most affected. Specifically, regarding the effect of hbx1 on SM, we found that genes in 44 SM gene clusters where upregulated while 49 were downregulated in the absence of hbx1, including genes in the SM clusters responsible for the synthesis of asparasone, piperazine and aflavarin, all known to be associated with sclerotia. In addition, our study revealed that hbx1 affects the expression of other transcription factor genes involved in development, including the conidiation central regulatory pathway and flb genes.
Keywords: Aspergillus flavus, hbx1, secondary metabolism, fungal development, transcriptome
The opportunistic phytopathogen, Aspergillus flavus, is often found colonizing oil seed crops such as peanut, corn, sorghum, tree nuts and cotton (Robens and Cardwell 2003). Dispersal of this fungus proceeds rapidly in the field through production of asexual spores termed conidia present on specialized structures denominated conidiophores. Once the fungus has colonized the crop it can survive in the field under harsh conditions for several years by forming resistant structures termed sclerotia (Horn et al., 2014). Upon colonization of the plant, A. flavus produces a number of mycotoxins, including the highly carcinogenic family of toxins known as aflatoxins (Bhatnagar et al., 2018). Contaminated crops are often destroyed or significantly reduced in value leading to substantial economic losses in the range of one billion US dollars annually during years of severe aflatoxin outbreaks (Robens & Cardwell 2003, & Wu et al. 2008). In developing nations where legislation is often not in place to regulate the allowable levels of aflatoxins in susceptible crops, consumption of aflatoxin-contaminated food can lead to immunosuppression, liver cancer, and in some cases death (Yard et al. 2013).
Successful control of aflatoxin contamination in crops will depend in part on research efforts directed toward understanding the regulatory mechanisms controlling A. flavus dissemination and survival, as well as mycotoxin biosynthesis and pathogenicity. It has been shown that A. flavus development is genetically linked to secondary metabolism, including the production of mycotoxins (Calvo and Cary 2015). Among several important regulators of fungal development and secondary metabolism is the light-responsive global regulator VeA. This fungal-specific protein has been shown to regulate asexual and sexual development as well as production multiple secondary metabolites in many fungal genera (Calvo et al., 2016), including Aspergillus (Kato et al., 2003, Duran et al. 2007, Dhingra et al. 2013, Lind et al. 2015). In A. flavus, loss of VeA results in increased conidiation, absence of sclerotia, and suppression of secondary metabolite production including aflatoxin, aflatrem, and cyclopiazonic acid (Duran et al. 2007). In addition to VeA, the arginine methyltransferase RmtA has been shown to be a positive regulator of both aflatoxin production and asexual development (Satterlee et al. 2016). Another example is RtfA, a homolog of a putative member of the Saccharomyces cerevisiae paf1 complex (Warner et al., 2007), that is also required for normal aflatoxin biosynthesis, sclerotial production and conidiation (Lohmar et al. 2016). Other examples are the genes encoding transcription factors mtfA (Zhuang et al. 2016) nsdC, and nsdD (Cary et al. 2012). The global regulator MtfA is a negative regulator of conidiation, required for normal maturation of sclerotia, and a positive regulator of aflatoxin production (Zhuang et al. 2016). Both nsdC and nsdD also demonstrated a role in the regulation of conidiophore development, and are essential for sclerotial formation, as well as influencing production of aflatoxin (Cary et al. 2012).
Recently homeobox domain transcription factor genes were identified in A. flavus, and disruption of the homeobox 1 (hbx1) gene abolished production of conidia and sclerotia as well as production of several mycotoxins (Cary et al. 2017). The hbx1 gene was also shown to regulate expression of several development regulators such as brlA (Cary et al., 2012), a keystone in the induction of conidiation (Adams et al. 1998). Alongside the effect on developmental regulators, expression of the aflatoxin specific transcription factor aflR and the global regulator veA were altered in the absence of hbx1 possibly contributing to the observed decrease in the production of several mycotoxins such as aflatrem, cyclopiazonic acid, and aflatoxin (Cary et al., 2017).
Based on the profound effect that hbx1 has on development and secondary metabolism in A. flavus, this gene represents a potential target for new strategies to control aflatoxin contamination of food and feed crops by A. flavus. To gain further insight into the regulatory scope of hbx1 we performed a transcriptome analysis. The impact of hbx1 on the gene expression profile of A. flavus was assessed over three-time points. Several thousand genes were under hbx1 control indicating that hbx1 is a global regulator, and its influence increased with time. An elevated number of transcription factors and developmental regulators were shown to be hbx1-dependent. Furthermore, a large numbers of secondary metabolite gene clusters are also affected by hbx1, among them seven are associated with known metabolites.
Materials & Methods
Strains used and growth conditions
Aspergillus flavus strains used in this study were the AF70 control, AF70 Δhbx1 and a genetically complemented Δhbx1 mutant (designated AF70 Δhbx1-COM) as described in Cary et al. (2017). Strains were point inoculated onto double strength 5/2 agar (50 mL V8 juice, 40 g agar, pH 5.2 per liter of medium (Chang et al. 1993) supplemented with 3.0 g ammonium sulfate and 1 mg/ml uracil (termed 2X V8 ASU) and incubated in the light at 30° for 6 days to promote conidiation. Conidia were collected from plates in 0.01% Triton X-100 and stored at 4°. Due to the inability of the Δhbx1 mutant to conidiate, cultures were maintained at -80° as glycerol stocks containing agar plugs of fungal mycelia.
Sequence analysis of plant homologs
Using the amino acid sequence for Hbx1 as query (XP_002380469.1) a BLASTp search was performed to identify possible homologs of Hbx1 in selected plant species. Species and sequences used were Arachis hypogaea (AKN10291.1), Zea mays (NP_001140916.1), Gossypium arboretum (XP_017643272.1) and Arabidopsis thaliana (AAA56907.1). A MAFFT multiple sequences alignment (https://mafft.cbrc.jp/alignment/software/) was performed to align the sequences and visualized using BoxShade (https://embnet.vital-it.ch/software/BOX_form.html).
RNA sequencing study
RNA preparation and sequencing:
Inoculated approximately 5 × 105 conidia/ml of the AF70 control and the Δhbx1-COM mutant into 500 ml peptone minimal salts (PMS; not conducive to aflatoxin production) (Buchanan and Lewis 1984) broth supplemented with 1 mg/ml uracil (PMSU) in 1 liter Ehrlenmeyer baffle flasks. Cultures were incubated at 30° in the dark with shaking at 250 rpm for 24 h. Mycelia of the AF70 Δhbx1 mutant were scraped from the surface of four 2X V8 ASU top agarose (0.5% agarose I, Amresco, Solon, OH) plates and placed in 25 mL 2X V8 ASU broth in a 50 ml Sarsteadt tube. Equal amounts of mycelia were macerated for 10 sec using a tissue grinder (Tissumizer SDT1810, Tekmar, Cincinnati, OH) then transferred into 500 ml PMSU broth in 1 liter Ehrlenmeyer baffle flask. Incubated at 30° in the dark with shaking at 250 rpm for 24 h. Collected mycelia from cultures of each of the three strains by filtering through sterile miracloth, transferred 0.5g wet weight into 25 ml of PDBU broth in 250 ml Ehrlenmeyer flasks (4 replicates) and incubated statically in the dark for 6 h (time point for initiation of aflatoxin gene expression), 24h and 48 h (approximate time points for initiation of conidia and sclerotia production, respectively). Cultures were filtered through sterile miracloth and the fungal tissue collected, frozen in liquid nitrogen and stored at -80°. The frozen mycelial samples were ground under liquid nitrogen with motar and pestle until powdered and transferred to a 50ml Sarstedt tube and stored frozen at -80 until ready for RNA extraction. RNA was isolated from 100-200 mg of the frozen ground mycelial samples using the TRI Reagent (Sigma T9424-100ML) and following the standard Direct-zol RNA MiniPrep kit (ZYMO Research, Irvine, CA) protocol using the double washes modification. RNA quality and quantity were determined using the Experion Automated Electrophoresis Station (Bio-Rad). Frozen RNA samples were shipped overnight on dry ice to North Carolina State University’s Genomics Sciences Laboratory for RNA sequencing. RNA libraries were prepared using the Ultra Directional RNA library prep kit from NEB using the manufacturer’s protocol for NEBNext PolyA mRNA magnetic isolation module. Sequencing was carried out by Illumina HiSeq 2500 at 125 bp single end reads.
RNA data analysis:
Read mapping
The single-end reads of three strains (Control, , -COM) each with three replicates at three time points (6 h, 24 h, and 48 h) were separately aligned to the reference genome (Nierman et al., 2015) using HISAT2 (Kim et al., 2015) version 2.0.5. The command used was hisat2 -x reference_genome_index –U fastq_file -S output_file.sam. HISAT2 utilizes Bowtie2 (Langmead & Salzberg 2012) and was run using software version 2.3.
Read counts
The mapped reads in SAM format were then analyzed using the feature Counts tool from the Subread package (Liao et al., 2013) version 1.6.0. This tool was employed to return a table of read counts for each gene. The command used was featureCounts -a reference_genome.gtf -p -s 2 -o output_file –primary input_file.sam. A bash script was used to combine all the separate read count files into one table.
Differentially expressed coding genes (DEGs)
The table of read counts was used as input for the R limma package (Ritchie et al., 2015). This package was used to determine DEGs by comparing read counts between two strains: Control vs. and Control vs. -COM. These comparisons were made at all three-time points: 6 h, 24 h, 48 h. The replicates of each condition at each time point were combined during this step of the analysis. The RPKM function in the R edgeR package (Robinson et al., 2009) determined the reads per kilobase per million (RPKM) values for all the genes.
Bash and Perl scripts were developed to parse the DEGs and RPKM data. An Excel file was created with the RPKM values for all genes across all conditions. FungiFun2 (Priebe et al., 2014) was used for FunCat term annotation of DEGs from Control vs. and Control vs. -COM. FungiDB (Basenko et al., 2018) was used for GO term annotation.
Functional annotation and generation of gene lists
Secondary Metabolite gene clusters (SMGs) were extracted from Ehrlich and Mack (2014). In addition, a list of transcription factors (TFs) were derived from the Fungal Transcription Factor Database (http://ftfd.snu.ac.kr/intro.php) (Park et al. 2008) for A. flavus and mapped to differentially expressed genes in A. flavus. Functional annotations of these transcription factors were obtained from NCBI. R (R Core Team 2017) version 3.4.1, specifically the ggplot2 package (Wickham 2009), was used to make statistical figures. The Venn diagrams were made using the R package VennDiagram (Chen & Boutros 2011). Fungal development-related genes from Aspergillus species were reviewed in Table S2 from Krijgsheld et al. (2013). FASTA sequences of these genes were used to search against the A. flavus genome to identify developmental genes.
The list of DEGs from the study performed by Dolezal and collaborators (Dolezal et al. 2013) was compared to the hbx1 DEGs to search for potentially hbx1–dependent virulence genes. Furthermore, we specifically looked for virulence-related secretory genes. The list of possible virulence-related hbx1 genes was compared to the A. flavus secretome-related genes in the FunSecKB2 database (Meinken et al., 2014). For higher confidence in results only the list of “curated secreted” and “highly likely secreted” genes in FunSecK2 were used.
Weighted gene network co-expression analysis
The gene co-expression network was made using WGCNA (Weighted Gene Network Co-expression Analysis) with a signed network, the biweight mid-correlation method, and a soft-thresholding power of 9. Variance stabilized counts from DESeq2 were used as input to WGNCA (Langfelder and Horvath 2008). Genes with missing values or zero variance were filtered out using the goodSamplesGenes function within the WGCNA package. Visualization of gene networks using wild-type A. flavus data were shown using Cytoscape v3.6.0 with the Edge-weighted Spring Embedded layout with minor manual adjustment. Relative edge weight values were calculated for the entire module containing hbx, and First Neighbor nodes were selected for additional analysis.
Data availability
Table S1 contains calculated expression values of sequenced RNA samples along with corresponding p-values. Table S2 contains a selected list of fungal developmental regulators that are shown to be hbx1-dependent. Table S3 is a subset of Table S1 that shows all known transcription factors in A. flavus and their corresponding expression pattern in regard to presence or absence of hbx1. The data are publicly available at NCBI’s SRA repository with the SRA Accession #: PRJNA494425. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7304252.
Results
Hbx1 is not conserved in plants species
To determine if the hbx1 product is conserved among plants and thus a possible viable target for control of A. flavus, a BLASTp search was performed to identify potential homologs. The best BLASTp hit for Hbx1 from Arachis hypogaea, Zea mays, Gossypium arboretum, and Arabidopsis thaliana were chosen for comparison to the A. flavus protein. Among these hits the query coverage of the results was very low (9–18%) localizing only around the homeobox domain located in Hbx1. Amino acid sequences of all species were then run through a MAFFT multiple sequence alignment. The results of the alignment are visualized in Figure 1. This result, together with the low percentage values, indicates that Hbx1 from Aspergillus is not conserved in these common plant hosts.
Figure 1.
- Multiple sequence alignment of Aspergillus flavus Hbx1 and possible closest predicted proteins in selected plant species. BLASTp search was carried out to find possible homologs in selected plant species and with the best hits. A MAFFT Sequence Alignment (https://www.ebi.ac.uk/Tools/msa/mafft/) was performed to show homology of amino acid sequences.
hbx1 is a global genetic regulator in A. flavus
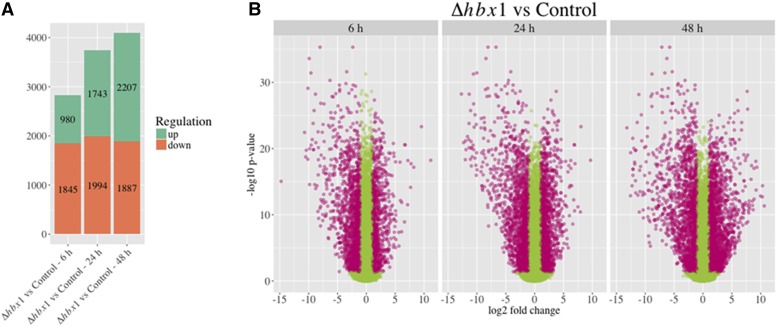
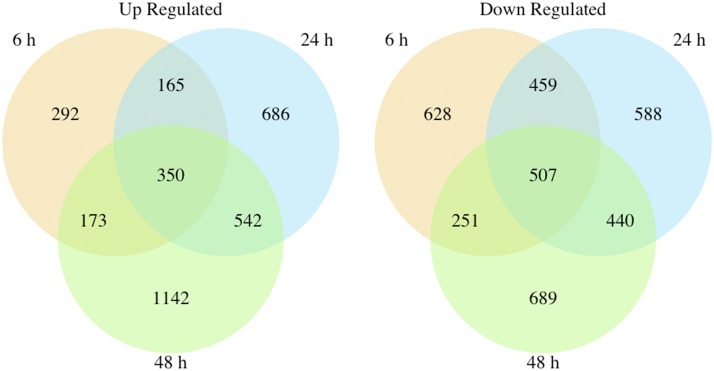
Across the time points assessed, absence of hbx1 caused a significant change in expression levels in more than 5000 genes in the A. flavus genome. Nearly 2000 genes were downregulated at each time point. In addition, while at the 6 h time point only 980 genes presented an increase in their expression, that number approximately doubled at the later time points (Figure 2, Table 1). Although a similar total amount of genes showed altered expression at each time point, there were not always the same DEGs at all three-time points. Only 350 genes of the entire genome were consistently upregulated by the loss of hbx1, while 507 genes experienced a significant decrease in expression at all three times points (Figure 3).
Figure 2.
Number of genes influenced by hbx1 A) Number of up-regulated (green) and down-regulated (orange) DEGs in vs. control at the 6 h, 24 h, and 48 h time points. B) Volcano plot of log2 folder change vs. log10 P-value of all the genes in vs. control at the 6 h, 24 h, and 48 h time points. DEGs are pink dots, other genes are shown as green dots. Pink dots with positive log2 fold change values are up-regulated DEGs. Pink dots with negative log2 fold change values are down-regulated DEGs. The x-axis represents the log2 of the fold change as determined by Limma. The y-axis is the log10 of the adjusted p value from Limma. The cut off-fold change value to determine differential expression is greater than 2 or less than 0.5. The cut off-adjusted p value to determine differential expression was greater than 0.05. Additional statistical representation of other comparison are in volcano plots located in Figure S1.
Table 1. – Percentage of A.flavus hbx1-dependent DEGs at each time point.
| 6 h | 24 h | 48 h | All 3 time points | ||
|---|---|---|---|---|---|
| Up regulated | Percent of total DEGs | 16.17% | 28.76% | 36.42% | 5.78% |
| Percent of total genome | 7.27% | 12.93% | 16.37% | 2.60% | |
| Down regulated | Percent of total DEGs | 30.45% | 32.90% | 31.14% | 8.37% |
| Percent of total genome | 13.68% | 14.79% | 13.99% | 3.76% |
Figure 3.
- Venn diagram visualizing the overlap of up regulated and down regulated genes in vs. Control at the 6 h, 24 h, and 48 h time points.
hbx1 is indispensable for normal secondary metabolism in A. flavus
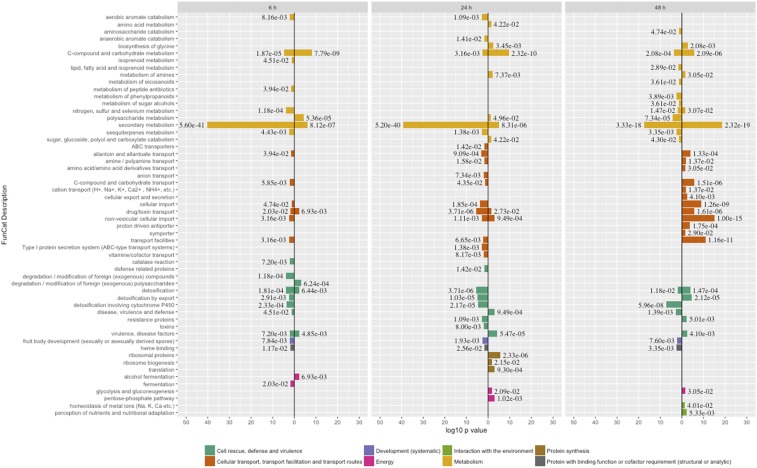
To elucidate the regulatory scope of hbx1 in A. flavus, a series of functional enrichment analyses were performed with the transcriptome data. Using the FungiFun2 platform we performed a Gene Ontology search using FunCat terms (Figure 4) (Priebe et al. 2014). The analysis revealed multiple enriched categories, with the largest one being related to metabolism, followed by cellular transport and cell rescue (the former particularly at 48 h) (Figure 4). Within this division of categories, metabolic genes involved in secondary metabolism were the largest group affected by loss of hbx1. At all-time points, most of the DEGs associated with secondary metabolism were downregulated in the absence of hbx1. GO terms were also used for functional analysis from FungiDB and are represented in Figure S2. This data also supports the pattern that secondary metabolism is the largest hbx1-affected category.
Figure 4.
- FunCat terms associated with DEGs found in vs. Control at 6 h, 24 h, and 48 h. The minus log10 of the p-value of DEGs in each term is proportional to the length of the bars. FunCat annotations and p-value as determined by FungiFun2(https://elbe.hki-jena.de/fungifun/fungifun.php): (i) metabolism is shown in orange, (ii) cellular transport, transport facilities and transport routes in light brown, (iii) cell rescue, defense & virulence in dark green, (iv) development in purple, (v) protein with binding function or cofactor requirement (structural or analytic) in black, (vi) protein synthesis in dark brown, (vii) energy in magenta, and (viii) interaction with the environment in light green. Down regulated genes are to the left of the origin and up regulated to the right.
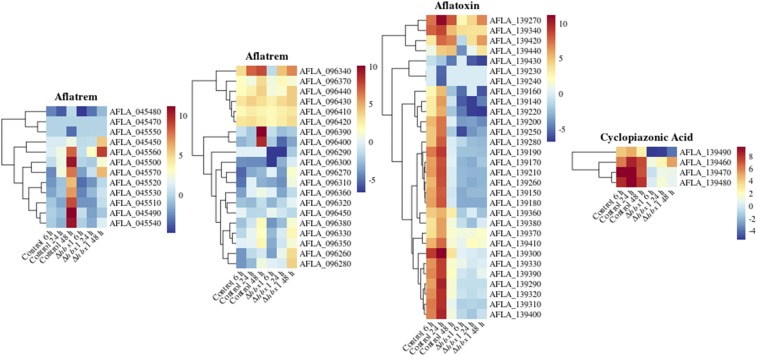
In a previous study we discovered that hbx1 is a positive regulator of aflatoxin, aflatrem, and cyclopiazonic acid biosynthesis (Cary et al. 2017). The current transcriptome analysis provides further insight into hbx1 regulation of biosynthetic gene clusters of those mycotoxins (Figure 5). In the aflatoxin cluster the majority of the genes are suppressed in the mutant strain. Previously mRNA transcripts from the aflatrem clusters were detected at approximately 48 h in the wild- type (Nicholson et al. 2009), coinciding with our observations (Figure 5), however such an increase was not observed in the hbx1 deletion mutant. In addition, all the genes in the cyclopiazonic acid genes cluster were down regulated in the absence of hbx1 (Figure 5).
Figure 5.
Heat map of RPKM values of genes on a log scale found in secondary metabolite gene clusters of aflatrem, aflatoxin, and cyclopiazonic acid (CPA). The RPKM value of each gene was calculated by averaging all the RPKM values of all replicates corresponding to that treatment at three different time points: 6 h, 24 h, and 48 h.
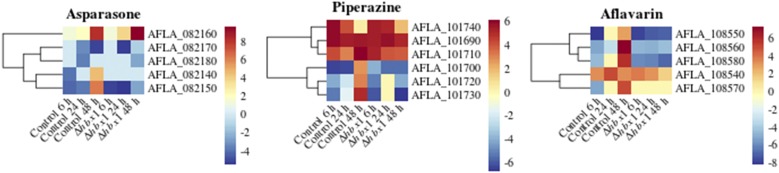
Other gene clusters involved in the production of secondary metabolites known to be associated with sclerotial development were also affected, particularly at the last time point, such as the asparasone, piperazine, and aflavarin gene clusters (Figure 6). In addition, other secondary metabolite gene clusters in A. flavus, without a described associated product (orphan clusters) were shown to be regulated by hbx1 (Figure S3). Other clusters were also affected but to a lesser extent (Table S1).
Figure 6.
Heat map of RPKM values of genes on a log scale found in sclerotia related -secondary metabolite gene clusters of asparasone, piperazine, and aflavarin. The RPKM value of each gene was calculated by averaging all the RPKM values of all replicates corresponding to that treatment at three different time points: 6 h, 24 h, and 48 h.
hbx1 is a master regulator of developmental regulatory genes and other transcription factors in A. flavus
The hbx1 gene is necessary for normal conidiation and sclerotial development in A. flavus (Cary et al., 2017), and it was shown to affect the expression of key developmental regulators, such as brlA, the master regulator of the asexual development (conidia) in Aspergillus. In addition, hbx1 also affected the expression of veA, which regulates multiple aspects of fungal development as well as influencing production of secondary metabolites (Calvo et al., 2016). In our current study, a list of selected A. flavus developmental regulatory genes (experimentally characterized in A. flavus and/or in other fungi), was applied to our data analysis to better understand the mechanism of action of hbx1 in A. flavus regulatory networks (Table S2). Our results revealed that several additional developmental transcription factors were also hbx1-dependent, such as the terminal gene in the central conidiation pathway, wetA (Wu et al., 2017) the fluffy genes flbA, flbC, flbD, and fluG (Purschwitz et al., 2008, Garzia et al., 2010, & Kwon et al., 2010, Chang et al. 2012), as well as the mating gene MAT1-1 (Ramirez-Prado et al., 2008).
Our hbx1 data analysis also extended to other A. flavus transcription factors. A list of A. flavus transcription factors were obtained from the Fungal Transcription Factor Database and used to search for DEGs. Annotations of the transcription factors were derived from NCBI. From these results, of the over six hundred transcription factors identified in this fungus, almost four hundred of them are influenced by hbx1 (Table S3). A subset of well-characterized genes from that list are also represented in Table 2.
Table 2. Annotated hbx1-dependent transcription factors. A list of A. flavus transcription factors was obtained from the Fungal Transcription Factor Database and compared to the list of hbx1 dependent DEGs. Annotations were retrieved from NCBI (full list is shown in Table S3). Expression values are those between the wild type (WT) and Δhbx1 at all time points assayed.
| Gene | AFLA ID | Description | 6 h | 24 h | 48 h |
|---|---|---|---|---|---|
| abp2 | AFLA_081210 | ARS binding protein Abp2, putative | −0.02695 | −1.05237 | −0.83165 |
| aflO | AFLA_139220 | aflO/ omtB/ dmtA/ O-methyltransferase B | −8.01939 | −10.9727 | −2.94881 |
| aflP | AFLA_139210 | aflP/ omtA/ omt-1/ O-methyltransferase A | −9.18684 | −11.8992 | −2.50201 |
| aflR | AFLA_139360 | aflR / apa-2 / afl-2 / transcription activator | −6.93292 | −5.75199 | −5.3899 |
| amdA | AFLA_048870 | C2H2 transcription factor (AmdA), putative | −1.27305 | 0.369593 | −0.66252 |
| amdR | AFLA_028560 | C6 transcription factor (AmdR), putative | −1.0392 | −0.75555 | −0.66391 |
| amdX | AFLA_002290 | C2H2 transcription factor (AmdX), putative | −1.02402 | −1.62432 | −0.7989 |
| amyR | AFLA_026160 | C6 transcription factor (AmyR), putative | 0.718895 | 1.399027 | 1.196918 |
| areA | AFLA_049870 | GATA transcriptional activator AreA | −0.76928 | −2.688 | 0.534173 |
| areB | AFLA_136100 | GATA transcription factor (AreB), putative | 0.136046 | −1.10235 | 0.413972 |
| azf1 | AFLA_054800 | C2H2 transcription factor (Azf1), putative | −0.59027 | −3.74734 | −3.02994 |
| brlA | AFLA_082850 | C2H2 type conidiation transcription factor BrlA | 0.391684 | −2.17902 | −2.37438 |
| cnjB | AFLA_051900 | zinc knuckle transcription factor (CnjB), putative | 3.893041 | 0.290236 | 2.115113 |
| creA | AFLA_134680 | C2H2 transcription factor (Crea), putative | −0.50862 | −0.56638 | −1.0953 |
| ctf1B | AFLA_012010 | C6 transcription factor (Ctf1B), putative | −0.04607 | 0.620485 | 1.55645 |
| erg2 | AFLA_069460 | C2H2 transcription factor (Egr2), putative | −1.55121 | −0.57147 | −1.05583 |
| flbC | AFLA_137320 | C2H2 conidiation transcription factor FlbC | −0.72449 | −2.36931 | −0.71241 |
| flbD | AFLA_080170 | MYB family conidiophore development protein FlbD, putative | −1.18991 | −2.3671 | −3.69073 |
| hpa3 | AFLA_131640 | HLH transcription factor (Hpa3), putative | −0.97532 | −1.5551 | −2.37946 |
| MAT-α-1 | AFLA_103210 | mating-type protein MAT alpha 1 | 0.301373 | 0.231333 | 3.258805 |
| mbf1 | AFLA_086430 | coactivator bridging factor 1 (Mbf1), putative | 0.637772 | 1.803474 | 0.237383 |
| nirA | AFLA_093040 | C6 transcription factor (NirA), putative | −0.36009 | −1.05781 | 0.124633 |
| nosA | AFLA_025720 | C6 transcription factor NosA | −5.37861 | −8.60563 | −3.24851 |
| nsdD | AFLA_020210 | sexual development transcription factor NsdD | −0.29566 | −1.18101 | −0.97544 |
| pcaG | AFLA_012100 | NDT80_PhoG domain protein PcaG | −1.3881 | −1.5446 | −0.30405 |
| regA | AFLA_073870 | C6 transcription factor RegA, putative | −0.25045 | 0.20309 | 1.643673 |
| rfeC | AFLA_044060 | C2H2 transcription factor (RfeC), putative | −0.42645 | −1.10049 | 0.597632 |
| rpn4 | AFLA_017640 | C2H2 transcription factor (Rpn4), putative | 0.491655 | 0.477596 | 3.176475 |
| seb1 | AFLA_110650 | C2H2 transcription factor (Seb1), putative | −0.18746 | −0.02585 | 1.021753 |
| sep1 | AFLA_048110 | forkhead transcription factor (Sep1), putative | −0.65362 | −1.43862 | −0.5192 |
| snt2 | AFLA_029990 | PHD finger and BAH domain protein (Snt2), putative | 0.07615 | −0.12486 | 1.163666 |
| srrA/skn7 | AFLA_034540 | stress response transcription factor SrrA/Skn7, putative | −0.77849 | −1.28158 | 0.119597 |
| ssb3 | AFLA_093820 | ssDNA binding protein Ssb3, putative | −0.00201 | 0.813483 | −1.02252 |
| steA | AFLA_048650 | sexual development transcription factor SteA | −0.53006 | −1.08547 | −0.48395 |
| stuA | AFLA_046990 | APSES transcription factor StuA | −1.62823 | −2.5412 | −1.79524 |
| swi5 | AFLA_031400 | C2H2 transcription factor Swi5 | −0.54244 | −1.11596 | −0.0182 |
Prediction of hbx1-dependent genes possibly involved in virulence
Previously Dolezal et al. (2013) analyzed the transcriptome of A. flavus during the infection of maize kernels. This study compared the gene expression profile of A. flavus during infection of viable kernels to that of non-viable. The data from this study was assessed into two groups, genes that were upregulated during infection of viable kernels and those downregulated. We compared these groups of genes to the hbx1-dependent transcriptome from our study. In total, 1125, 1451, and 1672 genes were differentially expressed in both studies at the 6, 24, and 48 h, respectively (Table S4). Table S4 also shows genes that exhibited the same trend over three-time points as well as genes that presented an expression pattern opposite that depicted in the Dolezal et al. virulence study compared to hbx1-dependent DEGs. Among them, 75 genes were found upregulated during infection of viable seeds but downregulated in the hbx1 mutant. Conversely 20 genes were found to present lower expression levels during infection while having increased expression in the hbx1mutant.
In the aforementioned study, genes encoding transcription factors, involved in secondary metabolism, as well as the fungal secretome were upregulated and predicted to be potential virulence factors (Dolezal et al., 2013). We further compared differentially expressed secretory genes from that study to the hbx1 transcriptome study. From this selected data set, 164, 196, and 235 secretory genes where differentially expressed in the absence of hbx1 at 6, 24, and 48 h respectively (Table S5).
Identification and visualization of gene regulatory networks correlated with hbx1 expression and knockout
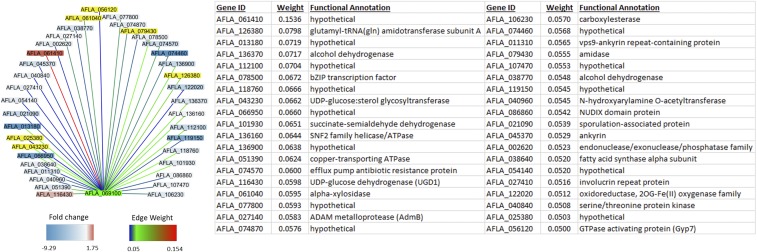
Expression values (variant stabilized read counts) from the RNA-seq data were subjected to WGCNA to identify networks of genes that are co-expressed with wild-type hbx1, and therefore potentially impacted by the hbx1 deletion. The homeobox transcription factor showed highest co-expression correlation with a hypothetical protein (AFLA_061410), (Figure 7), which was significantly upregulated in the hbx1 mutant (∼4 fold), and shared a high sequence similarity to a putative DNA methyltransferase. The putative hypothetical proteins down-regulated in the hbx1 mutant (AFLA_013180 and AFLA_066950) share a sequence similarity with an S-adenosyl-methionine dependent methyltransferase and a flavin adenine dinucleotide -binding proteins, respectively. These putative methyltranferases highly associated with hbx1 expression (and affected in the hbx1 mutant) are excellent candidate regulatory genes for near-downstream regulation by hbx1.
Figure 7.
Weighted Gene Co-expression Network Analysis was conducted using read counts from the isogenic control strain to identify genes co-expressed with hbx1 (AFLA_069100) and illustrated using Cytoscape (left). Edge coloration reflects the TOM value, indicating the relative significance of gene Co-expression (“Edge Weight”). The Node color reflects gene rlog2 fold changes in the 24 h sample of hbx1 knockout mutant relative to control (“Fold change”). Yellow indicates no change in relative expression levels. The functional annotation from Aspergillus flavus strain 3357 (Accession: GCA_000006275.2) is indicated for each AFLA gene Identifier (right).
Discussion
Previous studies showed that development is genetically associated with secondary metabolism, (Calvo et al. 2002; Calvo and Cary 2015). Recent published work also demonstrated that the hbx1 gene is one of those genetic links that is required for normal conidiation, sclerotial formation as well as secondary metabolism in the aflatoxin-producer and agriculturally important fungus A. flavus (Cary et al., 2017). In the current study we showed that while homeobox domains similar to that present in the Hbx1 protein are found throughout various phyla, the rest of the A. flavus Hbx1 amino acid sequence is not conserved in planta. This suggests that hbx1 could be a good target for strategies to control A. flavus infection that would not result in any off-target effects in agriculturally important crops susceptible to this opportunistic pathogen.
To gain insight into A. flavus hbx1 mechanism of action we investigated the extent of its regulatory scope performing a transcriptome study using RNA sequencing. Our analyses revealed a broad effect of hbx1 on the genome; in the absence of hbx1 more than 20% of the A. flavus genome presents changes in expression. In addition, we observed that the number of genes governed by hbx1 increases with time in the fungal culture. Based on our results, Hbx1 is a dynamic transcriptional regulator that, while it controls the expression of 857 genes at all time points assessed, a greater number of hbx1-dependent genes were affected at specific time points analyzed.
Our functional enrichment analysis indicated that while categories involved in cell rescue and defense, development and cellular transport were shown to be under the control of hbx1, the largest group of hbx1-dependent genes corresponds to the category of metabolism. Twelve secondary metabolite gene clusters, out of 56 clusters identified in A. flavus, including the kojic acid cluster (Ehrlich and Mack, 2014 & Ammar et al. 2017), were under hbx1 control. Of these 12 clusters, five of them were not associated with a known metabolic product, however the remaining clusters have already been characterized. Four of these clusters have been shown to be involved in the synthesis of potent mycotoxins, aflatoxin, aflatrem (split into two clusters), and cyclopiazonic acid (Yu et al., 2004, Chang et al., 2009, Nicholson et al., 2009). In addition, genes in the clusters involved in the production of asparasone, piperazine, and aflavarin were also shown to be suppressed in the absence of hbx1. These three metabolites are associated with sclerotial development (Calvo and Cary 2015). Both asparasone and aflavarin are specifically found within these structures (Cary et al. 2014, & Cary et al. 2015), and genes located in the piperazine cluster have been shown to affect their development (Forseth et al. 2012). Fungi concentrate secondary metabolites in reproductive structures for defense against herbivores and insects (Wicklow 1988, Gloer 1995, Gloer 1997, & Gloer 2007). Horn et al. (2009, 2014) reported ascospore-bearing ascocarps embedded within sclerotia of A. flavus and A. parasiticus. In these aflatoxin-producers, sclerotia play an important role as resting structures capable of surviving environmental extremes remaining viable after several years in the crop fields (Coley-Smith and Cooke 1971), and the hbx1-dependent secondary metabolites present in them contribute to their survival against biotic stress and possibly abiotic stresses. Since deletion of hbx1 results in abolishment of sclerotia in the fungus, it is possible that the effect of hbx1 on the expression of some of these secondary metabolite gene clusters specifically associated with a particular morphological structure could be indirect, by affecting developmental regulators that are repressed in the absence of hbx1. Whether the effect on these clusters is direct or indirect, these studies indicate an important role of hbx1 in A. flavus survival, promoting the formation of resistant structures and a chemical arsenal critical for defense against microbes, predators and other environmental insults.
The hbx1 gene is also necessary for conidiation. Our transcriptome analysis also indicated that the brlA central regulatory pathway is under hbx1 control, not only affecting brlA, but also wetA, a developmental regulator conserved in Aspergillus species (Wu et al., 2018). Furthermore, the aconidial phenotype of Δhbx1 resemble that of the fluffy mutants described in A. nidulans that revealed the flb regulatory pathway (reviewed by Ruger-Herreros et al. 2011; Krijgsheld et al. 2013). Indeed, A. flavus flbA, flbC, flbD, and flbE homologs (Chang et al., 2012) are down regulated in the absence of hbx1 while in the same strain at the early time point fluG had a significant increase of expression. This indicates that hbx1 is a regulator of these conidiophore biogenesis genes, and expression of some of these genes over time is significantly different from that observed in the control strain.
It is possible that Hbx1 might not bind directly to the promoters of the central regulatory pathway genes but affects their expression by controlling expression of genes upstream in the regulatory hierarchy. Examples of these might be genes like ppoC and stuA. Both of these genes have been shown to affect conidiophore development via brlA (Dutton et al., 1997, Tsitsigiannis et al., 2004, & Sheppard et al., 2005). In addition, other developmental genes were also under the influence of hbx1, for instance, the spore hydrophin gene rodA (Carrion et al., 2013) and also the nosA gene, encoding a putative Zn(II)(2)Cys(6) transcription factor previously described in several Aspergillus species (Vienken and Fischer 2006, Soukup et al., 2012, Zhao et al., 2017). In A. nidulans, nosA is necessary for cleistothecial primordium maturation (Vienken and Fischer 2006), and its homolog in A. flavus has been reported to be required for sclerotial production (Zhao et al., 2017). It is likely that the reduction in the expression of nosA in the A. flavus hbx1 mutant could contribute to prevent sclerotial formation in this strain.
Since hbx1 has a broad effect on A. flavus development and metabolism, we also investigated possible connections between hbx1 and virulence during corn infection based on the previous report by Dolezal et al. (2013). In our study, the DEGs identified from the hbx1-dependent transcriptome were compared to those identified by Dolezal et al. (2013) in viable and non-viable infected corn kernels. This allowed us to predict genes possibly involved in virulence that are controlled by hbx1. Genes identified as upregulated in the corn infection study but suppressed in the Δhbx1 transcriptome study could potentially be involved in virulence. Approximately 300 DEGs were identified at all time points that fit this description. Among them is the pes1 gene (AFLA_069330), that in Aspergillus fumigatus was found indispensable for virulence in the Galleria mellonella model (Reeves et al., 2006). Other genes in this group were shown to affect spore germination and secondary metabolism, such as sfk1. In Penicillium roqueforti, silencing of sfk1 alters condial germination and prevents production of roquefortine C, andrastin A, and mycophenolic acid (Torrent et al., 2017). Out of the mentioned group of 300 DEGs, 75 were consistently suppressed in the Δhbx1 mutant (Table S4). In this subgroup, beyond genes located in the aflatoxin gene cluster, most of these genes have not been investigated, and could be potential genes of interest in future studies to identify A. flavus virulence factors.
Secreted proteins such as hydrolytic enzymes are essential for successful infection of the host by the fungus (Lo Presti et al., 2015 -de Jonge et al., 2011 & Kale and Tyler 2011). With this in mind, we focused on analyzing components of the secretome regulated by hbx1, specifically those genes that may play a role in virulence. FunSecKB2 analysis revealed that among the genes in the Dolezal et al. (2013) study, approximately 50 secretome-related genes were upregulated during infection of viable seeds, while those same genes were downregulated in our transcriptome study of the hbx1 deletion mutant, at least at one time point, suggesting that this set of hbx1-dependent genes could be potentially be involved in virulence, for example genes encoding proteases (i.e.AFLA_057670), amylases (i.e., AFLA_123170) and other hydrolases (i.e., AFLA_065010, AFLA_088610, and AFLA_125970).
Weighted gene co-expression network analysis has been used to identify novel gene interactions by determining patterns of co-expression among several biological samples, which infers a functional relationship between genes. This process has been used to analyze RNA-seq data from Aspergillus species (Baltussen et al. 2018, Korani et al. 2018), and functional studies have demonstrated the validity of WGCNA (Calabrese et al., 2017, Wang et al. 2017). Here we identify the network of co-expressed genes using the isogenic control strain and identified several genes that are both significantly co-expressed with hbx1 and show altered expression patterns in the hbx1 knockout mutant. Three genes of particular interest that demonstrated altered regulation and relatively high correlation values are annotated as hypothetical proteins (AFLA_ 013180, AFLA_061410, and AFLA_066950). The impact of these hypothetical genes on A. flavus biology will be the focus of future studies of hbx1-dependent gene regulation.
We demonstrated that the expression of thousands of genes is affected by hbx1. In addition, we showed that hbx1-dependent regulation in A. flavus is dynamic in a time-dependent manner. The hbx1 gene is required for the production of structures needed for dissemination and survival of A. flavus and the production of detrimental secondary metabolites. This, together with the fact that Hbx1 is not conserved in other phyla suggest that this global regulator could be a target to develop novel methodologies to control the adverse health and economic impacts due to infection and aflatoxin contamination of many important crops by A. flavus.
Achnowledgements
This work was supported by USDA grant 58-6435-4-015 and the Department of Biological Sciences at Northern Illinois University. SE and YY are supported by NSF (DBI-1652164) and NIH (1R15GM114706) grants.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7304252.
Communicating editor: A. Rokas
Literature Cited
- Adams T., Boylan M., Timberlake W., 1998. BrlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 54: 353–362. pmid:3293800 [DOI] [PubMed]
- Ammar H. A., Srour A. Y., Ezzat S. M., Hoseny A. M., 2017. Identification and characterization of genes involved in kojic acid biosynthesis in Aspergillus flavus. Ann. Microbiol. 67: 691–702. 10.1007/s13213-017-1297-8 [DOI] [Google Scholar]
- Baltussen T. J. H., Coolen J. P. M., Zoll J., Verweij P. E., Melchers W. J. G., 2018. Gene co-expression analysis identifies gene clusters associated with isotropic and polarized growth in Aspergillus fumigatus conidia. Fungal Genet. Biol. 116: 62–72. 10.1016/j.fgb.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Basenko E., Pulman J., Shanmuga A., Harb O., Crouch K., et al. , 2018. FungiDB: an integrated bioinformatic resource for fungi and oomycetes. J. Fungi (Basel) 4: 39 10.3390/jof4010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar D., Rajasekaran K., Gilbert M., Cary J. W., Magan N., 2018. Advances in molecular and genomic research to safeguard food and feed supply from aflatoxin contamination. World Mycotoxin J. 11: 47–72. 10.3920/WMJ2017.2283 [DOI] [Google Scholar]
- Buchanan R. L., Lewis D. F., 1984. Regulation of aflatoxin biosynthesis: Effect of glucose on activities of various glycolytic enzymes. Appl. Environ. Microbiol. 48: 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese G. M., Mesner L. D., Stains J. P., Tommasini S. M., Horowitz M. C., et al. , 2017. Integrating GWAS and co-expression network data identifies bone mineral density genes SPTBN1 and MARK3 and an osteoblast functional module. Cell Syst. 4: 46–59.e4. 10.1016/j.cels.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A. M., Wilson R. A., Bok J. W., Keller N. P., 2002. Relationship between Secondary Metabolism and Fungal Development. Microbiology and Molecular Biology Reviews, Cell Syst. 66: 447–459. 10.1128/mmbr.66.3.447-459.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A. M., Cary J. W., 2015. Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol. 6 10.3389/fmicb.2015.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A. M., Lohmar J. M., Ibarra B., Satterlee T., 2016. Velvet Regulation of Fungal Development, Growth, Differentiation and Sexuality. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research), Vol. I, edited by Wendland J. Springer, Cham. [Google Scholar]
- Carrion S. D., Leal S. M., Ghannoum M. A., Aimanianda V., Latge J., et al. , 2013. The RodA hydrophobin on Aspergillus fumigatus spores masks dectin-1- and dectin-2-dependent responses and enhances fungal survival in vivo. J. Immunol. 191: 2581–2588. 10.4049/jimmunol.1300748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J., Harris-Coward P., Scharfenstein L., Mack B., Chang P. K., Wei Q., 2017. The Aspergillus flavus Homeobox Gene, hbx1, Is Required for Development and Aflatoxin Production. Toxins. 9: 315 10.3390/toxins9100315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J. W., Han Z., Yin Y., Lohmar J. M., Shantappa S., et al. , 2015. Transcriptome analysis of Aspergillus flavus Reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot. Cell 14: 983–997. 10.1128/EC.00092-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J. W., Harris-Coward P. Y., Ehrlich K. C., Mack B. M., Kale S. P., et al. , 2012. NsdC and NsdD Affect Aspergillus flavus Morphogenesis and Aflatoxin Production. Eukaryot. Cell 11: 1104–1111. 10.1128/EC.00069-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J. W., Harris-Coward P. Y., Ehrlich K. C., Mavungu J. D., Malysheva S. V., et al. , 2014. Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genet. Biol. 64: 25–35. 10.1016/j.fgb.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Chang P. K., Cary J. W., Bhatnagar D., Cleveland T. E., Bennett J. W., et al. , 1993. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 59: 3273–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P., Horn B. W., Dorner J. W., 2009. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet. Biol. 46: 176–182. 10.1016/j.fgb.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Chang P. K., Scharfenstein L. L., Mack B., Ehrlich K. C., 2012. Deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Appl. Environ. Microbiol. 78: 7557–7563. 10.1128/AEM.01241-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Boutros P. C., 2011. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley-Smith J. R., Cooke R. C., 1971. Survival and germination of fungal sclerotia. Annu. Rev. Phytopathol. 9: 65–92. 10.1146/annurev.py.09.090171.000433 [DOI] [Google Scholar]
- Dhingra S., Lind A. L., Lin H., Tang Y., Rokas A., et al. , 2013. The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS One 8: e77147 10.1371/journal.pone.0077147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal A. L., Obrian G. R., Nielsen D. M., Woloshuk C. P., Boston S. R., et al. , 2013. Localization, morphology and transcriptional profile of Aspergillus flavus during seed colonization. Mol. Plant Pathol. 14: 898–909. 10.1111/mpp.12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran R. M., Cary J. W., Calvo A. M., 2007. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73: 1158–1168. 10.1007/s00253-006-0581-5 [DOI] [PubMed] [Google Scholar]
- Dutton J. R., Johns S., Miller B. L., 1997. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 16: 5710–5721. 10.1093/emboj/16.18.5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich K., Mack B., 2014. Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins (Basel) 6: 1916–1928. 10.3390/toxins6061916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forseth R. R., Amaike S., Schwenk D., Affeldt K. J., Hoffmeister D., et al. , 2012. Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew. Chem. Int. Ed. 52: 1590–1594. 10.1002/anie.201207456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A., Etxebeste O., Herrero-García E., Ugalde U., Espeso E. A., 2010. The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Mol. Microbiol. 75: 1314–1324. 10.1111/j.1365-2958.2010.07063.x [DOI] [PubMed] [Google Scholar]
- Gloer J. B., 1995. Antiinsectan natural products from fungal sclerotia. Acc. Chem. Res. 28: 343–350. 10.1021/ar00056a004 [DOI] [Google Scholar]
- Gloer J. B., 1997, pp. 249–268 in “Applications of fungal ecology in the search for new bioactive natural products,” in The Mycota, Ed. 1st, edited by Wicklow D. T., Soderstrom B. Springer, Berlin. [Google Scholar]
- Gloer J. B., 2007. “Applications of fungal ecology in the search for new bioactive natural products,” in The Mycota, Vol. IV, Ed. 2nd, edited by Kubicek C. P., Druzhinina I. S. Springer-Verlag, New York, NY. [Google Scholar]
- Horn B. W., Geromy G., Carbone M., Carbone I., 2009. Sexual reproduction in Aspergillus flavus. Mycologia 101: 423–429. 10.3852/09-011 [DOI] [PubMed] [Google Scholar]
- Horn B. W., Sorensen R. B., Lamb M. C., Sobolev V. S., Olarte R. A., et al. , 2014. Sexual reproduction in Aspergillus flavus sclerotia naturally produced in corn. Phytopathology 104: 75–85. 10.1094/PHYTO-05-13-0129-R [DOI] [PubMed] [Google Scholar]
- de Jonge R., Bolton M. D., Thomma B. P., 2011. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Curr. Opin. Plant Biol. 14: 400–406. 10.1016/j.pbi.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Kale S. D., Tyler B. M., 2011. Entry of oomycete and fungal effectors into plant and animal host cells. Cell. Microbiol. 13: 1839–1848. 10.1111/j.1462-5822.2011.01659.x [DOI] [PubMed] [Google Scholar]
- Kato N., Brooks W., Calvo A. M., 2003. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans Is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2: 1178–1186. 10.1128/EC.2.6.1178-1186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L., 2015. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korani W., Chu Y., Holbrook C. C., Ozias-Akins P., 2018. Insight into genes regulating postharvest aflatoxin contamination of tetraploid peanut from transcriptional profiling. Genetics. 209143–56. Epub 2018/03/17. 10.1534/genetics.118.300478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijgsheld P, Bleichrodt R., van Veluw G. J., Wang F., Müller W. H., et al. , 2013. Development in Aspergillus. Stud Mycol. 74: 1–29. pmid:23450714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon N., Garzia A., Espeso E. A., Ugalde U., Yu J., 2010. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77: 1203–1219. 10.1111/j.1365-2958.2010.07282.x [DOI] [PubMed] [Google Scholar]
- Langfelder P., Horvath S., 2008. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W., 2013. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lind A. L., Wisecaver J. H., Smith T. D., Feng X., Calvo A. M., et al. , 2015. Examining the evolution of the regulatory circuit controlling secondary metabolism and development in the fungal genus Aspergillus. PLoS Genet. 11: e1005096 10.1371/journal.pgen.1005096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmar J. M., Harris-Coward P. Y., Cary J. W., Dhingra S., Calvo A. M., 2016. RtfA, a putative RNA-Pol II transcription elongation factor gene, is necessary for normal morphological and chemical development in Aspergillus flavus. Appl. Microbiol. Biotechnol. 100: 5029–5041. 10.1007/s00253-016-7418-7 [DOI] [PubMed] [Google Scholar]
- Meinken J., Asch D. K., Neizer-Ashun K. A., Chang G., Jr., Min X. J., 2014. FunSecKB2: A fungal protein subcellular location knowledgebase. Computational Molecular Biology, 4: 1–17. 10.5376/cmb.2014.04.0007 [DOI] [Google Scholar]
- Nicholson M. J., Koulman A., Monahan B. J., Pritchard B. L., Payne G. A., et al. , 2009. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 75: 7469–7481. 10.1128/AEM.02146-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman W. C., Yu J., Fedorova-Abrams N. D., Losada L., Cleveland T. E., et al. , 2015. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announc. 3: e00168-15 10.1128/genomeA.00168-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Park J., Jang S., Kim S., Kong S., et al. , 2008. FTFD: An informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24: 1024–1025. 10.1093/bioinformatics/btn058 [DOI] [PubMed] [Google Scholar]
- Lo Presti L., Lanver G., Schweizer S., Tanaka L., Liang L., et al. , 2015. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 66: 513–545. 10.1146/annurev-arplant-043014-114623 [DOI] [PubMed] [Google Scholar]
- Priebe S., Kreisel C., Horn F., Guthke R., Linde J., 2014. FungiFun2: A comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 31: 445–446. 10.1093/bioinformatics/btu627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purschwitz J., Müller S., Kastner C., Schöser M., Haas H., et al. , 2008. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 18: 255–259. 10.1016/j.cub.2008.01.061 [DOI] [PubMed] [Google Scholar]
- R Core Team , 2017. R: A Language and Environment for Statistical Computing. https://www.R-project.org/
- Ramirez-Prado J. H., Moore G. G., Horn B. W., Carbone I., 2008. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 45: 1292–1299. 10.1016/j.fgb.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Reeves E. P., Reiber K., Neville C., Scheibner O., Kavanagh K., et al. , 2006. A nonribosomal peptide synthetase (Pes1) confers protection against oxidative stress in Aspergillus fumigatus. FEBS J. 273: 3038–3053. 10.1111/j.1742-4658.2006.05315.x [DOI] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., et al. , 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robens J., Cardwell K., 2003. The costs of mycotoxin management to the USA: management of aflatoxins in the United States. J. Toxicol. Toxin Rev. 22: 139–152. 10.1081/TXR-120024089 [DOI] [Google Scholar]
- Robinson M. D., Mccarthy D. J., Smyth G. K., 2009. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruger-Herreros C., Rodríguez-Romero J., Fernández-Barranco R., Olmedo M., Fischer R., et al. , 2011. Regulation of conidiation by light in Aspergillus nidulans. Genetics 188: 809–822. 10.1534/genetics.111.130096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlee T., Cary J. W., Calvo A. M., 2016. RmtA, a putative arginine methyltransferase, regulates secondary metabolism and development in Aspergillus flavus. PLoS One 11: e0155575 10.1371/journal.pone.0155575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard D. C., Doedt T., Chiang L. Y., Kim H. S., Chen D., et al. , 2005. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 16: 5866–5879. 10.1091/mbc.e05-07-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup A. A., Farnoodian M., Berthier E., Keller N. P., 2012. NosA, a transcription factor important in Aspergillus fumigatus stress and developmental response, rescues the germination defect of a laeA deletion. Fungal Genet. Biol. 49: 857–865. 10.1016/j.fgb.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent C., Gil-Durán C., Rojas-Aedo J. F., Medina E., Vaca I., et al. , 2017. Role of sfk1 gene in the filamentous fungus Penicillium roqueforti. Front. Microbiol. 8: 2424 10.3389/fmicb.2017.02424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis D. I., Kowieski T. M., Zarnowski R., Keller N. P., 2004. Endogenous lipogenic regulators of spore balance in Aspergillus nidulans. Eukaryot. Cell 3: 1398–1411. 10.1128/EC.3.6.1398-1411.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vienken K., Fischer R., 2006. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol. Microbiol. 61: 544–554. 10.1111/j.1365-2958.2006.05257.x [DOI] [PubMed] [Google Scholar]
- Wang T., He X., Liu X., Liu Y., Zhang W., Huang Q., et al. , 2017. Weighted gene co-expression network analysis identifies FKBP11 as a key regulator in acute aortic dissection through a NF-kB dependent pathway. Frontiers in Physiology. 8:1010. 10.3389/fphys.2017.01010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner M. H., Roinick K. L., Arndt K. M., 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27: 6103–6115. 10.1128/MCB.00772-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., 2009. 2009 ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag, New York. [Google Scholar]
- Wicklow D. T., 1988, pp. 173–201 in “Metabolites in the coevolution of fungal chemical defence systems,” in Coevolution of Fungi with Plants and Animals, edited by Pirozynski K. A., Hawksworth D. Academic Press, New York, NY. [Google Scholar]
- Wu F., Liu Y., Bhatnagar D., 2008. Cost-Effectiveness Of Aflatoxin Control Methods: Economic Incentives. Toxin Rev. 27: 203–225. 10.1080/15569540802393690 [DOI] [Google Scholar]
- Wu M., Mead M. E., Kim S., Rokas A., Yu J., 2017. WetA bridges cellular and chemical development in Aspergillus flavus. PLoS One 12 10.1371/journal.pone.0179571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.-Y., Mead M. E., Lee M.-K., Ostrem Loss E. M., Kim S. C., et al. , 2018. Systematic dissection of the evolutionarily conserved WetA developmental regulator across a genus of filamentous fungi. mBio 9: e01130–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yard E. E., Daniel J. H., Lewis L. S., Rybak M. E., Paliakov E. M., et al. , 2013. Human aflatoxin exposure in Kenya, 2007: A cross-sectional study. Food Additives & Contaminants: Part A 30: 1322–1331. 10.1080/19440049.2013.789558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Chang P., Ehrlich K. C., Cary J. W., Bhatnagar D., et al. , 2004. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70: 1253–1262. 10.1128/AEM.70.3.1253-1262.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Spraker J.E., Bok J.W., Velk T., He Z.M., Keller N.P. 2017. A cellular fusion cascade regulated by LaeA is required for sclerotial development in Aspergillus flavus Front Microbiol. Oct 5:1925. eCollection 2017. 10.3389/fmicb.2017.01925 [DOI] [PMC free article] [PubMed]
- Zhuang Z., Lohmar J., Satterlee T., Cary J. W., Calvo A. M., 2016. The master transcription factor mtfA governs aflatoxin production, morphological development and pathogenicity in the fungus Aspergillus flavus. Toxins (Basel) 8: 29 10.3390/toxins8010029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Table S1 contains calculated expression values of sequenced RNA samples along with corresponding p-values. Table S2 contains a selected list of fungal developmental regulators that are shown to be hbx1-dependent. Table S3 is a subset of Table S1 that shows all known transcription factors in A. flavus and their corresponding expression pattern in regard to presence or absence of hbx1. The data are publicly available at NCBI’s SRA repository with the SRA Accession #: PRJNA494425. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7304252.