Abstract
The insect mushroom body (MB) is a conserved brain structure that plays key roles in a diverse array of behaviors. The Drosophila melanogaster MB is the primary invertebrate model of neural circuits related to memory formation and storage, and its development, morphology, wiring, and function has been extensively studied. MBs consist of intrinsic Kenyon Cells that are divided into three major neuron classes (γ, α′/β′ and α/β) and 7 cell subtypes (γd, γm, α′/β′ap, α′/β′m, α/βp, α/βs and α/βc) based on their birth order, morphology, and connectivity. These subtypes play distinct roles in memory processing, however the underlying transcriptional differences are unknown. Here, we used RNA sequencing (RNA-seq) to profile the nuclear transcriptomes of each MB neuronal cell subtypes. We identified 350 MB class- or subtype-specific genes, including the widely used α/β class marker Fas2 and the α′/β′ class marker trio. Immunostaining corroborates the RNA-seq measurements at the protein level for several cases. Importantly, our data provide a full accounting of the neurotransmitter receptors, transporters, neurotransmitter biosynthetic enzymes, neuropeptides, and neuropeptide receptors expressed within each of these cell types. This high-quality, cell type-level transcriptome catalog for the Drosophila MB provides a valuable resource for the fly neuroscience community.
Keywords: Drosophila, mushroom body, transcriptome, RNA-seq
Drosophila melanogaster is a powerful model system for behavioral neuroscience. The fly model takes advantage of a relatively simple brain that expresses homologous suites of genes and orchestrates a conserved yet highly diverse and elaborate suit of behaviors. Behavioral genetics in Drosophila affords the means to identify individual genes that function within identified neuronal cell types, whose connectivity and functional roles in behavior can be elucidated. The ability to form memories of past experience and to orchestrate adaptive and plastic changes in behavioral responses is an example of a fundamental field of behavioral neuroscience where Drosophila neurogenetics has made major contributions (Heisenberg 2003; Davis 2005; Margulies et al. 2005; Keene and Waddell 2007). Memory research in flies has led to the identification of fundamental cellular mechanisms of memory such as cAMP signaling and CREB-mediated gene transcription (Yin and Tully 1996; Heisenberg 2003; Davis 2005; Margulies et al. 2005; Keene and Waddell 2007), and also has contributed to our understanding of how memories are processed in a complex neural circuit. A primary site of associative learning in insects is the mushroom body (MB) (Strausfeld et al. 1998; Heisenberg 2003; Davis 2005; Margulies et al. 2005; Keene and Waddell 2007; Menzel 2012; Farris 2013), a paired brain structure that in Drosophila is comprised of approximately 2000 intrinsic Kenyon Cells (KCs) per hemisphere. MBs in fruit flies are critical sites of olfactory, visual and gustatory learning (Heisenberg 2003; Davis 2005; Margulies et al. 2005; Keene and Waddell 2007; Vogt et al. 2014; Masek and Keene 2016), and also play important roles in other behavioral contexts such as temperature preferences (Hong et al. 2008), sleep (Artiushin and Sehgal 2017) and responses to ethanol exposure (Kaun et al. 2011).
MB dependent plasticity is one of the most intensely studied aspects of invertebrate neurobiology. The morphology and developmental lineage of the neurons that populate the MB in Drosophila, as well as the identity and morphology of most of their neuronal inputs and outputs, have been fully characterized (Ito et al. 1998; Jefferis et al. 2002; Aso et al. 2014a; 2014b). Many functional manipulations of both neural activity and signaling pathways relevant to plasticity have been conducted within each of the identified neuronal cell types in this circuit (Connolly et al. 1996; Zars et al. 2000; Dubnau et al. 2001; McGuire et al. 2001; Isabel et al. 2004; Krashes et al. 2007; Blum et al. 2009; Trannoy et al. 2011; Qin et al. 2012; Huang et al. 2012; Cervantes-Sandoval et al. 2013; Perisse et al. 2013; Bouzaiane et al. 2015). Functional imaging studies have established neural activity correlates in behaving animals (Davis 2011). Together, these studies support the conclusion that the neurons of the MB play unique roles in memory acquisition, storage and retrieval. Moreover, memory storage over the course of minutes and hours after training relies on an evolving requirement for reverberating neural activity within a circuit that includes MB intrinsic neurons and the so-called extrinsic neurons that provide inputs and outputs (Dubnau and Chiang 2013; Cognigni et al. 2018). In contrast to the increasingly deep understanding of the development, connectivity and functional requirements of each cell type in this circuit, there is a surprising paucity of data on differences in their transcriptional profiles.
MB KCs can be divided into three major classes, γ, α′/β′ and α/β, based on the projection patterns of the axons (Crittenden et al. 1998; Lee et al. 1999). Extensive anatomical and functional characterization corroborates this classification as biologically relevant (Keene and Waddell 2007; Davis 2011; Dubnau and Chiang 2013). Gene expression differences between these three classes of MB KCs have been studied using microarray (Perrat et al. 2013), RNA sequencing (RNA-seq) (Crocker et al. 2016) and single-cell RNA-seq (Croset et al. 2018; Davie et al. 2018). However, it is known that the three KC classes can be further separated into seven subtypes: γd, γm, α′/β′ap, α′/β′m, α/βp, α/βs and α/βc KCs. The functional relevance of this further subdivision is supported by expression of split-GAL4 lines, analysis of the axonal projection patterns of individual neurons from each cell subtype (Luan et al. 2006; Aso et al. 2014a) and investigation of their functional roles in behavior (Perisse et al. 2013; Sitaraman et al. 2015a; Vogt et al. 2016; Yang et al. 2016). Until now, there have been no attempts to identify the unique transcriptional programs that control/establish the identity of each of these seven cell subtypes, while cell clustering using a resource of enormous single-cell transcriptional profiles showed only three MB KC clusters (Davie et al. 2018). But the availability of intersectional genetics approaches that make use of split-GAL4 provide the means to investigate each of the subtypes individually (Luan et al. 2006; Pfeiffer et al. 2008; Aso et al. 2014a).
Several methods have been developed to characterize the transcriptional programs of specific cell types in flies, such as TRAP (Heiman et al. 2008), TU-tagging (Miller et al. 2009), and EC-tagging (Hida et al. 2017). Here, we used an improved version of the INTACT method (Henry et al. 2012), called tandem affinity purification of intact nuclei and RNA-sequencing (TAPIN-seq; Davis et al. 2018), to profile the nuclear transcriptomes of all seven MB neuronal subtypes that constitute the MB (Aso et al. 2014a). These transcriptomes revealed ∼350 genes with either class- or subtype-specific expression, including several well-known and many new class or subtype markers. Moreover, our data provide a full accounting of the input-output signaling properties for each of these neuron subtypes including neurotransmitter biosynthetic machinery, neuropeptides and neurotransmitter and neuropeptide receptors.
Materials and Methods
Fly stocks
Because quantitative traits such as gene expression profiles can be sensitive to genetic background, we created a newly isogenic subline of w1118 (isoCJ1), which was itself derived from Canton-S wild type as an inbred line many years ago (Yin et al. 1994). To generate the isogenic strain, we used ten-generations of single male and female sibling crosses to generate 10 independent isogenic strains. MJ2 was selected based on its ability to form comparable olfactory short-term memory performance to the parental strain in the standard Pavlovian task (Supplemental Material, Figure S1). The nuclear envelope epitope tagged transgene P{5XUAS-unc84::2XGFP}attP40, the pJFRC28 strain P{10XUAS-IVS-GFP-p10}attP2 and each of the split-GAL4 inserts were backcrossed into this new MJ2 wild type strain for five generations to equilibrate each to this isogenic background. For each split-GAL4 combination, we separately backcrossed the GAL4 activating domain and DNA-binding domain components, and then combined the two hemi-drivers as a split-GAL4 line in the MJ2 background thereafter using standard balancer chromosomes that had themselves been equilibrated to the MJ2 strain. The UAS-WM P{5XUAS-myr::GFP-V5-p2A-His2B::mCherry-HA} reporter strain was generated using standard approaches (Chang et al. 2018). Flies were cultured on standard cornmeal food using the standard cornmeal recipe from Bloomington stock center. Food was supplemented with antibiotics.
For imaging to characterize expression with each split-GAL4 strain that had been reconstituted in the MJ2 background, we used 2 – 5 day old male flies. For RNA-seq sample preparation, each split-GAL4 line was crossed to the P{5XUAS-unc84::2XGFP}attP40. 2 – 5 day old adult progeny for each genotype were collected and frozen in liquid nitrogen between 10 am and 7 pm.
TAPIN purification of nuclei
Fly heads from a mixed population of male and female flies were first isolated with a customized sieve. 400 frozen heads were added to 6 mL of 20 mM sodium acetate pH 8.5, 2.5 mM MgCl2, 250 mM sucrose, 0.5% NP-40, 0.6 mM spermidine, 0.2 mM spermine, 1 mM DTT, 1× complete protease inhibitor (Sigma: 5056489001), 0.5 mg/mL torula RNA (ThermoFisher: AM7118), 0.6 mg/mL carboxyl coated Dynabeads (ThermoFisher: 14306D) and 1.6 μg anti-GFP antibody (ThermoFisher: G10362). Homogenization was carried out on ice by 50 tractions in a Dounce homogenizer using the tight pestle followed by filtration over either a 10 or 20 μm cup filter (Partec: 0400422314, 040042315).
Released chromatin and broken nuclei were adsorbed to carboxyl coated magnetic beads for 30 min at 4° with constant rotation. Unbound antibody was removed by incubating the sample on ice for 20 min with 100 μL of washed UNOsphere SUPra resin (Bio-Rad: 1560218). After the resin was removed on a 10 μm cup filter and the carboxyl beads on a magnet stand, the nuclei-containing supernatant was mixed with an equal volume of 500 mM sodium acetate pH 8.5, 250 mM sucrose, 6 mM EGTA, 6 mM EDTA, 0.6 mM spermidine, 0.2 mM spermidine, 1 mM DTT, 1× complete protease inhibitor, 0.25 mg/mL torula RNA and 30 μL Protein A Dynabeads (ThermoFisher: 10002D). A 2-hour incubation on ice with occasional agitation was used to recover tagged nuclei. Bead-bound nuclei were then recovered on a magnet stand and washed twice with 250 mM sodium acetate pH 8.5, 250 mM sucrose and 0.1% NP-40. Nuclei were then released at 37° for 1 hr by incubation in 50 μL of 10 mM Tris pH 7.5, 2.5 mM MgCl2, 0.5 mM CaCl2, 250 mM sucrose, 0.1% NP-40, 1 mg/mL torula RNA, 40 units RNAsin (Promega: N2515), 2 units DNAseI (NEB: M0303L), 320 units IdeZ protease (NEB: P0770S). The sample was diluted to 100 μL with 10 mM Tris pH 7.5, 2.5 mM MgCl2, 0.5 mM CaCl2, 250 mM sucrose and 0.1% NP-40, EGTA was added to 1 mM and the suspension was rapidly triturated 100 times. After returning the sample to a magnet stand, 90 μL of buffer containing released nuclei was removed and added to 1.5 μL of Protein G Dynabeads (ThermoFisher: 10004D) that were previously resuspended in 10 μL of 10 mM Tris pH 7.5, 2.5 mM MgCl2, 0.5 mM CaCl2, 250 mM sucrose and 0.1% NP-40. The second binding reaction was run for 1 – 3 hr on ice with occasional agitation, followed by 2× 250 μL washes in 10 mM Tris pH 7.5, 2.5 mM MgCl2, 0.5 mM CaCl2, 250 mM sucrose and 0.1% NP-40. Prior to the last wash a 5 μL aliquot was removed for quantitation and the remainder of the sample was solubilized in Arcturus Picopure RNA extraction buffer (ThermoFisher: KIT0204).
RNA-seq library construction
Nuclear RNA was DNAseI treated and purified using the Arcturus PicoPure system exactly as instructed by the supplier. Purified RNA was mixed with a 1:100,000 dilution of ERCC standard RNA (ThermoFisher: 4456740) and amplified using the Nugen Ovation v2 system (Nugen: 7102-32). cDNA was then blunted, ligated to barcoded linkers (Nugen: 0319-32, 0320-32) and sequenced in paired-end mode on an Illumina HiSeq2500 to 125 nt read lengths.
RNA-seq data analysis
We trimmed RNA-seq reads (5nt from the 5′ end of the forward read, using seqtk option “trim -b 5”; https://github.com/lh3/seqtk) to remove non-transcript sequences introduced by the NuGen Ovation kit and then pseudo-aligned these to the Drosophila transcriptome (ENSEMBL release 91, BDGP6) using kallisto (Bray et al. 2016) to estimate transcript abundances. We also included sequences for the synthetic ERCC spike-in species and TAPIN reporter in the transcriptome index. After pseudo-alignment, we removed ERCC, TAPIN reporter, and ribosomal RNA entries and renormalized the transcript abundance matrix to units of transcripts per million (TPM). To visualize TAPIN-seq signal across the genome, we also aligned trimmed reads to the whole genome (BDGP6, dm6) using STAR (Dobin et al. 2013), created bigWig genome tracks (deeptools; Ramírez et al. 2016), and visualized them in the IGV genome browser (Robinson et al. 2011).
To identify class- and subtype-enriched genes, we performed differential expression analysis using the estimated counts from kallisto as input to limma (Ritchie et al. 2015), voom (Law et al. 2014), and quantile normalizing the expression levels to account for differences in the number of genes detected in each sample (Table S1). We used criteria of at least 10 TPM abundance in one sample, at least twofold difference in expression, and 5% false discovery rate to identify differentially expressed genes.
Immunohistochemistry
Immunohistochemistry was performed essentially as in a previous report (Wu et al. 2013). Brains were dissected in isotonic PBS and immediately transferred to 4% paraformaldehyde in PBS for a 30-min fixation at room temperature. Fixed brain samples, were rinsed with isotonic PBS and incubated in PBS containing 2% Triton X-100, 10% normal goat serum (NGS; Penetration & Blocking Buffer) while being subjected to a degassing procedure (Chiang et al. 2011). Brain samples were agitated in the same buffer at 4° overnight. Brains were then transferred to primary antibodies diluted with PBS containing 0.25% Triton X-100, 1% NGS (Dilution Buffer) and agitated at 4° for 1–3 day. After primary antibody incubation, the brain samples were washed in PBS containing 1% Triton X-100, 3% NaCl (Washing Buffer) three times before they were moved to the 1:250 diluted secondary antibodies for one day agitation at 4°. For the biotin amplification staining (Figure 4, S3 & S4A), samples were washed three times and agitated in the 1:500 diluted Alexa Fluor 635 streptavidin (Thermo Fisher Scientific, USA: S-32364) at 4° for 1 day. Finally, the immunolabeled brain samples were washed three times, cleared and mounted in a drop of FocusClear (CelExplorer Labs, Taiwan: FC-101) between two coverslips separated by a spacer ring of ∼200 μm thickness, so the brain samples were not flattened. The Penetration & Blocking Buffer and Dilution Buffer contain additional 0.02% Sodium Azide as a preservative. For GAL4 line characterization, 1:100 dilution of mouse anti-dlg1 (4F3, deposited to the DSHB, USA by Goodman, C.) plus 1:250 dilution of rabbit anti-GFP (Thermo Fisher Scientific, USA: A-6455) primary antibody and 1:250 dilution of secondary antibody of Alexa Fluor 633-conjugated goat anti-mouse (Thermo Fisher Scientific, USA: A-21052) and Alexa Fluor 488-conjugated F(ab’)2-goat anti-rabbit (Thermo Fisher Scientific, USA: A-11070) were used. For the MB marker gene confirmation, a 1:4000 dilution of rabbit anti-sNPFp (Johard et al. 2008) or 1:20 dilution of mouse anti-Fas2 (1D4, deposited to the DSHB, USA by Goodman, C.) or 1:20 dilution of mouse anti-trio (9.4A, deposited to the DSHB, USA by Hama, C.) primary antibody, 1:250 dilution of secondary antibody of biotin-conjugated goat anti-rabbit (Thermo Fisher Scientific, USA: 65-6140) or biotin-conjugated goat anti-mouse (Thermo Fisher Scientific, USA: D-20691) were used. For the GABAergic identification staining, a 1:250 dilution of mouse anti-GFP (MilliporeSigma, USA: 11814460001) together with 1:250 dilution of rabbit anti-GABA (MilliporeSigma, USA: A2052) or 1:500 dilution of rabbit anti-Gad1 (Featherstone et al. 2000) or 1:400 dilution of rabbit anti-VGAT (Fei et al. 2010) primary antibodies, and 1:250 dilution of secondary antibody of Alexa Fluor 488-conjugated goat anti-mouse (Thermo Fisher Scientific, USA: A-11029) together with biotin-conjugated goat anti-rabbit or Alexa Fluor 647-conjugated goat anti-rabbit (Thermo Fisher Scientific, USA: A-21244) were used.
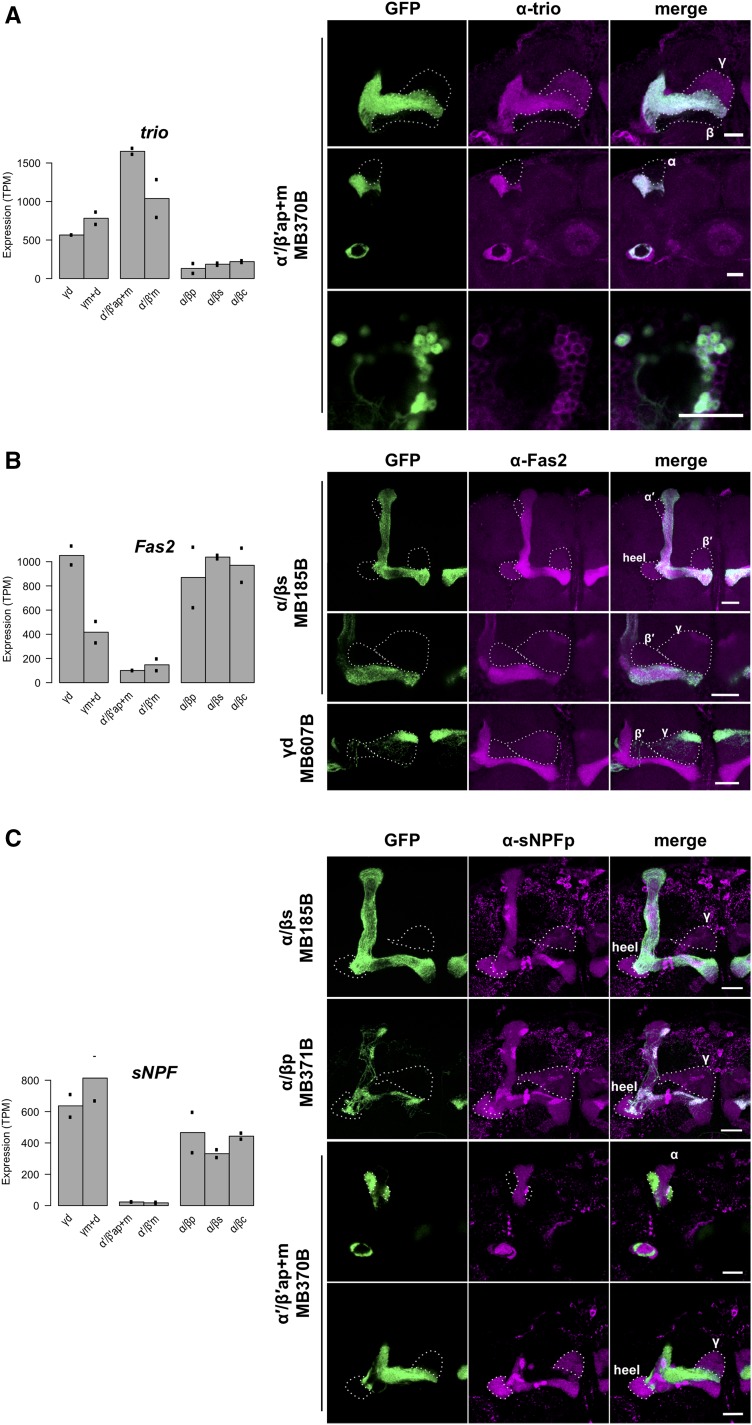
Figure 4.
(A) trio is depleted in α/β KCs. Whole mount anti-trio immunostaining confirmed strong signal in the MB α′/β′ lobes, moderate signal in the γ lobe, and no signal in the α/β lobes. The cell bodies of MB α′/β′ KC class also showed immunoreactivity. (B) Fas2 is depleted in MB α′/β′ KC class. Whole mount anti-Fas2 immunostaining confirmed stronger signal in the MB α/β and γd lobes. (C) sNPF is depleted in MB α′/β′ KC class. Whole mount anti-sNPF precursor immunostaining confirmed no detectable signal in the MB α′/β′ lobes. Among the immunoreactive α/β and γ lobes, the α/βp lobes showed the strongest signal. In each plot the bars represent the mean TPM, and the dots represent individual replicate values. Scale bars represent 20 μm. Expression patterns of the split-GAL4 lines were reported by P{10XUAS-IVS-GFP-p10}attP2.
Confocal imaging and post-imaging processing
Brain samples were imaged under a Zeiss LSM 800 confocal microscope with a 40X C-Apochromat water-immersion objective lens (N.A. value 1.2). The settings for scanning were manually adjusted. To overcome the limited field of view when imaging the GAL4 expression patterns, we scanned each brain in two parallel stacks of confocal images with some overlap between the two brain hemispheres, with a voxel size of 0.31 X 0.31 X 1.25 μm. We then stitched the two image stacks into a single data set with ‘Pairwise stitching’ function in Fiji (Preibisch et al. 2009; Schindelin et al. 2012), segmented the brain region based on the dlg1 staining channels with 3D Slicer (https://www.slicer.org; Kikinis et al. 2013), and made a ‘Z projection’ with Fiji (Schindelin et al. 2012). The MB subtype models were constructed from the GFP channel of the confocal images used for projections, by using the 3D Slicer to segment, show 3D, and conduct smoothing.
Cell counting
The confocal images for cell counting were acquired with a voxel size of 0.16 X 0.16 X 1.00 μm. The GFP channel was first used to identify KCs, and then the ‘Cell Counter’ plugin in Fiji was used to count all detectable nuclei in the mCherry channel (Preibisch et al. 2009). For each line, we counted three hemispheres from three different animals.
Data availability
All Drosophila strains are available upon request. Supplemental files are available at FigShare. Files S1 – S7 contain the expression pattern for each of the split-GAL4 drivers, while Files S8 – S14 contain one example of the cognate WM images used for cell counting. Figure S1 contains the olfactory short-term memory performance for 10 newly generated isogenic strains. Figure S2 contains overview of the workflow of TAPIN purification of nuclei and the molecular size distribution of amplified cDNA obtained from the 14 TAPIN-seq libraries. Figure S3 and S4 contains the view of the cell bodies of MB KCs in the confocal images that have double labeling for immunostaining and GAL4 expression. Table S1 contains the TAPIN-seq library statistics, including the numbers of raw reads, pseudoaligned reads and uniquely aligned reads, as well as the detected gene numbers. Table S2 contains the full list of genes enriched and depleted in individual MB classes and subtypes. The sequencing reads and processed data files, including the tables of estimated abundances, are available in NCBI GEO (GSE119629). All code used to analyze RNA-seq results and create the figures and tables in this manuscript are available in the GitHub repository (http://github.com/fredpdavis/mushroombody). Supplemental material available at Figshare: https://doi.org/10.25387/g3.7267481.
Results
TAPIN-seq profiling of MB neuronal cell subtypes
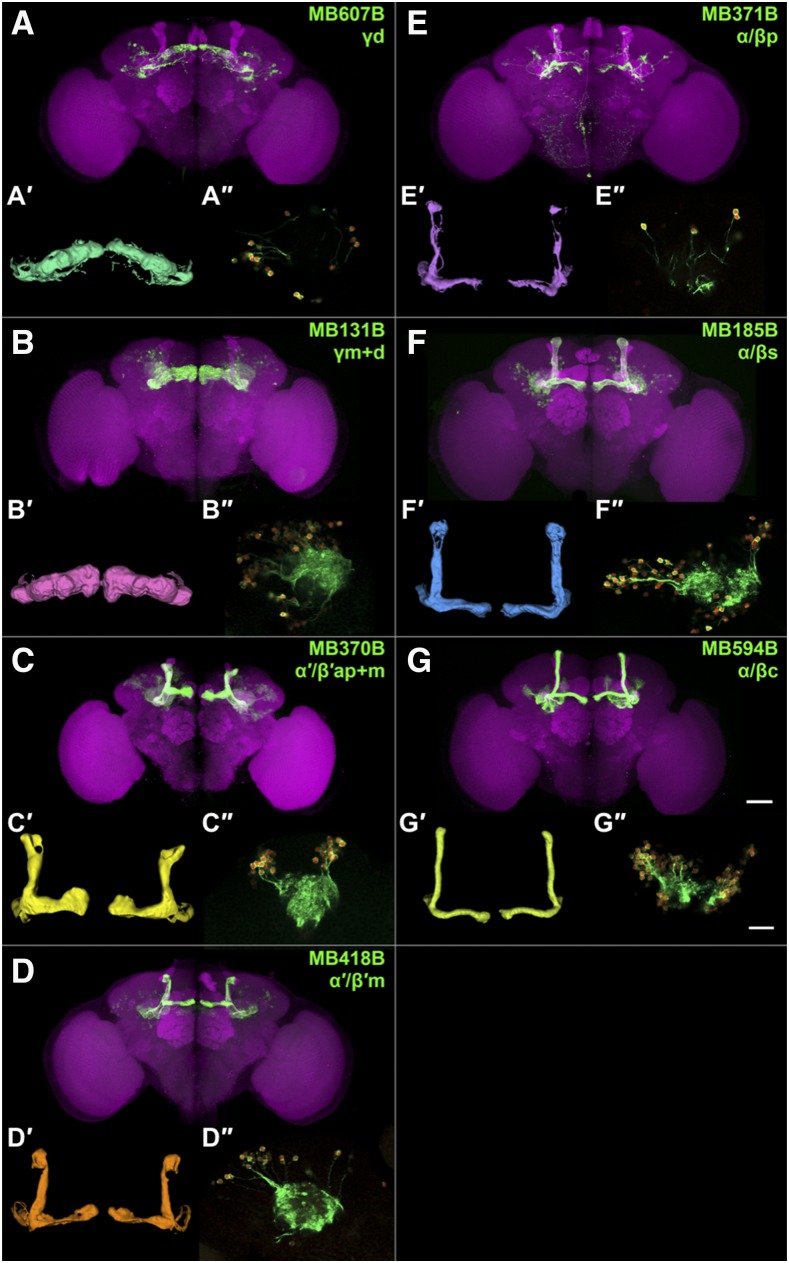
To label MB subtypes, we used seven split-GAL4 lines: MB607B (γd), MB131B (γm+d), MB370B (α′/β′ap+m), MB418B (α′/β′m), MB371B (α/βp), MB185B (α/βs) and MB594B (α/βc) (Aso et al. 2014a). We first backcrossed all the split GAL4 hemi-drivers and the nuclear-tag reporter (Henry et al. 2012) into MJ2, an isogenic Canton-S derivative (Methods). Because of the change in genetic background, we re-characterized the expression pattern of each split-GAL4 combination to confirm that the expected cell subtype-specific pattern had not been altered. We used a novel membrane-GFP-P2A-nuclear-mCherry dual label reporter (Watermelon or WM for short; see Methods) (Chang et al. 2018), which expresses membrane-tethered GFP and nuclear mCherry from a single transcript via a viral ribosome skip peptide (P2A; Daniels et al. 2014). The GFP label revealed neuronal morphology, thereby confirming MB subtype specificity, and the nuclear-mCherry marked each nucleus, which permitted an accurate cell count. Using WM labeling, we found that each split-GAL4 combination yielded limited expression in only small numbers of neurons outside the MB, but exhibited strong expression in the annotated MB cell subtype (Figures 1A – 1G, 1A′ – 1G′ & Files S1 – S7) (Aso et al. 2014a). Using high-resolution imaging (Figures 1A″–1G″ & Files S8 – S14), we were able to count the total number of MB KCs of each subtype that is labeled by a given spilt-GAL4 combination (Table 1). The numbers of labeled cells for each MB KC subtype are in general agreement with a previous report (Aso et al. 2014a) and it appears that most if not all of the neurons of each subtype are labeled with these Split-GAL4 combinations. In fact the total number of KCs labeled by five non-overlapping split-GAL4 lines – MB131B, MB370B, MB371B, MB185B and MB594B – is very close to the estimated total number of KCs (1,855 vs. 2,000; Aso et al. 2014a), making it unlikely that any major KC subtype is missed. Together, these results confirm the previously reported specificity and comprehensiveness of these split-GAL4 lines to label each of the MB KC neuronal subtypes.
Figure 1.
Characterizing mushroom body subtype drivers. (A-G) Expression pattern of the split-GAL4 driver lines used in this study. Green, the GFP plus anti-GFP immunoreactive signal; magenta, anti-dlg1 immunoreactive signal as a counterstain. The scale bar represents 50μm. (A′-G′) 3D model of the split-GAL4 expression pattern in MB. (A″-G″) Example of the high-resolution membrane-GFP-P2A-nuclear-mCherry dual label reporter (WM for short; see Methods) images used for cell counting. The scale bar represents 20μm. Genotype: MB607B > WM, R19B03-p65.AD/UAS-WM-2; R39A11-GAL4.DBD/+, MB131B > WM, R13F02-p65.AD/UAS-WM-2; R89B01-GAL4.DBD/+, MB370B > WM, R13F02-p65.AD/UAS-WM-2; R41C07-GAL4.DBD/+, MB418B > WM, R26E07-p65.AD/UAS-WM-2; R30F02-GAL4.DBD/+, MB371B > WM, R13F02-p65.AD/UAS-WM-2; R85D07-GAL4.DBD/+, MB185B > WM, R52H09-p65.AD/UAS-WM-2; R18F09-GAL4.DBD/+, and MB594B > WM, R13F02-p65.AD/UAS-WM-2; R58F02-GAL4.DBD/+.
Table 1. List of split-GAL4 lines labeling MB subtype (n = 3).
| DRIVER | MB SUBTYPE | NUMBER OF CELLS |
|---|---|---|
| MB607B | γd | 132.00 ± 6.03 |
| MB131B | γm+d | 574.33 ± 2.73 |
| MB370B | α′/β′ap+m | 405.33 ± 18.80 |
| MB418B | α′/β′m | 152.67 ± 3.71 |
| MB371B | α/βp | 77.33 ± 2.85 |
| MB185B | α/βs | 294.33 ± 16.68 |
| MB594B | α/βc | 505.33 ± 9.67 |
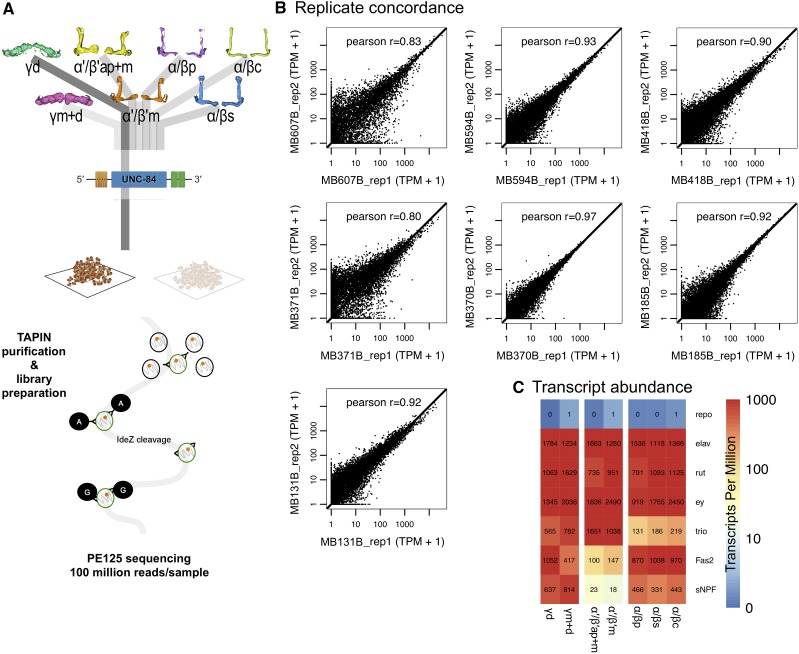
To profile the nuclear transcriptome of each MB subtype, we used TAPIN-seq (Davis et al. 2018), a modification of INTACT (Henry et al. 2012) that yields improved selectivity by use of a two-step purification (Figure S2A). The 7 characterized split-GAL4 combinations were used to express the nuclear membrane protein UNC84 fused with 2 copies of GFP (Figure 2A). For each split-GAL4 combination, tagged nuclei of a given MB KC subtype were purified from ∼400 fly heads. Nuclear RNA was then extracted and used to generate RNA-seq libraries (Figure S2B). We generated libraries from two independent biological replicates for each MB KC subtype, and paired-end sequenced them (Table S1). We estimated transcript abundances in each library using kallisto (Bray et al. 2016; see Methods). The sequencing reads and estimated abundances are available in NCBI GEO (GSE119629).
Figure 2.
TAPIN-seq profiling of MB subtypes. (A) Driver lines expressing in the seven Kenyon cell subtypes were crossed with the TAPIN-seq reporter, which labels the nuclei in each subtype. Nuclear RNA from each subtype was used to generate RNA-seq libraries, which were sequenced in paired-end mode. (B) We estimated reproducibility of the TAPIN-seq measurements by calculating the Pearson correlation between estimated transcript abundances (log2 transformed Transcripts Per Million + 1). (C) The TAPIN-seq transcriptomes recover the neuronal marker elav, while not detecting the glial marker repo. The transcriptomes also recover the expected expression patterns of the known pan-Kenyon cell markers ey and rut as well as class-enriched genes trio, Fas2, and sNPF.
Transcript abundances were well correlated between replicate libraries (Figure 2B). We observed strong expression of the neuron-specific marker elav in all subtypes (1,118 – 1,784 TPM), contrasting with low levels of the glial-specific gene repo (0.1 – 1.3 TPM; Figure 2C), consistent with a high-fidelity purification of TAPIN labeled nuclei. We also detected strong and broad expression of genes expected in all MB neuron subtypes, such as the transcription factor ey (919 – 2,490 TPM) and components of the cAMP signaling pathway such as rutabaga (735 – 1,629 TPM) which encodes the Ca2+/calmodulin-activated adenylyl cyclase (Figure 2C; Crittenden et al. 1998). The TAPIN-seq profiles also recovered the expected pattern of genes known to be enriched in individual classes, including trio, Fas2, and sNPF.
Genes enriched in MB neuronal classes and subtypes
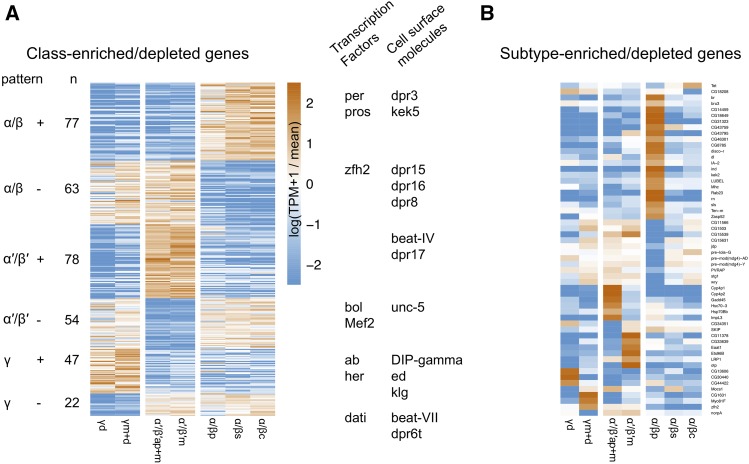
We next identified transcripts that are differentially expressed across MB neuron classes or subtypes using three criteria (Methods). We found 341 transcripts that are enriched or depleted in one of the three MB neuron classes (Figure 3A) and 57 that are enriched or depleted in one of the seven MB cell subtypes (Figure 3B; Table 2 for summary and Table S2 for the full gene list). To evaluate the accuracy of our differential expression analysis, we examined several genes encoding proteins reported to differentially label MB cell classes. Antibodies against trio, for example, are reported to label α′/β′ and γ classes (Awasaki et al. 2000). Indeed, the trio gene is identified in our analysis as an MB class-specific gene depleted in α/β KCs (average 179 TPM in α/β vs. 1,345 TPM α′/β′ and 674 TPM γ), and we confirm that anti-trio immunoreactivity is localized in the MB α′/β′ and γ lobes, and their cell bodies (Figure 4A). Fas2 has been reported to exhibit strong immunoreactive signal in the MB α/β lobe class, weak signal in the γ lobe class and no signal in the α′/β′ lobe class (Crittenden et al. 1998). Consistent with this protein distribution, we identify Fas2 transcripts as an MB class-specific gene depleted in α′/β′ KCs (124 TPM α′/β′ vs. 959 TPM α/β and 734 TPM γ). At the cell type level, Fas2 was enriched in in the γd subtype relative to γm+d (1,052 vs. 417 TPM, respectively). Using immunolabeling, we confirmed this pattern of anti-Fas2 immunoreactivity. As previously reported, we detect strong immunoreactive signal of Fas2 in the α/β lobe class of neurons, somewhat weaker signal in the γ lobe class and no signal in the α′/β′ lobe class neurons. We further demonstrate that Fas2 immunoreactivity appears weaker in the γm subtype compared to γd subtype KCs (Figure 4B), which mirrors the prediction from the TAPIN-seq described above.
Figure 3.
Differential expression of MB class-specific genes (A) and subtype-specific genes (B). Specific examples of up and downregulated genes for each subtype are indicated in (A).
Table 2. Number of genes enriched and depleted in individual MB classes and subtypes. Genes identified by limma/voom (q-value < 0.05, |fold change| > 2x).
| GROUPTYPE | GROUP | ENRICHED | DEPLETED |
|---|---|---|---|
| class | γ | 47 | 22 |
| class | α′/β′ | 78 | 54 |
| class | α/β | 77 | 63 |
| subtype | γd | 3 | 2 |
| subtype | γm+d | 3 | 1 |
| subtype | α′/β′ap+m | 6 | 2 |
| subtype | α′/β′m | 6 | 0 |
| subtype | α/βp | 21 | 11 |
| subtype | α/βs | 0 | 0 |
| subtype | α/βc | 1 | 1 |
We also examined the expression of the neuropeptide gene sNPF which we identified as an MB class-specific gene depleted in α′/β′ KCs (20 TPM in α′/β′ vs. 414 TPM α/β and 725 TPM γ). Immunolabeling with an antibody against the sNPF precursor confirmed a previous report and is consistent with the TAPIN-seq results. We observe no detectable signal in α′/β′ KC class neurons and strong signal in α/β and γ classes (Figure 4C; Johard et al. 2008). No noticeable signal was detected in the cell bodies of KCs (Figure S3), consistent with a previous description (Johard et al. 2008). At the protein level, we further noted elevated immunoreactivity in the MB α/βp subtype, which was not reflected by the TAPIN-seq results (Figure 4C). This discrepancy points either to limitations of TAPIN mediated profiling with these split-GAL4 lines and/or the importance of post-transcriptional regulatory mechanisms in determining the accumulation of the neuropeptide. Taken together, the identification of known differentially expressed transcripts and the immunostaining data for three identified examples broadly corroborate the fidelity of the TAPIN-seq results.
The differentially expressed genes also include several transcriptional regulators with class- and subtype-specific patterns (Figure 3). For example, we identify transcriptional regulators enriched in the γm+d (zfh2), α′/β′m (Ets96B, otp), α/βp (br, bru3, dl, disco-r, ind, rn), and α/βc (Tet) subtypes. Although the abundance and significance of DNA methylation in Drosophila is unclear, enrichment of the Tet DNA methyltransferase is intriguing given the role of DNA methylation in memory formation in other insects (Biergans et al. 2012) and mammals (Day and Sweatt 2010). The subtype-enriched expression could reflect a remnant of a functional expression pattern from an ancestral species with DNA methylation.
Several genes encoding cell-surface molecules were also differentially expressed across the subtypes. These genes included members of gene families previously implicated in specifying synaptic connectivity, including the defective proboscis extension response (Dpr) as well as the Dpr-interacting protein (DIP). Interactions between proteins from these families have been previously documented and shown to underlie synaptic connectivity in circuits including the visual system (Özkan et al. 2013; Carrillo et al. 2015). The expression patterns we observed for several cell-surface molecules suggest they might be involved in specifying class- and subtype-specific connectivity.
Neurotransmitter output
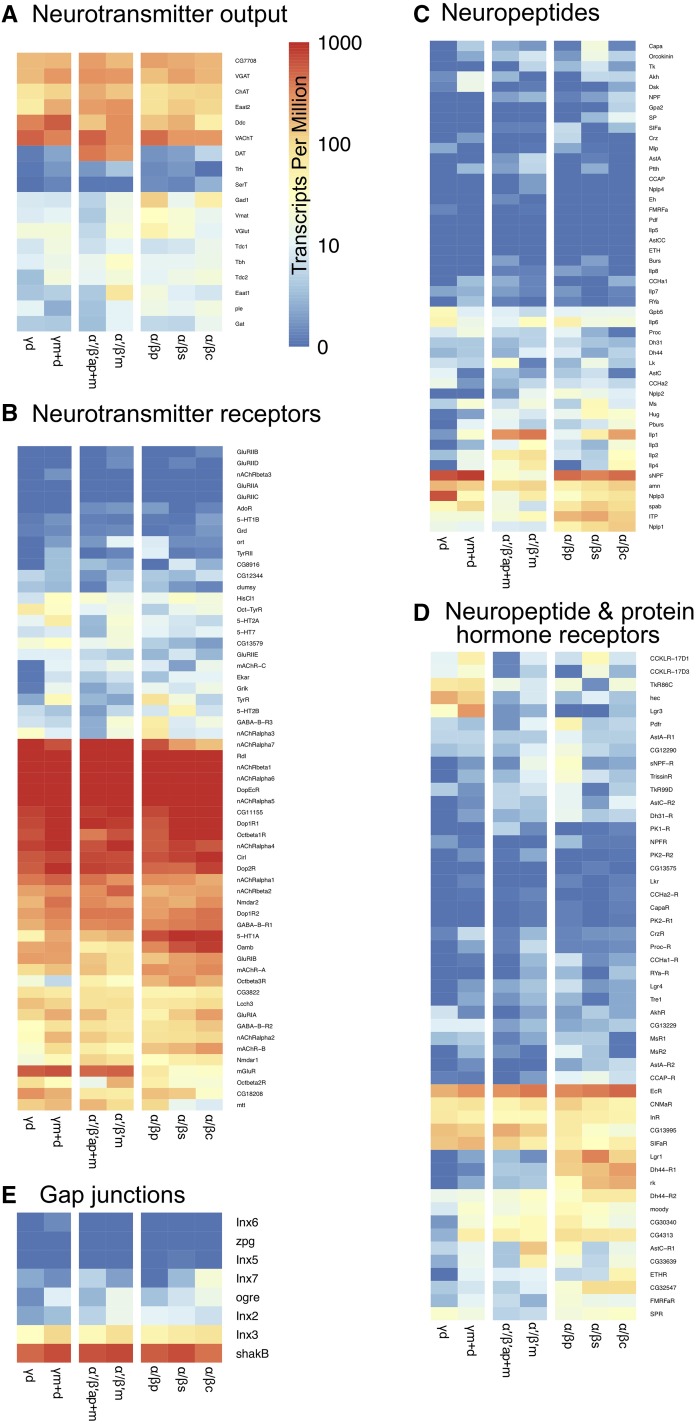
To explore the functional utility of our TAPIN-seq measurements, we next focused on genes related to the input and output properties of the MB cell subtypes. Specifically, we examined genes encoding neurotransmitter biosynthetic enzymes, neurotransmitter transporters (Figure 5A), neurotransmitter receptors (Figure 5B), neuropeptides (Figure 5C), neuropeptide and protein hormone receptors (Figure 5D), and gap junction components (Figure 5E). A recent report has established that the primary neurotransmitter in the MB is acetylcholine (Barnstedt et al. 2016). Because our method profiled each of the seven MB neuronal subtypes, we re-assessed this conclusion at a higher resolution. We confirmed that all seven MB cell subtypes express high levels of both choline acetyltransferase (ChAT; 75 – 198 TPM) and vesicular acetylcholine transporter (VAChT; 257 – 517 TPM).
Figure 5.
TAPIN-seq profiles of genes related to neurotransmitter biosynthetic enzymes, neurotransmitter transporters (A), neurotransmitter receptors (B), neuropeptides (C), neuropeptide and protein hormone receptors (D), and functional components of gap junctions (E).
We next asked whether one or more KC subtypes might co-release other small molecule neurotransmitters, but found no strong argument to support such a conclusion. In a few cases, we see moderate expression of biosynthetic enzymes for other neurotransmitters including GABA, serotonin and dopamine. But in each case, there are findings that undermine the conclusion that these neurotransmitters are consistently produced/released by any of the 7 KC subtype. For example, we do see strong expression in all MB cell subtypes of vesicular GABA transporter (VGAT; 152 – 279 TPM), which is an essential transporter that is responsible for packaging the neurotransmitter GABA into synaptic vesicles (Fei et al. 2010). On the other hand, the gene for GABA biosynthetic enzyme, Gad1, is moderately expressed in only the α/βp (121 TPM) and α/βc subtypes of KCs (54 TPM) and at lower levels in the remaining cell subtypes (4 – 14 TPM). In principle, moderate Gad1 expression might be consistent with the hypothesis that GABA is released from a fraction of α/βp and/or α/βc subtype neurons. To examine this possibility, we conducted immunofluorescence experiments with antibodies against GABA, Gad1, and VGAT but found no marked immunoreactivity in any α/βp KC subtype cell bodies (Figure S4). This result suggests that the most likely explanation for our observed Gad1 pattern is that the split-GAL4 lines we used for α/βp and α/βc KC subtypes drive low levels of expression in some subset of GABAergic neurons outside the MB (Figure S4A).
A similar set of findings are apparent with Dopa decarboxylase (Ddc), a commonly used marker for dopaminergic or serotonergic neurons. Ddc catalyzes the decarboxylation of dopa to dopamine and 5-hydroxytryptophan to serotonin but not tyrosine to tyramine (Gramates et al. 2017). We see fairly strong Ddc expression in all MB cell types (55 – 574 TPM), especially the γm+d (574 TPM) and γd (339 TPM) KCs. However, we do not see expression of Tryptophan hydroxylase (Trh; 0 – 3 TPM), which provides the first and rate-limiting step in the synthesis of serotonin. Nor do we detect serotonin transporter (SerT; 0 – 2 TPM; Giang et al. 2011) in any of the 7 cell subtypes (Figure 5A). Pale (ple), which encodes tyrosine hydroxylase (TH) for dopamine synthesis, also is not highly expressed in any MB cell subtype (2 – 16 TPM; Figure 5A). Thus, we conclude that all MB KC subtypes likely release acetylcholine (Barnstedt et al. 2016; Crocker et al. 2016) and no other small molecule neurotransmitters.
Neurotransmitter receptors
We next examined expression profiles of small molecule neurotransmitter receptors. Dopamine has been established as a key input to MB for many behaviors including aversive and appetitive olfactory learning (Kim et al. 2007; Claridge-Chang et al. 2009; Qin et al. 2012; Liu et al. 2012; Burke et al. 2012), modulation of motivational state (Krashes et al. 2009), regulated forgetting (Berry et al. 2012; Shuai et al. 2015), sleep (Sitaraman et al. 2015b), courtship behaviors (Kuo et al. 2015; Lim et al. 2018), and temperature preference (Bang et al. 2011). A network of dopaminergic neurons innervates all MB cell subtypes (Aso et al. 2014a), and all four dopamine receptors, Dop1R1, Dop1R2, Dop2R and DopEcR, are expressed in all 7 MB KC subtypes (594 – 1152, 222 – 408, 444 – 1059, 3284 – 8335 TPM, respectively; Figure 5B) (Han et al. 1996; Kim et al. 2007; Qin et al. 2012; Ishimoto et al. 2013; cf Draper et al. 2007). We confirmed that the Dopamine transporter (DAT) is preferentially expressed in both MB α′/β′ap and α′/β′m cell subtypes (397 and 262 TPM in α′/β′ and α′/β′m, respectively, vs. 0.2 – 4.3 TPM in other subtypes; Figure 5A) (Croset et al. 2018; Davie et al. 2018), suggesting that precise temporal control of dopaminergic signaling may be at play in α′/β′ KCs. We also observe equivalently high levels of expression in all MB KC subtypes for all six GABA receptor genes. Among the three GABAA receptors, Resistance to dieldrin (Rdl) is strongly expressed (5,021 – 9,548 TPM), consistent with previous findings (Harrison et al. 1996; Liu et al. 2007), while Ligand-gated chloride channel homolog 3 (Lcch3) is moderately expressed (70 – 153 TPM), and Drosophila Glycine receptor (Grd) is not detected (0.1 – 0.8 TPM). All the GABAB receptors are broadly expressed in MB except for GABA-B-R3 (1 – 61 TPM; Figure 5B). These findings are consistent with a report that establishes the importance of GABAergic feedback from the anterior paired lateral neurons to MB KCs (Lin et al. 2014), mediated by both ionotropic GABAA and metabotropic GABAB receptors (Inada et al. 2017).
Serotonergic signaling in the MB also is involved in olfactory memory formation, sleep regulation and stress response modulation (Yuan et al. 2006; Lee et al. 2011; Haynes et al. 2015; Ries et al. 2017). We found that among five serotonin receptors, only 5-HT1A is expressed in the MB, with an enrichment in the α/β lobe KC class (Figure 5B). This is consistent with the finding that dorsal paired medial (DPM) neurons release serotonin onto 5-HT1A receptors expressed in α/β KCs to support anesthesia-resistant memory formation (Lee et al. 2011). A previous report observed 5-HT1B-GAL4 expression and 5-HT1B immunoreactivity in the γ KC class (Yuan et al. 2005). We did not detect nuclear 5-HT1B transcripts by TAPIN-seq (0.1 – 1.0 TPM), but we cannot rule out the possibility that higher levels of transcripts and protein are present in the cytoplasm, beyond detection in this nuclear transcriptome.
MB KCs receive nicotinic acetylcholine receptor (nAChR)-mediated synaptic transmission from antennal lobe projection neurons (Gu and O’Dowd 2006). We found the nAChR subunits nAChRalpha1, nAChRalpha4, nAChRalpha5, nAChRalpha6, nAChRalpha7, nAChRbeta1 and nAChRbeta2 are strongly expressed in all seven MB cell subtypes, but nAChRalpha3 and nAChRbeta3 transcripts are absent or undetectable (Figure 5B). Two muscarinic acetylcholine receptors (mAChRs), mAChR-A and mAChR-B, are also expressed in the MB with an enrichment in α/β lobe KC class (Figure 5B & Table S2).
We also examined the expression of the six known octopamine receptors (Gramates et al. 2017). We found that Oamb and CG18208 (recently characterized as Octα2R; Qi et al. 2017), which are α-adrenergic-like receptors, are class- and subtype-specific genes, respectively (Table S2). Oamb TAPIN-seq signal is strongly detected in α/β class KCs and far lower levels are seen in α′/β′ class KCs (Figure 5B) (Crittenden et al. 1998; Kim et al. 2013). The three β-adrenergic-like receptors — Octβ1R, Octβ2R and Octβ3R — are all detected in the MB with variable levels across cell subtypes (cf Wu et al. 2013 for Octβ2R immunolabeling).
Although transient glutamate immunoreactivity has been shown in α/βc lobe KC class of young adult, VGlut expression has never been observed in the MB KCs (Daniels et al. 2008; Sinakevitch et al. 2010). Consistent with this conclusion, we observe low VGlut transcript abundance (5 – 50 TPM; Figure 5A). We also confirmed that the NMDA receptors, both Nmdar1 and Nmdar2, are broadly expressed in the MB (Figure 5B) (Xia et al. 2005; Ueno et al. 2017); ionotropic receptors GluRIA and GluRIB are also expressed (Figure 5B). Flies also have a unique metabotropic glutamate receptor called mGluR, which has been previously observed by immunolabeling throughout the adult brain, but minimally in the MB lobes (Devaud et al. 2008). Here, with the cell type resolution of our dataset, we identified mGluR as an MB class-specific gene that is expressed in a subset of KCs — mGluR is depleted in α/β class KCs (40 TPM; Table S2) relative to both α′/β′ and γ classes of KCs (504 and 612 TPM, respectively; Figure 5B).
Neuropeptides
In addition to small neurotransmitter systems, we also examined expression of neuropeptides and neuropeptide receptors. We consistently detect expression of a small group of well-characterized and putative neuropeptides. Unexpectedly, this includes amnesiac (amn), which is detected at fairly robust levels in all 7 MB KC subtypes (52 – 175 TPM) — although this small gene resides in the intron of another gene, Hers, which is highly expressed (218 – 239 TPM), thus complicating the accurate estimation of amn abundance. Previous work has established a requirement during memory formation for amn expression in a single pair of DPM neurons outside MB (Waddell et al. 2000). sNPF expression appears as a class-specific transcript, with high levels in α/β and γ KC classes and very little expression in the α′/β′ KC class (414, 725, 20 TPM, respectively; Figure 4C). This is consistent with previous reports (Johard et al. 2008) and we further confirm this expression pattern by immunostaining (Figure 4C). Another notable class-specific neuropeptide is Drosophila Insulin-like peptide 1 (Ilp1; Liu et al. 2016b), expressed at high levels in α′/β′ class of KCs (284, 382 TPM in α′/β′, α′/β′m, respectively) and α/βc subtype (220 TPM) but not in most other MB KC subtypes (0.8 – 55 TPM; Figure 5C). As with neuropeptides, we also detect a panel of neuropeptide and protein hormone receptors, some of which are robustly expressed in all MB KC subtypes (e.g., Ecdysone receptor), some of which are class specific (e.g., Dh44-R1 and hector), and some of which are enriched or depleted in one or more subtypes of neurons (e.g., AstC-R1 and ETHR; Figure 5D).
Finally, because of a report that gap junctions may form between MB KCs of different classes and play a role in visual learning (Liu et al. 2016a), we examined the expression levels of the eight gap junction genes in MB cell subtypes. shakB is strongly expressed (433 – 751 TPM), and Inx3 moderately (29 – 107 TPM), in all MB cell subtypes (Figure 5E). Although Inx5 and Inx6 are reportedly required in the α/β and α′/β′ KC classes for visual learning and memory (Liu et al. 2016a), we do not detect either gene in any of the 7 MB KC subtypes (0 – 0.2 TPM, 0 – 0.5 TPM, respectively; Figure 5E). We cannot rule out the possibility that functionally relevant levels of expression are below our detection limit or that cytoplasmic RNA levels are higher.
Discussion
Our results establish a high-quality, neuronal cell type-level transcriptome for Drosophila MB. We identified 350 differentially expressed genes, which includes most of the previously reported MB lobe (class specific) markers and many novel class-specific or cell subtype-specific profiles of expression. In addition to the subtype level resolution of our experimental design, the TAPIN approach that we used also offers several advantages and technical differences with these prior approaches. First, because TAPIN is compatible with flash frozen tissue as the input, the method introduces minimal disturbance to the endogenous transcriptome as compared to more lengthy procedures for purification of neurons for expression profiling. Second, it may be relevant that TAPIN explicitly profiles nuclear RNAs, likely enriching for actively transcribed/nascent transcripts vs. abundant ones that are stably maintained in the cytoplasm. Thus, it would be attractive to apply this method to profile transcriptional response to behavioral perturbations.
Several previous studies have used genome-wide methods to profile expression in the Drosophila MB (Perrat et al. 2013; Crocker et al. 2016; Croset et al. 2018; Davie et al. 2018; Jones et al. 2018). Perrat et al. used a microarray-based approach to profile expression of each of the three major classes of MB KCs (purified by flow cytometry from dissociated brains) and compared these profiles with expression in the rest of the brain. They first focused on the expression of transposons (Perrat et al. 2013), and subsequently used the same transcriptome dataset to discover that MB KCs are cholinergic (Barnstedt et al. 2016) based on expression of biosynthetic enzymes. Crocker et al. used an RNA-seq-based approach to profile expression in relatively small pools of physically isolated α/β and γ class neurons to search for memory-related changes in gene expression (Crocker et al. 2016). Most recently, Croset et al. and Davies et al. used droplet-based single cell sequencing to profile the Drosophila brain, and by clustering the single cells they were able to identify the three MB classes, but not the further sub-division into neuronal subtypes (Croset et al. 2018; Davie et al. 2018).
Although we used a different profiling method and resolved transcriptomes at the cell subtype rather than class level, our findings are broadly compatible with prior reports (Perrat et al. 2013; Barnstedt et al. 2016; Crocker et al. 2016; Croset et al. 2018; Davie et al. 2018). Our dataset reveals strong expression of both ChAT and VAChT, consistent with the conclusion that MBs are cholinergic (Barnstedt et al. 2016). Our findings further support the conclusion that all of the individual MB KC subtypes are cholinergic. The datasets also are consistent in the expression of known class-specific markers. One notable difference is that Crocker et al. reported high levels of expression of the 5-HT1B receptor in both α/β and γ classes of KCs (Crocker et al. 2016), and Davie et al. also observed 5-HT1B expression in α/β and γ KCs single cell clusters (Davie et al. 2018). In contrast, we see no evidence for expression of this receptor in our TAPIN-seq profiles (0.1 – 1 TPM). This difference could reflect methodology: Crocker and Davie both measured 5-HT1B receptor transcripts in the cytoplasm while we measured the levels that are actively transcribed or present in the nucleus. This technical difference could be especially relevant for neurotransmitter receptors, some of which can be translated locally at dendrites (Steward and Banker 1992).
Our dataset is the first to profile expression in this brain region at neuronal subtype resolution. This level of resolution is critical given the wealth of data on the functional differences of each MB KC subtype in Drosophila behaviors. Our dataset provides a full accounting within each of the MB KC subtypes of the profiles of expression of the cellular machinery to produce and receive neurotransmission, including small molecule transmitters and their receptors, neuropeptides and neuropeptide receptors and subunits of gap junctions. It is noteworthy that the TAPIN expression dataset supports the conclusions that all the adult MB KC subtypes are cholinergic, and that none of the subtypes express genes that would suggest the co-release of GABA, dopamine, glutamate, or serotonin. On the other hand, we detect expression of a spectrum of neuropeptides and their receptors. This observation is consistent with the hypothesis that MB KCs may co-release both acetylcholine and several neuropeptides (Takemura et al. 2017).
In addition to these findings with regards to the inputs and outputs, we identified 350 differentially expressed genes including many that distinguish MB KC classes or even individual cell subtypes. MB α/βp subtype showed 21 enriched genes and 11 depleted genes, contrasting with two other subtypes in the α/β class and two other classes. This uniqueness is supported by its unique odor responses (Perisse et al. 2013) and connectivity (Chen et al. 2012). Despite the limitation in the methodology that we profile the MB γm subtype using the spilt-GAL4 line MB131B that has minor expression in γd, and profile the α′/β′ap subtype using MB370B that has minor expression in α′/β′m (Figure 1 & Table 1), we still identified distinct sets of enriched/depleted genes (Figure 3 & Table S2), indicating the differences between two subtypes in MB γ or α′/β′ classes.
Our dataset provides a valuable resource for the fly neuroscience community to conduct functional studies. For example, our data provide a list of previously unknown class specific and sub-type specific transcripts, whose impact on the functional differences between these neurons are not known (Figure 3; Table S2). An arsenal of genetic tools to manipulate any gene’s function within each of these cell subtypes already exists (Caygill and Brand 2016; Kaya-Çopur and Schnorrer 2016). In addition to olfactory associative memory, MBs also play fundamental roles in other forms of memory including visual and gustatory (Vogt et al. 2014; Masek and Keene 2016), temperature preference (Hong et al. 2008), courtship behaviors (Kuo et al. 2015; Lim et al. 2018), stress response (Ries et al. 2017), food-seeking (Tsao et al. 2018), sleep (Artiushin and Sehgal 2017) and responses to ethanol (Kaun et al. 2011). This dataset will facilitate the discovery of neural mechanisms for each of these conserved behaviors.
Acknowledgments
We thank Y. Aso and G. Rubin (Janelia Research Campus) for the split-GAL4 lines and pJFRC28 reporter, J. Veenstra (Université de Bordeaux 1, France) for the rabbit anti-sNPFp antibody, D. Krantz (University of California, Los Angeles) for the rabbit anti-VGAT antibody, F. Jackson (Tufts University) for the rabbit anti-Gad1 antibody and the Developmental Studies Hybridoma Bank (The University of Iowa), created by the NICHD, for the mouse monoclonal antibodies against dlg1, Fas2 and trio. We thank W.-W. Liao (Washington University in St. Louis) for technical assistance in analysis of the alignment files. We also are grateful to J. Azpurua, J. Beshel, Y.-H. Chang, R. Keegan, S. Krupp, L. Talbot for helpful discussions or comments on the manuscript. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). This work was supported by US National Institutes on Deafness and Other Communication Disorders [5R01DC013071-06 to J.D.]; DART NeuroScience LLC [to J.D.]; Taiwan National Science Council [102-2917-I-564-004 to M.-F.M.S.]; and US National Institute of Arthritis and Musculoskeletal and Skin Diseases [Intramural Research Program to F.P.D.].
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7267481.
Communicating editor: M. Arbeitman
These authors contributed equally in this work.
Literature Cited
- Artiushin G., Sehgal A., 2017. The Drosophila circuitry of sleep-wake regulation. Curr. Opin. Neurobiol. 44: 243–250. 10.1016/j.conb.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Hattori D., Yu Y., Johnston R. M., Iyer N. A., et al. , 2014a The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3: e04577 10.7554/eLife.04577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Sitaraman D., Ichinose T., Kaun K. R., Vogt K., et al. , 2014b Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife 3: e04580 10.7554/eLife.04580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T., Saito M., Sone M., Suzuki E., Sakai R., et al. , 2000. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron 26: 119–131. 10.1016/S0896-6273(00)81143-5 [DOI] [PubMed] [Google Scholar]
- Bang S., Hyun S., Hong S.-T., Kang J., Jeong K., et al. , 2011. Dopamine signalling in mushroom bodies regulates temperature-preference behaviour in Drosophila. PLoS Genet 7: e1001346 10.1371/journal.pgen.1001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnstedt O., Owald D., Felsenberg J., Brain R., Moszynski J.-P., et al. , 2016. Memory-relevant mushroom body output synapses are cholinergic. Neuron 89: 1237–1247. 10.1016/j.neuron.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J. A., Cervantes-Sandoval I., Nicholas E. P., Davis R. L., 2012. Dopamine is required for learning and forgetting in Drosophila. Neuron 74: 530–542. 10.1016/j.neuron.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biergans S. D., Jones J. C., Treiber N., Galizia C. G., Szyszka P., 2012. DNA methylation mediates the discriminatory power of associative long-term memory in honeybees. PLoS ONE 7: e39349 10.1371/journal.pone.0039349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A. L., Li W., Cressy M., Dubnau J., 2009. Short- and long-term memory in Drosophila require cAMP signaling in distinct neuron types. Curr. Biol. 19: 1341–1350. 10.1016/j.cub.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzaiane E., Trannoy S., Scheunemann L., Plaçais P.-Y., Preat T., 2015. Two independent mushroom body output circuits retrieve the six discrete components of Drosophila aversive memory. Cell Reports 11: 1280–1292. 10.1016/j.celrep.2015.04.044 [DOI] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L., 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527 (erratum: Nat. Biotechnol. 34: 888) 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Burke C. J., Huetteroth W., Owald D., Perisse E., Krashes M. J., et al. , 2012. Layered reward signalling through octopamine and dopamine in Drosophila. Nature 492: 433–437. 10.1038/nature11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo R. A., Özkan E., Menon K. P., Nagarkar-Jaiswal S., Lee P.-T., et al. , 2015. Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins. Cell 163: 1770–1782. 10.1016/j.cell.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill E. E., Brand A. H., 2016. The GAL4 system: a versatile system for the manipulation and analysis of gene expression. Methods Mol. Biol. 1478: 33–52. 10.1007/978-1-4939-6371-3_2 [DOI] [PubMed] [Google Scholar]
- Cervantes-Sandoval I., Martin-Peña A., Berry J. A., Davis R. L., 2013. System-like consolidation of olfactory memories in Drosophila. J. Neurosci. 33: 9846–9854. 10.1523/JNEUROSCI.0451-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-H., Keegan R. M., Prazak L., Dubnau J., 2018. Cellular labeling of endogenous virus replication (CLEVR) reveals de novo insertions of the gypsy endogenous retrovirus in cell culture and in both neurons and glial cells of aging fruit flies. bioRxiv. 10.1101/445221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-C., Wu J.-K., Lin H.-W., Pai T.-P., Fu T.-F., et al. , 2012. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science 335: 678–685. 10.1126/science.1212735 [DOI] [PubMed] [Google Scholar]
- Chiang A.-S., Lin C.-Y., Chuang C.-C., Chang H.-M., Hsieh C.-H., et al. , 2011. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr. Biol. 21: 1–11. 10.1016/j.cub.2010.11.056 [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A., Roorda R. D., Vrontou E., Sjulson L., Li H., et al. , 2009. Writing memories with light-addressable reinforcement circuitry. Cell 139: 405–415. 10.1016/j.cell.2009.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognigni P., Felsenberg J., Waddell S., 2018. Do the right thing: neural network mechanisms of memory formation, expression and update in Drosophila. Curr. Opin. Neurobiol. 49: 51–58. 10.1016/j.conb.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly J. B., Roberts I. J., Armstrong J. D., Kaiser K., Forte M., et al. , 1996. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science 274: 2104–2107. 10.1126/science.274.5295.2104 [DOI] [PubMed] [Google Scholar]
- Crittenden J. R., Skoulakis E. M., Han K.-A., Kalderon D., Davis R. L., 1998. Tripartite mushroom body architecture revealed by antigenic markers. Learn. Mem. 5: 38–51. [PMC free article] [PubMed] [Google Scholar]
- Crocker A., Guan X.-J., Murphy C. T., Murthy M., 2016. Cell-type-specific transcriptome analysis in the Drosophila mushroom body reveals memory-related changes in gene expression. Cell Reports 15: 1580–1596. 10.1016/j.celrep.2016.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V., Treiber C. D., Waddell S., 2018. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. eLife 7: e34550 10.7554/eLife.34550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R. W., Gelfand M. V., Collins C. A., DiAntonio A., 2008. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J. Comp. Neurol. 508: 131–152. 10.1002/cne.21670 [DOI] [PubMed] [Google Scholar]
- Daniels R. W., Rossano A. J., Macleod G. T., Ganetzky B., 2014. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS One 9: e100637 10.1371/journal.pone.0100637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., et al. , 2018. A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174: 982–998.e20. 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F. P., Nern A., Picard S., Reiser M. B., Rubin G. M., et al. , 2018. A genetic, genomic, and computational resource for exploring neural circuit function. bioRxiv. 10.1101/385476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., 2005. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu. Rev. Neurosci. 28: 275–302. 10.1146/annurev.neuro.28.061604.135651 [DOI] [PubMed] [Google Scholar]
- Davis R. L., 2011. Traces of Drosophila memory. Neuron 70: 8–19. 10.1016/j.neuron.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J. J., Sweatt J. D., 2010. DNA methylation and memory formation. Nat. Neurosci. 13: 1319–1323. 10.1038/nn.2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaud J.-M., Clouet-Redt C., Bockaert J., Grau Y., Parmentier M.-L., 2008. Widespread brain distribution of the Drosophila metabotropic glutamate receptor. Neuroreport 19: 367–371. 10.1097/WNR.0b013e3282f524c7 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper I., Kurshan P. T., McBride E., Jackson F. R., Kopin A. S., 2007. Locomotor activity is regulated by D2-like receptors in Drosophila: an anatomic and functional analysis. Dev. Neurobiol. 67: 378–393. 10.1002/dneu.20355 [DOI] [PubMed] [Google Scholar]
- Dubnau J., Chiang A.-S., 2013. Systems memory consolidation in Drosophila. Curr. Opin. Neurobiol. 23: 84–91. 10.1016/j.conb.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Dubnau J., Grady L., Kitamoto T., Tully T., 2001. Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature 411: 476–480. 10.1038/35078077 [DOI] [PubMed] [Google Scholar]
- Farris S. M., 2013. Evolution of complex higher brain centers and behaviors: behavioral correlates of mushroom body elaboration in insects. Brain Behav. Evol. 82: 9–18. 10.1159/000352057 [DOI] [PubMed] [Google Scholar]
- Featherstone D. E., Rushton E. M., Hilderbrand-Chae M., Phillips A. M., Jackson F. R., et al. , 2000. Presynaptic glutamic acid decarboxylase is required for induction of the postsynaptic receptor field at a glutamatergic synapse. Neuron 27: 71–84. 10.1016/S0896-6273(00)00010-6 [DOI] [PubMed] [Google Scholar]
- Fei H., Chow D. M., Chen A., Romero-Calderón R., Ong W. S., et al. , 2010. Mutation of the Drosophila vesicular GABA transporter disrupts visual figure detection. J. Exp. Biol. 213: 1717–1730. 10.1242/jeb.036053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giang T., Ritze Y., Rauchfuss S., Ogueta M., Scholz H., 2011. The serotonin transporter expression in Drosophila melanogaster. J. Neurogenet. 25: 17–26. 10.3109/01677063.2011.553002 [DOI] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Santos G. D., Urbano J.-M., Antonazzo G., et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. 10.1093/nar/gkw1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., O’Dowd D. K., 2006. Cholinergic synaptic transmission in adult Drosophila Kenyon cells in situ. J. Neurosci. 26: 265–272. 10.1523/JNEUROSCI.4109-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K.-A., Millar N. S., Grotewiel M. S., Davis R. L., 1996. DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron 16: 1127–1135. 10.1016/S0896-6273(00)80139-7 [DOI] [PubMed] [Google Scholar]
- Harrison J. B., Chen H. H., Sattelle E., Barker P. J., Huskisson N. S., et al. , 1996. Immunocytochemical mapping of a C-terminus anti-peptide antibody to the GABA receptor subunit, RDL in the nervous system in Drosophila melanogaster. Cell Tissue Res. 284: 269–278. 10.1007/s004410050587 [DOI] [PubMed] [Google Scholar]
- Haynes P. R., Christmann B. L., Griffith L. C., 2015. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. eLife 4: e03868 10.7554/eLife.03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M., Schaefer A., Gong S., Peterson J. D., Day M., et al. , 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135: 738–748. 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M., 2003. Mushroom body memoir: from maps to models. Nat. Rev. Neurosci. 4: 266–275. 10.1038/nrn1074 [DOI] [PubMed] [Google Scholar]
- Henry G. L., Davis F. P., Picard S., Eddy S. R., 2012. Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 40: 9691–9704. 10.1093/nar/gks671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida N., Aboukilila M. Y., Burow D. A., Paul R., Greenberg M. M., et al. , 2017. EC-tagging allows cell type-specific RNA analysis. Nucleic Acids Res. 45: e138–e138. 10.1093/nar/gkx551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.-T., Bang S., Hyun S., Kang J., Jeong K., et al. , 2008. cAMP signalling in mushroom bodies modulates temperature preference behaviour in Drosophila. Nature 454: 771–775. 10.1038/nature07090 [DOI] [PubMed] [Google Scholar]
- Huang C., Zheng X., Zhao H., Li M., Wang P., et al. , 2012. A permissive role of mushroom body α/β core neurons in long-term memory consolidation in Drosophila. Curr. Biol. 22: 1981–1989. 10.1016/j.cub.2012.08.048 [DOI] [PubMed] [Google Scholar]
- Inada K., Tsuchimoto Y., Kazama H., 2017. Origins of cell-type-specific olfactory processing in the Drosophila mushroom body circuit. Neuron 95: 357–367.e4. 10.1016/j.neuron.2017.06.039 [DOI] [PubMed] [Google Scholar]
- Isabel G., Pascual A., Preat T., 2004. Exclusive consolidated memory phases in Drosophila. Science 304: 1024–1027. 10.1126/science.1094932 [DOI] [PubMed] [Google Scholar]
- Ishimoto H., Wang Z., Rao Y., Wu C.-F., Kitamoto T., 2013. A novel role for ecdysone in Drosophila conditioned behavior: linking GPCR-mediated non-canonical steroid action to cAMP signaling in the adult brain. PLoS Genet. 9: e1003843 10.1371/journal.pgen.1003843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Suzuki K., Estes P., Ramaswami M., Yamamoto D., et al. , 1998. The organization of extrinsic neurons and their implications in the functional roles of the mushroom bodies in Drosophila melanogaster Meigen. Learn. Mem. 5: 52–77. [PMC free article] [PubMed] [Google Scholar]
- Jefferis G. S. X. E., Marin E. C., Watts R. J., Luo L., 2002. Development of neuronal connectivity in Drosophila antennal lobes and mushroom bodies. Curr. Opin. Neurobiol. 12: 80–86. 10.1016/S0959-4388(02)00293-3 [DOI] [PubMed] [Google Scholar]
- Johard H. A. D., Enell L. E., Gustafsson E., Trifilieff P., Veenstra J. A., et al. , 2008. Intrinsic neurons of Drosophila mushroom bodies express short neuropeptide F: relations to extrinsic neurons expressing different neurotransmitters. J. Comp. Neurol. 507: 1479–1496. 10.1002/cne.21636 [DOI] [PubMed] [Google Scholar]
- Jones S. G., Nixon K. C. J., Chubak M. C., Kramer J. M., 2018. Mushroom body specific transcriptome analysis reveals dynamic regulation of learning and memory genes after acquisition of long-term courtship memory in Drosophila. G3 (Bethesda) 8: 3433–3446. 10.1534/g3.118.200560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun K. R., Azanchi R., Maung Z., Hirsh J., Heberlein U., 2011. A Drosophila model for alcohol reward. Nat. Neurosci. 14: 612–619. 10.1038/nn.2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Çopur A., Schnorrer F., 2016. A Guide to Genome-Wide In Vivo RNAi Applications in Drosophila. Methods Mol. Biol. 1478: 117–143. 10.1007/978-1-4939-6371-3_6 [DOI] [PubMed] [Google Scholar]
- Keene A. C., Waddell S., 2007. Drosophila olfactory memory: single genes to complex neural circuits. Nat. Rev. Neurosci. 8: 341–354. 10.1038/nrn2098 [DOI] [PubMed] [Google Scholar]
- Kikinis R., Pieper S. D., Vosburgh K. G., 2013. 3D Slicer: a platform for subject-specific image analysis, visualization, and clinical support, pp. 277–289 in Intraoperative Imaging and Image-Guided Therapy, Springer New York, New York, NY. [Google Scholar]
- Kim Y.-C., Lee H.-G., Han K.-A., 2007. D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J. Neurosci. 27: 7640–7647. 10.1523/JNEUROSCI.1167-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-C., Lee H.-G., Lim J., Han K.-A., 2013. Appetitive learning requires the alpha1-like octopamine receptor OAMB in the Drosophila mushroom body neurons. J. Neurosci. 33: 1672–1677. 10.1523/JNEUROSCI.3042-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes M. J., DasGupta S., Vreede A., White B., Armstrong J. D., et al. , 2009. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell 139: 416–427. 10.1016/j.cell.2009.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes M. J., Keene A. C., Leung B., Armstrong J. D., Waddell S., 2007. Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron 53: 103–115. 10.1016/j.neuron.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S.-Y., Wu C.-L., Hsieh M.-Y., Lin C.-T., Wen R.-K., et al. , 2015. PPL2ab neurons restore sexual responses in aged Drosophila males through dopamine. Nat. Commun. 6: 7490 10.1038/ncomms8490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K., 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15: R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.-T., Lin H.-W., Chang Y.-H., Fu T.-F., Dubnau J., et al. , 2011. Serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila. Proc. Natl. Acad. Sci. USA 108: 13794–13799. 10.1073/pnas.1019483108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Lee A., Luo L., 1999. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126: 4065–4076. [DOI] [PubMed] [Google Scholar]
- Lim J., Fernandez A. I., Hinojos S. J., Aranda G. P., James J., et al. , 2018. The mushroom body D1 dopamine receptor controls innate courtship drive. Genes Brain Behav. 17: 158–167. 10.1111/gbb.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A. C., Bygrave A. M., de Calignon A., Lee T., Miesenböck G., 2014. Sparse, decorrelated odor coding in the mushroom body enhances learned odor discrimination. Nat. Neurosci. 17: 559–568. 10.1038/nn.3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Plaçais P.-Y., Yamagata N., Pfeiffer B. D., Aso Y., et al. , 2012. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488: 512–516. 10.1038/nature11304 [DOI] [PubMed] [Google Scholar]
- Liu Q., Yang X., Tian J., Gao Z., Wang M., et al. , 2016a Gap junction networks in mushroom bodies participate in visual learning and memory in Drosophila. eLife 5: e13238 10.7554/eLife.13238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Krause W. C., Davis R. L., 2007. GABAA receptor RDL inhibits Drosophila olfactory associative learning. Neuron 56: 1090–1102. 10.1016/j.neuron.2007.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liao S., Veenstra J. A., Nässel D. R., 2016b Drosophila insulin-like peptide 1 (DILP1) is transiently expressed during non-feeding stages and reproductive dormancy. Sci. Rep. 6: 26620 10.1038/srep26620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H., Peabody N. C., Vinson C. R., White B. H., 2006. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52: 425–436. 10.1016/j.neuron.2006.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies C., Tully T., Dubnau J., 2005. Deconstructing memory in Drosophila. Curr. Biol. 15: R700–R713. 10.1016/j.cub.2005.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P., Keene A. C., 2016. Gustatory processing and taste memory in Drosophila. J. Neurogenet. 30: 112–121. 10.1080/01677063.2016.1185104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Davis R. L., 2001. The role of Drosophila mushroom body signaling in olfactory memory. Science 293: 1330–1333. 10.1126/science.1062622 [DOI] [PubMed] [Google Scholar]
- Menzel R., 2012. The honeybee as a model for understanding the basis of cognition. Nat. Rev. Neurosci. 13: 758–768. 10.1038/nrn3357 [DOI] [PubMed] [Google Scholar]
- Miller M. R., Robinson K. J., Cleary M. D., Doe C. Q., 2009. TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat. Methods 6: 439–441. 10.1038/nmeth.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan E., Carrillo R. A., Eastman C. L., Weiszmann R., Waghray D., et al. , 2013. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154: 228–239. 10.1016/j.cell.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisse E., Yin Y., Lin A. C., Lin S., Huetteroth W., et al. , 2013. Different kenyon cell populations drive learned approach and avoidance in Drosophila. Neuron 79: 945–956. 10.1016/j.neuron.2013.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrat P. N., DasGupta S., Wang J., Theurkauf W., Weng Z., et al. , 2013. Transposition-driven genomic heterogeneity in the Drosophila brain. Science 340: 91–95. 10.1126/science.1231965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T.-T. B., Misra S., et al. , 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105: 9715–9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S., Saalfeld S., Tomancak P., 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25: 1463–1465. 10.1093/bioinformatics/btp184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y.-X., Xu G., Gu G.-X., Mao F., Ye G.-Y., et al. , 2017. A new Drosophila octopamine receptor responds to serotonin. Insect Biochem. Mol. Biol. 90: 61–70. 10.1016/j.ibmb.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Qin H., Cressy M., Li W., Coravos J. S., Izzi S. A., et al. , 2012. Gamma neurons mediate dopaminergic input during aversive olfactory memory formation in Drosophila. Curr. Biol. 22: 608–614. 10.1016/j.cub.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., et al. , 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries A.-S., Hermanns T., Poeck B., Strauss R., 2017. Serotonin modulates a depression-like state in Drosophila responsive to lithium treatment. Nat. Commun. 8: 15738 10.1038/ncomms15738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., et al. , 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai Y., Hirokawa A., Ai Y., Zhang M., Li W., et al. , 2015. Dissecting neural pathways for forgetting in Drosophila olfactory aversive memory. Proc. Natl. Acad. Sci. USA 112: E6663–E6672. 10.1073/pnas.1512792112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinakevitch I., Grau Y., Strausfeld N. J., Birman S., 2010. Dynamics of glutamatergic signaling in the mushroom body of young adult Drosophila. Neural Dev. 5: 10 10.1186/1749-8104-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D., Aso Y., Jin X., Chen N., Felix M., et al. , 2015a Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr. Biol. 25: 2915–2927. 10.1016/j.cub.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D., Aso Y., Rubin G. M., Nitabach M. N., 2015b Control of sleep by dopaminergic inputs to the Drosophila mushroom Body. Front. Neural Circuits 9: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O., Banker G. A., 1992. Getting the message from the gene to the synapse: sorting and intracellular transport of RNA in neurons. Trends Neurosci. 15: 180–186. 10.1016/0166-2236(92)90170-D [DOI] [PubMed] [Google Scholar]
- Strausfeld N. J., Hansen L., Li Y., Gomez R. S., Ito K., 1998. Evolution, discovery, and interpretations of arthropod mushroom bodies. Learn. Mem. 5: 11–37. [PMC free article] [PubMed] [Google Scholar]
- Takemura S.-Y., Aso Y., Hige T., Wong A., Lu Z., et al. , 2017. A connectome of a learning and memory center in the adult Drosophila brain. eLife 6: e26975 10.7554/eLife.26975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trannoy S., Redt-Clouet C., Dura J.-M., Preat T., 2011. Parallel processing of appetitive short- and long-term memories in Drosophila. Curr. Biol. 21: 1647–1653. 10.1016/j.cub.2011.08.032 [DOI] [PubMed] [Google Scholar]
- Tsao C.-H., Chen C.-C., Lin C.-H., Yang H.-Y., Lin S., 2018. Drosophila mushroom bodies integrate hunger and satiety signals to control innate food-seeking behavior. eLife 7: e35264 10.7554/eLife.35264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K., Suzuki E., Naganos S., Ofusa K., Horiuchi J., et al. , 2017. Coincident postsynaptic activity gates presynaptic dopamine release to induce plasticity in Drosophila mushroom bodies. eLife 6: e21076 10.7554/eLife.21076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt K., Aso Y., Hige T., Knapek S., Ichinose T., et al. , 2016. Direct neural pathways convey distinct visual information to Drosophila mushroom bodies. eLife 5: e14009 10.7554/eLife.14009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt K., Schnaitmann C., Dylla K. V., Knapek S., Aso Y., et al. , 2014. Shared mushroom body circuits underlie visual and olfactory memories in Drosophila. eLife 3: e02395 10.7554/eLife.02395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S., Armstrong J. D., Kitamoto T., Kaiser K., Quinn W. G., 2000. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell 103: 805–813. 10.1016/S0092-8674(00)00183-5 [DOI] [PubMed] [Google Scholar]
- Wu C.-L., Shih M.-F. M., Lee P.-T., Chiang A.-S., 2013. An octopamine-mushroom body circuit modulates the formation of anesthesia-resistant memory in Drosophila. Curr. Biol. 23: 2346–2354. 10.1016/j.cub.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Xia S., Miyashita T., Fu T.-F., Lin W.-Y., Wu C.-L., et al. , 2005. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr. Biol. 15: 603–615. 10.1016/j.cub.2005.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-H., Shih M.-F. M., Chang C.-C., Chiang M.-H., Shih H.-W., et al. , 2016. Additive expression of consolidated memory through Drosophila mushroom body subsets. PLoS Genet. 12: e1006061 10.1371/journal.pgen.1006061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. C., Tully T., 1996. CREB and the formation of long-term memory. Curr. Opin. Neurobiol. 6: 264–268. 10.1016/S0959-4388(96)80082-1 [DOI] [PubMed] [Google Scholar]
- Yin J. C., Wallach J. S., Del Vecchio M., Wilder E. L., Zhou H., et al. , 1994. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79: 49–58. 10.1016/0092-8674(94)90399-9 [DOI] [PubMed] [Google Scholar]
- Yuan Q., Joiner W. J., Sehgal A., 2006. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 16: 1051–1062. 10.1016/j.cub.2006.04.032 [DOI] [PubMed] [Google Scholar]
- Yuan Q., Lin F., Zheng X., Sehgal A., 2005. Serotonin modulates circadian entrainment in Drosophila. Neuron 47: 115–127. 10.1016/j.neuron.2005.05.027 [DOI] [PubMed] [Google Scholar]
- Zars T., Fischer M., Schulz R., Heisenberg M., 2000. Localization of a short-term memory in Drosophila. Science 288: 672–675. 10.1126/science.288.5466.672 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All Drosophila strains are available upon request. Supplemental files are available at FigShare. Files S1 – S7 contain the expression pattern for each of the split-GAL4 drivers, while Files S8 – S14 contain one example of the cognate WM images used for cell counting. Figure S1 contains the olfactory short-term memory performance for 10 newly generated isogenic strains. Figure S2 contains overview of the workflow of TAPIN purification of nuclei and the molecular size distribution of amplified cDNA obtained from the 14 TAPIN-seq libraries. Figure S3 and S4 contains the view of the cell bodies of MB KCs in the confocal images that have double labeling for immunostaining and GAL4 expression. Table S1 contains the TAPIN-seq library statistics, including the numbers of raw reads, pseudoaligned reads and uniquely aligned reads, as well as the detected gene numbers. Table S2 contains the full list of genes enriched and depleted in individual MB classes and subtypes. The sequencing reads and processed data files, including the tables of estimated abundances, are available in NCBI GEO (GSE119629). All code used to analyze RNA-seq results and create the figures and tables in this manuscript are available in the GitHub repository (http://github.com/fredpdavis/mushroombody). Supplemental material available at Figshare: https://doi.org/10.25387/g3.7267481.