Abstract
Southern Leaf Blight (SLB), Northern Leaf Blight (NLB), and Gray Leaf Spot (GLS) caused by Cochliobolus heterostrophus, Setosphaeria turcica, and Cercospora zeae-maydis respectively, are among the most important diseases of corn worldwide. Previously, moderately high and significantly positive genetic correlations between resistance levels to each of these diseases were identified in a panel of 253 diverse maize inbred lines. The goal of this study was to identify loci underlying disease resistance in some of the most multiple disease resistant (MDR) lines by the creation of chromosome segment substitution line (CSSL) populations in multiple disease susceptible (MDS) backgrounds. Four MDR lines (NC304, NC344, Ki3, NC262) were used as donor parents and two MDS lines (Oh7B, H100) were used as recurrent parents to produce eight BC3F4:5 CSSL populations comprising 1,611 lines in total. Each population was genotyped and assessed for each disease in replicated trials in two environments. Moderate to high heritabilities on an entry mean basis were observed (0.32 to 0.83). Several lines in each population were significantly more resistant than the MDS parental lines for each disease. Multiple quantitative trait loci (QTL) for disease resistance were detected for each disease in most of the populations. Seventeen QTL were associated with variation in resistance to more than one disease (SLB/NLB: 2; SLB/GLS: 7; NLB/GLS: 2 and 6 to all three diseases). For most populations and most disease combinations, significant correlations were observed between disease scores and also between marker effects for each disease. The number of lines that were resistant to more than one disease was significantly higher than would be expected by chance. Using the results from individual QTL analyses, a composite statistic based on Mahalanobis distance (Md) was used to identify joint marker associations with multiple diseases. Across all populations and diseases, 246 markers had significant Md values. However further analysis revealed that most of these associations were due to strong QTL effects on a single disease. Together, these findings reinforce our previous conclusions that loci associated with resistance to different diseases are clustered in the genome more often than would be expected by chance. Nevertheless true MDR loci which have significant effects on more than one disease are still much rarer than loci with single disease effects.
Keywords: Maize disease resistance, Multiple disease resistance, QTL
Genetic resistance is the most cost-effective and environment-friendly method for reducing losses in yield and quality in agricultural crops caused by plant disease. Disease resistance is often described in the literature as being either qualitative or quantitative. Qualitative disease resistance typically confers high levels of resistance and is generally controlled by a single or a few genes with major effects (Bent and Mackey 2007; Ross 1986; Van der Plank 1965). Quantitative disease resistance (QDR) typically confers partial resistance, is usually based on the effect of multiple genes and is generally believed to be much more durable in the field than qualitative resistance (Poland et al. 2009, Niks et al. 2015).
Multiple disease resistance (MDR) can refer to a host plant with resistance to more than one disease or to a gene or allele that confers resistance to more than one disease (Wiesner-Hanks and Nelson 2016). The MDR principle can be traced back at least to 1902 where a cowpea cultivar was described to be resistant to wilt and root-knot, Neocosmospora vasinfecta and Heterodera radicicola, respectively (Orto 1902). Since then, many authors have reported on successful development of lines with MDR in a great number of crops such as wheat, barley and potato (Jansky and Rouse 2003; Mitchell-Olds et al., 1995; Risk et al., 2013; Schnurbusch et al., 2004). Multiple-disease resistant lines may contain several genes acting independently to confer resistance to multiple diseases, or single genes conferring multiple resistance to several diseases. Several examples of MDR genes are described in the literature. For example, Mi-1 confers resistance to both aphids and nematodes in tomato (Vos et al., 1998) and Lr34/Yr18 confers resistance to leaf rust, stripe rust, steam rust, and powdery mildew in wheat (Spielmeyer et al. 2013). Other genes conferring MDR in wheat include Lr46/Yr29, Lr67, and Yr30 (Bariana et al., 2007; Rosewarne et al., 2008; Spielmeyer et al., 2013; William et al., 2007). In this study, the genetic basis of resistance to three diseases of maize was investigated: Gray leaf spot (GLS); Northern leaf blight (NLB); and Southern leaf blight (SLB). Each of these diseases are caused by an ascomycete fungal pathogen that infects leaves of maize.
SLB is caused by the necrotrophic fungus Cochliobolus heterostrophus (anamorph Bipolaris maydis) and often occurs where maize is grown under hot and humid conditions. Irregularly shaped lesions initially from the lower to upper canopy of the plant and expand until they coalesce within leaves to cover large sections or entire leaves (White 1999). In the early 1970s, C. heterostrophus race T, which produces the so-called T-toxin, caused an important epidemic in the U.S., causing overall losses of approximately 12% -15% (Ullstrup 1972). After the epidemic, farmers in the U.S. reverted to growing lines that were not susceptible to C. heterostrophus race T. Currently, the most common race of C. heterostrophus in the U.S. is race O, which can cause yield losses of greater than 40% in inoculated experimental fields (Byrnes and Pataky, 1989, Fisher, Hooker, et al., 1976, Gregory, Ayers, et al., 1979), although losses are normally much lower due to the use of resistant hybrids. Resistance to C. heterostrophus race O is quantitatively inherited with primarily additive or partially dominant gene action (Holley and Goodman 1989). Numerous QTL studies have been conducted to map loci associated with SLB resistance (Balint-Kurti et al., 2008b; Balint-Kurti et al., 2006; Balint-Kurti et al., 2007; Bian et al., 2014; Carson et al., 2004; Kump et al., 2011; Negeri et al., 2011; Zwonitzer et al., 2009; Zwonitzer et al., 2010).
NLB caused by the hemibiotrophic fungus Setosphaeria turcica (anamorph Exserohilum turcicum) is found throughout the world in humid areas with moderate temperatures. Like SLB, the disease initially develops on the lower canopy and spreads to leaves of the upper canopy, producing expansive cigar-shaped lesions. In some U.S. regions, yield loses can be greater than 50% (Raymundo and Hooker 1981; Tefferi et al., 1996). Both qualitative and quantitative forms of phenotypic variation in resistance to NLB have been characterized. The qualitative resistance genes Htl, Ht2, Ht3 confer resistance to specific races of NLB (Bentolila et al., 1991; Hooker 1963; Simcox and Bennetzen 1993; Yin et al., 2003). One of these race-specific genes, Htnl, was recently identified as a wall-associated receptor-like kinase (Hurni et al., 2015). Because qualitative resistance is generally less durable in the field, some research has focused on quantitative disease resistance, revealing a polygenic architecture composed primarily of additive effects (Balint-Kurti et al., 2010; Poland et al., 2011; Chung et al., 2010, 2011, Jamann et al. 2016).
GLS is caused by the related species Cercospora zeae-maydis and C. zeina (Meisel et al. 2009; Ward et al. 1999) which both have a necrotrophic lifestyle. Most reports of GLS describe its occurrence in the United States (Ward et al., 1997, Tehon and Daniels 1925) and Africa (Dunkle and Levy 2000), with some reports of GLS in China and Latin America (Ward et al., 1999). Disease development is favored in temperate, humid conditions. Similar to SLB and NLB, symptoms are first observed on the lower leaves as a mixture of small, irregularly shaped spots and semi-rectangular lesions which spread up the plant while lesions expand parallel to the veins of the leaf (Ward et al., 1999). Losses due to GLS can range from 10 to 25% annually, but in severe cases this value can reach 50% (Freppon et al., 1996; Ward et al., 1999). Residues from continuous cropping managed under conservation tillage also promote the development of the disease (Ward et al., 1999). Phenotypic variation in resistance to GLS in maize is generally quantitative, with a polygenic architecture primarily composed of additive allele effects (Lyimo et al., 2011; Zhang et al., 2012 Balint-Kurti et al., 2008a; Derera et al., 2008; Gordon et al., 2006; Juliatti et al., 2009; Lennon 2014; Zwonitzer et al., 2010, Benson et al. 2015).
While the infection processes of the pathogens causing SLB, NLB, and GLS have several distinguishing features, they share some aspects (Beckman and Payne 1982; Jennings and Ullstrup 1957). For instance, they all penetrate the leaf and, in early stages, grow in living tissue, but ultimately derive their nutrition from dead tissue. Gene(s) affecting some of these shared pathogenesis processes might be expected to confer MDR. Indeed, using multivariate statistical analysis of data on SLB, NLB, and GLS resistance scored in a panel of maize inbred lines developed for high-resolution association mapping (Flint-Garcia et al. 2005), Wisser et al. (2011) found moderately high positive genetic correlations between resistances in all pairwise combinations. Because linkage disequilibrium (LD) in that panel is low (Flint-Garcia et al. 2005), the authors inferred that a component of the variation in MDR was attributable to alleles with pleiotropic effects. Single nucleotide polymorphisms (SNPs) within a glutathione S-transferase gene on chromosome 7 were associated with MDR.
The goal of the current study was to produce chromosome segment substitution lines (CSSL) in which a whole genome tiling path of introgressions from MDR lines is captured in MDS genomic backgrounds and to use these populations to determine if loci underlying MDR could be identified. The rationale for choosing this approach was twofold; First we reasoned that the effects of some resistance loci might be best observed in a susceptible background and second, we wanted to identify MDR NIL lines that could be used directly in subsequent studies probing the mechanisms of resistance. Four top-ranking MDR lines (NC304, NC344, Ki3 and NC262) identified in Wisser et al. (2011) were used as donor parents and two low-ranking MDS lines (Oh7B, H100) were used as recurrent parents to produce eight CSSL populations.
Materials and Methods
Plant materials, populations
Six inbred maize lines were used to produce eight different populations. The parental materials were: inbred line H100 (developed by Indiana Agricultural Experiment Station from N28 x H91; Dudley et al., 1991), inbred line Ki3 (developed from Suwan-1(S) C4 (Thailand); Liu et al., 2003), inbred line NC262 (developed by North Carolina State University from McNair 14 x McNair 18; Nelson et al., 2016), inbred line NC304 (developed by North Carolina State University from (Pioneer X105A x H5) x H101; Nelson et al., 2016), inbred line NC344 (developed by North Carolina State University from (McNair 14x18)^2 x [(NC246 x NC248) x C103]; Nelson et al., 2016), and inbred line Oh7B (developed in Ohio from (Oh07 × 38-11) x Oh07; Liu et al., 2003).
Eight chromosome segment substitution populations were created by crossing four MDR donor lines, Ki3, NC262, NC304, and NC344 as females, with two multiple disease susceptible recurrent lines, H100 and Oh7B as male. After the F1 was obtained, three generations of backcrosses with H100 or Oh7B as males were performed, followed by four generations of self-pollination via single-seed descent to obtain BC3F4:5 lines (Figure 1). The F4:5 lines were subsequently increased by sib-mating within each line. An identification code was ascribed to each line, starting with the prefix DRIL (for “Disease Resistance Introgression Line”) followed by a population code for each cross based on line codes: (H100 = 2, Ki3 = 3, NC262 = 5, NC304 = 6, NC344 = 7, Oh7B = 8); where 32, 52, 62, and 72 correspond to Ki3/H100, NC262/H100, NC304/H100, and NC344/H100, respectively, and 38, 58, 68, and 78 correspond to Ki3/OH7B, NC262/OH7B, NC304/OH7B, and NC344/OH7B, respectively. In each case the code number for the donor (female) parent is listed first and the number for the recurrent (male) parent is listed second, followed by a numerical identifier for each line (e.g., DRIL32-001, DRIL32-002, etc.).
Figure 1.
Scheme used to produce all chromosome segment substitution lines populations in this study.
Generation of genotypic data
Young leaf tips from six plants per line were collected and lyophilized for DNA extraction and genotyping. DNA extraction was performed using the commercially available Gentra Puregene Tissue Kit. The final DRIL populations were genotyped with the Pioneer Illumina publicplex platform using 765 single nucleotide polymorphism (SNP) markers (Jones et al., 2009). The genetic map locations used were based on the IBM4 map (Fu et al., 2006). Files S1-S8 have details of all the markers used in each population.
On average 3.1% of the marker data were missing. Missing data imputation was performed using a set of simple algorithms in Microsoft Excel based on flanking markers. For each line, if a data point was missing for a marker (X), and if the parental genotypes for the two flanking markers were identical, then the missing data were imputed to be the same parental genotype as the flanking markers. This imputation criteria was also used for up to two consecutive missing marker values (e.g., consecutive marker calls AA, XX, XX, AA becomes AA AA AA AA, where X is missing data and A is a parental allele). If the flanking markers differed, then the missing markers were imputed as the recurrent parental allele.
The BC3F4:5 lines are expected to carry 6.25% introgressed DNA derived from the donor genome, and 78% of that introgressed DNA is expected to be present as homozygous introgressions (Figure 1). The genotypic data were systematically evaluated to identify and eliminate lines that substantially deviated from these expectations. In these cases, it was likely that the lines had been the product of one or more contaminant pollinations.
To eliminate lines that were not likely the product of the given crossing scheme, DRILs were eliminated if they exceeded two standard deviations from the median value of either: (i) the total percentage of markers in a line that were heterozygous and the number of introgression segments in a line; (ii) the percentage of donor parent alleles detected for each line; or (iii) any combination of (i) and (ii).
A similar process was performed to identify markers with anomalous scores. For each DRIL population, any given marker locus would be expected to carry donor parent alleles at approximately 6.25% of loci assessed and 78% of these donor alleles would be expected to be present in the homozygous state. Additionally, marker loci would be expected to have similar genotypes at genetically linked marker loci most of the time. After anomalous lines had been eliminated, two parameters were calculated from the remaining lines in each population: the percentage of lines that were heterozygous for a particular marker and the percentage in each population that a marker was different from its flanking marker (we refer to this metric as percent unlinked marker). If a marker presented a value in any of these two parameters that was higher than the median plus two standard deviations, it was eliminated. The exceptions to this were when the marker was the final marker on a chromosome and when a high percentage of unlinked markers was caused by the anomalous behavior of the neighboring marker. To determine this, the most anomalous markers (top third) were eliminated first and then the analysis was performed again to reevaluate the remaining markers each time.
Inoculum Preparation and Inoculation Procedure
Sorghum grains were soaked from 3 to 4 days in water, placed in 1 L flasks, and autoclaved for one hour (120 PSI and 121°). Autoclaved grain was inoculated with either C. heterostrophus, S. turcica, or C. zeae-maydis. The fungus grew at room temperature (23-25°) for approximately 10, 14 or 21 days for C. heterostrophus, S. turcica, or C. zeae-maydis respectively until the sorghum was colonized with the fungus. The sorghum was air-dried and stored at 4°. Each isolate of each fungus was grown and dried separately. The dried sorghum infested with isolates of the same species was thoroughly mixed just prior to inoculation. The same mixture was used to inoculate the entire trial. Twenty-five to thirty-day old maize plants were inoculated by adding 6 to 10 infested sorghum kernels placed into the whorl of each maize plant.
Disease evaluations
All trials used an augmented lattice design. The number of entries varied between each population and are shown in Table 1. Two environments were used for every assessment and two replications were used in every environment except for the case of NC262/H100 where only one replication was planted in one of the years in the GLS evaluation. The number of blocks within each replication varied depending on the number of lines in the population but was at least 10 in every case. A recurrent parent check was included in each block and the resistant donor parent was also planted several times in each replication.
Table 1. Number of lines and markers and associated parameters for each population.
| Population | Ki3/H100 | Ki3/Oh7B | NC262/H100 | NC262/Oh7B |
|---|---|---|---|---|
| Lines | 265 | 204 | 195 | 111 |
| Markers | 245 | 239 | 271 | 209 |
| 1Percent Recurrent Parent | 93.5 | 92.7 | 91.3 | 85.2 |
| 2Percent Donor Parent | 6.5 | 7.3 | 8.7 | 14.8 |
| 3Percent Heterozygous | 0.8 | 1.2 | 1.6 | 3.1 |
| 4Percent Segregating Introgressions | 12.2 | 16.7 | 19.2 | 19.8 |
| 5Number of Introgressions | 6.3 | 6.9 | 7.8 | 10.5 |
| 6Percent Donor Genome | 100 | 98 | 100 | 100 |
| Population | NC304/H100 | NC304/Oh7B | NC344/H100 | NC304/Oh7B |
|---|---|---|---|---|
| Lines | 252 | 108 | 258 | 218 |
| Markers | 270 | 254 | 270 | 241 |
| Percent Homozygous Recurrent Parent | 93.1 | 91 | 93.2 | 92.05 |
| Percent Homozygous Donor Parent | 6.9 | 9 | 6.8 | 7.5 |
| Percent Heterozygous | 1.6 | 1.2 | 1.3 | 1 |
| Percent Segregating Introgressions | 19.9 | 12.4 | 19.6 | 13.7 |
| Number of Introgressions | 6.7 | 7.6 | 6.4 | 5.9 |
| Percent Donor Genome Represented | 100 | 100 | 100 | 100 |
The total number of markers scored as homozygous for the recurrent parent allele plus half of the total number of markers scored as heterozygous, divided by the total number of markers used for the population, multiplied by 100.
The total number of markers scored as homozygous for the donor parent allele plus half of the total number of markers scored as heterozygous, divided by the total number of markers used for the population multiplied by 100.
Number of markers scored as heterozygous divided by the total number of markers multiplied by 100.
Number of markers scored as heterozygous divided by the total number of markers scored as homozygous or heterozygous for the donor parent allele multiplied by 100.
Number of segments that contain homozygous or heterozygous donor parental allele.
Number of markers scored as homozygous or heterozygous for the donor parental allele contained within the population, divided by the total number of markers multiplied by 100. The population mean of these statistics are shown. For % of introgression the range is also shown.
Field evaluations for SLB disease were performed during the summers of 2012 and 2013 for the NC262/H100 and NC262/Oh7B populations, and in 2014 and 2015 for the Ki3/H100, NC304/H100, NC344/H100, Ki3/Oh7B, NC304/Oh7B, and NC344/Oh7B populations. All SLB phenotypic evaluations were performed at Central Crops Research Station (CCRS) in Clayton, NC. Experiments were planted in 1.8 m single rows with a 0.9 m row width using 10 seeds per plot. To assure high disease pressure, SLB inoculations were carried out 30 days after planting using a mixture of three isolates of C. heterostrophus including 2-16Bm, Hm540 (Carson 1998), and an unnamed isolate provided by Syngenta. Visual scores of the disease were taken three or four times at intervals of eight to ten days starting two weeks after anthesis (when 50% of the plants in a row were shedding pollen) on a scale 1 to 9 where 1 = 100% leaf area affected by the pathogen and 9 = no disease. For each plot, days from planting to anthesis, plant height, and ear height were recorded.
Field evaluations for NLB disease were performed during summer of 2013 for NC262/H100 and NC262/OH7B population at CCRS (environment 1) and at Aurora, NY, Cornell University NY (environment 2). Ki3/Oh7B, NC304/Oh7B, and NC344/Oh7B populations were evaluated for NLB at CCRS in 2014 and 2015. Ki3/H100, NC304/H100, and NC344/H100 were evaluated in 2014 at CCRS. At CCRS trials were planted as described above. In Aurora NY, trials were planted in 2.4 m single rows with a 1 m row width using 10 seeds per plot. To assure high disease pressure, NLB inoculations were performed using several S. turica isolates (ET238A, ET471A-1, ET30A, ET3A, ET28A, ET257A, Cairo05, 235A, race 0 from Syngenta, Race1 from Syngenta, ET252A, ET28A, ET30A, ET222A) 30 to 45 days after planting in Clayton. In New York, inoculations were performed using a local isolate NY001 (race 1). At Clayton, three to four visual scores of the disease were taken at intervals of eight to ten days starting two weeks after anthesis. At NY, scores were taken two times, at an interval of one week, starting two weeks after anthesis. For each plot, days to anthesis was also recorded. At each location, the percentage of diseased leaf area was used to score the disease. For the purpose of correlation analysis, NLB disease scores (from 0 to 100%, where 0 is the most resistant) were converted to a scale similar to the SLB and GLS phenotyping system. Field evaluations for GLS disease were performed during the summers of 2012 and 2013 at Andrews, NC for the NC262/H100 and NC262/OH7B populations, and at Andrews, NC and at the Virginia Tech Kentland Farm, near Blacksburg, VA, during the summer of 2014 for the Ki3/H100, NC304/H100 and NC344/H100 populations. The Ki3/OH7B, NC304/OH7B, and NC344/OH7B populations underwent GLS phenotypic evaluations during summers of 2014 and 2015 at Andrews NC and Blacksburg VA respectively. Trials were planted in 4 m single rows with a 1 m row width using 16 seeds per plot. The fields used in Andrews contained infected plant debris from the previous season and therefore had a very high inoculum disease pressure and artificial inoculation was not required. In Virginia, inoculations were performed using a mixture of 10 field-derived C. zeae-maydis isolates 30 to 45 days after planting. Visual scores of the disease were taken twice at an interval of 10 days starting two weeks after anthesis. Ratings ranged from 1 to 9 where 1 = 100% leaf area affected by the pathogen and 9 = no disease. Days toanthesis was also recorded in Andrews for the NC262/H100 and NC262/OH7B populations.
The symptoms of SLB, NLB and GLS are distinct from each other. In every case in every experiment we observed almost exclusively the symptoms expected of the targeted disease until late in the season (after completion of data collection when the plants had started to senesce).
Statistical Analysis
Statistical analysis of phenotypic data were performed using R (R core development team 2008). Q-Q, residual, and distribution plots were also produced to determine normality and variance homogeneity. The standardized area under disease progress curve (sAUDPC, a quantitative summary of disease severity over time; Simko and Piepho 2012) was calculated for each genotype and each disease based on a minimum of two and maximum of four sequential ratings. Using lme4 (Bates et al., 2015), a univariate linear mixed model was fit to the data for each disease:
where, is the response variable as described above, genotype () is a fixed effect, block , replication , environment () and genotype-by-environment () is a random effect and is a random residual effect. Least square means (LSMeans) for each genotype within each disease were obtained and the lines that had LSMeans significantly different from the recurrent check H100 or Oh7B were identified using a multiple comparison test, mulcomp/glht in R, in which a multiple test correction due to heterogeneous or unequal variances among groups was accounted.
Heritability was estimated on an entry mean basis (Holland et al. 2003). This was determined for each population-disease combination. Variance components used to compute hertitability were estimated from a model similar to the one described above, except that genotype was fit as a random effect in order to also estimate the genotypic variance.
Pairwise Pearson correlations between LSMeans for each disease within each population were calculated. A Chi square test was used to show test whether the number of multiple disease resistant lines identified in each genetic background (H100 and Oh7B) was higher than would be expected due to random chance. Fisher’s exact test was used to test the same thing for individual populations as Chi square does not deal properly with smaller numbers (<5) (Bearden et al. 1982).
QTL analyses
For each DRIL population and disease, QTL analysis was performed using the QTL IciMapping software (Meng, Li, et al., 2015), employing the option for non-idealized CSSL QTL analysis (CSSL with more than one introgression). For this analysis, all recurrent homozygous genotypes, heterozygous genotypes and missing genotypes were coded as 0 and homozygous donor genotypes were coded as 2. The program allows for marker multicollinearity control, and for this a “by condition number” of 1000. A RSTEP-LRT method, which is a likelihood ratio test based on stepwise regression, was used for QTL mapping. Stepwise regression was used to select the most significant markers and a likelihood ratio test was used to calculate the LOD scores for each marker. The probability threshold used for stepwise regression was 0.01 while LOD thresholds were determined by permutation testing (1000 iterations) with a type I error of 0.05.
Outputs of QTL analysis included LOD values, percentage of variance explained, and additive effects of the marker. Additive effects of NLB were converted to the same SLB/GLS scale. Pairwise correlations between the marker additive effects were calculated. The IBM4 genetic map was used as a guide for placing the markers on the maize genetic map (Fu et al., 2006; Jones et al., 2009).
Composite Statistic
Following Rousseeuw and Zomeren (1990), Rousseeuw (1985) and Lotterhous et al. (2017), a composite statistic based on Mahalanobis distance (Md) was used to identify marker associations that represent multivariate outliers in three-dimensional space (i.e., three traits). For each trait, LOD scores were converted into p-values: (Nyholt 2000). Using the negative log10 of the p-values for the three traits, minimum covariance determinant was used to compute the trait-wise covariance matrix for robust detection of outliers (Lotterhos et al. 2017). The analysis was performed using the OutlierMahdist function of the rrcovHD package v. 1.4-4 (Todorov and Filzmoser 2009).
Md2 values follow a distribution with degrees of freedom equal to the number of dimensions (Rousseeuw and Zomeren 1990), so the probability of Md2 for each marker was computed. The “BH” method (Benjamini and Hochberg 1995) of the p.adjust function in R was used to adjust for multiple marker tests per population. Significant multivariate outliers were declared at a 1% false discovery rate. In addition, because a number of significant outliers were identified despite having sub-threshold LOD scores for all three traits, an additional higher threshold for significance was set based on whether the Md2 value was significant and the marker LOD score from QTL mapping was greater than the LOD threshold for at least one of the three traits. Note: the LOD threshold varied, as it was computed by permutation for each population-disease combination (see above). Custom R scripts were used to make figures for the results from analyses using the composite statistic.
Statement of data and reagent availability
All lines are available on request form the corresponding authors. The populations are currently being deposited at the Maize Genetics Stock Center (http://maizecoop.cropsci.uiuc.edu/). All genotypic and phenotypic information used in this work is included in the supplementary files. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7265756.
Results and Discussion
Eight different CSSL DRIL populations were created (Figure 1) in which four multiple disease resistant (MDR) donor parent maize lines (Ki3, NC262, NC304, and NC344) were crossed as females with two multiple disease susceptible (MDS) recurrent parent lines (H100 and Oh7B) as males.
Each population was assessed in disease-specific nurseries for resistance to SLB, NLB, and GLS in two environments with two replications per environment. The phenotypic distributions were, as expected, centered around the recurrent parent phenotype in each case, with the distributions skewed slightly toward resistance (Figure S1). Each DRIL was genotyped using 765 SNP markers. After quality control to eliminate anomalous DRILs, 1611 DRILs from all populations were maintained for further analysis (Table 1). Following quality control of the marker data, 209 to 271 polymorphic markers were retained in each population (Table 1). Based on imputed genotypes, the entire donor parent genome was captured by introgressions within each population except for the Ki3/Oh7B population in which 98% of the Ki3 genome was represented (Table 1).
Exploratory visual data analysis of Q-Q plots, residual plots and spatial field plots of phenotypic values did not reveal any obvious deviations from normality in the data distribution or obvious spatial field effects (not shown); a linear mixed model with normally distributed random variables and without spatial effects was fit to the data. Plotting LSMean estimates for each population-disease-environment combination indicated that donor parental lines were, as expected, highly resistant for all diseases and that the recurrent parental lines were always close to the mean of the entire population. In three out of 48 population-disease-environment combinations assessed, namely the NC304/H100, NC344/H100, and Ki3/H100 populations evaluated for NLB in Aurora NY, the data were deemed unusable based on inconsistent phenotypes for the repeated parental checks. In these cases, we relied on data from just one environment with two replications.
Significant differences were always observed between the donor and recurrent parents for each disease and for each population (Table 2). In every population, some DRILs were significantly more resistant than the recurrent parent for each disease (File S9). However, no significant transgressive segregation was identified. In other words, we did not identify any DRILs that were significantly more resistant than the resistant parent or significantly more susceptible than the susceptible parent (Figure S1, File S9).
Table 2. LSMeans for donor and recurrent parents for each population.
| 1Population | Ki3/H100 | Ki3/Oh7B | NC262/H100 | NC262/Oh7B |
|---|---|---|---|---|
| SLB Don | 8.0*** | 8.0*** | 7.8*** | 6.9*** |
| SLB Rec | 4.6 | 7.8 | 4.4 | 5.7 |
| NLB Don | 12.1*** | 12.8*** | 9.9*** | 15.5*** |
| NLB Rec | 53.4 | 52.4 | 68.4 | 58.9 |
| GLS Don | 8.1*** | 8.2*** | 7.7*** | 6.9*** |
| GLS Rec | 6 | 5.9 | 4.3 | 4.6 |
| Population | NC304/H100 | NC304/Oh7B | NC344/H100 | NC344/Oh7B |
|---|---|---|---|---|
| SLB Don | 8.0*** | 7.9*** | 8.0*** | 7.9*** |
| SLB Rec | 4.8 | 5.8 | 4.6 | 5.8 |
| NLB Don | 10.4*** | 12.7*** | 6.6*** | 9.9*** |
| NLB Rec | 61.8 | 50.5 | 47.7 | 55.6 |
| GLS Don | 8.1*** | 8.3*** | 8.0*** | 8.0*** |
| GLS Rec | 5.9 | 5.9 | 6 | 5.7 |
Don = donor, Rec = recurrent. Significant differences between donor and recurrent parent are represented by *** at a level of 0.001. In each population, first line is the donor and the second is the recurrent parent. SLB and GLS scored on a 1-9 scale with 9 being resistant. NLB scored on a 0–100% scale with 0 being resistant.
Pearson correlation coefficients between replications within environments varied from 0.17 to 0.73 for SLB, 0.23 to 0.63 for NLB, and 0.25 to 0.62 for GLS (Table S1). Similarly, correlations between environments were moderate but significant and varied between 0.35-0.74, 0.39-0.65, and 0.33-0.66 for all SLB, NLB, and GLS experiments respectively (Table S1). It should be noted that CSSL populations display relatively low levels of line-to-line genetically-determined phenotypic variation by design, since the DRILs were on average ≈94% similar to each other. This lack of phenotypic variation leads to lower levels of correlation and heritability than would be expected in other similarly-sized types of bi-parental mapping populations (e.g., recombinant inbred lines). Nevertheless, heritability on an entry mean basis was relatively high for most of the traits: among all populations, heritability for SLB ranged between 0.65 and 0.85; heritability for NLB ranged between 0.67 and 0.83; and heritability for GLS ranged between 0.59 and 0.82 (Table 3).
Table 3. Heritabilities on an entry mean basis for each population and disease.
| Population/ Disease | Ki3/ | Ki3/ | NC262/ | NC262/ | NC304/ | NC304/ | NC344/ | NC344/ |
|---|---|---|---|---|---|---|---|---|
| H100 | Oh7B | H100 | Oh7B | H100 | Oh7B | H100 | Oh7B | |
| SLB | 0.85 | 0.80 | 0.65 | 0.82 | 0.85 | 0.81 | 0.83 | 0.76 |
| NLB | 0.67 | 0.76 | 0.74 | 0.71 | 0.83 | 0.77 | 0.75 | 0.75 |
| GLS | 0.63 | 0.61 | 0.76 | 0.82 | 0.73 | 0.65 | 0.64 | 0.59 |
Pairwise correlation coefficients between LSMeans for different diseases measured in each population were significant in most of the cases. Among the 24 comparisons made (three pairwise comparisons in each of eight populations), 21 were significant at a P < 0.05 (Table 4). Significant correlation coefficients between SLB and NLB ranged from 0.15 to 0.35 among eight populations. Significant correlations coefficients between SLB and GLS ranged from 0.17 to 0.53 among six populations. Significant correlation coefficients between NLB and GLS ranged from 0.18 to 0.47 among seven populations. These results support the hypothesis that the same genes or linked genes contribute to MDR.
Table 4. Pairwise correlations coefficients and p-values for combination of diseases LSMeans scores.
| Population | 1Ki3/H100 (N = 265) | Ki3/Oh7B (N = 207) | NC262/H100 (N = 194) | NC262/Oh7B (N = 111) | ||||
|---|---|---|---|---|---|---|---|---|
| Disease | NLB | GLS | NLB | GLS | NLB | GLS | NLB | GLS |
| SLB | 0.24*** | 0.53*** | 0.35*** | 0.37*** | 0.16* | 0.40*** | 0.21* | 0.17* |
| NLB | . | 0.23*** | . | 0.47*** | . | 0.09 | . | 0.27** |
| Population | NC304/H100 (N = 251) | NC304/Oh7B (N = 108) | NC344/H100 (N = 258) | NC344/Oh7B (N = 216) | ||||
|---|---|---|---|---|---|---|---|---|
| Disease | NLB | GLS | NLB | GLS | NLB | GLS | NLB | GLS |
| SLB | 0.35*** | 0.53*** | 0.24* | 0.35*** | 0.15* | 0.41*** | 0.21** | 0.11 |
| NLB | . | 0.18** | . | 0.30** | . | 0.18** | . | 0.18** |
Significant p-values are represented by ***, **, *, and * at a level of 0.001, 0.01, 0.05, and 0.1 respectively.
Using a 253-line diverse association mapping population, Wisser et al. (2011) reported significant pairwise correlation coefficients between these same three diseases that ranged from 0.55 to 0.67. Those correlations are somewhat higher than those obtained in the present study, probably due to the fact that the panel was much more phenotypically diverse than the populations used in this study. Lennon (2014) used several CSSL populations derived from the commonly used maize lines B73 and several teosinte accessions that were constructed in a similar way to the populations used in the present study. Similar to our findings using CSSL populations, correlations between LSMeans for resistance to SLB and GLS ranged between 0.23 and 0.51, with all of them being significant at level of 0.10.
Fisher’s exact test was performed to determine whether the number of MDR lines (i.e., lines that were significantly more resistant than the recurrent parent for more than one disease) in each population was higher than would be expected by chance. In other words, we tested the hypothesis that resistance to one disease is genetically independent of resistance to another disease. In summary, among 24 tests for combinations of two diseases, six were significant at P < 0.01, seven at P < 0.1, and 11 were not significant (Table 5). Similarly, among eight tests for MDR to three diseases, six were significant with P < 0.0001, and two were not significant. Chi-square tests were conducted on the frequencies of MDR lines within all the four DRIL populations derived from H100 and also from the four derived from Oh7B inbred lines. For the H100 set, all tests were significant. For the Oh7B set, three out of four tests were significant (Table 5). These results suggest that, in most cases, a line that is resistant to one disease has an increased likelihood of being resistant to another disease. Such multiple disease resistance could be caused by shared genomic regions contributing to resistance via coupling phase linkage of donor alleles conferring resistance to different diseases or via the pleiotropic effects of the same sequence variant.
Table 5. Results for Fisher exact test and Chi square test to determine whether multiply disease resistant lines are present at higher levels than would be predicted given the frequencies of lines resistant to each single disease.
| Population | Ki3/ | Ki3/ | NC262/ | NC262/ | NC304/ | NC304/ | NC344/ | NC344/ | H100_ | Oh7B_ |
|---|---|---|---|---|---|---|---|---|---|---|
| H100 | Oh7B | H100 | Oh7B | H100 | Oh7B | H100 | Oh7B | Pooled | Pooled | |
| 1Pop Size | 265 | 207 | 194 | 111 | 251 | 108 | 258 | 216 | 968 | 642 |
| 2SLB # Sig_lines | 74 | 40 | 6 | 37 | 101 | 38 | 49 | 23 | 230 | 138 |
| NLB # Sig_lines | 17 | 27 | 2 | 2 | 9 | 16 | 12 | 6 | 40 | 51 |
| GLS # Sig_lines | 17 | 4 | 42 | 38 | 32 | 1 | 12 | 10 | 103 | 53 |
| 3SLB/NLB | 49* | 16*** | 1* | 2 | 7* | 9* | 3 | 0 | 20*** | 27*** |
| SLB/GLS | 13*** | 4** | 6*** | 14 | 28*** | 1 | 6* | 1 | 53*** | 20** |
| NLB/GLS | 2 | 3** | 2* | 2 | 4* | 0 | 1 | 1 | 9* | 6 |
| SLB/NLB/GLS | 2*** | 3*** | 1*** | 2*** | 4*** | 0 | 1*** | 0 | 8*** | 5*** |
Number of lines in the population.
Number of lines that were statistically different than the recurrent parent for the indicated disease.
Number of lines that were statistically different than the recurrent parent for the indicated combination of diseases.
Significant p-values are represented by ***, **, *, and * at a level of 0.001, 0.01, 0.05, and 0.1 respectively. For combinations of diseases, significance implies that there were more lines significantly resistant to multiple diseases than was expected based on the proportions that were significantly resistant to each single disease.
The DRIL populations were used to map loci associated with each disease, and a composite statistic was used to identify potential MDR loci. Based on single trait analysis, a total of 56, 20, and 28 QTL of small to moderate effect were detected for SLB, NLB, and GLS respectively over all the populations (Table S2, Figure 2). The number of QTL observed among populations varied between 2 and 18. The maximum number of QTL were detected in the NC344/H100 population (18 QTL: 7, 4, and 7 for SLB, NLB, and GLS respectively), followed by the NC304/H100 population (17: 11, 0, and 6); the lowest numbers of QTL were detected in the NC262/Oh7B (2: 0, 2, and 0) population, likely due in part to the relatively small number of lines in this population.
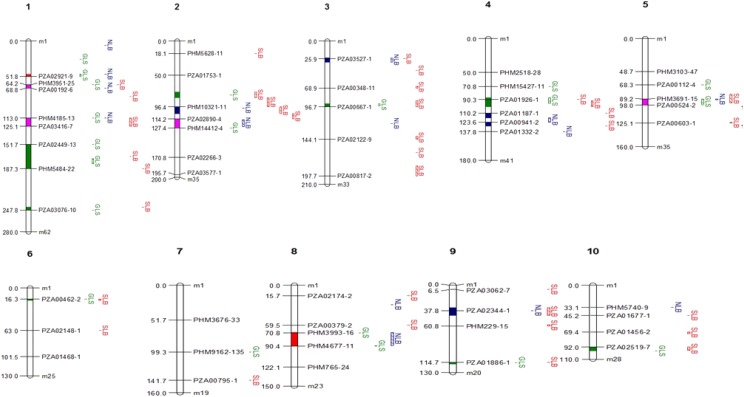
Figure 2.
QTL detected for single, pairwise or threewise disease resistance (chromosomes 1,2, and 3). Colored segments on chromosomes represent regions associated with more than one disease though not necessarily in the same population (red = NLB/GLS, blue = SLB/NLB, green = SLB/GLS, and fuchsia = threewise). Detailed parameters for these QTL are available in Table S4.
For SLB, QTL were detected on all chromosomes, with additive effects ranging between -0.32 and 0.46 (on our 1-9 scale, with a positive value indicating that donor allele increases resistance and vice-versa). For NLB, QTL were detected on all chromosomes except 6 and 7, and additive effects were small to moderate and varied between -6.15 and 3.92 (on a scale from 0–100% where 0 is the most resistant with a negative value indicating that donor allele increases resistance). For GLS, QTL were detected on all chromosomes, with additive effects ranging between -0.25 and 0.35 (on a scale from 1-9, the same as SLB).
Pearson correlations between marker additive effects for different diseases were calculated. From 24 correlations (from 8 populations and 3 diseases), 13 were significant at a P < 0.05 (Table 6). These results are again congruent with the hypothesis that there exists a shared component of the genetic basis of resistance to these three diseases. Indeed, four markers exceeded the corresponding LOD threshold for two diseases in four different populations (Ki3/H100 [NLB-GLS]; Ki3/Oh7B [NLB-GLS]; NC304/H100 [SLB-GLS]; NC344/H100 [SLB-GLS]), but none exceeded the threshold for all three. In some cases, however, the LOD score of a marker was above the significance threshold for one disease, but just below the threshold for another disease. An example of this can be observed in the Ki3/H100 population on chromosome 9 at 114.68 cM where the marker had a LOD score for SLB of 5.6, well above the threshold (3.2), and not far below for NLB (threshold = 3.7, markers LOD = 2.9) and GLS (threshold = 3.03, markers LOD = 2.3).
Table 6. Pairwise correlations coefficients between marker additive effects for combination of diseases.
| Population | Ki3/H100 1(N = 213) | Ki3/Oh7B N = 212) | NC262/H100 (N = 243) | NC262/Oh7B (N = 192) | ||||
|---|---|---|---|---|---|---|---|---|
| Disease | NLB | GLS | NLB | GLS | NLB | GLS | NLB | GLS |
| SLB | 0.12* | 0.37*** | 0.00 | 0.06 | 0.14* | 0.17* | −0.09 | 0.14* |
| NLB | . | 0.19** | . | 0.22** | . | 0.18** | . | −0.02 |
| Population | NC304/H100 (N = 239) | NC304/Oh7B (N = 209) | NC344/H100 (N = 246) | NC344/Oh7B (N = 210) | ||||
|---|---|---|---|---|---|---|---|---|
| Disease | NLB | GLS | NLB | GLS | NLB | GLS | NLB | GLS |
| SLB | 0.05 | 0.23*** | 0.19** | 0.15* | 0.08 | 0.26*** | 0.06 | 0 |
| NLB | . | 0.06 | . | 0.17 | . | 0.14* | . | 0.28*** |
Number of markers segregating in each population (N). Correlations between marker effects were estimated using all the markers selected by the stepwise regression procedure and used in the QTL analysis. Significant p-values are represented by ***, **, *, and * at a level of 0.001, 0.01, 0.05, and 0.1 respectively.
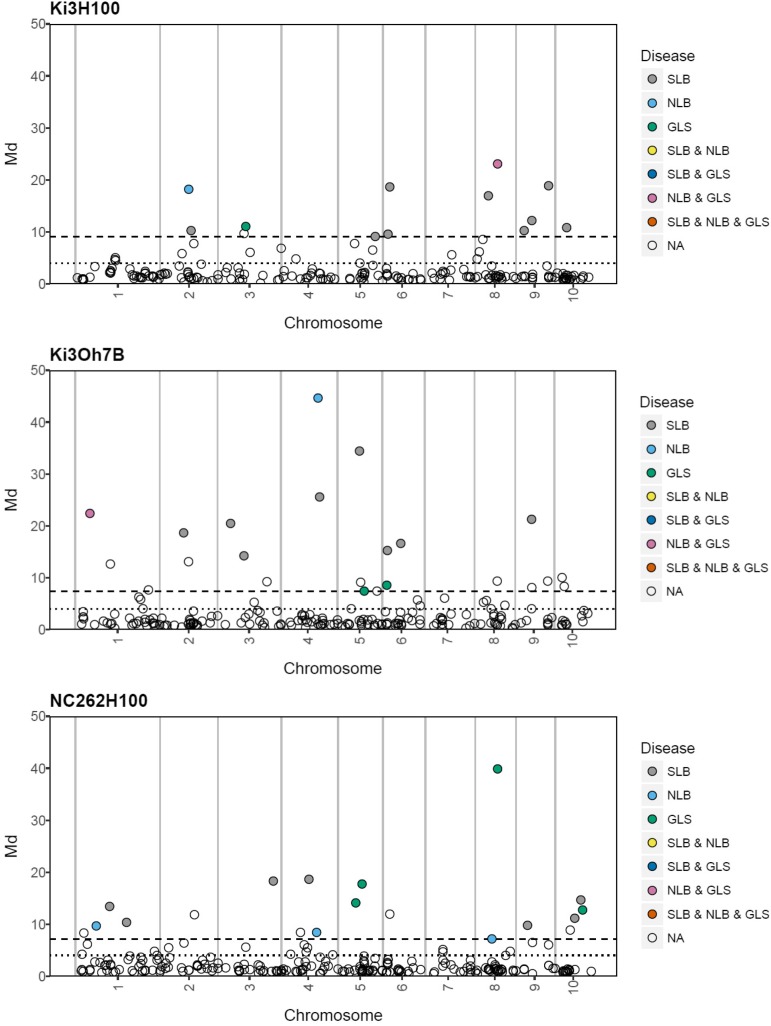
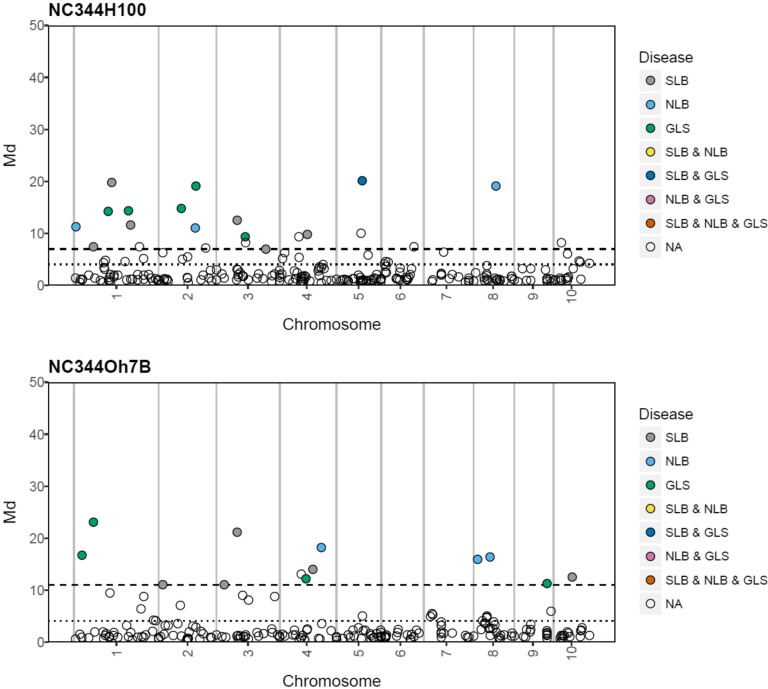
In order to identify additional markers potentially associated with MDR, a composite statistic, Md (Rousseeuw 1985, Rousseeuw and Zomeren, 1990; Lotterhous et al. 2017), which accounts for covariance between the QTL test results for each trait was used (the Md statistic was used because the IciMapping software does not include a multivariate QTL test statistic). Similar to a standard multivariate test, significant multi-trait marker associations can occur when a marker for any one trait or multiple traits deviate. Indeed, all of the significant marker associations identified by single disease QTL analysis (Figure 2) were also declared significant according to the Md statistic (1% FDR). However, several additional markers with relatively low LOD scores for individual traits had significantly large Md values (Figure 3). In total, 246 of the 1652 computed marker Md values were significant (File S10). Across all populations, the additive effects of these markers were correlated as expected for MDR between SLB and GLS (0.18) and NLB and GLS (0.15), but the correlation was essentially zero (-0.01) between SLB and NLB (instead, specific populations were positively correlated and others were negatively correlated). Therefore, Md helped highlight potential MDR loci, but inspection of the individual test statistic results and more highly replicated validation studies are needed to confirm the association of specific loci with MDR. Moreover, due to intricacies in QTL detection that may result in neighboring markers being declared significant despite the same variant being causal, or because MDR is due to linkage and not pleiotropic effects, loci with closely linked marker associations also represent good candidates for further investigation. Therefore, the co-localization of QTL for different diseases within and across populations was also examined.
Figure 3.
Multivariate outlier markers detected by the Md composite statistic. The dotted line corresponds to a 1% FDR for the Md value. Points are color coded according to whether the marker LOD score exceeded the disease-specific permutation threshold from QTL analysis for one or more diseases. The dashed line marks the Md value at which the minimum LOD threshold for a single disease exceeds the lowest threshold (thresholds were specific to each disease).
QTL for each disease were identified at the same loci in different populations (Table S3). For instance, fourteen QTL for SLB co-localized in at least two populations, and two of them were detected in four of ten populations (on chromosome 3, at 66.37-68.94 cM, and on chromosome 9 at 59.01-60.93). For NLB, six QTL co-localized in two populations; and for GLS, six QTL co-localized in at least two populations (Table S4).
When QTL were localized to maize bins (Davis et al. 1999, Table S4), several genomic regions were associated with resistance to more than one disease in the same population. QTL for SLB and GLS co-localized in bins 1.05, 1.06, 3.04, 4.05, 6.01, and 10.04. For SLB and NLB, three QTL co-localized in bins 1.05, 2.05, and 4.05. For NLB and GLS, three QTL co-localized in bins 1.03, 2.05, and 8.03. One QTL in bin 5.04 (chromosome 5 at 90.96-95.23 cM) was associated with resistance to all three diseases in population NC344/H100; in this case, the allele derived from the MDR resistant donor increased susceptibility rather than resistance (Table S4). These results reinforce prior findings (e.g., Tanksley et al., 1996, Balint-Kurti et al., 2008b, Balint-Kurti, et al., 2007, Kump et al. 2011, Benson et al. 2015) that favorable alleles can be found in lines with unfavorable phenotypes. In addition, we noted numerous QTL associated with one disease in one population and with a second or third disease in a different population (Table S4).
Previous studies reported MDR QTL in bins 1.06, 9.02-9.03 for all three diseases, in bins 1.08-1.09, 2.04, 3.04, and 10.05 for SLB and GLS, in 1.04 and 2.02 for NLB and GLS and in 6.01and 8.05 for NLB and SLB (Balint-Kurti et al., 2010; Belcher et al., 2012; Chung et al., 2011; Zwonitzer et al., 2010). Also, a GST gene in bin 7.02 was associated with variation in MDR (Wisser et al., 2011). In this study, some QTL co-localized with those reported previously, but the association with all three diseases was not observed. The one at bin 1.06, reported as MDR for three diseases, was identified for SLB and GLS in this study. The QTL at 9.03, also reported previously to confer resistance to three diseases, was identified in this study for SLB and NLB. QTL previously identified in bins 1.08-1.09 and 3.04 for SLB and GLS, were also identified in this study to confer resistance to SLB and GLS. Most of MDR QTL (for three or two diseases) detected in this study are novel.
In summary, using eight populations comprising more than 1611 lines, we identified a large number of single disease resistance QTL, and several genomic regions with effects on multiple diseases (Figures 2 and 3). Using several approaches, we demonstrated that alleles associated with resistances to these three diseases tend to co-localize in the genome and confer MDR (allelic effects consistently increase resistance). It is likely that in some cases these are due to the co-localization of alleles with disease specific effects and in others to the pleiotropic effects of single alleles. These populations will likely be of utility in mapping other traits including disease resistance. One recent study used some of these populations to map loci for resistance to Goss’s Wilt (Cooper et al. 2018). It will also be helpful to develop higher density genotypic datasets for these populations. We are currently working on fine mapping some of these loci to distinguish between these possibilities.
Acknowledgments
Funding for the work was provided by USDA-ARS, the Corn Growers’ Association of North Carolina and by NSF grant #1127076 to RJW and PBK. LLZ was supported by a grant from Fulbright Colombia and COLCIENCIAS. We thank Cathy Herring and the staff at Central crops for excellent field support, Shannon Sermons, David Rhyne and Greg Marshall for their technical support. We thank Daniel Gorman, David Bubeck and DuPont-Pioneer for their assistance with field trials in Andrews NC. We thank Jim Holland and Marc Cubeta for their advice and for reviewing the manuscript.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7265756.
Communicating editor: D. Zamir
Literature Cited
- Balint-Kurti P. J., Wisser R., Zwonitzer J. C., 2008a. Use of an advanced intercross line population for precise mapping of quantitative trait loci for gray leaf spot resistance in maize. Crop Sci. 48: 1696–1704. 10.2135/cropsci2007.12.0679 [DOI] [Google Scholar]
- Balint-Kurti P. J., Yang J. Y., Van Esbroeck G., Jung J., Smith M. E., 2010. Use of a Maize Advanced Intercross Line for Mapping of QTL for Northern leaf blight resistance and multiple disease resistance. Crop Sci. 50: 458–466. 10.2135/cropsci2009.02.0066 [DOI] [Google Scholar]
- Balint-Kurti P. J., Zwonitzer J. C., Pe M. E., Pea G., Lee M., et al. , 2008b. Identification of quantitative trait loci for resistance to southern leaf blight and days to anthesis in two maize recombinant inbred line populations. Phytopathology 98: 315–320. 10.1094/PHYTO-98-3-0315 [DOI] [PubMed] [Google Scholar]
- Balint-Kurti P. J., Krakowsky M. D., Jines M. P., Robertson L. A., Molnar T. L., et al. , 2006. Identification of quantitative trait loci for resistance to southern leaf blight and days to anthesis in a maize recombinant inbred line population. Phytopathology 96: 1067–1071. 10.1094/PHYTO-96-1067 [DOI] [PubMed] [Google Scholar]
- Balint-Kurti P. J., Zwonitzer J. C., Wisser R. J., Carson M. L., Oropeza-Rosas M. A., et al. , 2007. Precise mapping of quantitative trait loci for resistance to southern leaf blight, caused by Cochliobolus heterostrophus race O, and flowering time using advanced intercross maize lines. Genetics 176: 645–657. 10.1534/genetics.106.067892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariana H. S., Miah H., Brown G. N., Willey N., Lehmensiek A., 2007. Molecular mapping of durable rust resistance in wheat and its implication in breeding. Wheat Production in Stressed Environments 12: 723–728. 10.1007/1-4020-5497-1_88 [DOI] [Google Scholar]
- Bates D., Machler M., Bolker B., Walker S., 2015. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 67: 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Bearden W. O., Sharma S., Teel J. E., 1982. Sample size effects on chi square and other statistics used in evaluating causal models. J. Mark. Res. 19: 425–430. 10.2307/3151716 [DOI] [Google Scholar]
- Beckman P. M., Payne G. A., 1982. External growth, penetration, and development of Cercospora zeae-maydis in corn leaves. Phytopathology 72: 810–815. 10.1094/Phyto-72-810 [DOI] [Google Scholar]
- Belcher A. R., Zwonitzer J. C., Cruz J. S., Krakowsky M. D., Chung C. L., et al. , 2012. Analysis of quantitative disease resistance to southern leaf blight and of multiple disease resistance in maize, using near-isogenic lines. Theor. Appl. Genet. 124: 433–445. 10.1007/s00122-011-1718-1 [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. [Google Scholar]
- Bent A. F., Mackey D., 2007. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436. 10.1146/annurev.phyto.45.062806.094427 [DOI] [PubMed] [Google Scholar]
- Bentolila S., Guitton C., Bouvet N., Sailland A., Nykaza S., et al. , 1991. Identification of an RFLP marker tightly linked to the Ht1 Gene in maize. Theor. Appl. Genet. 82: 393–398. 10.1007/BF00588588 [DOI] [PubMed] [Google Scholar]
- Benson J. M., Poland J. A., Benson B. M., Stromberg E. L., Nelson R. J., 2015. Resistance to gray leaf spot of maize: genetic architecture and mechanisms elucidated through nested association mapping and near-isogenic line analysis. PLoS Genet. 11: e1005045 10.1371/journal.pgen.1005045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y., Yang Q., Balint-Kurti P. J., Wisser R. J., Holland J. B., 2014. Limits on the reproducibility of marker associations with southern leaf blight resistance in the maize nested association mapping population. BMC Genomics 15: 1068 10.1186/1471-2164-15-1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes K. J., Pataky J. K., 1989. Relationships between yield of three maize hybrids and severity of southern leaf blight caused by race O of Bipolaris maydis. Plant Dis. 73: 834–840. 10.1094/PD-73-0834 [DOI] [Google Scholar]
- Carson M. L., 1998. Aggressiveness and perennation of isolates of Cochliobolus heterostrophus from North Carolina. Plant Dis. 82: 1043–1047. 10.1094/PDIS.1998.82.9.1043 [DOI] [PubMed] [Google Scholar]
- Carson M. L., Stuber C. W., Senior M. L., 2004. Identification and mapping of quantitative trait loci conditioning resistance to southern leaf blight of maize caused by Cochliobolus heterostrophus race O. Phytopathology 94: 862–867. 10.1094/PHYTO.2004.94.8.862 [DOI] [PubMed] [Google Scholar]
- Chung C.-L., Jamann T., Longfellow J., Nelson R., 2010. Characterization and fine-mapping of a resistance locus for northern leaf blight in maize bin 8.06. Theor. Appl. Genet. 121: 205–227. 10.1007/s00122-010-1303-z [DOI] [PubMed] [Google Scholar]
- Chung C. L., Poland J., Kump K., Benson J., Longfellow J., et al. , 2011. Targeted discovery of quantitative trait loci for resistance to northern leaf blight and other diseases of maize. Theor. Appl. Genet. 123: 307–326. 10.1007/s00122-011-1585-9 [DOI] [PubMed] [Google Scholar]
- Cooper J. S., Balint-Kurti P. J., Jamann T. M., 2018. Identification of Quantitative Trait Loci for Goss’s Wilt of Maize. Crop Sci. 58: 1192–1200. 10.2135/cropsci2017.10.0618 [DOI] [Google Scholar]
- Davis G. L., McMullen M. D., Baysdorfer C., Musket T., Grant D., et al. , 1999. A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics 152: 1137–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derera J., Tongoona P., Pixley K. V., Vivek B., Laing M. D., et al. , 2008. Gene action controlling gray leaf spot resistance in southern African maize germplasm. Crop Sci. 48: 93–98. 10.2135/cropsci2007.04.0185 [DOI] [Google Scholar]
- Dudley J. W., Maroof M. A. S., Rufener G. K., 1991. Molecular markers and grouping of parents in maize breeding programs. Crop Sci. 31: 718–723. 10.2135/cropsci1991.0011183X003100030036x [DOI] [Google Scholar]
- Dunkle L. D., Levy M., 2000. Genetic relatedness of African and United States populations of Cercospora zeae-maydis. Phytopathology 90: 486–490. 10.1094/PHYTO.2000.90.5.486 [DOI] [PubMed] [Google Scholar]
- Fisher D. E., Hooker A. L., Lim S. M., Smith D. R., 1976. Leaf infection and yield loss caused by four Helminthosporium leaf diseases of corn. Phytopathology 66: 942–944. 10.1094/Phyto-66-942 [DOI] [Google Scholar]
- Flint-Garcia S. A., Thuillet A. C., Yu J. M., Pressoir G., Romero S. M., et al. , 2005. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 44: 1054–1064. 10.1111/j.1365-313X.2005.02591.x [DOI] [PubMed] [Google Scholar]
- Freppon J. T., Pratt R. C., Lipps P. E., 1996. Chlorotic lesion response of maize to Cercospora zeae-maydis and its effect on gray leaf spot disease. Phytopathology 86: 733–738. 10.1094/Phyto-86-733 [DOI] [Google Scholar]
- Fu Y., Wen T. J., Ronin Y. I., Chen H. D., Guo L., et al. , 2006. Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics 174: 1671–1683. 10.1534/genetics.106.060376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. G., Lipps P. E., Pratt R. C., 2006. Heritability and components of resistance to Cercospora zeae-maydis derived from maize inbred VO613Y. Phytopathology 96: 593–598. 10.1094/PHYTO-96-0593 [DOI] [PubMed] [Google Scholar]
- Gregory L. V., Ayers J. E., Nelson R. R., 1979. The influence of cultivar and location on yield loss in corn (Zea mays) due to southern corn leaf blight Helminthosporium maydis. Plant Dis. Rep. 63: 891–895. [Google Scholar]
- Holland J. B., Nyquist W. E., Cervantes-Martinez C. T., 2003. Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 22: 9–112. [Google Scholar]
- Holley R. N., Goodman M. M., 1989. New sources of resistance to southern corn leaf-blight from tropical hybrid maize derivatives. Plant Dis. 73: 562–564. 10.1094/PD-73-0562 [DOI] [Google Scholar]
- Hooker A.L. 1963. Inheritance of Chlorotic-Lesion Resistance to Helminthosporium Turcicum in Seedling Corn. Phytopathology 53:660-&.
- Hurni S., Scheuermann D., Krattinger S. G., Kessel B., Wicker T., et al. , 2015. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. USA 112: 8780–8785. 10.1073/pnas.1502522112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamann T. M., Luo X., Morales L., Kolkman J., Chung C.-L., et al. , 2016. A remorin gene is implicated in quantitative disease resistance in maize. Theor. Appl. Genet. 129: 591–602. 10.1007/s00122-015-2650-6 [DOI] [PubMed] [Google Scholar]
- Jansky S. H., Rouse D. I., 2003. Multiple disease resistance in interspecific hybrids of potato. Plant Dis. 87: 266–272. 10.1094/PDIS.2003.87.3.266 [DOI] [PubMed] [Google Scholar]
- Jennings P. R., Ullstrup A. J., 1957. A histological study of 3 Helminthosporium leaf blights of corn. Phytopathology 47: 707–714. [Google Scholar]
- Jones E., Chu W. C., Ayele M., Ho J., Bruggeman E., et al. , 2009. Development of single nucleotide polymorphism (SNP) markers for use in commercial maize (Zea mays L.) germplasm. Mol. Breed. 24: 165–176. 10.1007/s11032-009-9281-z [DOI] [Google Scholar]
- Juliatti F. C., Pedrosa M. G., Silva H. D., da Silva J. V. C., 2009. Genetic mapping for resistance to gray leaf spot in maize. Euphytica 169: 227–238. 10.1007/s10681-009-9943-2 [DOI] [Google Scholar]
- Kump K. L., Bradbury P. J., Wisser R. J., Buckler E. S., Belcher A. R., et al. , 2011. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 43: 163–168. 10.1038/ng.747 [DOI] [PubMed] [Google Scholar]
- Lennon J. R., 2014. Making the most of Corn: The identification and characterization of exotic genes and germplasm for maize improvement, North Carolina State University, Raleigh. [Google Scholar]
- Liu K. J., Goodman M., Muse S., Smith J. S., Buckler E., et al. , 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotterhos K. E., Card D. C., Schaal S. M., Wang L., Collins C., Verity B., 2017. Composite measures of selection can improve the signal-to-noise ratio in genome scans. 8: 717–727. 10.1111/2041-210X.12774 [DOI] [Google Scholar]
- Lyimo H. J. F., Pratt R. C., Mnyuku R. S. O. W., 2011. Heritability and gene effect estimates for components of partial resistance to grey leaf spot of maize by generation mean analysis. Plant Breed. 130: 633–639. 10.1111/j.1439-0523.2011.01890.x [DOI] [Google Scholar]
- Meisel B., Korsman J., Kloppers F. J., Berger D. K., 2009. Cercospora zeina is the causal agent of grey leaf spot disease of maize in southern Africa. Eur. J. Plant Pathol. 124: 577–583. 10.1007/s10658-009-9443-1 [DOI] [Google Scholar]
- Meng L., Li H., Zhang L., Wang J., 2015. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3: 269–283. 10.1016/j.cj.2015.01.001 [DOI] [Google Scholar]
- Mitchell-Olds T., James R. V., Palmer M. J., Williams P. H., 1995. Genetics of Brassica rapa (syn. campestris). 2. Multiple disease resistance to three fungal pathogens: Peronospora parasitica, Albugo candida and Leptosphaeria maculans. Heredity 75: 362–369. 10.1038/hdy.1995.147 [DOI] [PubMed] [Google Scholar]
- Negeri A. T., Coles N. D., Holland J. B., Balint-Kurti P. J., 2011. Mapping QTL controlling southern leaf blight resistance by joint analysis of three related recombinant inbred line populations. Crop Sci. 51: 1571–1579. 10.2135/cropsci2010.12.0672 [DOI] [Google Scholar]
- Nelson P. T., Krakowsky M. D., Coles N. D., Holland J. B., Bubeck D. M., et al. , 2016. Genetic characterization of the north carolina state university maize lines. Crop Sci. 56: 259–275. 10.2135/cropsci2015.09.0532 [DOI] [Google Scholar]
- Niks R. E., Qi X., Marcel C., 2015. Quantitative resistance to biotrophic filamentous plant pathogens: Concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol. 53: 445–470. 10.1146/annurev-phyto-080614-115928 [DOI] [PubMed] [Google Scholar]
- Nyholt D. R., 2000. All LODs Are Not Created Equal. Am. J. Hum. Genet. 67: 282–288. 10.1086/303029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orto W. A., 1902. The wilt disease of the cowpea and its control US Dep. Agric. Bur. Plant Industries Bull 17: 9–20. [Google Scholar]
- Poland J. A., Bradbury P. J., Buckler E. S., Nelson R. J., 2011. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. USA 108: 6893–6898. 10.1073/pnas.1010894108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J. A., Balint-Kurti P. J., Wisser R. J., Pratt R. C., Nelson R. J., 2009. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14: 21–29. 10.1016/j.tplants.2008.10.006 [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2011 R: A Language and Environment for Statistical Computing. Vienna, Austria : the R Foundation for Statistical Computing. ISBN: 3-900051-07-0. Available online at http://www.R-project.org/
- R core development team , D.C.T. 2008. R: A language and environment for statistical computing Vienna, Austria : the R Foundation for Statistical Computing. ISBN: 3-900051-07-0. Available online at http://www.R-project.org/
- Raymundo A. D., Hooker A. L., 1981. Measuring the relationship between northern corn leaf blight and yield losses. Plant Dis. 65: 325–327. 10.1094/PD-65-325 [DOI] [Google Scholar]
- Risk J. M., Selter L. L., Chauhan H., Krattinger S. G., Kumlehn J., et al. , 2013. The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol. J. 11: 847–854. 10.1111/pbi.12077 [DOI] [PubMed] [Google Scholar]
- Rosewarne G. M., Singh R. P., Huerta-Espino J., Rebetzke G. J., 2008. Quantitative trait loci for slow-rusting resistance in wheat to leaf rust and stripe rust identified with multi-environment analysis. Theor. Appl. Genet. 116: 1027–1034. 10.1007/s00122-008-0736-0 [DOI] [PubMed] [Google Scholar]
- Ross H., 1986. Potato breeding. Problems and perspectives. Adv. Plant Breed 13: 5–68. [Google Scholar]
- Rousseeuw, P. J., and B. C. van Zomeren, 1990 Unmasking multivariate outliers and leverage points. J. Am. Stat. Assoc. 85: 633–639. 10.2307/2289995 [DOI] [Google Scholar]
- Rousseeuw, P.J., 1985 Multivariate estimation with high breakdown point. In W. Grossmann, G. Pflug, I. Vincze, W. Wertz (eds.), Mathematical Statistics and Applications, pp. 283–297. Reidel Publishing, Dordrecht. [Google Scholar]
- Schnurbusch T., Paillard S., Schori A., Messmer M., Schachermayr G., et al. , 2004. Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor. Appl. Genet. 108: 477–484. 10.1007/s00122-003-1444-4 [DOI] [PubMed] [Google Scholar]
- Simcox K. D., Bennetzen J. L., 1993. The use of molecular markers to study Setosphaeria turcica resistance in maize. Phytopathology 83: 1326–1330. 10.1094/Phyto-83-1326 [DOI] [Google Scholar]
- Simko I., Piepho H.-P., 2012. The area under the disease progress stairs: Calculation, advantage, and application. Phytopathology 102: 381–389. 10.1094/PHYTO-07-11-0216 [DOI] [PubMed] [Google Scholar]
- Spielmeyer W., Mago R., Wellings C., Ayliffe M., 2013. Lr67 and Lr34 rust resistance genes have much in common - they confer broad spectrum resistance to multiple pathogens in wheat. BMC Plant Biol. 13: 96 10.1186/1471-2229-13-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S. D., Grandillo S., Fulton T. M., Zamir D., Eshed Y., et al. , 1996. Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 92: 213–224. 10.1007/BF00223378 [DOI] [PubMed] [Google Scholar]
- Tefferi A., Hulluka M., Welz H. G., 1996. Assessment of damage and grain yield loss in maize caused by northern leaf blight in western Ethiopia. J. Plant Dis. Prot. 103: 353–363. [Google Scholar]
- Tehon L. R., Daniels E., 1925. A note on the brown leaf-spot of alfalfa. Phytopathology 15: 714–719. [Google Scholar]
- Todorov, V., and P. Filzmoser, 2009 An object-oriented framework for robust multivariate analysis. J. Stat. Softw. 32: 1–47. 10.18637/jss.v032.i03 [DOI] [Google Scholar]
- Ullstrup A. J., 1972. Impacts of southern corn leaf blight epidemics of 1970–1971. Annu. Rev. Phytopathol. 10: 37–50. 10.1146/annurev.py.10.090172.000345 [DOI] [Google Scholar]
- Van der Plank J. E., 1965. Plant Diseases: Epidemics and Control. Trails. Brit. mycol. Soc. 48: 157–160. [Google Scholar]
- Vos P., Simons G., Jesse T., Wijbrandi J., Heinen L., et al. , 1998. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 16: 1365–1369. 10.1038/4350 [DOI] [PubMed] [Google Scholar]
- Ward J. M. J., Laing M. D., Rijkenberg F. H. J., 1997. Frequency and timing of fungicide applications for the control of gray leaf spot in maize. Plant Dis. 81: 41–48. 10.1094/PDIS.1997.81.1.41 [DOI] [PubMed] [Google Scholar]
- Ward J. M. J., Stromberg E. L., Nowell D. C., Nutter F. W., 1999. Gray leaf spot - A disease of global importance in maize production. Plant Dis. 83: 884–895. 10.1094/PDIS.1999.83.10.884 [DOI] [PubMed] [Google Scholar]
- Wiesner-Hanks T., Nelson R., 2016. Multiple disease resistance in plants. Annu. Rev. Phytopathol. 54: 229–252. 10.1146/annurev-phyto-080615-100037 [DOI] [PubMed] [Google Scholar]
- White D. G., 1999. Compendium of corn diseases, Ed. 3rd APS Press, American Phytopathological Society, St. Paul, Minnesota. [Google Scholar]
- William H. M., Singh R. P., Huerta-Espino J., Rosewarne G., 2007. Characterization of genes for durable resistance to leaf rust and yellow rust in CIMMYT spring wheats. Wheat Production in Stressed Environments 12: 65–70. 10.1007/1-4020-5497-1_7 [DOI] [Google Scholar]
- Wisser R. J., Kolkman J. M., Patzoldt M. E., Holland J. B., Yu J. M., et al. , 2011. Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis and implicates a GST gene. Proc. Natl. Acad. Sci. USA 108: 7339–7344. 10.1073/pnas.1011739108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X. Y., Wang Q. H., Yang J. L., Jin D. M., Wang F., et al. , 2003. Fine mapping of the Ht2 (Helminthosporium turcicum resistance 2) gene in maize. Chin. Sci. Bull. 48: 165–169. 10.1360/03tb9034 [DOI] [Google Scholar]
- Zhang Y., Xu L., Fan X. M., Tan J., Chen W., et al. , 2012. QTL mapping of resistance to gray leaf spot in maize. Theor. Appl. Genet. 125: 1797–1808. 10.1007/s00122-012-1954-z [DOI] [PubMed] [Google Scholar]
- Zwonitzer J. C., Bubeck D. M., Bhattramakki D., Goodman M. M., Arellano C., et al. , 2009. Use of selection with recurrent backcrossing and QTL mapping to identify loci contributing to southern leaf blight resistance in a highly resistant maize line. Theor. Appl. Genet. 118: 911–925. 10.1007/s00122-008-0949-2 [DOI] [PubMed] [Google Scholar]
- Zwonitzer J. C., Coles N. D., Krakowsky M. D., Arellano C., Holland J. B., et al. , 2010. Mapping resistance quantitative trait loci for three foliar diseases in a maize recombinant inbred line population-evidence for multiple disease resistance? Phytopathology 100: 72–79. 10.1094/PHYTO-100-1-0072 [DOI] [PubMed] [Google Scholar]