Abstract
The Janus Kinase/Signal Transducer and Activator of Transcription (JAK/STAT) and epidermal growth factor receptor (EGFR) signaling pathways are conserved regulators of tissue patterning, morphogenesis, and other cell biological processes. During Drosophila oogenesis, these pathways determine the fates of epithelial follicle cells (FCs). JAK/STAT and EGFR together specify a population of cells called the posterior follicle cells (PFCs), which signal to the oocyte to establish the embryonic axes. In this study, whole genome expression analysis was performed to identify genes activated by JAK/STAT and/or EGFR. We observed that 317 genes were transcriptionally upregulated in egg chambers with ectopic JAK/STAT and EGFR activity in the FCs. The list was enriched for genes encoding extracellular matrix (ECM) components and ECM-associated proteins. We tested 69 candidates for a role in axis establishment using RNAi knockdown in the FCs. We report that the signaling protein Semaphorin 1b becomes enriched in the PFCs in response to JAK/STAT and EGFR. We also identified ADAM metallopeptidase with thrombospondin type 1 motif A (AdamTS-A) as a novel target of JAK/STAT in the FCs that regulates egg chamber shape. AdamTS-A mRNA becomes enriched at the anterior and posterior poles of the egg chamber at stages 6 to 7 and is regulated by JAK/STAT. Altering AdamTS-A expression in the poles or middle of the egg chamber produces rounder egg chambers. We propose that AdamTS-A regulates egg shape by remodeling the basement membrane.
Keywords: EGFR, JAK/STAT, oogenesis, AdamTS-A, Drosophila, Sema1b
An important biological question is how signaling pathways promote tissue patterning and morphogenetic change. Many pathways carry out these functions by transcriptionally activating or repressing target gene expression. In many developmental and disease contexts, specific signaling pathways have been implicated, but it is not yet understood which transcriptional targets they act on to affect these processes, or how these transcriptional changes are translated into changes in cell and tissue morphology. The goal of this study was to identify novel transcriptional targets of JAK/STAT and EGFR/Ras/MAPK signaling. These conserved signaling pathways regulate normal cell differentiation and proliferation, but can also promote oncogenic transformation, tumor development, and metastasis in disease contexts (Voldborg et al. 1997; Hou et al. 2002; Arbouzova and Zeidler 2006). Both pathways contribute to the development of certain cancers, such as primary intestinal T-cell lymphomas (Nicolae et al. 2016) and hepatocellular carcinomas (Calvisi et al. 2006) and it is thought that combined pathway inhibition may therefore be more effective than inhibiting either pathway on its own in some disease contexts (Winter et al. 2014). These pathways can also function synergistically to regulate differentiation and morphogenesis during normal cell differentiation and morphogenesis.
The Drosophila melanogaster egg chamber is a well-characterized system for studying how signaling pathways specify cell fates and effect morphogenetic change (Horne-Badovinac and Bilder 2005). Egg chambers undergo a highly stereotyped developmental progression that is divided into 14 stages (Spradling 1993). Egg chambers contain two main cell types: somatic epithelial cells, called follicle cells (FCs), which surround the egg chamber in a monolayer; and germline cells, which generate the future egg. Egg chambers originate from a structure called the germarium, which contains the germline stem cells and follicle stem cells. Germline stem cells divide asymmetrically to produce daughter cells called cystoblasts. These divide four times to give rise to a cluster of 16 cells, 15 of which will become nurse cells and one of which will differentiate into the oocyte. Once the 16-cell cluster is formed, follicle cells surround the cluster in a monolayered epithelium to generate the egg chamber (Spradling 1993). Egg chambers are connected to each other by special follicle cells called stalk cells as they grow. A string of egg chambers, surrounded by a muscle sheath, is collectively termed an ovariole and can be thought of as an assembly line that produces mature eggs.
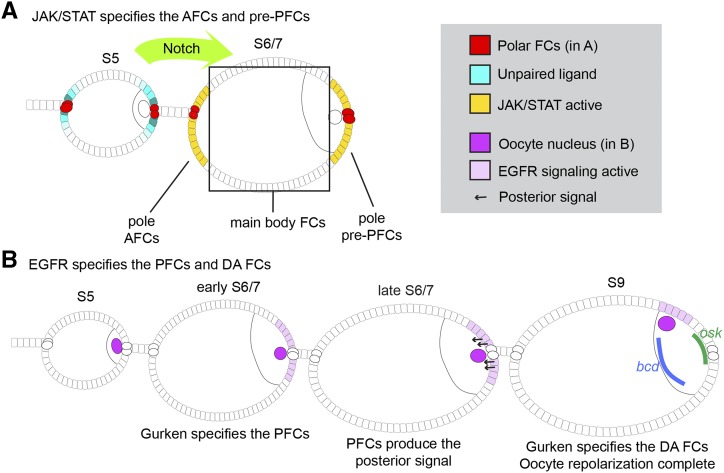
During FC development, signaling generates FC sub-populations with particular functions during oogenesis. If these cell types are not properly specified spatially and temporally, morphogenesis and/or patterning are disrupted in the egg chamber, and later, in the embryo (Berg 2005). The JAK/STAT and EGFR pathways play crucial roles in the patterning of the FCs. Special follicle cells called the polar cells, which can be thought of as signaling hubs and are located at the anterior and posterior ends of each egg chamber, secrete the JAK/STAT signaling ligand Unpaired (Upd; McGregor et al. 2002; Figure 1A). During stages 3 to 6, JAK/STAT signaling becomes activated in a gradient in the FCs at the egg chamber poles. In the anterior, JAK/STAT activity specifies the anterior follicle cell (AFC) fate. The future posterior follicle cells (PFCs) are exposed to Upd from the polar cells as well as Gurken, the active ligand for the EGFR, from the oocyte (Figure 1A,B). Together, JAK/STAT and EGFR activity produce the posterior follicle cell (PFC) fate (Xi et al. 2003). Previous work has demonstrated that by removing JAK/STAT or EGFR it is possible to disrupt PFC formation, and that ectopic activity of both pathways in the follicle cells can induce ectopic PFCs (Xi et al. 2003; Fregoso Lomas et al. 2016). In the Drosophila egg chamber, EGFR signaling activates the conserved MAPK/ERK signaling cascade (Schnorr and Berg 1996); consequently, disrupting the components of this cascade can also interfere with PFC specification.
Figure 1.
JAK/STAT and EGFR signaling pattern the follicle cells and establish the A/P axis of the developing Drosophila egg. A) The polar cells (red) secrete the JAK/STAT signaling ligand Unpaired (cyan). Unpaired activates JAK/STAT signaling activity in the follicle cell (FC) poles (yellow) by stage 5 (S5) to specify the AFCs and presumptive PFCs (pre-PFCs). Main body FCs are indicated. FCs become competent to respond to the JAK/STAT signal at stage 6, following Notch signaling (light green). B) Gurken/EGFR signaling (light purple) specifies the PFCs at stage 6/7 (S6/7). Once specified, the PFCs signal (arrows) to direct oocyte repolarization. During repolarization, the oocyte nucleus (magenta) migrates to the dorsoanterior (DA) corner, bicoid RNA (bcd, blue) accumulates at the anterior, and oskar RNA (osk, green) localizes to the posterior. At stage 9 (S9), Gurken/EGFR signaling specifies the DA follicle cells.
The PFCs play a key role in the establishment of the embryonic body axes. Prior to stage 6/7, the oocyte microtubules (MTs) are organized with the minus ends at the posterior cortex (Theurkauf et al. 1992). At stage 6/7, shortly after the PFCs are specified, they signal to the oocyte to trigger the reorganization of the oocyte cytoskeleton (González-Reyes et al. 1995; Roth et al. 1995; Figure 1B). This event is known as oocyte repolarization, and the signaling event that precedes it is referred to as posterior signaling. If posterior signaling occurs normally, the oocyte microtubules become polarized by stage 9 such that the minus ends are oriented toward the anterior of the egg chamber (Clark et al. 1994; Cha et al. 2002). During this process, microtubules push the oocyte nucleus anteriorly (Zhao et al. 2012). Oocyte repolarization has important implications for the development of the embryo because it directs the localization of the anterior determinant, bicoid (bcd), and the posterior determinant, oskar (osk) (González-Reyes et al. 1995; Roth et al. 1995; Figure 1B). These RNAs encode proteins that pattern the head and tail structures in the embryo, and accordingly, if they are not localized properly, the head or tail structures do not form correctly (Frohnhöfer and Nüsslein-Volhard 1986; Lehmann and Nüsslein-Volhard 1986; Berleth et al. 1988; Kim-Ha et al. 1991).
In addition to specifying posterior follicle cell fates, JAK/STAT and EGFR regulate additional cell and tissue morphologies in the egg chamber. EGFR signaling specifies the dorsoanterior (DA) follicle cells (Figure 1B), which migrate and change shape during late oogenesis to form the tubular dorsal appendages (Price et al. 1989; James et al. 2002; Osterfield et al. 2013). JAK/STAT signaling is required for the specification and proper morphology of the stalk cells, for specification and migration of the border cells, and was recently found to regulate the overall shape of the egg chamber (Silver and Montell 2001; McGregor et al. 2002; Baksa et al. 2002; Crest et al. 2017).
To understand how the JAK/STAT and EGFR pathways pattern the egg chamber and in particular, how they promote posterior signaling, we used gene expression profiling to identify genes transcribed downstream of these pathways. We used genetic tools to ectopically express the JAK/STAT signaling ligand Upd (UAS-upd; Tulina and Matunis 2001) and a constitutively active (CA) version of EGFR (UAS-λtop; Queenan et al. 1997) in the FCs to generate ectopic PFCs. Using microarrays, we identified 317 genes whose expression was enriched under these conditions. Candidate genes that encode cell-surface expressed or secreted (CSS proteins), transcription factors, signaling proteins, ECM proteins and some proteins of unknown function were selected for further investigation. RNAi knockdown was used to test 69 of these candidates for a role in oocyte repolarization. By this approach, we identified a new PFC-enriched protein, Sema1b, which requires both JAK/STAT and EGFR activity for its accumulation in the PFCs. The microarray dataset was also enriched for extracellular matrix (ECM) proteins; therefore, we used RNAi knockdown to study the consequences of disrupting several ECM proteins in the FCs. By this approach, we discovered a novel role for the metalloprotease AdamTS-A during oogenesis. Our results show that AdamTS-A is expressed in the AFCs and PFCs downstream of JAK/STAT signaling and is required for egg chamber elongation in a manner that appears to require its protease activity. To our knowledge, our work is the first to describe a metalloprotease that regulates Drosophila egg chamber shape. We propose that AdamTS-A helps to remodel the basement membrane at the egg chamber poles to promote elongation.
Materials and Methods
Fly strains and genetics
The following stocks were obtained from the Bloomington Drosophila Stock Center (BDSC): tub:Gal80TS (BL-7019), sema-1aP1 (BL-11097), and AdamTS-AMI14156 (BL-61726). AdamTS-A RNAi lines (VDRC-33347 and VDRC-110157) were ordered from the Vienna Drosophila Stock Center (VDRC). Sema1b-VenusCPTI003971 (DGGR-115463) and Sema1b-GAL4NP1166 (DGGR-103911) are from the Kyoto Stock Center. UAS-upd was a gift from Doug Harrison. UAS-λtop4.4 is described in Queenan et al. 1997. The hop RNAi line used in this study (BL-32966; Figures 3F, 4C, 6D and 6J-L) is described in Recasens-Alvarez et al. (2017). The UAS-upd, UAS-λtop recombinant chromosome was generated by meiotic recombination. mirr-GAL4; tub:GAL80TS and fru-GAL4; tub:GAL80TS (Crest et al. 2017), as well as tj-GAL4 were gifts from David Bilder. GR1-GAL4 is described in Gupta and Schüpbach, 2003. UASp-eGFP-48C was a gift from Elizabeth Gavis. AdamTS-AKO was a gift from Afshan Ismat (Ismat et al. 2013). AdamTS-Arnwy1 was a gift from James Skeath (Skeath et al. 2017). staufen-GFP (Martin et al. 2003) was a gift from Daniel St Johnston; the original insertion was mobilized to generate an insertion on the X-chromosome using Δ2-3 transposase. Oregon R (OreR) flies were used as wild type (WT). RNAi lines used in the screen were from the BDSC or VDRC and their stock numbers are provided in Supplemental Material, File S5.
Figure 3.
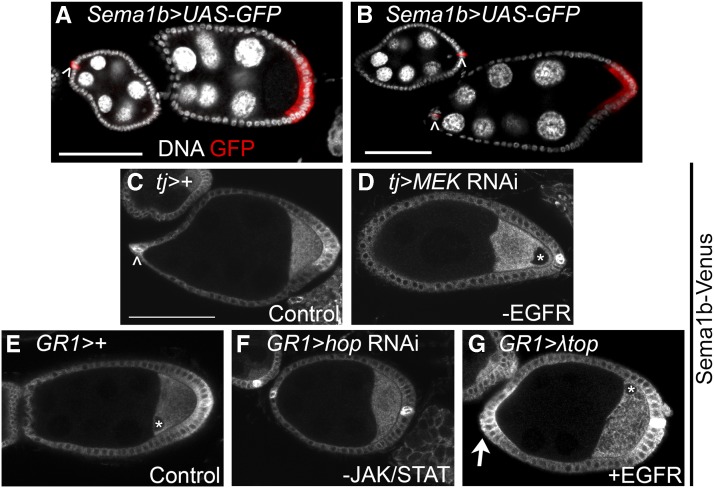
Sema1b RNA and protein are enriched in the PFCs and polar FCs. Sema1b protein accumulates in FCs with both JAK/STAT and EGFR signaling. A,B) To visualize Sema1b expression during oogenesis, Sema1b-GAL4 was used to drive a UASp-eGFP reporter. Sema1b was observed in the polar follicle cells (arrowheads) and in the PFCs beginning at stage 7 (red). Polar cells outside of this z-plane also express the reporter. C,E) Sema1b-Venus protein trap was used as a reporter for Sema1b protein localization during oogenesis. Sema1b-Venus was detected in the polar follicle cells (arrowhead) and PFCs. Stage 8 is pictured in panels C-E. C) Control (GAL4 only) for comparison to D. D) When MEK RNAi knockdown was very effective in disrupting PFC differentiation, as evidenced by the loss of oocyte nuclear migration (*), Sema1b-Venus enrichment in the PFCs was also disrupted. E) Control (GAL4 only) for comparison to F and G. F) Disruption of JAK/STAT signaling using hop knockdown disrupts Sema1b-Venus accumulation in the PFCs. G) Ectopic expression of constitutively active EGFR (λtop) produces ectopic Sema1b-Venus accumulation in the AFCs (arrow) and occasionally causes a loss of Sema1b enrichment in the PFCs (data not shown). Asterisks (*) mark visible oocyte nuclei. Scale bars represent 50 μm. Table S1 includes quantification of Sema1b-Venus accumulation in the AFC and PFCs and oocyte nuclear migration defects to accompany panels C-G. An anti-GFP antibody was used to detect UAS-GFP in panels A&B and Sema1b-Venus in panels C-G.
Figure 4.
Sema1b knockdown disrupts oocyte repolarization; however, Sema1b mutants do not appear to have oocyte repolarization defects. A-A’’) Stau-GFP (green) and Vasa (red) both localize to the oocyte posterior in Control (GAL4 only) stage 10 egg chambers. F-actin is in white. B-B’’) Sema1b RNAi produces mislocalization of both Stau-GFP and Vasa to the center of the oocyte at stage 10. C&D) Egg chambers heterozygous or homozygous for Sema1bKO (a deletion allele) do not show oocyte repolarization defects by Vasa (red) immunostaining. F-actin is in white. E) Sema1aP1 follicle cell clones were generated in a Sema1bKO mutant background. All FCs in this egg chamber are doubly mutant (a total FC clone is pictured). Vasa localization in these egg chambers is normal. All scale bars represent 50 μm.
Figure 6.
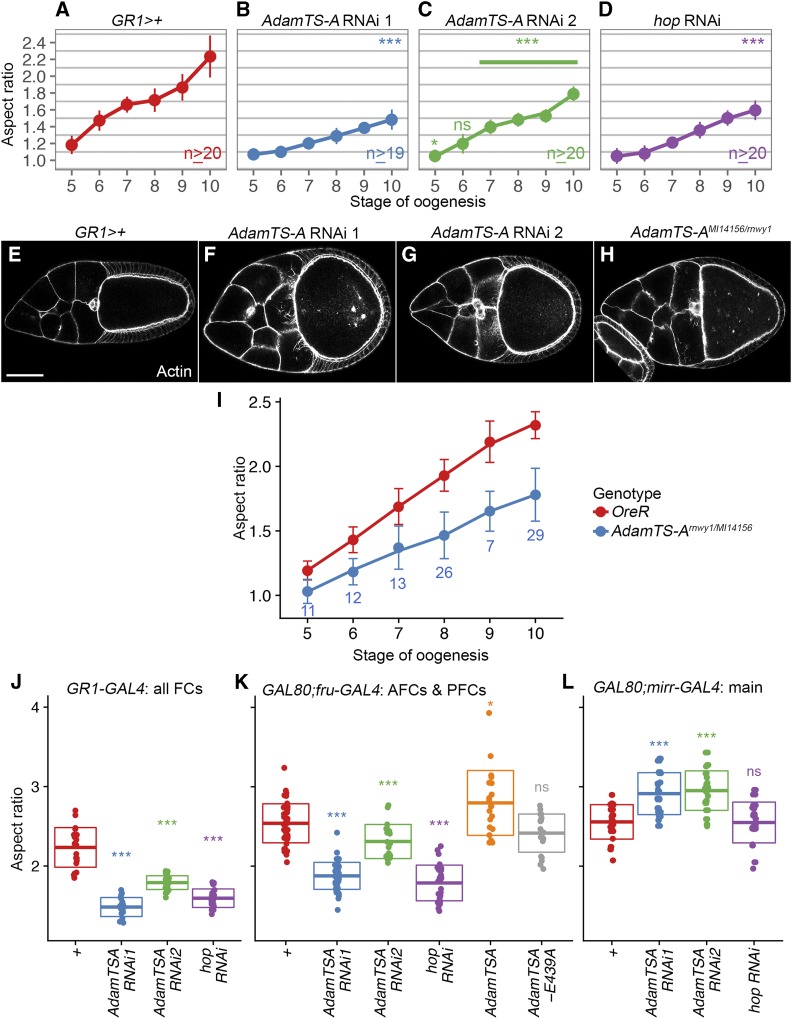
Disrupting AdamTS-A produces round egg chambers. A-D) AdamTS-A and hop RNAi knockdown delay egg chamber elongation beginning at stage five. n values reflect sample sizes for each data point. E-H) Disrupting AdamTS-A results in a pronounced round egg chamber phenotype at stage 10. Confocal micrographs of stage 10 egg chambers of following genotypes E) GR1>+ (GAL4 only control for comparison to F&G), F) AdamTS-A RNAi 1, G) AdamTS-A RNAi 2, and H) AdamTS-AMI14156/rnwy1. Scale bar represents 50 μm. I) AdamTS-AMI14156/rnwy1 egg chambers are significantly rounder than OreR controls beginning at stage 5. For all OreR data points, n ≥ 19. For AdamTS-AMI14156/rnwy1, n values are indicated in blue. J-L) Quantification of egg chamber aspect ratios at stage 10 demonstrates that J) RNAi knockdown of AdamTS-A or JAK/hop ubiquitously in the follicle cells using the GR1-GAL4 driver, or disrupting AdamTS-A or JAK/hop in the terminal follicle cells using (K) fru-GAL4 and GAL80 is sufficient to produce a significantly rounder egg at stage 10. Overexpression of AdamTS-A in the terminal follicle cells using a fru-GAL4 and GAL80 (J) or RNAi knockdown of AdamTS-A in the main body follicle cells using a mirr-GAL4 and GAL80 (L) cause hyperelongation. However, expression of a protease-dead form of AdamTS-A (AdamTS-AE439A) in the terminal follicle cells (K) does not produce hyperelongation. hop RNAi knockdown in the poles produces a significant elongation defect (K); however, knockdown in the main body follicle cells does not affect the aspect ratio. All error bars represent standard deviation of means. For all panels, * = P < 0.05; *** = P < 0.0005, ns = not significant.
Full experimental genotypes and incubation temperatures are reported in File S7.
Microarray preparation and data analysis
Ovaries from females fed yeast for two days were dissected in PBS and flash frozen in liquid nitrogen. Ovaries from WT (OreR) females were dissected in three batches, frozen and pooled to generate a reference pool of RNA. 5-10 ovary pairs from 5-8 day old GAL80TS; GR1-GAL4/UAS-upd, UAS-λtop and GAL80TS; GR1-GAL4/TM6 egg chambers were pooled for each preparation; this minimizes biological variation and ensured a sufficient quantity of RNA was collected for microarray analysis. RNA was extracted using TRIzol reagent (ThermoFisher), cleaned using an RNeasy kit (Qiagen), and DNase-treated with the Turbo DNA-free kit (ThermoFisher) according to the manufacturer’s instructions. RNA concentrations were then measured by a NanoDrop spectrophotometer. To confirm that RNA was free of degradation, RNA used for microarray analysis was analyzed by non-denaturing agarose gel electrophoresis.
Each labeling reaction was performed using 322 ng of RNA. Reference RNA samples were labeled with Cy3-CTP and all other samples were always labeled with Cy5-CTP. Labeling and hybridization were performed using a Quick Amp two-color labeling kit (Agilent), and a labeling protocol optimized by Maitreya Dunham and Bing He (He et al. 2012). Labeled cRNA was purified with a RNeasy column (Qiagen) and hybridized at 65° for 17 h, and then washed using Agilent’s proprietary buffers according to standard protocols. The microarray slides used were 4X44K Drosophila oligo arrays (print 021791), based on V2 of the Drosophila genome. Slides (6 arrays on 3 slides) were scanned immediately following washing to minimize sample degradation. Scans were made at a resolution of 5μ/pixel using an Agilent G2505C scanner and Agilent Feature Extraction version 11.0.11. Raw data were stored at Princeton’s PUMA database (http://puma.princeton.edu/). Bioconductor’s Limma package for R (www.bioconductor.org) was used to Loess normalize, background correct (by the normexp method), and quantile normalize, and finally, to identify differentially expressed probes. False discovery rate (FDR) estimates were calculated in Limma using the Benjamini-Hotchberg method (Benjamini and Hotchberg 1995).
To generate a list of all genes upregulated in GAL80TS; GR1-GAL4/UAS-upd, UAS-λtop ovaries as compared to control GAL80TS; GR1-GAL4/TM6 ovaries, expression thresholds and a p-value threshold were selected. Transcripts included in the final 317 gene dataset were upregulated at least twofold with a false discovery rate threshold of 0.05. If more than one probe was present per gene in the arrays, or if duplicate probes were present in the arrays, values corresponding to the most highly significant data were included in Tables 1-2. Raw and normalized microarray data have been deposited at GEO.
Table 1. Microarray Validation. PFC-expressed genes are enriched in the microarray dataset. Genes that are known transcriptional targets of the JAK/STAT or EGFR pathways are indicated. Fold enrichment and adjusted P-values for differential expression are also provided.
| Gene ID | Fold change | Adj. P- value | JAK/ STAT | EGFR | Reference(s) |
|---|---|---|---|---|---|
| aos | *1.54 | 2.60E-03 | x | Queenan et al. 1997, Zhao and Bownes 1999 | |
| bib | 2.98 | 7.07E-05 | Ruohola et al. 1991 | ||
| CG11275 | 17.2 | 5.03E-06 | Jambor et al. 2015 | ||
| dome | 2.37 | 8.44E-05 | x | Ghiglione et al. 2002, Xi et al. 2003 | |
| H15 | 7.45 | †8.57E-01 | x | x | Fregoso Lomas et al. 2013, Fregoso Lomas et al. 2016 |
| ImpL2 | 5.78 | 1.56E-05 | x | x | Jordan et al. 2005 |
| jim | 2.98 | 1.48E-03 | x | Doerflinger et al. 1999 | |
| kek1 | 2.01 | 1.38E-03 | x | Musacchio and Perrimon, 1996, Queenan et al. 1997 | |
| mid | 2.09 | 1.75E-03 | x | x | Fregoso Lomas et al. 2013, Fregoso Lomas et al. 2016 |
| pnt | 2.58 | 2.93E-04 | x | x | Morimoto et al. 1996, Xi et al. 2003, Flaherty et al. 2009 |
| Socs36E | 15.5 | 3.38E-05 | x | Rawlings et al. 2004 | |
| Stat92E | *1.99 | 1.34E-03 | x | Xi et al. 2003, Assa-Kunik et al. 2007 | |
| sty | 4.23 | 3.32E-05 | x | Reich et al. 1999 | |
| tsl | 3.86 | 3.91E-04 | x | Savant-Bhonsale and Montell 1993, Furriols et al. 2007 |
Below the twofold expression threshold but above the P-value threshold.
Below the P-value threshold but above the twofold expression threshold.
Table 2. Genes encoding ECM and ECM-associated proteins identified in the microarray dataset. Each protein’s role or function is briefly summarized, and fold changes and adjusted P-values for differential expression are provided.
| Gene | Fold change | Adj. P value | Role |
|---|---|---|---|
| upd | 63.1 | 3.99E-08 | Ligand |
| Mmp2 | 5.54 | 1.36E-04 | Protease |
| SPARC | 3.34 | 7.30E-05 | Collagen-binding |
| AdamTS-A | 2.84 | 8.82E-05 | Protease |
| Ppn | 2.76 | 8.82E-05 | Protease |
| scb | 2.49 | 1.65E-04 | Integrin |
| trol | 2.49 | 3.34E-05 | HSPG |
| m | 2.45 | 2.40E-04 | Secreted or ECM |
| LanB1 | 2.27 | 1.92E-03 | Laminin |
| Fas1 | 2.26 | 5.63E-03 | Adhesion |
| Mp | 2.23 | 6.55E-04 | Collagen XVIII |
| Col4a1 | 2.18 | 8.67E-03 | Collagen IV |
| LanB2 | 2.02 | 2.58E-03 | Laminin |
Immunofluorescence
Females were fed yeast for two days, dissected in room temperature PBS, fixed in 4% PFA for 20 min at room temperature, and washed in PBST (1X PBS with 0.1% Triton X-100; Sigma-Aldrich). Primary antibodies used were rabbit anti-GFP (AB3080P; 1:500; Millipore), guinea pig anti-Midline (1:200, a gift from Laura Nilson; Fregoso Lomas et al. 2013) and goat anti-Vasa (dN-13; 1:1000; Santa Cruz Biotechnology). Secondary antibodies used were Alexa Fluor 488, 555, 568 or 647-conjugated (1:400; Life Technologies). DNA was stained with Hoechst (10 μg/mL; Life Technologies). Alexa Fluor 546 Phalloidin (1:500; Life Technologies) or Alexa Fluor 647 Phalloidin (1:250; Life Technologies) were used to stain F-actin. For all experiments, egg chambers were mounted in Aqua-Poly/Mount (Polysciences). All micrographs in this study were taken using a Nikon A1 laser scanning confocal microscope. Images were acquired using a 40X, 1.30 NA Nikon Apo Plan Fluor or a 60X, 1.40 NA Nikon Plan objective. Images were prepared using Fiji and Adobe Photoshop CS6.
Fluorescent in situ hybridization (FISH)
Ovaries were prepared for FISH as described in Abbaszadeh and Gavis (2016) with two deviations: ovaries were incubated in MeOH at -20° for 15 min and mounted in Aqua-Poly/Mount. FISH probes conjugated to Quasar 570 were ordered from LGC Biosearch Technologies. 5 nmol of lyophilized probes were resuspended in 200 μL of TE Buffer (pH 8.0), and 1 μL of resuspended probe was used for each 100 μL hybridization reaction. A full list of probes used for FISH can be found in File S6.
RNAi knockdown
To test candidate genes for a role in oocyte repolarization, stau-GFP; tj-GAL4; UAS-Dcr2/TM6 virgins were crossed to UAS-RNAi males. Crosses were carried out at 25°, and once F1 females hatched, they were incubated at 29° with yeast for two days prior to dissection. Egg chambers were stained with Alexa Fluor 546 Phalloidin and Hoechst. The penetrance of Stau-GFP mislocalization was scored using an EVOS FL epifluorescence microscope (ThermoFisher) at stages 9 and 10 of oogenesis. Egg chambers that appeared to be dying or that could not be accurately staged were not scored. In control egg chambers, Stau-GFP localizes strongly to the posterior of the oocyte at stage 10 of oogenesis. If Stau accumulated at the center of the oocyte, appeared dispersed or in blobs, or only accumulated weakly at the posterior, we considered Stau “mislocalized.” If other oogenesis phenotypes were observed, these were recorded as well. RNAi lines were considered ‘hits’ if they produced ≥50% Stau-GFP mislocalization at stage 10.
A full list of RNAi knockdown reagents is included in File S5, which also reports the penetrance of Stau-GFP mislocalization for each line tested. When possible, we tested multiple RNAi lines for each candidate gene.
Generation of the Sema1bKO allele
We designed a scheme to generate a functional null Sema1b allele by replacing a large portion of the Sema1b coding region with a dsRed-containing cassette. ∼1 kb homology arms were designed to flank the gRNA cut site, and cloned into the pHD-dsRed-attP vector. 100 ng/μL of each gRNA plasmid and 500 ng/μL of the HDR plasmid were injected into w; FRT42D; nos-Cas9 embryos by Rainbow Transgenic Flies. Mutagenized flies were identified by expression of 3XP3::dsRed in the adult eye. Sequencing was performed to confirm integration of the dsRed-containing cassette at the intended genomic location with removal of the targeted region. We also confirmed by sequencing that the vector was not integrated at the Sema1b locus and that the flanking regions were not altered.
Generating Sema1aP1 mutant clones in a Sema1bKO mutant background
Negatively-marked mitotic clones were generated in the FCs by the Flp-FRT system (Golic and Lindquist 1989; Xu and Rubin 1993). The following stocks were crossed to produce clones in a Sema1bKO mutant background: w, hsFlp; Sema1aP1, FRT40A, Sema1bKO/CyO and ubiGFP, FRT40A, Sema1bKO.
Egg chamber aspect ratio calculation
Egg chambers stained with Hoechst and Alexa Fluor 546 Phalloidin were mounted onto glass slides with a coverslip and photographed using an EVOS FL epifluorescence microscope. ImageJ was used to measure egg chambers along the A/P and D/V axes. The aspect ratio is the ratio of the length along the A/P axis to the width, measured along the D/V axis. Egg chambers were only included in this analysis if there were no gaps in the follicular epithelium and if the egg chamber did not appear to be degenerating. Welch’s two-sample t-tests were used to compare the aspect ratios, lengths and widths of two groups.
Data availability
All data necessary to confirm the conclusions presented in this article are represented fully within it and the associated materials. Strains are available upon request. Raw and normalized microarray data were deposited at the Gene Expression Omnibus (GEO) under accession number GSE118881. File S1 contains a list of all microarray probes that pass the fold change and P-value thresholds. The genes in this list are sorted into categories and their function, if known, is summarized. Raw P-values and adjusted P-values are also reported in File S1. Files S2, S3 and S4 contain the result of GO statistical overrepresentation analysis of the microarray dataset using Panther, LAGO, and DAVID, respectively. File S5 contains a list of the genes selected for the RNAi screen as well as Stau-GFP scoring (at stage 10) for each RNAi line tested. File S6 lists the probe sequences used for AdamTS-A FISH. File S7 contains a full list of experimental genotypes and incubation temperatures. Table S1 contains quantification associated with Figure 3. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7209809.
Results
This study aimed to identify targets of the JAK/STAT and EGFR pathways during Drosophila oogenesis. The research that established the instructive role of JAK/STAT and EGFR in FC patterning showed that modulating these pathways can interconvert certain sub-populations of FCs. For instance, ectopically expressing EGFR in the anterior is sufficient to convert AFCs into PFC-like cells that morphologically resemble PFCs and express PFC-specific markers (Lee and Montell 1997; Keller Larkin et al. 1999; Xi et al. 2003). Similarly, if EGFR is lost from the PFCs, they assume an AFC fate and express AFC markers (Roth et al. 1995; González-Reyes et al. 1995, Xi et al. 2003). If Upd is ectopically expressed in the main body follicle cells, they assume an AFC fate (Xi et al. 2003).
However, few downstream effectors for these pathways have been identified. To identify genes transcriptionally activated by JAK/STAT and EGFR in the FCs, we used the GAL4-UAS system (Brand and Perrimon 1993) to ectopically express JAK/STAT and EGFR pathway components throughout the follicular epithelium. Using the GR1-GAL4 driver, we ubiquitously expressed the JAK/STAT ligand Unpaired (UAS-upd) in the FCs to activate JAK/STAT (Figure S1 shows the GR1-GAL4 driver’s expression pattern). To activate EGFR signaling in the FCs, we expressed a dominant active EGFR using UAS-λtop (Queenan et al. 1997). Previous work has shown that ectopic expression of these two signaling components is sufficient to convert AFCs and main body follicle cells into PFCs (Fregoso Lomas et al. 2016). However, to confirm that this was the case, we stained control and ectopic expression egg chambers using an antibody against Midline (Mid), which is expressed specifically in the PFCs (Fregoso Lomas et al. 2013; Figure 2A). Ectopic JAK/STAT and EGFR activity successfully converted all FCs into PFC-like cells with detectable Mid protein expression (Figure 2B). Microarrays were then used to identify transcripts expressed in response to ectopic EGFR and/or JAK/STAT signaling.
Figure 2.
Ectopic expression of JAK/STAT and EGFR in the follicular epithelium of the Drosophila egg chamber. A) Midline (Mid, white) protein is expressed specifically in the PFCs in a control (GAL4 and GAL80 only) stage 7 egg chamber. F-actin is shown in red. Scale bar is 10 μm. B) Ectopic expression of upd (JAK/STAT ligand) and λtop (constitutively active EGFR) results in Mid expression in all FCs. A&B both show stage 7/8 egg chambers. The egg chamber in B is not as elongated as the one depicted in A because ectopic expression of upd in the FCs disrupts egg chamber elongation (Alégot et al. 2018). C) Cellular component gene ontology (GO) terms enriched in egg chambers with ectopic EGFR and JAK/STAT activity. Analysis of the microarray dataset was performed with PANTHER (http://www.pantherdb.org/). Extracellular matrix (ECM) proteins (blue) are overrepresented.
Ectopic expression of EGFR and JAK/STAT signaling components identifies both known and novel transcriptional targets
We found 589 microarray probes representing 317 unique genes to be upregulated at least twofold in ectopic expression ovaries using an adjusted P-value threshold of 0.05 (File S1). Of the 14 genes that are known to date to be transcriptionally enriched in the PFCs, 12 are found in the 317 gene dataset with significant upregulation (Table 1). The other two genes, argos (aos) and Stat92E, were upregulated and passed the FDR threshold, but fell below the twofold expression threshold. We also identified a number of other transcripts that are known JAK/STAT and EGFR signaling targets in other contexts. Several transcripts identified by microarray analysis are known to be required in the PFCs for oocyte repolarization but are not known JAK/STAT or EGFR targets. These include alpha- and beta-spectrin (Lee et al. 1997; Wong et al. 2015) and mbs (Sun et al. 2011). The fact that the microarray dataset includes essentially all of the known PFC-expressed genes validates our microarray dataset and strongly suggests that it should include other, as yet unidentified PFC-expressed transcripts.
To test if the microarray “hit” list was enriched for transcripts encoding proteins involved in particular biological processes or localized to certain parts of the cell, we used the PANTHER online database (www.pantherdb.org/) for statistical overrepresentation tests (Mi et al. 2013). Notably, extracellular matrix (ECM) proteins are 10.31-fold statistically overrepresented in the microarray dataset (FDR = 3.41x10−3, Fisher’s exact test; Figure 2C). We also performed complementary gene ontology (GO) analysis using LAGO and DAVID, which also suggested an enrichment of ECM proteins. GO analysis data can be found in the supplement (Files S2-S4). ECM proteins present in the 317 transcript dataset include SPARC, Collagen IV (Col4a1), Terribly reduced optic lobes (Trol, the Drosophila homolog of Perlecan), the collagen Multiplexin (Mp); the laminins Laminin B1 (LanB1) and Laminin B2 (LanB2); Fasciclin 1 (Fas1) and the αPS3 integrin Scab (Scb), Kekkon-1 (Kek1), CG7702, the matrix-modifying protein Sulf1, and the proteases Papilin (Ppn), Matrix metalloprotease 2 (Mmp2), and AdamTS-A (Table 2).
We next tested 69 candidate genes from the microarray for a requirement in posterior signaling during oogenesis. RNAi knockdown was used to disrupt candidate gene expression in the FCs, and a staufen-GFP transgene (stau-GFP; Martin et al. 2003) was used to visualize oocyte polarity at stages 9-10 of oogenesis. We focused on genes encoding cell surface and secreted (CSS; Kurusu et al. 2008) proteins, ECM proteins, cytoskeleton interactors and transcription factors because these classes of proteins seemed the most likely to be involved in the differentiation of the PFCs or their function in signaling to repolarize the oocyte. We generated the stock stau-GFP; tj-GAL4; UAS-Dcr2 and crossed virgins to males carrying UAS-RNAi constructs or RNAi controls. tj-GAL4 and UAS-Dcr2 facilitate candidate gene knockdown in the FCs, and the stau-GFP transgene is a useful readout for oocyte repolarization (Martin et al. 2003; Dietzl et al. 2007). traffic jam-GAL4 (tj-GAL4) has been used successfully in previous RNAi screens in the FCs (Handler et al. 2013; Berns et al. 2014) and it produces expression in the FCs throughout their development (Olivieri et al. 2010). An RNAi line was considered a “hit” if it produced a Stau-GFP mislocalization phenotype of ≥50% penetrance at stage 10. 11 RNAi lines targeting nine different genes were counted as “hits” (Table 3). We tested ten RNAi lines targeting seven known posterior signaling genes as positive controls (File S5). Of these ten lines, three disrupted oogenesis before stage 10 (lines targeting ERK, EGFR and α-Spec), and two produced a Stau-GFP mislocalization phenotype (an α-Spec RNAi line and a MEK RNAi line; see Table 3). The remainder of the positive control lines tested fell below the 50% mislocalization cutoff (for stage 10) and were not scored as ‘hits’. As an additional positive control, we performed a cross with a dominant negative EGFR transgene (UAS-DN-DER; Buff et al. 1998). This positive control successfully resulted in Stau mislocalization. Based on the results of our control experiments, we expected false negatives in the RNAi knockdown screen and that some posterior signaling genes might cause a premature termination of normal oogenesis before stage 10.
Table 3. RNAi knockdown and accompanying control lines characterized as ‘hits.’ Lines where >50% of stage 10 egg chambers showed Stau-GFP mislocalization were considered ‘hits’.
| Gene | Stock | % Stau misloc S10 | Cellular function |
|---|---|---|---|
| Negative control lines | |||
| N/A | OreR | 3 | N/A |
| N/A | V60101 | 0 | KK library control |
| Positive control lines | |||
| EGFR | UAS-DN-DER | 100 | EGFR pathway |
| α-spec | BL42801 | 74 | Spectrin |
| MEK | BL32920 | 68 | EGFR pathway |
| RNAi screen ‘hits’ | |||
| CG6340 | V34160 | 100 | Unknown |
| Csk | V109813 | 98 | Kinase activity |
| Csk | V32877 | 96 | Kinase activity |
| CG13510 | V28434 | 95 | Unknown |
| CG8547 | V110523 | 92 | Unknown |
| ImpL3 | V110190 | 92 | Lactate dehydrogenase |
| Sema1b | V107233 | 88 | Signaling ligand |
| pico | V16371 | 86 | Actin regulator |
| Sema1b | BL28588 | 71 | Signaling ligand |
| CG13506 | V14127 | 70 | Unknown |
| Ppn | V16523 | 55 | ECM protease |
Only one gene encoding a CSS or signaling protein was identified as a ‘hit’ in the RNAi knockdown screen: the putative signaling ligand Semaphorin 1b (Sema1b; Table 3). Posterior signaling defects were also seen for several RNAi knockdown lines targeting genes of unknown function, including Ecdysone-induced protein L3 (ImpL3), which encodes the metabolic enzyme lactate dehydrogenase, pico, an actin-regulator, and Papilin (Ppn), a matrix metalloprotease. C-terminal Src kinase (Csk), encodes a tyrosine kinase that is best studied for its role in inhibiting the activity of the two Drosophila Src kinases, Src42A and Src64B (Pedraza et al. 2004). Csk has been shown to genetically interact with components of JAK/STAT, Hippo and EGFR/Ras signaling pathways in Drosophila (Stewart et al. 2003; Read et al. 2004; Hirabayashi et al. 2013; Kwon et al. 2015). We also fortuitously found that two RNAi lines targeting AdamTS-A resulted in the production of abnormally round egg chambers. AdamTS-A is a member of the AdamTS family of metalloproteases, and it has been reported to be expressed in a number of migratory tissues in the Drosophila embryo, including the caudal visceral mesoderm, trachea, and hemocytes. Proteolytic targets of AdamTS-A have not yet been definitively reported, but it is thought to modify ECM components in the larval CNS and to regulate cell-matrix adhesion in the developing salivary gland (Ismat et al. 2013; Skeath et al. 2017). We selected Sema1b and AdamTS-A for further study based on their RNAi knockdown phenotypes, expression patterns (see below), and putative functions.
sema1b mRNA and Sema1b protein are enriched in the polar cells and PFCs
Microarray analysis demonstrated a 3.6-fold increase in Sema1b transcript in ovaries with ectopic PFCs. To visualize the expression pattern of Sema1b mRNA in the ovary, we used the GAL4 driver Sema1bNP1166 (Figure S2) to drive the expression of a GFP reporter (UASp-EGFP). GFP expression was largely confined to the polar follicle cells and PFCs (Figure 3A,B). To test whether Sema1b protein localizes to the PFCs, as the previous result suggests, we used the protein trap line Sema1bCPTI003971, where Venus is spliced into the Sema1b protein between exons 2 and 3 (Figure S2). We observed enrichment of Venus in the polar FCs and PFCs at mid-oogenesis (Figure 3C). Together these data suggest that Sema1b is enriched in the PFCs and polar cells at both the mRNA and protein levels.
Sema1b protein enrichment in the PFCs requires JAK/STAT and EGFR signaling
Semaphorins are signaling proteins that can signal by physically interacting with other Semaphorins or with a closely-related family of signaling proteins called Plexins. Semaphorins and Plexins share an approximately 500 amino acid domain called a Sema domain, which they use to interact (Kolodkin et al. 1993). Semaphorins come in transmembrane, secreted, and membrane-linked (GPI-anchored) forms, and all Plexins are transmembrane proteins (Yu and Kolodkin 1999). There are five known Semaphorins and two Plexins encoded in the fly genome. To test whether Sema1b expression is regulated by JAK/STAT or EGFR signaling, we examined the levels of Sema1b-Venus in different knockdown conditions. MEK RNAi knockdown, which was used to reduce EGFR signaling, disrupted Sema1b-Venus enrichment in the PFCs. A strong disruption of EGFR signaling interferes with the migration of the oocyte nucleus to the dorsoanterior corner (Neuman-Silberberg and Schüpbach 1996; Figure 3D). The GAL4 system used to express the MEK RNAi construct produced a variable effect; some egg chambers displayed a strong phenotype and produced a failure of oocyte nuclear movement, and others did not. In egg chambers where the oocyte nucleus failed to move, Sema1b-Venus levels were substantially reduced in the PFCs (Figure 3D). RNAi knockdown of the kinase JAK/hopscotch (hop) also strongly disrupted Sema1b-Venus in the PFCs (compare Figure 3E&F). Finally, we observed that if EGFR was ectopically activated in the FCs by expressing UAS-λtop, Sema1b-Venus was observed in the AFCs, where endogenous JAK/STAT signaling is present, but not in the main body follicle cells (Figure 3G). Taken together, these data indicate that both JAK/STAT and EGFR signaling promote Sema1b expression in the PFCs. The penetrance of Sema1b localization and nuclear migration defects for each genetic condition in Figure 3C-G is reported in Table S1.
Sema1b RNAi knockdown produces oocyte repolarization defects (Figure 4A,B)
Using Stau-GFP as an oocyte polarity marker, we observed posterior signaling defects for two overlapping Sema1b RNAi knockdown lines (VDRC-107233/line 1 and BL-32877/line 2). Sema1b RNAi line 1 produced Stau-GFP mislocalization with 89% penetrance (n = 116) and line 2 produced Stau-GFP mislocalization with 71% penetrance (n = 24) at stage 10. Control egg chambers, where Stau-GFP; tj-GAL4; UAS-Dcr2 were crossed to WT resulted in 3% Stau-GFP mislocalization (n = 98), and the RNAi (KK) library control (VDRC-60101) produced 3% Stau-GFP mislocalization (n = 75). Since Sema1b RNAi line 1 produced a stronger phenotype, we further characterized the Sema1b RNAi knockdown phenotype using this line. To confirm the Stau-GFP mislocalization phenotype, we performed immunostaining for a second marker of oocyte polarity: the protein Vasa, which localizes to the oocyte posterior at stage 10 in response to posterior signaling (Figure 4A; Hay et al. 1988). Sema1b RNAi knockdown causes Vasa mislocalization (Figure 4B). As a final readout of oocyte polarity in Sema1b RNAi knockdown egg chambers, we used a kinesin-lacZ transgenic reporter. In control stage 9 egg chambers, MTs were correctly polarized and Kinesin-βgal always localized to the posterior of the egg chamber (100%, n = 19; Figure S3A). By contrast, when Sema1b was knocked down in the FCs, Kinesin-βgal was frequently mislocalized (78% mislocalization; n = 32; Figure S3B).
Egg chambers mutant for Sema1b do not display oocyte repolarization defects
To further test whether Sema1b is required for oocyte repolarization, we generated a Sema1b mutant allele using CRISPR and homology driven repair (HDR). Our approach replaced a large portion of the Sema1b coding region, including the conserved Semaphorin domain, with a dsRed cassette (Figure S2; Gratz et al. 2014). The allele was verified by sequencing. Because the signaling domain was removed in the mutant and a frameshift was generated that produced a premature stop codon, the allele is likely a functional, if not protein, null. Given the strong oocyte repolarization defects we observed by RNAi knockdown, we were surprised to find that the allele generated by CRISPR, Sema1bKO, was homozygous viable and females were fertile. Ovaries from homozygous mutant females appeared normal and Vasa protein localized properly to the posterior (compare Figure 4C&D). Two independently derived Sema1bKO lines were examined, and neither displayed a Vasa mislocalization phenotype. We also did not observe Vasa mislocalization defects in egg chambers with FC clones mutant for Sema-1bKO (data not shown). To determine if functional redundancy between the two known type I Semaphorins might account for the lack of defects observed in the Sema1bKO mutant allele, we generated mutant clones for a lethal Sema1a null allele (Sema1aP1) in a Sema1bKO homozygous mutant background. We again did not detect any posterior signaling defects; Vasa localized correctly when Sema1a and Sema1b were both disrupted in the follicle cells (Figure 4E). As discussed later, it is possible that other related proteins, such as other Sema-domain containing proteins, could compensate for the loss of Sema1b in the Sema1bKO mutant.
AdamTS-A is transcriptionally activated by JAK/STAT in the follicle cells
We fortuitously observed that RNAi knockdown of the metalloprotease AdamTS-A strongly disrupts egg chamber elongation, a process that is regulated by JAK/STAT signaling (Crest et al. 2017; Alégot et al. 2018). To determine where AdamTS-A is expressed, we used fluorescent in situ hybridization (FISH) to examine AdamTS-A expression (Figure 5A&D; see File S6 for FISH probes). AdamTS-A transcripts are present in FCs early in oogenesis in the germarium. Additionally, by stages 6/7, AdamTS-A transcript levels become more enriched in the terminal follicle cells (the AFCs and PFCs) than in the main body follicle cells.
Figure 5.
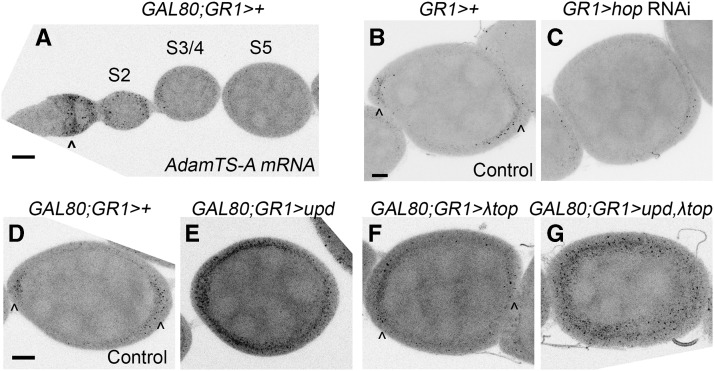
AdamTS-A expression is regulated by JAK/STAT signaling. A) Fluorescent in situ hybridization (FISH) of control (GAL4 and GAL80 only) egg chambers shows AdamTS-A expression in the germarium (arrowhead). B-G show stage 6/7 egg chambers. B) Control (GAL4 only) egg chamber for comparison with C, showing AdamTS-A expression in the anterior and posterior follicle cells (AFCs and PFCs) beginning at mid-oogenesis (arrowheads). AdamTS-A is present at reduced levels in the main body follicle cells. C) hop RNAi in the follicle cells causes a reduction in AdamTS-A expression. D) Control (GAL4 and GAL80 only) egg chamber for comparison with E-G. Arrowheads indicate AFC and PFC enrichment. E) Ectopic upd expression causes increased AdamTS-A expression in the FCs. Increased AdamTS-A expression in the main body follicle cells is particularly notable. F) Egg chambers expressing constitutively active EGFR (λtop) resemble controls. G) Egg chambers co-expressing upd and λtop show ectopic AdamTS-A expression. Panels A-G are maximum intensity projections generated from 21 z-slices at 1 μm intervals except for F, which is a projection of 20 z-slices. Scale bars represent 10 μm.
To test whether AdamTS-A expression is activated by JAK/STAT or EGFR, we ubiquitously activated both pathways in the FCs and then used FISH to detect any resulting changes in AdamTS-A expression. UAS-upd expression in the follicle cells caused strong ectopic AdamTS-A expression in the main-body follicle cells at stage 6/7 (Figure 5E). UAS-λtop expression does not appear to affect AdamTS-A in the main body follicle cells (Figure 5F). Coexpression of UAS-upd and UAS-λtop resembles the UAS-Upd-only condition (Figure 5G). If JAK/STAT signaling activates AdamTS-A expression in the poles of the egg chamber, then disrupting JAK/STAT should reduce AdamTS-A expression. Consistent with this prediction, hop RNAi knockdown causes a reduction in AdamTS-A expression in the egg chamber poles at stage 6/7 (compare Figure 5B&C). Together, these data suggest that JAK/STAT regulates AdamTS-A expression in the FCs.
AdamTS-A is required for egg chamber elongation
To quantify the elongation defect observed in AdamTS-A RNAi knockdown egg chambers, the aspect ratio (length:width ratio) was measured at stages 5 to 10 (Figure 6A-C). When AdamTS-A RNAi 1 was expressed under the control of the GR1-GAL4 driver (Gupta and Schüpbach 2003), which produces expression in the FCs starting at stage 4 of oogenesis (Figure S1), elongation defects were already evident at stage 5 of oogenesis (Figure 6B&F). AdamTS-A RNAi 2 produced a milder phenotype, but also significantly reduced egg chamber aspect ratios at most stages of oogenesis (Figure 6C&G). Disrupting JAK/STAT signaling is also sufficient to reduce egg chamber elongation beginning at stage 5 (hop RNAi; Figure 6D), which is consistent with previously published findings (Alégot et al. 2018).
To confirm the phenotypes observed by RNAi knockdown, we tested several different combinations of AdamTS-A alleles to find a transheterozygous mutant condition where adults survive and females produce viable ovaries. AdamTS-AMI14156 homozygotes survive, but females usually have no detectable ovaries or very small malformed ovaries with severe packaging defects; consequently, these flies never lay eggs. The AdamTS-Arnwy1 allele is reportedly viable and sub-fertile (Skeath et al. 2017), but we were not able to obtain homozygous adult flies, which suggests the allele may have developed a secondary lethal mutation. However, heteroallelic AdamTS-AMI14156/AdamTS-Arnwy1 females produce ovaries of a more normal size and morphology, and while they have reduced fertility, they occasionally lay eggs. Stage 10 AdamTS-AMI14156/AdamTS-Arnwy1 egg chambers, like AdamTS-A RNAi egg chambers, have an abnormal shape (Figure 6H). We quantified egg chamber elongation in AdamTS-AMI14156/AdamTS-Arnwy1 egg chambers and found that, consistent with our RNAi knockdown studies, heteroallelic egg chambers are also significantly rounder than controls at stages 5 to 10 (Figure 6I, P < 0.0005).
Since FISH showed that AdamTS-A is enriched at the egg chamber poles as compared to the middle, we wanted to test whether altering AdamTS-A expression in the poles – or in the main body follicle cells - changes egg chamber shape. fru-GAL4 drives expression specifically in the egg chamber terminal follicle cells (the poles; see Figure 1A; Barth et al. 2012; Borensztejn et al. 2013; Crest et al. 2017). mirr-GAL4 has a complementary expression pattern and is expressed in the main body follicle cells beginning at stage 7 (Isabella and Horne-Badovinac 2015; Crest et al. 2017). To validate these drivers, we used them to disrupt JAK/STAT signaling by expressing a hop/JAK RNAi line. Consistent with prior studies, disrupting JAK/STAT by knocking down hop/JAK ubiquitously (GR1-GAL4) or in the terminal follicle cells (fru-GAL4) produced rounder stage 10 egg chambers (Figure 6J&K), but knockdown in the main body follicle cells (mirr-GAL4), where JAK/STAT is not normally active, did not alter egg chamber shape (Figure 6L).
Interestingly, AdamTS-A RNAi knockdown in the terminal follicle cells produced hypoelongation (Figure 6K), but knockdown in the main body follicle cells resulted in hyperelongation (Figure 6L). We also found that ectopic expression of AdamTS-A (UAS-AdamTS-A; Ismat et al. 2013) in the poles produced a modest but significant hyperelongation (Figure 6K). Taken together, these results suggest that proper egg chamber shape requires differential AdamTS-A expression along the A/P axis, such that it is enriched in the poles and reduced in the middle. To test whether AdamTS-A’s protease activity is required for its function in egg chamber elongation, the fru-GAL4 driver was used to express the protease dead form of the protein in the poles (UAS-AdamTS-AE439A; Ismat et al. 2013). Unlike the wildtype form of the protein, the protease dead form did not produce hyperelongation (Figure 6K). This suggests that the protease activity of AdamTS-A promotes egg elongation.
Discussion
Signaling pathways play a critical role during animal development. They organize the body plan, and also sculpt the tissues and organs of the developing animal. Establishing the embryonic body axes is one of the first and most important events in an animal’s development. In Drosophila melanogaster this process begins during oogenesis. In developing egg chambers, special follicle cells called the PFCs signal to establish the A/P axis. These cells are specified by a combination of JAK/STAT and EGFR signaling, but it is unclear which targets of JAK/STAT and EGFR signaling mediate their differentiation or signaling behavior. The experiments described herein aimed to identify target genes activated by these two pathways during development, and to screen potential targets for a function in A/P axis establishment or in other aspects of oogenesis.
We used a gene expression approach to identify established and novel transcripts regulated by JAK/STAT and/or EGFR signaling in the follicle cells. Most known PFC-expressed transcripts were present in the microarray dataset, which demonstrates that our gene expression approach was able to successfully identify PFC-expressed RNAs. Surprisingly, transcripts encoding ECM proteins were overrepresented in our microarray screen dataset, which indicates that JAK/STAT and/or EGFR may regulate the ECM at a transcriptional level. We were also surprised to find genes (mbs, α-spec and β-spec) in the microarray dataset that encode proteins involved in posterior signaling but which are not known to be differentially expressed in the PFCs or regulated by the JAK/STAT or EGFR pathways. Using RNAi, we identified new functions and expression patterns for two genes during Drosophila oogenesis. Sema-1b is a novel PFC-expressed transcript, and AdamTS-A becomes enriched in the AFCs and PFCs and plays a role in egg chamber elongation.
It is important to consider the limitations of the approaches used in this study. From the microarray data we have collected, it is not possible to predict whether particular candidate genes are targets of JAK/STAT and/or EGFR signaling. A second important consideration is that the RNAi knockdown reagents used in our screen may not be ideal tools for disrupting candidate oocyte polarization genes in the FCs. It is well-established that many RNAi lines do not effectively reduce target gene expression, which can produce false negatives (Dietzl et al. 2007; Perkins et al. 2015). Consistent with this observation, several control RNAi lines directed against genes known to be required for posterior signaling (i.e., ERK, MEK, EGFR, and PP1β, see File S5) were not classified as ‘hits’ using the criteria we established. False positives are also a recurrent problem in RNAi screens. Long RNAi reagents can produce off-target gene knockdown in a sequence-dependent manner by cross-hybridizing to transcripts resembling the desired target. It has also been proposed that some RNAi reagents can produce off-target effects in a sequence-independent manner. It is therefore also important to consider that a few false-positives may be present among the RNAi screen ‘hits’.
Sema1b is expressed in the PFCs and is regulated by JAK/STAT and EGFR
Experiments using transcriptional and translational reporters both suggest that the signaling protein Sema1b is enriched in the PFCs. Using a translational reporter, we observed that disrupting JAK/STAT or EGFR signaling was sufficient to eliminate Sema1b enrichment in the PFCs. Additionally, when EGFR signaling was ectopically activated, Sema1b became strongly enriched in the anterior FCs (AFCs), where JAK/STAT is active, but not in the main body follicle cells, where JAK/STAT is inactive. This suggests that these pathways act together to promote Sema1b expression in the FCS. We did not test whether JAK/STAT and EGFR function to directly regulate Sema1b transcription. It is also not clear whether Sema1b has a functional role in the FCs.
Sema1b RNAi knockdown produced a strong oocyte polarization phenotype; however, results obtained with our Sema1bKO CRISPR-generated allele suggest that Sema1b is not essential for oocyte polarity. It is possible that the Sema1b RNAi knockdown phenotype is the result of off-target gene knockdown, however, there are no predicted off-target genes that would easily explain the Sema1b knockdown phenotype. The Sema1b RNAi reagents do have a 17 bp region of homology that matches Sema1a; however, disrupting both Sema1a and Sema1b using mutants did not produce a posterior signaling phenotype. There are a number of other Sema domain-containing proteins in the Drosophila genome, and it was not feasible to disrupt additional ones in this study. It is also possible that the RNAi pathway or certain RNAi reagents used in our study might function systemically to disrupt oocyte repolarization; however, we did not see a significant polarity problem in our controls or in most RNAi lines that we tested. It is possible that the genetic background used for RNAi sensitizes the oocyte for polarity defects, where knockdown of a gene that is peripherally involved in the process could generate a phenotype that it might not show in a different genetic background. These possibilities could be investigated by generating additional Sema1b RNAi knockdown reagents and by testing different genetic backgrounds in the presence of the Sema1bKO mutation.
JAK/STAT signaling activates AdamTS-A expression in follicle cells
We showed that AdamTS-A is enriched at the poles of the egg chamber at stage 6/7. Ectopic JAK/STAT pathway activity in the FCs substantially increases AdamTS-A expression in the main body follicle cells, where it is normally present at lower levels. Furthermore, disruption of JAK/STAT signaling using hop RNAi knockdown suggests that JAK/STAT signaling is required to establish the normal pattern of AdamTS-A expression during oogenesis.
Our analysis does not permit us to distinguish whether AdamTS-A is a direct or indirect target of JAK/STAT signaling in the FCs. Confirmed direct targets of JAK/STAT signaling are bound, often at multiple sites, by Stat92E, the sole Drosophila STAT protein (Yan et al. 1996). The AdamTS-A genomic locus contains a number of predicted Stat92E binding sites. Our analysis of a recent ChIP-Seq dataset (Kudron et al. 2018; epic.gs.washington.edu/modERN/) revealed that in the 0-12 h Drosophila embryo, eGFP-Stat92E binds at three sites in the first intron and promoter of AdamTS-A (eGFP-Stat92E ChIP peaks at the AdamTS-A locus are marked in Figure S6). It will also be interesting to investigate whether AdamTS-A is regulated by JAK/STAT signaling in the other tissues where it is known to be expressed, such as the caudal visceral mesoderm (CVM), salivary gland, hemocytes, and central nervous system glia (Ismat et al. 2013; Skeath et al. 2017).
AdamTS-A promotes egg chamber elongation
We found that AdamTS-A RNA is expressed strongly in the terminal follicle cells and more weakly in the main body follicle cells. Disrupting AdamTS-A throughout the follicular epithelium or in the terminal FCs should reduce the difference in the expression of AdamTS-A along the length of the egg chamber. In contrast, knockdown specifically in the main body FCS or ectopic AdamTS-A expression in the terminal FCs should enhance AdamTS-A’s enrichment in the egg chamber poles.
In our study, any perturbations that should enhance differential AdamTS-A expression along the A/P axis promote hyperelongation and those that reduce the difference produce hypoelongation, leading to a round egg chamber phenotype. By examining AdamTS-A RNAi knockdown and hypomorphic mutant egg chambers, we found that AdamTS-A is required during the early stages of egg chamber elongation, which is also when JAK/STAT signaling is required. This leads us to propose a model for AdamTS-A function in egg chamber elongation.
The basement membrane (BM), which forms at the basal side of the follicular epithelium, plays an important role in regulating signaling, growth and egg chamber shape. Egg chambers initially grow isotropically, or evenly, in all directions. However, they begin to elongate at stage 5. Live imaging studies revealed that egg chambers rotate perpendicular to the A/P axis throughout much of oogenesis with rotation occurring most rapidly during stages 5 to 9 of oogenesis (Haigo and Bilder 2011). This timing also corresponds to the most rapid phases of egg chamber elongation (Haigo and Bilder 2011; Cetera et al. 2014).
The observation that elongation and rotation coincide led to the proposal that egg chamber rotation promotes elongation (Haigo and Bilder 2011). Consistent with this hypothesis, all genetic perturbations reported to disrupt rotation or to cause off-axis rotation also affect elongation. Proteins involved in rotation include the atypical cadherin Fat2, the ECM receptor Lar, Laminins, Misshapen (Msn; a kinase that regulates integrins), and βPS-integrin/Myospheroid (Mys) (Lewellyn et al. 2013; Haigo and Bilder 2011; Viktorinová and Dahmann 2013; Barlan et al. 2017; Díaz de la Loza et al. 2017.) During rotation, the FCs deposit ColIagen IV (Viking; Vkg) to the BM in fibrils oriented perpendicular to the axis of elongation. Collagen is believed to function as a molecular corset during elongation (Haigo and Bilder 2011). If collagen is disrupted, for example by ColIV RNAi knockdown or collagenase treatment, egg chamber elongation is also disrupted. However, it is possible to disrupt elongation without disrupting rotation. For example, ectopic SPARC prevents collagen deposition and disrupts elongation but does not interfere with rotation (Isabella and Horne-Badovinac 2015).
Elongation requires several distinct processes that are temporally overlapping (reviewed in Gates 2012; Cetera and Horne Badovinac 2015). The published literature on AdamTS-A suggests a possible mechanism by which it could direct the growth of the egg chamber. In the central nervous system (CNS), AdamTS-A modifies the basement membrane to constrain and direct tissue growth. In this context. AdamTS-A and Perlecan (Pcan/Trol) act together to promote tissue softness, and act in opposition to Collagen IV and βPS-integrin (Mys), which promote tissue stiffness (Skeath et al. 2017). Recent work shows that the second phase of egg chamber elongation requires an asymmetric gradient of basement membrane stiffness, with stiffer matrix at the middle and softer matrix at the poles, and this stiffness matrix appears to require JAK/STAT pathway activity (Crest et al. 2017). It is therefore attractive to propose that AdamTS-A expression is induced by JAK/STAT signaling at the egg chamber poles, and functions to soften the basement membrane at the poles. All our manipulations that led to an increased difference of AdamTS-A at the poles vs. the middle of the egg chambers resulted in hyperelongation, whereas manipulations that decreased this difference resulted in hypoelongation. Varying AdamTS-A levels affects Collagen IV levels in the central nervous system, and Collagen IV has been proposed as a possible target for AdamTS-A’s protease activity in the CNS (Skeath et al. 2017). The results we obtained by modulating AdamTS-A expression in the FCs are consistent with it functioning to promote tissue softness, and ColIV is an attractive possible target for AdamTS-A in the egg chamber; however, further studies are warranted to test this hypothesis.
In summary, we used differential expression analysis to generate a list of 317 potential PFC-enriched genes. Genes in this dataset may also be transcriptionally activated by JAK/STAT and/or EGFR signaling. We have investigated some of these genes for a role in oocyte polarization, but others remain to be tested. Our work suggests that Sema1b is expressed in the PFCs, but it remains unclear whether Sema1b plays a functional role in oogenesis. Our work with AdamTS-A demonstrates that our screen also has the potential to uncover novel JAK/STAT and/or EGFR target genes with key functional roles in oogenesis.
Acknowledgments
The authors thank Julie Merkle and Olivier Devergne for reading and offering feedback on this manuscript and Bing He and Colin Watson for their assistance in designing the microarray experiments. We thank the genomics core facility at Princeton, especially John Matese, for their assistance with data management, storage and experimental design. We thank Mahrose Mehdi for assisting in the characterization of AdamTS-A alleles. We also thank the other members of the Schüpbach and Wieschaus labs for their helpful suggestions and Gary Laevsky for sharing his expertise in confocal microscopy. This work was supported by National Institute of General Medicine (grant R01GM077620 to T.S.) and by the Howard Hughes Medical Institute.
Author contributions: J.W. performed the experiments described in this study. J.W. and T.S. designed the experiments. J.W. analyzed the data, assembled figures, and drafted the manuscript with T.S. The authors declare no conflicts of interest in this study.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7209809.
Communicating editor: M. Wolfner
Literature Cited
- Abbaszadeh E. K., Gavis E. R., 2016. Fixed and live visualization of RNAs in Drosophila oocytes and embryo. Methods 98: 34–41. 10.1016/j.ymeth.2016.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alégot H., Pouchin P., Bardot O., Mirouse V., 2018. Jak-Stat pathway induces Drosophila follicle elongation by a gradient of apical contractility. eLife 7: 1–21. 10.7554/eLife.32943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assa-Kunik E., Torres I. L., Schejter E., St Johnston D., Shilo B.-Z., 2007. Drosophila follicle cells are patterned by multiple levels of Notch signaling and antagonism between the Notch and JAK/STAT pathways. Dev. 134: 1161–1169. 10.1242/dev.02800 [DOI] [PubMed] [Google Scholar]
- Arbouzova N. I., Zeidler M. P., 2006. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Dev. 133: 2605–2616. 10.1242/dev.02411 [DOI] [PubMed] [Google Scholar]
- Baksa K., Parke T., Dobens L. L., Dearolf C. R., 2002. The Drosophila STAT protein, Stat92E, Regulates Follicle Cell Differentiation during Oogenesis. Dev. Biol. 243: 166–175. 10.1006/dbio.2001.0539 [DOI] [PubMed] [Google Scholar]
- Barlan K., Cetera M., Horne-Badovinac S., 2017. Fat2 and Lar Define a Basally Localized Planar Signaling System Controlling Collective Cell Migration. Dev. Cell 40: 467–477. 10.1016/j.devcel.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth J., Hafen E., Köhler K., 2012. The lack of autophagy triggers precocious activation of Notch signaling during Drosophila oogenesis. BMC Dev. Biol. 12: 35 10.1186/1471-213X-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300. [Google Scholar]
- Berg C. A., 2005. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 21: 346–355. 10.1016/j.tig.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Reichstein S., et al. , 1988. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO 7: 1749–1756. 10.1002/j.1460-2075.1988.tb03004.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns N., Woichansky I., Friedrich S., Kraft N., Riechmann V., 2014. A genome-scale in vivo RNAi analysis of epithelial development in Drosophila identifies new proliferation domains outside of the stem cell niche. J. Cell Sci. 127: 2736–2748. 10.1242/jcs.144519 [DOI] [PubMed] [Google Scholar]
- Borensztejn A., Boissoneau E., Fernandez G., Agnès F., Pret A. M., 2013. JAK/STAT autocontrol of ligand-producing cell number through apoptosis. Dev. 140: 195–204. 10.1242/dev.079046 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Dev. 118: 401–415. http://dev.biologists.org/content/118/2/401 [DOI] [PubMed] [Google Scholar]
- Buff E., Carmena A., Gisselbrecht S., Jimenez F., Michelson A. M., 1998. Signalling by the Drosophila epidermal growth factor receptor is required for the specification and diversification of embryonic muscle progenitors. Dev. 125: 2075–2086. http://dev.biologists.org/content/125/11/2075 [DOI] [PubMed] [Google Scholar]
- Calvisi D. F., Ladu S., Gorden A., Farina M., Conner E. A., et al. , 2006. Ubiquitous Activation of Ras and Jak/Stat Pathways in Human HCC. Gastroenterology 130: 1117–1128. 10.1053/j.gastro.2006.01.006 [DOI] [PubMed] [Google Scholar]
- Cetera M., Horne-Badovinac S., 2015. Round and round gets you somewhere: collective cell migration and planar polarity in elongating Drosophila egg chambers. Curr. Opin. Genet. Dev. 32: 10–15. 10.1016/j.gde.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M., Ramirez-San Juan G. R., Oakes P. W., Lewellyn L., Fairchild M. J., et al. , 2014. Epithelial rotation promotes the global alignment of contractile actin bundles during Drosophila egg chamber elongation. Nat. Commun. 5: 5511 10.1038/ncomms6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha B.-J., Serbus L. R., Koppetsch B. S., Theurkauf W. E., 2002. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat. Cell Biol. 4: 592–598. 10.1038/ncb832 [DOI] [PubMed] [Google Scholar]
- Clark I., Giniger E., Ruohola-Baker H., Jan L. Y., Jan Y. N., 1994. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 4: 289–300. 10.1016/S0960-9822(00)00068-3 [DOI] [PubMed] [Google Scholar]
- Crest J., Diz-Muñoz A., Chen D.-Y., Fletcher D. A., Bilder D., 2017. Organ sculpting by patterned extracellular matrix stiffness. eLife 6: 1–16. 10.7554/eLife.24958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz de la Loza M. C., Díaz-Torres A., Zurita F., Rosalez-Nieves A. E., Moeendarbary E., et al. , 2017. Laminin Levels Regulate Tissue Migration and Anterior-Posterior Polarity during Egg Morphogenesis in Drosophila. Cell Reports 20: 211–223. 10.1016/j.celrep.2017.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Doerflinger H., Lepesant J.-A., Yanicostas C., 1999. Differential expression of the Drosophila zinc finger gene jim in the follicular epithelium. Mech. Dev. 86: 177–182. 10.1016/S0925-4773(99)00109-4 [DOI] [PubMed] [Google Scholar]
- Flaherty M. S., Zavadil J., Ekas L. A., Bach E. A., 2009. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev. Dyn. 238: 2235–2253. 10.1002/dvdy.21989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregoso Lomas M., Hails F., Boisclair Lachance J.-F., Nilson L. A., 2013. Response to the Dorsal Anterior Gradient of EGFR Signaling in Drosophila Oogenesis Is Prepatterned by Earlier Posterior EGFR Activation. Cell Reports 4: 791–802. 10.1016/j.celrep.2013.07.038 [DOI] [PubMed] [Google Scholar]
- Fregoso Lomas M., De Vito S., Boisclair Lachance J.-F., Houde J., Nilson L. A., 2016. Determination of EGFR Signaling Output by Opposing Gradients of BMP and JAK/STAT Activity. Curr. Biol. 26: 2572–2582. 10.1016/j.cub.2016.07.073 [DOI] [PubMed] [Google Scholar]
- Frohnhöfer H. G., Nüsslein-Volhard C., 1986. Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature 324: 120–125. 10.1038/324120a0 [DOI] [Google Scholar]
- Furriols M., Ventura G., Casanova J., 2007. Two distinct but convergent groups of cells trigger Torso receptor tyrosine kinase activation by independently expressing torso-like. Proc. Natl. Acad. Sci. USA 104: 11660–11665. 10.1073/pnas.0700991104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J., 2012. Drosophila egg chamber elongation: Insights into how tissues and organs are shaped. Fly (Austin) 6: 213–227. 10.4161/fly.21969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C., Bach E. A., Paraiso Y., Carraway K. L., III, Noselli S., et al. , 2002. Mechanism of activation of the Drosophila EGF Receptor by the TGFα ligand Gurken during oogenesis. Dev. 129: 175–186. http://dev.biologists.org/content/129/1/175 [DOI] [PubMed] [Google Scholar]
- Golic K., Lindquist S., 1989. The FLP Recombinase of Yeast Catalyzes Site-Specific Recombination in the Drosophila Genome. Cell 59: 499–509. 10.1016/0092-8674(89)90033-0 [DOI] [PubMed] [Google Scholar]
- González-Reyes A., Elliott H., St Johnston D., 1995. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature 375: 654–658. 10.1038/375654a0 [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubenstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly Specific and Efficient CRISPR/Cas9-Catalyzed Homology-Directed Repair in Drosophila. Genetics 196: 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta T., Schüpbach T., 2003. Cct1, a phosphatidylcholine biosynthesis enzyme, is required for Drosophila oogenesis and ovarian morphogenesis. Dev. 130: 6075–6087. 10.1242/dev.00817 [DOI] [PubMed] [Google Scholar]
- Haigo S., Bilder D., 2011. Global Tissue Revolutions in a Morphogenetic Movement Controlling Elongation. Science 331: 1071–1074. 10.1126/science.1199424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D., Meixner K., Pizka M., Lauss K. C., Schmied C., et al. , 2013. The Genetic Makeup of the Drosophila piRNA Pathway. Mol. Cell 50: 762–777. 10.1016/j.molcel.2013.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B., Ackerman L., Barbel S., Jan L. Y., Jan Y. N., 1988. Identification of a component of Drosophila polar granules. Dev. 103: 625–640. http://dev.biologists.org/content/103/4/625 [DOI] [PubMed] [Google Scholar]
- He B., Caudy A., Parsons L., Rosebrock A., Pane A., et al. , 2012. Mapping the pericentric heterochromatin by comparative genomic hybridization analysis and chromosome deletions in Drosophila melanogaster. Genet. Res. 22: 2507–2519. 10.1101/gr.137406.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S., Baranski T., Cagan R., 2013. Transformed Drosophila Cells Evade Diet-Mediated Insulin Resistance through Wingless Signaling. Cell 154: 664–675. 10.1016/j.cell.2013.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne-Badovinac S., Bilder D., 2005. Mass Transit: Epithelial Morphogenesis in the Drosophila egg chamber. Dev. Dyn. 232: 559–574. 10.1002/dvdy.20286 [DOI] [PubMed] [Google Scholar]
- Hou S. X., Zheng Z., Chen X., Perrimon N., 2002. The JAK/STAT Pathway in Model Organisms: Emerging Roles in Cell Movement. Dev. Cell 3: 765–778. 10.1016/S1534-5807(02)00376-3 [DOI] [PubMed] [Google Scholar]
- Isabella A. J., Horne-Badovinac S., 2015. Dynamic regulation of basement membrane protein levels promotes egg chamber elongation in Drosophila. Dev. Biol. 406: 212–221. 10.1016/j.ydbio.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismat A., Cheshire A. M., Andrew D. J., 2013. The secreted AdamTS-A metalloprotease is required for collective cell migration. Dev. 140: 1981–1993. 10.1242/dev.087908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H., Surendranath V., Kalinka A. T., Mejstrik P., Saalfeld S., et al. , 2015. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. eLife 4: 1–22. 10.7554/eLife.05003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K. E., Dorman J. B., Berg C. A., 2002. Mosaic analyses reveal the function of Drosophila Ras in embryonic dorsoventral patterning and dorsal follicle cell morphogenesis. Dev. 129: 2209–2222. http://dev.biologists.org/content/129/9/2209 [DOI] [PubMed] [Google Scholar]
- Jordan K. C., Hatfield S. D., Tworoger M., Ward E. J., Fischer K. A., et al. , 2005. Genome wide analysis of transcript levels after perturbation of the EGFR pathway in the Drosophila ovary. Dev. Dyn. 232: 709–724. 10.1002/dvdy.20318 [DOI] [PubMed] [Google Scholar]
- Keller Larkin M., Deng W.-M., Holder K., Tworoger M., Clegg N., et al. , 1999. Role of Notch pathway in terminal follicle cell differentiation during Drosophila oogenesis. Dev. Genes Evol. 209: 301–311. 10.1007/s004270050256 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Smith J. L., Macdonald P. M., 1991. oskar mRNA Is Localized to the Posterior Pole of the Drosophila Oocyte. Cell 66: 23–35. 10.1016/0092-8674(91)90136-M [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Matthes D. J., Goodman C. S., 1993. The semaphorin Genes Encode a Family of Transmembrane and Secreted Growth Cone Guidance Molecules. Cell 75: 1389–1399. 10.1016/0092-8674(93)90625-Z [DOI] [PubMed] [Google Scholar]
- Kudron M. M., Victorsen A., Gevirtzman L., Hillier L. W., Fisher W. W., et al. , 2018. The ModERN Resource: Genome-Wide Binding Profiles for Hundreds of Drosophila and Caenorhabditis elegans Transcription Factors. Genetics 208: 937–949. 10.1534/genetics.117.300657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu M., Cording A., Taniguchi M., Menon K., Suzuki E., et al. , 2008. A Screen of Cell-Surface Molecules Identifies Leucine-Rich Repeat Proteins as Key Mediators of Synaptic Target Selection. Neuron 59: 972–985. 10.1016/j.neuron.2008.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H. J., Waghmare I., Verghese S., Singh A., Singh A., et al. , 2015. Drosophila C-Terminal Src kinase regulates growth via the Hippo signaling pathway. Dev. Biol. 397: 67–76. 10.1016/j.ydbio.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Lee T., Montell D. J., 1997. Multiple Ras Signals Pattern the Drosophila Ovarian Follicle Cells. Dev. Biol. 185: 25–33. 10.1006/dbio.1997.8537 [DOI] [PubMed] [Google Scholar]
- Lee J. K., Brandin E., Branton D., Goldstein L. S. B., 1997. α-Spectrin is required for ovarian follicle monolayer integrity in Drosophila melanogaster. Dev. 124: 353–362. http://dev.biologists.org/content/124/2/353 [DOI] [PubMed] [Google Scholar]
- Lehmann R., Nüsslein-Volhard C., 1986. Abdominal Segmentation, Pole Cell Formation, and Embryonic Polarity Require the Localized Activity of oskar, a Maternal Gene in Drosophila. Cell 47: 141–152. 10.1016/0092-8674(86)90375-2 [DOI] [PubMed] [Google Scholar]
- Lewellyn L., Cetera M., Horne-Badovinac S., 2013. Misshapen decreases integrin levels to promote epithelial motility and planar polarity in Drosophila. J. Cell Biol. 200: 721–729. 10.1083/jcb.201209129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. G., Leclerc V., Smith-Litière K., St Johnston D., 2003. The identification of novel genes required for Drosophila anteroposterior axis formation in a germline clone screen using GFP-Staufen. Dev. 130: 4201–4215. 10.1242/dev.00630 [DOI] [PubMed] [Google Scholar]
- McGregor J. R., Xi R., Harrison D. A., 2002. JAK signaling is somatically required for follicle cell differentiation in Drosophila. Dev. 129: 705–717. http://dev.biologists.org/content/129/3/705 [DOI] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Thomas P. D., 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41: D377–D386. 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto A. M., Jordan K. C., Tietze J. K., Britton J. S., O’Neill E. M., et al. , 1996. Pointed, an ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Dev. 122: 3745–3754. http://dev.biologists.org/content/122/12/3745 [DOI] [PubMed] [Google Scholar]
- Musacchio M., Perrimon N., 1996. The Drosophila kekkon Genes: Novel Members of both the Leucine-Rich Repeat and Immunoglobulin Superfamilies Expressed in the CNS. Dev. Biol. 178: 63–76. 10.1006/dbio.1996.0198 [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S., Schüpbach T., 1996. The Drosophila TGF-α-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech. Dev. 59: 105–113. 10.1016/0925-4773(96)00567-9 [DOI] [PubMed] [Google Scholar]
- Nicolae A., Xi L., Pham T. H., Navarro W., Meeker H. G., et al. , 2016. Mutations in the JAK/STAT and RAS signaling pathways are common in intestinal T-cell lymphomas. Leukemia 30: 2245–2247. 10.1038/leu.2016.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D., Sykora M. M., Sachidanandam R., Mechtler K., Brennecke J., 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO 29: 3301–3317. 10.1038/emboj.2010.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterfield M., Du X., Schüpbach T., Wieshaus E., Shvartsman S. Y., 2013. Three-Dimensional Epithelial Morphogenesis in the Developing Drosophila Egg. Dev. Cell 24: 400–410. 10.1016/j.devcel.2013.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza L. G., Stewart R. A., Li D.-M., Xu T., 2004. Drosophila Src-family kinases function with Csk to regulate cell proliferation and apoptosis. Oncogene 23: 4754–4762. 10.1038/sj.onc.1207635 [DOI] [PubMed] [Google Scholar]
- Perkins L. A., Holderbaum L., Tao R., Hu Y., Sopko R., et al. , 2015. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 201: 843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. V., Clifford R. J., Schüpbach T., 1989. The Maternal Ventralizing Locus torpedo Is Allelic to faint little ball, an Embryonic Lethal, and Encodes the Drosophila EGF Receptor Homolog. Cell 56: 1085–1092. 10.1016/0092-8674(89)90641-7 [DOI] [PubMed] [Google Scholar]
- Queenan A. M., Ghabrial A., Schüpbach T., 1997. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Dev. 124: 3871–3880. http://dev.biologists.org/content/124/19/3871 [DOI] [PubMed] [Google Scholar]
- Rawlings J. S., Rennebeck G., Harrison S. M., Xi R., Harrison D. A., 2004. Two Drosophila suppressors of cytokine signaling (SOCS) differentially regulate JAK and EGFR pathway activities. BMC Cell Biol. 5: 38 10.1186/1471-2121-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R. D., Bach E. A., Cagan R. L., 2004. Drosophila C-terminal Src kinase negatively regulates organ growth and cell proliferation through inhibition of the Src, Jun N-terminal kinase, and STAT pathways. Mol. Cell. Biol. 24: 6676–6689. 10.1128/MCB.24.15.6676-6689.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recasens-Alvarez C., Ferreira A., Milán M., 2017. JAK/STAT controls organ size and fate specification by regulating morphogen production and signalling. Nat. Commun. 8: 13815 10.1038/ncomms13815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich A., Sapir A., Shilo B., 1999. Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Dev. 126: 4139–4147. http://dev.biologists.org/content/126/18/4139 [DOI] [PubMed] [Google Scholar]
- Roth S., Neuman-Silberberg F. S., Barcelo G., Schüpbach T., 1995. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell 81: 967–978. 10.1016/0092-8674(95)90016-0 [DOI] [PubMed] [Google Scholar]
- Ruohola H., Bremer K. A., Baker D., Swedlow J. R., Jan L. Y., et al. , 1991. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell 66: 433–449. 10.1016/0092-8674(81)90008-8 [DOI] [PubMed] [Google Scholar]
- Savant-Bhonsale S., Montell D. J., 1993. torso-like encodes the localized determinant of Drosophila terminal pattern formation. Genes Dev. 7: 2548–2555. 10.1101/gad.7.12b.2548 [DOI] [PubMed] [Google Scholar]
- Schnorr J. D., Berg C. A., 1996. Differential activity of Ras1 during patterning of the Drosophila Dorsoventral Axis. Genetics 144: 1545–1557. http://www.genetics.org/content/144/4/1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D. L., Montell D. J., 2001. Paracrine Signaling through the JAK/STAT Pathway Activates Invasive Behavior of Ovarian Epithelial Cells in Drosophila. Cell 107: 831–841. 10.1016/S0092-8674(01)00607-9 [DOI] [PubMed] [Google Scholar]
- Skeath J. B., Wilson B. A., Romero S. E., Snee M. J., Zhu Y., et al. , 2017. The extracellular metalloprotease AdamTS-A anchors neural lineages in place within and preserves the architecture of the central nervous system. Dev. 144: 3102–3113. 10.1242/dev.145854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R. A., Li D.-M., Huang H., Xu T., 2003. A genetic screen for modifiers of the lats tumor suppressor gene identifies C-terminal Src kinase as a regulator of cell proliferation in Drosophila. Oncogene 22: 6436–6444. 10.1038/sj.onc.1206820 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., 1993. Developmental Genetics of oogenesis, pp. 1–70 in The Development of Drosophila melanogaster, edited by Bate M., Martinez Arias A. Cold Spring Harbor Laboratory Press, Plainview. [Google Scholar]
- Sun Y., Yan Y., Denef N., Schüpbach T., 2011. Regulation of somatic myosin activity by Protein Phosphatase 1β controls Drosophila oocyte polarization. Dev. 138: 1991–2001. 10.1242/dev.062190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W. E., Smiley S., Wong M. L., Alberts B. M., 1992. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Dev. 115: 923–936. http://dev.biologists.org/content/115/4/923 [DOI] [PubMed] [Google Scholar]
- Tulina N., Matunis E., 2001. Control of Stem Cell Self-Renewal in Drosophila Spermatogenesis by JAK-STAT Signaling. Science 294: 2546–2549. 10.1126/science.1066700 [DOI] [PubMed] [Google Scholar]
- Viktorinová I., Dahmann C., 2013. Microtubule Polarity Predicts Direction of Egg Chamber Rotation in Drosophila. Curr. Biol. 23: 1472–1477. 10.1016/j.cub.2013.06.014 [DOI] [PubMed] [Google Scholar]
- Voldborg B. R., Damstrup L., Spand-Thomsen M., Skovgaard Poulsen H., 1997. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann. Oncol. 8: 1197–1206. 10.1023/A:1008209720526 [DOI] [PubMed] [Google Scholar]
- Winter P. S., Sarosiek K. A., Lin K. H., Meggendorfer M., Schnittger S., et al. , 2014. RAS signaling promotes resistance to JAK inhibitors by suppressing BAD-mediated apoptosis. Sci. Signal. 7: ra122 10.1126/scisignal.2005301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. K. L., Li W., An Y., Duan Y., Li Z., et al. , 2015. β-Spectrin Regulates the Hippo Signaling Pathway and Modulates the Basal Actin Network. J. Biol. Chem. 290: 6397–6407. 10.1074/jbc.M114.629493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi R., McGregor J. R., Harrison D. A., 2003. A Gradient of JAK Pathway Activity Patterns the Anterior-Posterior Axis of the Follicular Epithelium. Dev. Cell 4: 167–177. 10.1016/S1534-5807(02)00412-4 [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G. M., 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Dev. 117: 1223–1237. http://dev.biologists.org/content/117/4/1223 [DOI] [PubMed] [Google Scholar]
- Yan R., Luo H., Darnell J. E., Jr., Dearolf C. R., 1996. A JAK-STAT pathway regulates wing vein formation in Drosophila. Proc. Natl. Acad. Sci. USA 93: 5842–5847. 10.1073/pnas.93.12.5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.-H., Kolodkin A. L., 1999. Semaphorin Signaling: A Little Less Per-Plexin. Neuron 22: 11–14. 10.1016/S0896-6273(00)80672-8 [DOI] [PubMed] [Google Scholar]
- Zhao D., Bownes M., 1999. Misexpression of argos, an inhibitor of EGFR signaling in oogenesis, leads to the production of bicephalic, ventralized, and lateralized Drosophila melanogaster eggs. Genesis 25: 375–386. [DOI] [PubMed] [Google Scholar]
- Zhao T., Graham O. S., Raposo A., St Johnston D., 2012. Growing microtubules push the oocyte nucleus to polarise the Drosophila dorsal-ventral axis. Science 336: 999–1003. 10.1126/science.1219147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data necessary to confirm the conclusions presented in this article are represented fully within it and the associated materials. Strains are available upon request. Raw and normalized microarray data were deposited at the Gene Expression Omnibus (GEO) under accession number GSE118881. File S1 contains a list of all microarray probes that pass the fold change and P-value thresholds. The genes in this list are sorted into categories and their function, if known, is summarized. Raw P-values and adjusted P-values are also reported in File S1. Files S2, S3 and S4 contain the result of GO statistical overrepresentation analysis of the microarray dataset using Panther, LAGO, and DAVID, respectively. File S5 contains a list of the genes selected for the RNAi screen as well as Stau-GFP scoring (at stage 10) for each RNAi line tested. File S6 lists the probe sequences used for AdamTS-A FISH. File S7 contains a full list of experimental genotypes and incubation temperatures. Table S1 contains quantification associated with Figure 3. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7209809.