Abstract
Ohno’s hypothesis predicts that the expression of the single X chromosome in males needs compensatory upregulation to balance its dosage with that of the diploid autosomes. Additionally, X chromosome inactivation ensures that quadruple expression of the two X chromosomes is avoided in females. These mechanisms have been actively studied in mice and humans but lag behind in domestic species. Using RNA sequencing data, we analyzed the X chromosome upregulation in sheep fetal tissues from day 135 of gestation under control, over or restricted maternal diets (100%, 140% and 60% of National Research Council Total Digestible Nutrients), and in conceptuses, juvenile, and adult somatic tissues. By computing the mean expression ratio of all X-linked genes to all autosomal genes (X:A), we found that all samples displayed some levels of X chromosome upregulation. The degrees of X upregulation were not significant (P-value = 0.74) between ovine females and males in the same somatic tissues. Brain, however, displayed complete X upregulation. Interestingly, the male and female reproduction-related tissues exhibited divergent X dosage upregulation. Moreover, expression upregulation of the X chromosome in fetal tissues was not affected by maternal diets. Maternal nutrition, however, did change expression levels of several X-linked genes, such as sex determination genes SOX3 and NR0B1. In summary, our results showed that X chromosome upregulation occurred in nearly all sheep somatic tissues analyzed, thus support Ohno’s hypothesis in a new species. However, the levels of upregulation differed by different subgroups of genes such as those that are house-keeping and “dosage-sensitive”.
Keywords: Ohno’s hypothesis, X chromosome upregulation, Maternal nutrition, Ovine
In mammals, deviation from diploidy may induce detrimental consequences (Birchler et al. 2005). For example, gene duplications or deletions can induce cancer (Giam and Rancati 2015), and chromosome monosomy or trisomy usually causes fetal lethality (Burgess et al. 2014). Mammalian males, however, are monosomic for the X chromosome, yet do not suffer from the deleterious effects of X monosomy (Chen et al. 2014). This is likely the result of a still debated mechanism of X chromosome dosage compensation (Pessia et al. 2014; Mank et al. 2014; Veitia et al. 2015; Graves 2016). Susumu Ohno hypothesized that upregulation of the X-linked genes in the heterogametic sex (XY) would be necessary to maintain their expression to the levels of the diploid autosomes (Ohno 1966). This solved the dosage imbalance of X-linked genes in males, yet subjected females to quadruple levels of X expression. Another mechanism, X chromosome inactivation (XCI), the inactivation of one of the two X chromosomes in every cell of the female, balances the X chromosome gene dosage between males and females (Lyon 1961). Both X chromosome upregulation and XCI are necessary components of the X chromosome dosage compensation in mammals (Ohno 1966).
Although XCI has been characterized in many species (Goto and Monk 1998; Xue et al. 2002; Deakin et al. 2009; Lee 2011; Livernois et al. 2012; Sahakyan et al. 2018), verification of X chromosome upregulation has not been conducted until chromosome-wide expression analysis became possible (Pessia et al. 2014). X chromosome upregulation has been studied using data from microarray (Gupta et al. 2006; Nguyen and Disteche 2006) and RNA sequencing (RNA-seq) (Xiong et al. 2010; Deng et al. 2011; Lin et al. 2007, 2011, 2012; Pessia et al. 2012) by computing the mean or median expression ratio of all X-linked genes to all autosomal genes (X:A). An X:A ratio of 1.0 or greater implies the doubling of X-linked gene transcription from the single active X, indicating complete compensatory upregulation. An X:A ratio of 0.5 indicates that expression levels of X-linked genes is half of those of the autosome pairs, suggesting no compensatory upregulation. When an X:A ratio falls between 0.5 and 1, it is termed partial compensation (Deng et al. 2011). A full compensatory upregulation of X-linked genes has been recently observed only in “dosage-sensitive” genes in eutherian mammals (Julien et al., 2012; Lin et al., 2012; Pessia et al., 2012). These genes usually code for proteins complexes with structural, regulatory and housekeeping functions (Kondrashov and Koonin 2004; Birchler 2012; Pessia et al. 2012).
XCI and X chromosome upregulation have been studied in mice and humans, but such investigations lag behind in domestic species (Disteche 2012). XCI has been shown in sheep (Luciani et al. 1979), but little is known about its onset and no information is available on X dosage compensation. With completion of the ovine genome sequencing (Jiang et al. 2014) and the advancement of RNA-seq technology (Wolf and Bryk 2011), a number of RNA-seq datasets in sheep somatic tissues are now available for X chromosome upregulation studies.
The sheep has been frequently used as a model for human pregnancy and fetal development (Barry and Anthony 2008). Poor maternal nutrition, either over- or restricted-feeding, has been shown to alter gene expression in fetal tissues (Du et al. 2011; Pillai et al. 2016; Duan et al. 2018). Changes in DNA methylation is likely involved because restrictedly nourished ewes carried fetuses with altered DNA methyltransferase in the hypothalamus (Begum et al. 2012). Similarly, human metastable epialleles, which are variably expressed in genetically identical individuals, have also been persistently changed epigenetically by maternal nutrition in early pregnancy (Dominguez-Salas et al. 2014). These findings that maternal diet alters fetal epigenetics are of particular interest because XCI and X chromosome upregulation are epigenetically regulated processes (Goto and Monk 1998). However, the effects of maternal nutrition on X chromosome dosage compensation and X-linked gene expression have yet to be studied in the sheep, an important species for both agriculture and human medicine.
Using data generated by us (GSE111306) (Duan et al. 2018) and two additional RNA-seq datasets, PRJEB6169 (Jiang et al. 2014) and PRJNA254105 (Brooks et al. 2015), we were able to achieve the first comprehensive evaluation of X chromosome upregulation in sheep. Furthermore, we also investigated the effects of different maternal diets on the expression of X-linked genes. Our hypothesis was that X chromosome upregulation in the sheep would be partial, similar to that in the bovine as reported by us (Duan et al. 2016) and others (Ka et al. 2016). We further hypothesized that different maternal diets would alter the expression levels of X-linked genes in ovine fetal tissues.
Materials and Methods
Experimental design and RNA sequencing
Animal protocols, tissues collection, and RNA sequencing library preparation were described in Pillai et al. (2017) and Duan et al. (2018). Briefly, 12 pregnant ewes were individually housed and randomly assigned to control- (100% NRC requirement, Con, n = 4), overfed- (140%, Over, n = 4) or restricted- (60%, Res, n = 4) diets calculated by the National Research Council requirement for total digestible nutrients for a ewe pregnant with twins (Pillai et al. 2016). The ewes remained on their respective diets until day 135 of gestation when they were killed. Fifteen fetuses, control (n = 7), overfed (n = 4) and restricted (n = 4) were included in this study. Full organ of brain, kidney, and lung were collected, flash-frozen in liquid nitrogen and stored at -80° until RNA extraction.
RNA was extracted from fetal brain, kidney, and lung using TRIzol (Invitrogen, Grand Island, NY) according to the manufacturer’s instructions. Library preparation was carried out using TruSeq RNA library prep kit (Illumina, RS-122-2001, RS-122-2002) and quantified using real-time PCR. Agilent 2100 Bioanalyzer (Agilent) was used to assess the size distribution and to determine the RNA integrity number (RIN). All RNA samples for sequencing had RIN values greater or equal to 7 (Duan et al. 2018). The sequencing was performed on Nextseq 500 System (Illumina) with 75 bp paired-end reads in three sequencing runs. Overall, we obtained 2,149 million raw sequencing reads that passed filtering from three sequencing runs of 45 fetal tissue samples. The raw read dataset has been uploaded to GEO database with the accession number GSE111306.
Additional RNA-seq datasets
In addition to the RNA-seq data described above, two additional RNA-seq datasets were downloaded from Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under the accession numbers PRJNA254105 (Brooks et al. 2015) and PRJEB6169 (Jiang et al. 2014). PRJNA254105 included whole conceptuses at day 14 of gestation. PRJEB6169 contained data from adult and juvenile (6-10 months) heart, brain, liver, biceps femoris, rumen, female and male specific tissues, including the cervix, ovarian follicles, ovary, uterus, corpus luteum, testes, and placenta and membranes.
RNA-seq data trimming, mapping and assembly
Sequence adapter and quality trimming were conducted using Sickle v1.33 (Joshi and Fass 2011) with the parameters Q score ≥ 30 and length ≥ 20 (-q30, -l20). RNA-seq reads were checked using FastQC v0.11.3 (Andrews 2010) for quality control. Filtered RNA-seq reads from fetal tissues of day 135 of gestation were aligned to the sheep reference genome Oar_v4.0 using Hisat2 v2.0.5 (Kim et al. 2015). The mapping rates of all datasets are summarized in Table S1. The average mapping rate for our data are 90% with 19,846,496 reads mapped to the genome, whereas the additional datasets had an averaged 75% mapping rate with 12,854,507 reads aligned.
Aligned reads for each tissue from all three datasets were assembled using IsoEM v1.1.4 (Nicolae et al. 2011). The mRNA level of each gene was estimated by log2-transformed transcripts per kilobase million (TPM) within each dataset and quantified using IsoEM (version 1.1.4; Nicolae et al., 2011). TPM normalizes for gene length first and then for sequencing depth. This was preferred to RPKM/FPKM because it normalizes among transcriptome sizes of different samples and allows more appropriate comparisons of gene expression across samples (Soneson et al. 2015). Gene expression levels in TPM were log2-transformed to minimize variations. Expressed genes were defined as TPM ≥ 1 (Clark et al. 2017). A total of 7,166 genes were expressed among all tissues and defined as “dosage sensitive” genes (Sangrithi et al. 2017). Genes in the pseudoautosomal regions (PARs) of the sex chromosomes were obtained from Ruminant PARs annotation by Raudsepp and Chowdhary (2015).
Dosage compensation calculation
A total of 20,519 genes are in the sheep genome and assigned to each chromosome. The X:A ratio was calculated as the Relative X Expression (RXE); the difference between the log2-transformed mean TPM values of the X chromosome and autosomes (A), using the formula below, where X-linked and autosomal genes were expressed as x and a, respectively:
An RXE ≥ 0 represents a full up-regulation of X. An RXE between 0 and -1 indicates partial X chromosome upregulation. An RXE of -1 indicates a lack of X chromosome upregulation.
We also calculated the relative expression of each autosome pair (RGE) over all other chromosomes (excluding mitochondria and the Y chromosome which are not annotated in sheep). The RGE value was used to determine if the expression of the X chromosome deviated from the normal range of expression by the autosomes and if a particular autosome pair is more/less expressed than the rest of the chromosomes. The RGE was calculated using the following formula:
Where i represents a particular pair of autosomes, n represents all autosomes. n-i represents all autosomes excluding the autosome i. If the RGE of an autosome pair was greater than or equal to 0, it represents upregulation of that autosome pair. An RGE between 0 and -1 indicates downregulation.
The boxplots of RXE and RGE were generated in R (R Development Core Team 2008) using ggplot2 package (Wickham 2009). In these plots the lower and upper hinges encompass the 25th and 75th percentile of the data. The distance between the hinges is the inter-quartile range (IQR). The lower and upper whiskers extending from the hinges represent values no further than 1.fivefold of the IQR or within 95% confidence interval. Outliers were plotted individually beyond the end of whiskers (McGill et al. 1978) and labeled with numbers of the corresponding chromosomes.
Differentially expressed X-linked genes across maternal nutrition
Differentially expressed genes (DEGs) between Con and Over or Con and Res were determined using IsoDE version 2 (Al Seesi et al. 2014; Mandric et al. 2017). IsoDE2 is based on 200 bootstrap replicates where sampling from the original data were performed with replacement and stratified by the group variables (Al Seesi et al. 2014). Bootstrapping (Efron and Tibshirani 1994) is advantageous because it offers a reliable solution to the lack/low replicates and allows distinction between biological differences and technical variability or noise. Bootstrapping method is simple to apply and does not require any distribution assumptions. In each comparison, a gene was deemed differentially expressed if it showed log2 fold change (FC) > 1 between two treatments and significantly different (P-value ≤ 0.05). The X-linked DEGs (Table S2) were a sub-group of the total DEGs from all chromosomes.
Gene ontology analysis
A Gene Ontology (GO) classification was conducted using DAVID 6.8 (Huang et al. 2009b, 2009a). GO categories with P-value ≤ 0.05 were considered significantly overrepresented. The pie plot of “dosage sensitive” genes categorized by protein functions was made in PANTHER classification system (Mi et al. 2013).
Data Availability Statement
The datasets analyzed during the current study are available in the Gene Expression Omnibus and BioProject:
GSE111306: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111306
PRJNA254105: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA254105
PRJEB6169: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB6169
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7221467.
Results
X chromosome upregulation in ovine major organs and reproductive tissues
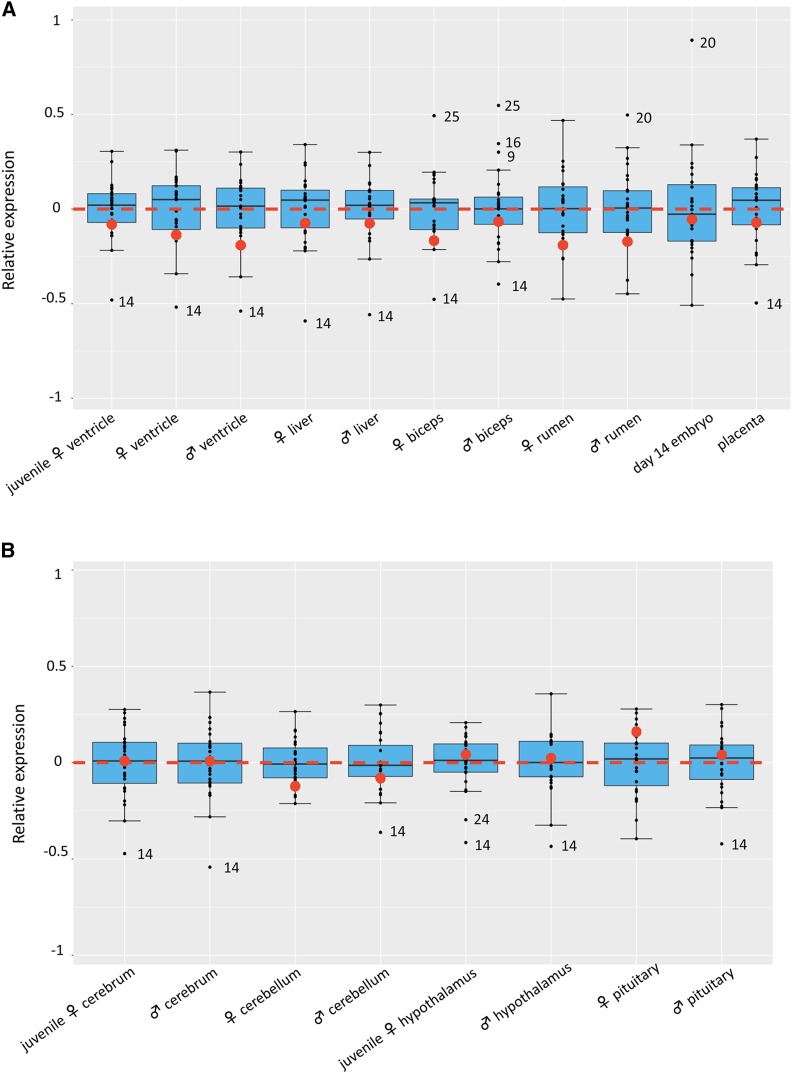
We found that the liver, muscle, rumen, heart of juveniles and adults, conceptuses and placenta and membrane at day 14 of gestation displayed partial X chromosome upregulation with RXE values ranging from -0.19 to -0.05, and an overall average RXE of -0.12 (Figure 1A). Interestingly, all RXE were much closer to 0 than to -1, indicating a substantial amount of dosage compensation across the entire X chromosome. The upregulation, however, appeared to be more pronounced in the brain. The RXE ranged from -0.12 to 0.16 in the cerebrum, cerebellum, hypothalamus, and pituitary (Figure 1B), suggesting complete X chromosome upregulation with the exception of the cerebellum. No significant difference (P-value = 0.74, by student t-test) was observed between males and females in the same tissue, demonstrating that X chromosome upregulation occurred similarly in both sexes, despite of the difference in the number of X chromosomes. Moreover, the RXE values fell within 1.5 times of the interquartile ranges (25–75% of the data) of RGEs of autosome pairs for all examined tissues (Figure 1 A and 1B), suggesting the single active X chromosome in somatic tissues balanced its gene transcription outputs with those of the autosome pairs.
Figure 1.
Boxplots of log2-transformed relative expression of the X chromosome (RXE) and each autosome pair (RGE) in major tissues (A) and brain (B) of juvenile and adult sheep. Red dots: mean RXEs for all replicates within a tissue type. Black dots: mean RGEs for each autosome pair. Numbers by black dots: autosomes whose RGEs fell outside of the expression quartiles for the tissue. Red dotted line: the border for complete (above line) and incomplete (below line) dosage compensation. The X:A ratio was calculated as the Relative X expression, RXE = log2 (X) − log2 (A), the difference between the log2-transformed mean TPM values of X and A. An RXE value of 0 means the expression of X and autosome is equal, suggesting X dosage compensation. Positive and negative RXE values indicate complete and incomplete dosage compensation, respectively. An RXE of -1, however, represents the lack of X dosage compensation. RGE of each autosome pair over all other chromosomes was used to evaluate the deviation of X expression to autosomes.
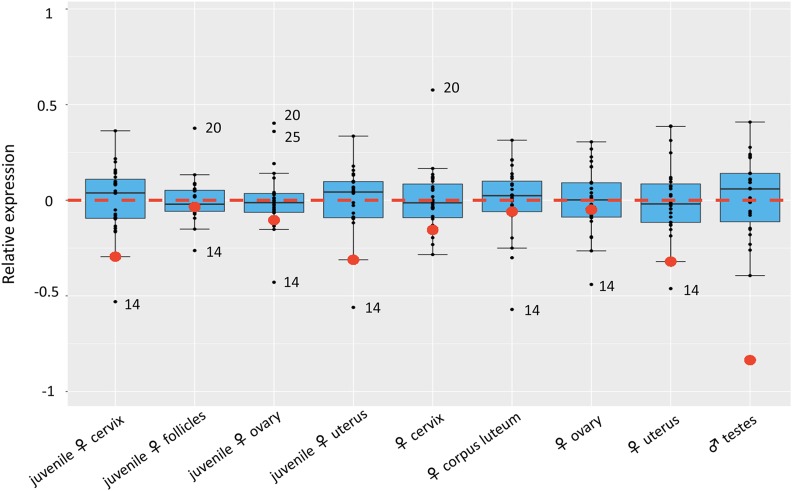
Partial X chromosome upregulation was also observed in juvenile and adult female reproduction-related tissues. These included the cervix, ovarian follicle, ovary, uterus, and corpus luteum. The overall averaged RXE was -0.19 and -0.15 for juvenile and adult female tissues, respectively (Figure 2). Among these, juvenile follicles, adult ovaries and corpora lutea had the greatest RXE values. However, a different pattern was observed in the male specific reproductive tissues studied. The testes exhibited an average RXE of -0.84, or a near lack of X chromosome upregulation (i.e., RXE = -1; Figure 2).
Figure 2.
Boxplots of log2-transformed relative expression of the X chromosome (RXE) and each autosome pair (RGE) in female- and male-specific tissues. Red dots: RXEs for all replicates within a tissue type. Black dots: RGEs for each autosome pair. Numbers by black dots: autosomes whose RGEs fell outside of the expression quartiles for the tissue. Red dotted line: the border for complete (above line) and incomplete (below line) dosage compensation. The X:A ratio was calculated as the Relative X expression, RXE = log2 (X) − log2 (A), the difference between the log2-transformed mean TPM values of X and A. An RXE value of 0 means the expression of X and autosome is equal, suggesting X dosage compensation. Positive and negative RXE values indicate complete and incomplete dosage compensation, respectively. An RXE of -1, however, represents the lack of X dosage compensation. RGE of each autosome pair over all other chromosomes was used to evaluate the deviation of X expression to autosomes.
We also observed that a number of autosome pairs had either greater than or less than the overall averaged gene expression. Chromosome 14, for example, was very “quiet” in gene expression at the chromosomal level, falling outside of the 1.5 interquartile ranges of RGEs of the other autosome pairs in many tissues (Figure 1 A). Conversely, Chromosomes 20 and 25 were “active” in many tissues with high RGE values (greater than 0.5). Interestingly, the expression activity was negatively correlated (r = -0.94) with the numbers of expressed genes (TPM ≥ 1) on these chromosomes. With an averaged 751 expressed genes, Chromosome 14 was less active (average RGE=-0.44) than Chromosomes 20 (average RGE = 0.22) and 25 (average RGE = 0.25) with 416 and 202 expressed genes, respectively. Rather, expression activity of these chromosomes may be related to the functions of genes that they contain. We therefore analyzed the gene ontology (GO) terms of lowly (1≤ TPM < 50) expressed genes on Chromosomes 14, 20, and 25. The GO terms for Chromosome 14 included regulation of DNA-templated transcription, which corresponds to the reduced expression of transcription factors in most of tissues (Vaquerizas et al. 2009). On the other hand, the major GO categories of highly expressed genes (TPM > 100) on Chromosomes 20 and 25 were enriched in nucleosome assembly and sarcomere organization (Table S3). These terms corresponded to greater activities of Chromosome 20 in early conceptuses and Chromosome 25 in bicep muscles (Figure 1A).
X chromosome upregulation in ovine fetuses under different maternal nutrition
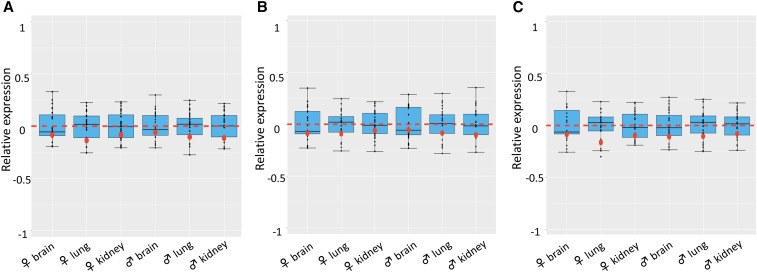
X chromosome upregulation in ovine fetuses at day 135 of gestation was not affected by maternal diet, either over- or restricted-nutrition (P-value = 0.59, 0.70, respectively, by Wilcoxon Rank Sums tests). Tissues (brain, kidney, and lung) of both male and female fetuses from mothers of all three treatment groups displayed partial dosage compensation. The RXEs of the three tissues ranged from -0.13 to -0.05, (Figure 3A), from -0.14 to -0.05 (Figure 3B), and from -0.16 to -0.08 (Figure 3C) for the Con-, Over- and Res-fed group, respectively. All RXE values fell within the RGE ranges, suggesting the single X upregulated on its expression to levels close to the autosome pairs. These observations indicate that different maternal nutrition did not affect the upregulation of the X chromosome in fetal tissues.
Figure 3.
Boxplots of log2-transformed expression of the X chromosome (RXE) and each autosome pair by fetal tissues from mothers under different nutritional treatments: Control (A), Overfed (B) and Restricted (C). Red dots: RXEs for all replicates within a treatment group. Black dots: RGEs for each autosome pair. Numbers by black dots: autosomes whose RGEs fell outside of the expression quartiles for the tissue. Red dotted line: the border for complete (above line) and incomplete (below line) dosage compensation. The X:A ratio was calculated as the Relative X expression, RXE = log2 (X) − log2 (A), the difference between the log2-transformed mean TPM values of X and A. An RXE value of 0 means the expression of X and autosome is equal, suggesting X dosage compensation. Positive and negative RXE values indicate complete and incomplete dosage compensation, respectively. An RXE of -1, however, represents the lack of X dosage compensation. RGE of each autosome pair over all other chromosomes was used to evaluate the deviation of X expression to autosomes.
X chromosome upregulation in different gene subgroups
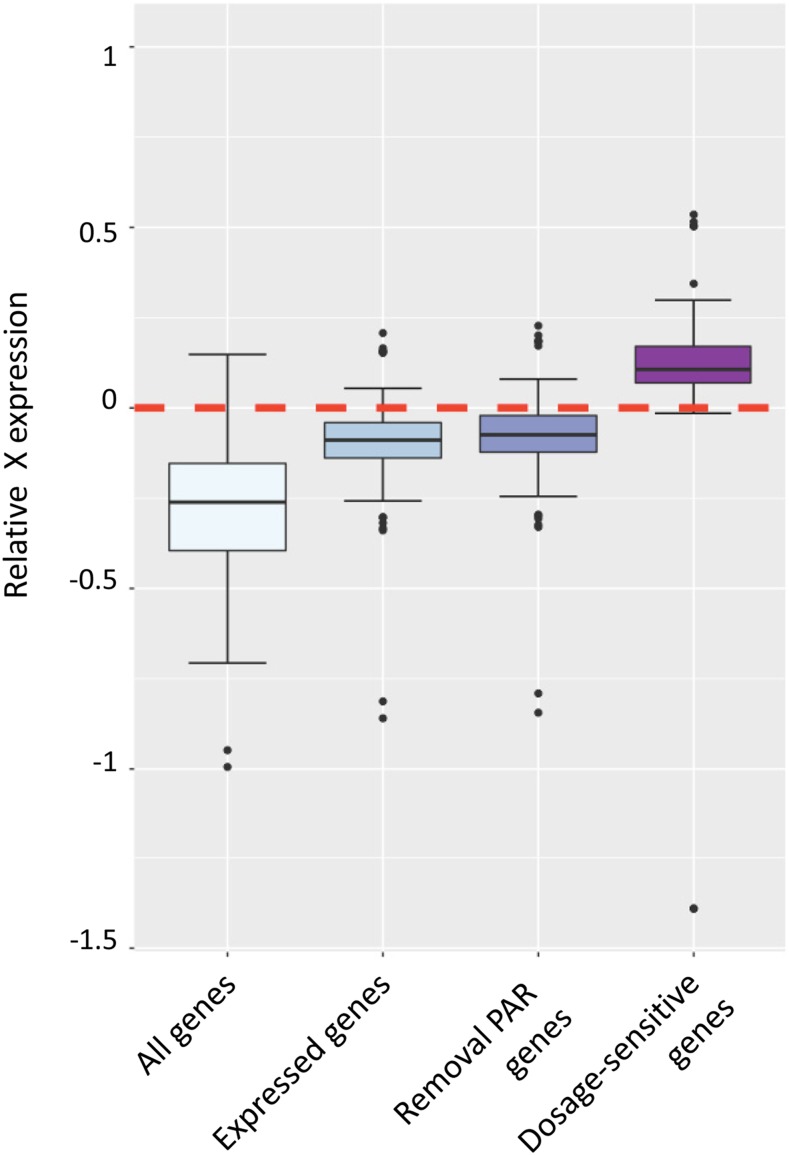
We calculated the RXE values in the gene categories of “All genes”, “Expressed genes”, “Genes subject to XCI (removal of PAR genes)” and “Dosage sensitive genes” (Figure 4). “All genes” included low- and non-expressed genes (TPM < 1), and had the lowest RXE values among the four subgroups of genes. The median of RXE in the “All genes” category was close to -0.5, indicating that when all X-linked genes were considered (including those with leaky expression), there was nearly no upregulation of the X chromosome. “Expressed genes” gave a partial X chromosome upregulation with a median RXE value close to 0, suggesting that this group contained genes of the X chromosome that were not subjected to X dosage compensation. Therefore, we removed the 14 genes located in ovine PAR. These genes have a homologous copy on the Y chromosome, and are not subjected to XCI. The RXE values without PAR were slightly increased, indicating that the PAR genes had lower expression. Moreover, we characterized another category - “Dosage sensitive genes”, which were ubiquitously expressed across all samples in our study and were mostly housing-keeping genes such as those involved in nucleic acid binding, cytoskeletal proteins and transferase (Figure S1 and Table S4). These “Dosage sensitive genes” had the highest median RXEs of greater than 0, corresponding to a full X upregulation. Taken together, our analysis of the four gene categories suggests that dosage regulation is highly related to gene functions.
Figure 4.
Boxplot of RXE values in the categories of “All genes”, “Expressed genes”, “Genes subject to XCI (removal of genes in PAR)” and “Dosage sensitive genes”. Red dotted line: the border for complete (above line) and incomplete (below line) dosage compensation.
Effects of maternal nutrition on the expression of X-linked genes in ovine fetal tissues
A total of 1,228 X-linked genes were annotated in the current ovine genome (Jiang et al. 2014). Among these, 625 genes were expressed (TPM ≥ 1) by the three fetal tissues combined (Table S2). The mean number of expressed X-linked genes by each fetal tissue was calculated for each maternal nutrition group (Table 1). On average, 518.2 ± 14.7 out of 625 X-linked genes were expressed in the three organs. Specifically, the brain expressed the most genes (536.1 ± 14.3; control group), followed by the kidney (517.3 ± 9.2) and the lung expressed the fewest (506 ± 3.7). The numbers of the expressed X-linked genes were not significantly different (P-value > 0.05, by one-way ANOVA) across the maternal nutrition treatments (Table 1). However, the levels of expression of the X-linked genes were affected by maternal nutrition. A total of 57 X-linked genes were differentially expressed among treatment groups. For example, two genes related to sex determination- SOX3 and NR0B1-were down-regulated in fetal brains of the Over group (Table S5). The changes in sex-linked genes may provide a mechanism for the highly debated observation that skewed sex ratio was related to maternal nutrition (Mathews et al. 2008). The top eight X-linked DEGs (PAGE4, S100G, SOX3, KCNE5, CLDN2, DUSP21, LOC105610402, and SLC6A14) were summarized in Table 2 and were all expressed 8X (> 3 log2-Fold Change) more than that of the controls. Taken together, these expression data clearly demonstrated an effect of poor maternal nutrition on gene expression during fetal development.
Table 1. Mean numbers of expressed (TPM ≥ 1) X-linked genes in tissues of day 135 fetuses from control (n = 7), overfed (n = 4) and restricted (n = 4) mothers.
| Treatments | ||||
|---|---|---|---|---|
| Control | Overfed | Restricted | P-value | |
| Brain | 536.1 ± 15.4 | 531.8 ± 8.7 | 530.3 ± 9.2 | 0.73 |
| Kidney | 517.3 ± 9.9 | 514.5 ± 9.1 | 518.8 ± 2.2 | 0.77 |
| Lung | 506.0 ± 4.0 | 503.3 ± 7.4 | 502.0 ± 4.2 | 0.44 |
Table 2. Differentially expressed X-linked genes by tissues of ovine fetuses from mothers under control, overfed and restricted nutrition treatments.
| Comparison | Tissue | Gene | Expression in controls (TPM) | Expression in treated (TPM) | Log2 FC* |
|---|---|---|---|---|---|
| Con vs. Over | Brain | PAGE4 | 99.80 | 0.18 | -∞ |
| Brain | S100G | 33.62 | 0.46 | -∞ | |
| Brain | SOX3 | 1.23 | 0.15 | -∞ | |
| Kidney | KCNE5 | 1.71 | 0.27 | -∞ | |
| Kidney | PAGE4 | 1.46 | 0.86 | −3.23 | |
| Lung | CLDN2 | 0.26 | 6.36 | 4.71 | |
| Con vs. Res | Brain | DUSP21 | 0.08 | 1.04 | 5.21 |
| Brain | LOC105610402 | 1.04 | 0.00 | -∞ | |
| Brain | S100G | 33.62 | 1.27 | −5.20 | |
| Lung | SLC6A14 | 1.57 | 0.10 | −5.43 |
Log2 FC: calculated by using bootstrapping; FC: fold change.
∞: infinity;
PAGE4: PAGE family member 4; S100G: S100 calcium binding protein G; SOX3: SRY-box 3; KCNE5: potassium voltage-gated channel subfamily E regulatory subunit 5; CLDN2: claudin 2; DUSP21: dual specificity phosphatase 21; LOC105610402: 60S ribosomal protein L17; SLC6A14: solute carrier family 6 member 14.
Discussion
To our knowledge, this is the first study of X chromosome compensatory expression upregulation in sheep. We conclude that X chromosome upregulation was present, but largely partial. Additionally, X chromosome upregulation in fetal organs was not affected by the different maternal diets. While a number of species, both invertebrates and vertebrates, have been examined for their X:A ratios, whether X expression is globally upregulated is still highly debated [reviewed in (Gu and Walters 2017)]. Recent studies in therian mammals, including the human, mouse, bovine, and non-human primates mostly support the partial X chromosome upregulation conclusion with X:A ratio being close to 1 (Gu and Walters 2017; Duan et al., 2016; Ka et al., 2016). Our findings here contribute to the consensus of partial X chromosome upregulation in a new species.
The estimation of X:A ratios differs when different gene subgroups and different tissues are analyzed, thus resulting in completely different conclusions over Ohno’s hypothesis (Sangrithi and Turner 2018). Some of the low- and non-expressed genes in somatic tissues were found to be highly expressed in testis. These genes are more enriched on the X chromosome than on autosomes (Rice 1996; Deng et al. 2011; Disteche 2016). Therefore, when the analysis included low- and non-expressed genes, the estimation of X chromosome upregulation is biased. Our result showed that RXE was closer to -0.5 when “all genes” were included, while RXE was close to 0 when only expressed genes were used. These two different types of gene categorization and inclusion corresponded to the opposite findings by Xiong et al. (2010) and Deng et al. (2011). Additionally, dosage compensation requires both X chromosome upregulation and XCI. Not all genes on the inactive X, however, are silenced. A group of X-linked genes escape inactivation (Disteche et al. 2002; Berletch et al. 2011; Al Nadaf et al. 2012; Balaton and Brown 2016). These include all genes in PARs (Helena Mangs and Morris 2007) and a few in non-PAR regions of X (Tukiainen et al. 2017). As the homologous region of the mammalian sex chromosomes, genes on PARs are expressed from both the X and Y chromosomes (Vermeesch et al. 1997). However, we are not able to exclude any non-PAR genes from the group “subject to XCI” due to the lack of information on non-PAR genes that escape XCI from these regions in the ovine, we were only able to exclude PAR genes in the group of “Subject to XCI”. Of the 20 annotated genes in ovine PAR (Figure S2) (Raudsepp and Chowdhary 2015), 14 were expressed in our study. They were P2RY8, DHRSX, ZBED1, CD99, XG, GYG2, ARSE, MXRA5, PRKX, NLGN4X, STS, PNPLA4, TBL1X, and GPR143. They had relatively low expression levels, ranging in TPM from 1-50 while the average expression level of X-linked genes was 78.5 in TPM. Not much change in RXE values was found when the PAR genes were removed from the expressed group; possibly due their small number. Furthermore, our analysis showed a full compensatory upregulation of “dosage-sensitive” X-linked genes in sheep. This is in agreement with previous findings in the mouse and human by Ramsköld et al. (2009) and Sangrithi et al. (2017) who suggested that ubiquitous gene expression corresponded to the housekeeping function of dosage sensitive genes. It is likely that in order not to create limiting effects, gene products of this subgroup must be generated at comparable levels to those of the same pathways yet encoded by autosome pairs. Therefore, ubiquitously expressed genes are much more upregulated compared to other genes on the X chromosome.
There are a few exceptions to the general finding that ovine tissues underwent partial X chromosome upregulation. One exception is the brain, which had the greatest overall RXE values among all somatic tissues (RXE ranged from -0.12 to 0.16). This greater degree of X chromosome upregulation has also been observed in other species, including the human, mouse (Nguyen and Disteche 2006), old world monkeys, opossum, platypus, and chicken (Julien et al. 2012). The higher X chromosome upregulation is likely the result of both greater levels as well as numbers of expression of X-linked genes in the brain (Table1). During evolution, the X chromosome accumulated an excess of sex- and reproduction-related genes (Saifi and Chandra 1999). Greater expression of X-linked genes in the brain has been described as “the large X-chromosome effect” (Wu and Davis 1993), which was hypothesized to influence general cognitive ability, female mating choices and contribute to species diversification (Zechner et al. 2001). Therefore, it is expected that the brain would have a higher RXE.
Another exception to the overall X chromosome expression upregulation was seen in the sheep male reproduction-related tissues. The RXE was extremely low in sheep testes, corresponding to the observation of low X:A ratio in both the testes and spermatids in mice, indicating an X-specific partial repression in these cells (Nguyen and Disteche 2006). It was reported that the X:A ratio remained low in spermatogonia (Nguyen and Disteche 2006; Sangrithi and Turner 2018). Subsequently both the X and Y chromosomes become inactivated by meiotic sex chromosome inactivation during spermatogenesis (Manterola et al. 2009). This suppression of the X chromosome is likely the cause for the low X:A ratio.
Day 14 whole embryos, on the other hand, had an RXE value of -0.05 which was very close to full dosage compensation (RXE = 0). High X chromosome upregulation in early embryos could be a rebound after the release of repression of sex chromosomes in sperm. In the early embryos this release is necessary for X upregulation initiation (Wang et al. 2016). The expression of X chromosome was reported to be upregulated after the blastocyst stage which continued during 6.5 to 10.5 days post coitum development in mice (Nguyen and Disteche 2006). Mouse embryonic stem cells from both XX and XY embryos were also found to undergo X upregulation (Lin et al. 2011). Although XCI is known to operate in sheep fetuses (Luciani et al. 1979), little is known about its onset and regulation. In the bovine conceptuses, the onset of random XCI is found to have been established before day 14 (Bermejo-Alvarez et al. 2011). In the ovine, it is very likely that XCI has occurred by day 14 due to its shorter gestation (King et al. 1985; Stevens et al. 1990). It is therefore highly possible that the Day 14 ovine conceptuses had only one active X chromosome. The greater RXE value in Day 14 conceptuses thus may imply that the single active X chromosome just started its compensation process. This is consistent with the greater RXE values observed in Day 10-19 conceptuses in the bovine (Duan et al. 2017).
In summary, our comprehensive analyses of X chromosome dosage compensation suggest upregulation of gene expression from the single active X chromosome in most ovine tissues of both sexes.
Acknowledgments
The authors thank Zoetis for donating the controlled intravaginal drug release devices (CIDRs) used for estrus synchronization and the UConn Livestock staff, Dr. Thomas Hoagland, Victor Delaire and the animal science undergraduate students for the animal care during this experiment.This work was supported by the USDA-ARS grant: 1265-31000-091-02S, the USDA Multi-state regional grant: W3171, National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number 2013–01919, Department of Education of Xinjiang Uygur Autonomous Region Scholarship: 2016-3-0036 and Studying Abroad program for Excellent Ph.D. Students of Guangxi Zhuang Autonomous Region: 2014-2. XCT, JD, KF, MZ and ZJ designed the study; AJ, SP, MH, SR, KG and SZ preformed animal breeding, feeding and care, necropsy, and tissue sample collection; KF, MZ and HJ preformed the RNA-seq experiment; JD, NJ, SA, IM and RO analyzed the data; KF, JD and XCT wrote the manuscript, NJ, AJ, RO, SZ, and KG edited the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7221467.
Communicating editor: J. Birchler
Literature Cited
- Al Nadaf S., Deakin J. E., Gilbert C., Robinson T. J., Graves J. A. M., et al. , 2012. A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosoma 121: 71–78. 10.1007/s00412-011-0343-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Seesi S., Tiagueu Y. T., Zelikovsky A., Măndoiu I. I., 2014. Bootstrap-based differential gene expression analysis for RNA-Seq data with and without replicates. BMC Genomics 15: S2 10.1186/1471-2164-15-S8-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S., 2010. FastQC A Quality Control tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Balaton B. P., Brown C. J., 2016. Escape Artists of the X Chromosome. Trends Genet. TIG 32: 348–359. 10.1016/j.tig.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Barry J. S., Anthony R. V., 2008. The Pregnant Sheep as a Model for Human Pregnancy. Theriogenology 69: 55–67. 10.1016/j.theriogenology.2007.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum G., Stevens A., Smith E. B., Connor K., Challis J. R. G., et al. , 2012. Epigenetic changes in fetal hypothalamic energy regulating pathways are associated with maternal undernutrition and twinning. FASEB J. 26: 1694–1703. 10.1096/fj.11-198762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch J. B., Yang F., Xu J., Carrel L., Disteche C. M., 2011. Genes that escape from X inactivation. Hum. Genet. 130: 237–245. 10.1007/s00439-011-1011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Alvarez P., Rizos D., Lonergan P., Gutierrez-Adan A., 2011. Transcriptional sexual dimorphism in elongating bovine embryos: implications for XCI and sex determination genes. Reproduction 141: 801–808. 10.1530/REP-11-0006 [DOI] [PubMed] [Google Scholar]
- Birchler J. A., 2012. Claims and counterclaims of X-chromosome compensation. Nat. Struct. Mol. Biol. 19: 3–5. 10.1038/nsmb.2218 [DOI] [PubMed] [Google Scholar]
- Birchler J. A., Riddle N. C., Auger D. L., Veitia R. A., 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21: 219–226. 10.1016/j.tig.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Brooks K. E., Burns G. W., Spencer T. E., 2015. Peroxisome proliferator activator receptor gamma (PPARG) regulates conceptus elongation in sheep. Biol. Reprod. 92: 42 10.1095/biolreprod.114.123877 [DOI] [PubMed] [Google Scholar]
- Burgess T., Downie L., Pertile M. D., Francis D., Glass M., et al. , 2014. Monosomy 21 Seen in Live Born Is Unlikely to Represent True Monosomy 21: A Case Report and Review of the Literature. Case Rep. Genet. 2014: 965401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-X., Golovnina K., Sultana H., Kumar S., Oliver B., 2014. Transcriptional effects of gene dose reduction. Biol. Sex Differ. 5: 5 10.1186/2042-6410-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E. L., Bush S. J., McCulloch M. E. B., Farquhar I. L., Young R., et al. , 2017. A high resolution atlas of gene expression in the domestic sheep (Ovis aries). PLoS Genet. 13: e1006997 10.1371/journal.pgen.1006997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin J. E., Chaumeil J., Hore T. A., Graves J. A. M., 2009. Unravelling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes. Chromosome Res. 17: 671–685. 10.1007/s10577-009-9058-6 [DOI] [PubMed] [Google Scholar]
- Deng X., Hiatt J. B., Nguyen D. K., Ercan S., Sturgill D., et al. , 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43: 1179–1185. 10.1038/ng.948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C. M., 2012. Dosage compensation of the sex chromosomes. Annu. Rev. Genet. 46: 537–560. 10.1146/annurev-genet-110711-155454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C. M., 2016. Dosage compensation of the sex chromosomes and autosomes. Semin. Cell Dev. Biol. 56: 9–18. 10.1016/j.semcdb.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C. M., Filippova G. N., Tsuchiya K. D., 2002. Escape from X inactivation. Cytogenet. Genome Res. 99: 36–43. 10.1159/000071572 [DOI] [PubMed] [Google Scholar]
- Dominguez-Salas P., Moore S. E., Baker M. S., Bergen A. W., Cox S. E., et al. , 2014. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat. Commun. 5: 3746 10.1038/ncomms4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., Zhao J. X., Yan X., Huang Y., Nicodemus L. V., et al. , 2011. Fetal muscle development, mesenchymal multipotent cell differentiation, and associated signaling pathways. J. Anim. Sci. 89: 583–590. 10.2527/jas.2010-3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Jue N. K., Jiang Z., O’Neill R., Wolf E., et al. , 2017. 125 incomplete compensatory up-regulation of x–linked genes in bovine germline, early embryos, and somatic tissues. Reprod. Fertil. Dev. 29: 171 10.1071/RDv29n1Ab125 [DOI] [Google Scholar]
- Duan J. E., Jue N. K., Jiang Z., O’Neill R., Wolf E., et al. , 2016. 144 dosage compensation and x–linked gene expression in bovine in vivo and in vitro embryos. Reprod. Fertil. Dev. 28: 202 10.1071/RDv28n2Ab144 [DOI] [Google Scholar]
- Duan J., Zhang M., Flock K., Seesi S. A., Mandoiu I., et al. , 2018. Effects of Maternal Nutrition on the Expression of Genomic Imprinted Genes in Ovine Fetuses. Epigenetics 13: 793–807. 10.1080/15592294.2018.1503489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B., Tibshirani R. J., 1994. An Introduction to the Bootstrap, CRC Press, Boca Raton, FL. [Google Scholar]
- Giam M., Rancati G., 2015. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 10: 3 10.1186/s13008-015-0009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T., Monk M., 1998. Regulation of X–Chromosome Inactivation in Development in Mice and Humans. Microbiol. Mol. Biol. Rev. 62: 362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. A. M., 2016. Did sex chromosome turnover promote divergence of the major mammal groups? BioEssays 38: 734–743. 10.1002/bies.201600019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Walters J. R., 2017. Evolution of Sex Chromosome Dosage Compensation in Animals: A Beautiful Theory, Undermined by Facts and Bedeviled by Details. Genome Biol. Evol. 9: 2461–2476. 10.1093/gbe/evx154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Parisi M., Sturgill D., Nuttall R., Doctolero M., et al. , 2006. Global analysis of X-chromosome dosage compensation. J. Biol. 5: 3 10.1186/jbiol30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helena Mangs A., Morris B. J., 2007. The Human Pseudoautosomal Region (PAR): Origin, Function and Future. Curr. Genomics 8: 129–136. 10.2174/138920207780368141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Xie M., Chen W., Talbot R., Maddox J. F., et al. , 2014. The Sheep Genome Illuminates Biology of the Rumen and Lipid Metabolism. Science 344: 1168–1173. 10.1126/science.1252806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N., Fass J., 2011. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files.
- Julien P., Brawand D., Soumillon M., Necsulea A., Liechti A., et al. , 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10: e1001328 10.1371/journal.pbio.1001328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka S., Ahn H., Seo M., Kim H., Kim J. N., et al. , 2016. Status of dosage compensation of X chromosome in bovine genome. Genetica 144: 435–444. 10.1007/s10709-016-9912-3 [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K. K., Seidel G. E., Elsden R. P., 1985. Bovine Embryo Transfer Pregnancies. II. Lengths of Gestation. J. Anim. Sci. 61: 758–762. 10.2527/jas1985.614758x [DOI] [PubMed] [Google Scholar]
- Kondrashov F. A., Koonin E. V., 2004. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 20: 287–290. 10.1016/j.tig.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Lee J. T., 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 12: 815–826. 10.1038/nrm3231 [DOI] [PubMed] [Google Scholar]
- Lin H., Gupta V., Vermilyea M. D., Falciani F., Lee J. T., et al. , 2007. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 5: e326 10.1371/journal.pbio.0050326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Halsall J. A., Antczak P., O’Neill L. P., Falciani F., et al. , 2011. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno’s hypothesis. Nat. Genet. 43: 1169–1170, author reply 1171–1172. 10.1038/ng.992 [DOI] [PubMed] [Google Scholar]
- Lin F., Xing K., Zhang J., He X., 2012. Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc. Natl. Acad. Sci. USA 109: 11752–11757. 10.1073/pnas.1201816109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livernois A. M., Graves J. M., Waters P. D., 2012. The origin and evolution of vertebrate sex chromosomes and dosage compensation. Heredity 108: 50–58. 10.1038/hdy.2011.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani, J., J. Bézard, M. Devictor-Vuillet, and P. Mauléon, 1979 3H-thymidine labelling pattern of preleptotene chromosome condensation stages in the fetal sheep ovary. Ann. Biol. Anim. Biochim. Biophys. 19: 1241–1250.
- Lyon M. F., 1961. Gene Action in the X-chromosome of the Mouse (Mus musculus L.). Nature 190: 372–373. 10.1038/190372a0 [DOI] [PubMed] [Google Scholar]
- Mandric I., Temate-Tiagueu Y., Shcheglova T., Al Seesi S., Zelikovsky A., et al. , 2017. Fast bootstrapping-based estimation of confidence intervals of expression levels and differential expression from RNA-Seq data. Bioinformatics 33: 3302–3304. 10.1093/bioinformatics/btx365 [DOI] [PubMed] [Google Scholar]
- Mank J. E., Hosken D. J., Wedell N., 2014. Conflict on the Sex Chromosomes: Cause, Effect, and Complexity. Cold Spring Harb. Perspect. Biol. 6: a017715 10.1101/cshperspect.a017715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola M., Page J., Vasco C., Berríos S., Parra M. T., et al. , 2009. A High Incidence of Meiotic Silencing of Unsynapsed Chromatin Is Not Associated with Substantial Pachytene Loss in Heterozygous Male Mice Carrying Multiple Simple Robertsonian Translocations. PLoS Genet. 5: e1000625 10.1371/journal.pgen.1000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews F., Johnson P. J., Neil A., 2008. You are what your mother eats: evidence for maternal preconception diet influencing foetal sex in humans. Proc. Biol. Sci. 275: 1661–1668. 10.1098/rspb.2008.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill R., Tukey J. W., Larsen W. A., 1978. Variations of Box Plots. Am. Stat. 32: 12–16. [Google Scholar]
- Mi H., Muruganujan A., Casagrande J. T., Thomas P. D., 2013. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8: 1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. K., Disteche C. M., 2006. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 38: 47–53. 10.1038/ng1705 [DOI] [PubMed] [Google Scholar]
- Nicolae M., Mangul S., Măndoiu I. I., Zelikovsky A., 2011. Estimation of alternative splicing isoform frequencies from RNA-Seq data. Algorithms Mol. Biol. 6: 9 10.1186/1748-7188-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1966. Sex Chromosomes and Sex-Linked Genes, Springer-Verlag, Berlin, Heidelberg: 10.1007/978-3-662-35113-0 [DOI] [Google Scholar]
- Pessia E., Engelstädter J., Marais G. A. B., 2014. The evolution of X chromosome inactivation in mammals: the demise of Ohno’s hypothesis? Cell. Mol. Life Sci. CMLS 71: 1383–1394. 10.1007/s00018-013-1499-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessia E., Makino T., Bailly-Bechet M., McLysaght A., Marais G. A. B., 2012. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc. Natl. Acad. Sci. USA 109: 5346–5351. 10.1073/pnas.1116763109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S. M., Sereda N. H., Hoffman M. L., Valley E. V., Crenshaw T. D., et al. , 2016. Effects of Poor Maternal Nutrition during Gestation on Bone Development and Mesenchymal Stem Cell Activity in Offspring. PLoS One 11: e0168382 10.1371/journal.pone.0168382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S. M., Jones A. K., Hoffman M. L., McFadden K. K., Reed S. A., et al. , 2017. Fetal and organ development at gestational days 45, 90, 135 and at birth of lambs exposed to under- or over-nutrition during gestation. Trans. Anim. Sci 1: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2008. R: A language and environment for statistical computing.
- Ramsköld D., Wang E. T., Burge C. B., Sandberg R., 2009. An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data. PLOS Comput. Biol. 5: e1000598 10.1371/journal.pcbi.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T., Chowdhary B. P., 2015. The Eutherian Pseudoautosomal Region. Cytogenet. Genome Res. 147: 81–94. 10.1159/000443157 [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1996. Evolution of the Y Sex Chromosome in Animals. Bioscience 46: 331–343. 10.2307/1312947 [DOI] [Google Scholar]
- Sahakyan, A., Y. Yang, and K. Plath, 2018 The Role of Xist in X–Chromosome Dosage Compensation. Trends Cell Biol. S0962–8924: 30100–4. [DOI] [PMC free article] [PubMed]
- Saifi G. M., Chandra H. S., 1999. An apparent excess of sex- and reproduction-related genes on the human X chromosome. Proc. Biol. Sci. 266: 203–209. 10.1098/rspb.1999.0623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi M. N., Royo H., Mahadevaiah S. K., Ojarikre O., Bhaw L., et al. , 2017. Non-Canonical and Sexually Dimorphic X Dosage Compensation States in the Mouse and Human Germline. Dev. Cell 40: 289–301.e3. 10.1016/j.devcel.2016.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi M. N., Turner J. M. A., 2018. Mammalian X Chromosome Dosage Compensation: Perspectives From the Germ Line. BioEssays News Rev. Mol. Cell. Dev. Biol. 40: e1800024. [DOI] [PubMed] [Google Scholar]
- Soneson C., Love M. I., Robinson M. D., 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000 Res. 4: 1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D., Alexander G., Bell A. W., 1990. Effect of prolonged glucose infusion into fetal sheep on body growth, fat deposition and gestation length. J. Dev. Physiol. 13: 277–281. [PubMed] [Google Scholar]
- Tukiainen T., Villani A.-C., Yen A., Rivas M. A., Marshall J. L., et al. , 2017. Landscape of X chromosome inactivation across human tissues. Nature 550: 244–248. 10.1038/nature24265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas J. M., Kummerfeld S. K., Teichmann S. A., Luscombe N. M., 2009. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 10: 252–263. 10.1038/nrg2538 [DOI] [PubMed] [Google Scholar]
- Veitia R. A., Veyrunes F., Bottani S., Birchler J. A., 2015. X chromosome inactivation and active X upregulation in therian mammals: facts, questions, and hypotheses. J. Mol. Cell Biol. 7: 2–11. 10.1093/jmcb/mjv001 [DOI] [PubMed] [Google Scholar]
- Vermeesch J. R., Petit P., Kermouni A., Renauld J.-C., Van Den Berghe H., et al. , 1997. The IL-9 Receptor Gene, Located in the Xq/Yq Pseudoautosomal Region, Has an Autosomal Origin, Escapes X Inactivation and Is Expressed from the Y. Hum. Mol. Genet. 6: 1–8. 10.1093/hmg/6.1.1 [DOI] [PubMed] [Google Scholar]
- Wang F., Shin J., Shea J. M., Yu J., Bošković A., et al. , 2016. Regulation of X-linked gene expression during early mouse development by Rlim. eLife 5 10.7554/eLife.19127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., 2009. Ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- Wolf J. B., Bryk J., 2011. General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics 12: 91 10.1186/1471-2164-12-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. I., Davis A. W., 1993. Evolution of postmating reproductive isolation: the composite nature of Haldane’s rule and its genetic bases. Am. Nat. 142: 187–212. 10.1086/285534 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Chen X., Chen Z., Wang X., Shi S., et al. , 2010. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 42: 1043–1047. 10.1038/ng.711 [DOI] [PubMed] [Google Scholar]
- Xue F., Tian X. C., Du F., Kubota C., Taneja M., et al. , 2002. Aberrant patterns of X chromosome inactivation in bovine clones. Nat. Genet. 31: 216–220. 10.1038/ng900 [DOI] [PubMed] [Google Scholar]
- Zechner U., Wilda M., Kehrer-Sawatzki H., Vogel W., Fundele R., et al. , 2001. A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution? Trends Genet. TIG 17: 697–701. 10.1016/S0168-9525(01)02446-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available in the Gene Expression Omnibus and BioProject:
GSE111306: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111306
PRJNA254105: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA254105
PRJEB6169: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB6169
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7221467.