Abstract
In covariate (sub)models of population pharmacokinetic models, most covariates are normalized to the median value; however, for body weight, normalization to 70 kg or 1 kg is often applied. In this article, we illustrate the impact of normalization weight on the precision of population clearance (CLpop) parameter estimates. The influence of normalization weight (70, 1 kg or median weight) on the precision of the CLpop estimate, expressed as relative standard error (RSE), was illustrated using data from a pharmacokinetic study in neonates with a median weight of 2.7 kg. In addition, a simulation study was performed to show the impact of normalization to 70 kg in pharmacokinetic studies with paediatric or obese patients. The RSE of the CLpop parameter estimate in the neonatal dataset was lowest with normalization to median weight (8.1%), compared with normalization to 1 kg (10.5%) or 70 kg (48.8%). Typical clearance (CL) predictions were independent of the normalization weight used. Simulations showed that the increase in RSE of the CLpop estimate with 70 kg normalization was highest in studies with a narrow weight range and a geometric mean weight away from 70 kg. When, instead of normalizing with median weight, a weight outside the observed range is used, the RSE of the CLpop estimate will be inflated, and should therefore not be used for model selection. Instead, established mathematical principles can be used to calculate the RSE of the typical CL (CLTV) at a relevant weight to evaluate the precision of CL predictions.
Electronic supplementary material
The online version of this article (10.1007/s40262-018-0652-7) contains supplementary material, which is available to authorized users.
Key Points
| Normalization to a weight outside the observed weight range (e.g. 70 kg normalization in a paediatric study) can increase the uncertainty of parameter estimates in pharmacokinetic covariate models. |
| The predictive performance of pharmacokinetic models and their covariate submodels is unaffected by weight normalization. |
| When normalizing outside the observed covariate range, the RSEs of the corresponding population estimates should generally not be used for model evaluation. The RSE of the typical parameter at a relevant covariate value can be used instead. |
Introduction
In population pharmacokinetic modelling, covariate models are built to describe between-subject variability in pharmacokinetic parameters (e.g. clearance [CL]) based on patient information (e.g. weight) [1, 2]. These covariate models can then be used to support personalized pharmacotherapy [3]. One of the most commonly identified covariates in population pharmacokinetic models is the body weight of the patient [4–8]. The relationship between drug CL and weight is often described using a power function:
| 1 |
| 2 |
where CLTV represents the predicted CL for a typical individual with weight equal to WTi, CLpop represents the population estimate of CL for an individual with a weight equal to the normalization weight (WTnorm), WTi represents the individual’s weight, EXPWT represents the exponent that characterizes the influence of weight on CL, CLi represents the individual post hoc predictions of CL for individual i, and eETA represents the post hoc estimate of the deviation of individual i from the predicted CL of a typical individual.
EXPWT in this weight-based Eq. 1 can be fixed a priori (for instance to 1, 0.75 or 0.67) or estimated [9–12]. For continuous covariates other than weight, it is common to normalize to the mean or the median covariate value of the covariate in the dataset, whereas for weight, normalization to 70 kg is often chosen [2, 13–15]. The rationale for this approach is that the estimate of CLpop will then represent the value of a typical 70 kg adult, which can easily be compared with other (adult) studies [2, 5, 10]. For similar reasons, a WTnorm of 1 kg can be chosen or, when no explicit normalization is performed, this normalization is implicitly chosen [9]. Recently, Mahmood and Tegenge investigated the impact of 70 and 1 kg weight normalization on CLTV predictions and concluded that weight normalization has no impact on CLTV predictions [9].
The concept of normalization has been extensively studied in linear regression [16, 17]. When normalizing the data to the mean, the relative standard error (RSE) of the intercept term is minimized, while normalizing outside of the data range can result in estimates with poor precision [16]. In this context, the power function in Eq. 1 can be considered a linear model in the log domain, with an intercept of log CLpop [13]. If the same concepts apply, we might expect the RSE of the estimate of CLpop to be minimal when normalizing weight to the geometric mean. Alternatively, normalizing to 1 kg or 70 kg might result in high RSE of CLpop, especially in populations with high or low weights.
As the impact of 70 kg normalization on the RSE of CLpop could be considerable, especially when analysing data from neonatal or morbidly obese patients, insight into the statistical consequences of the selected normalization weight seems important among those involved in population pharmacokinetic modelling. In this article, we provide mathematical derivations of the phenomena and illustrate the impact of weight normalization on the precision of the parameter estimate of CLpop. For this, we used an existing neonatal pharmacokinetic dataset, as well as simulated data of various paediatric and obese populations.
Methods
Case Study
A dataset from a previously published population pharmacokinetic analysis of phenobarbital in term and preterm neonates was used [18]. This dataset contained phenobarbital plasma concentrations collected during therapeutic drug monitoring from 53 neonates up to 80 h after the last phenobarbital dose. The weight of these neonates ranged from 0.45 to 4.5 kg and had a median value of 2.7 kg [18].
These data were modelled using a one-compartment model with interindividual variability on CL and volume of distribution (V) and a proportional error model. The covariate model consisted of a linear model (exponent fixed to a value of 1) for weight on V and a power model with an estimated exponent for weight on CL (Eq. 1). For three different values of WTnorm (1, 2.7, and 70 kg), parameter estimates were obtained using NONMEM 7.3 [19]. The collinearity of the parameter estimates was assessed using the condition number, which is defined as the square root of the ratio between the largest and smallest eigenvalue of the correlation matrix. RSEs of the parameter estimates (i.e., CLpop and EXPWT) in NONMEM were obtained using two different methods: calculation from the estimated variance–covariance matrix in NONMEM, and calculation calculated from 1000 bootstrap runs, which were performed using PsN 4.2.0. for each of the three models [20].
Furthermore, from these 1000 bootstrap runs, the estimates of CLpop and EXPWT (see Eq. 1) were used to calculate the predicted function of CLTV over weight for each of the bootstrap runs over a weight range of 0.5–200 kg. For each of the three weight-normalized models, we used these functions of CLTV over weight to obtain the 95% confidence interval and RSE of CLTV predictions over the weight range of 0.5–200 kg for all three values of WTnorm.
RSE of CLTV predictions over weight were also calculated using the variance–covariance matrix obtained from NONMEM [13]. For this, we log-transformed the power function of Eq. 1, which resulted in the following linear model:
| 3 |
where LogCLpop represents the estimate of the natural logarithm of CL of an individual whose weight is equal to the normalization weight. With a linear model, we can use principles from linear regression to calculate the RSE of the predictions of CLTV at an arbitrary weight WTi:
| 4 |
| 5 |
where VARLogCLpop represents the variance of the estimate of LogCLpop, represents the covariance between the estimates for LogCLpop and EXPWT, and VAREXPwt represents the variance of the estimate of EXPWT. Mathematical derivation of Eqs. 4 and 5 are supplied in the electronic supplementary material.
Impact of WTnorm on the Relative Standard Error of CLpop for Different Weight Distributions
To further study the impact of WTnorm in a covariate function on the RSE of the estimate of CLpop with different weight distributions, we generated pharmacokinetic datasets of patient populations with six different weight distributions in R version 3.3.2. We then fitted a one-compartmental pharmacokinetic model described in the previous subsection, re-estimating all parameters.
For this purpose, three paediatric weight distributions (PEDIAT1, PEDIAT2 and PEDIAT3) and three weight distributions of adult populations including obese patients (OBESE1, OBESE2 and OBESE3) were used to generate simulated datasets (Table 2).
Table 2.
Characteristics of the different weight distributions and summary of the simulation results
| Distribution | Geometric mean (kg) | SD on log-scale | Distance between geometric mean and 70 kg (in SD on log-scale) | Median RSE ratio CLpop (Eq. 6) for 70 kg normalization | Covariance step successful with 70 kg normalization (%) | Covariance step successful with geometric mean normalization (%) | Covariate step successful in both normalizations (%) |
|---|---|---|---|---|---|---|---|
| PEDIAT1 Log-normal |
20 | 0.25 | 5.0 | 4.3 | 72 | 57 | 54 |
| PEDIAT2 Uniform 10–32 kg |
20 | 0.33 | 3.9 | 3.5 | 79 | 63 | 59 |
| PEDIAT3 Uniform 1–51 kg |
20 | 0.83 | 1.5 | 1.7 | 85 | 85 | 84 |
| OBESE1 Uniform 110–220 |
162 | 0.2 | − 4.2 | 3.5 | 31 | 32 | 28 |
| OBESE2 Uniform 80–160 |
118 | 0.2 | − 2.6 | 2.3 | 47 | 48 | 45 |
| OBESE3 Uniform 45–160 |
97 | 0.35 | − 0.9 | 1.1 | 52 | 53 | 50 |
RSE relative standard error, SD standard deviation, CLpop population clearance
For each weight distribution, 250 datasets consisting of 50 patients were randomly sampled from the distributions using R software. For each patient, concentrations were simulated for 24, 72 and 120 h after a single 10 mg/kg dose. The simulated datasets were fitted with the models described in Sect. 2.1 and weight was normalized to either 70 kg or the expected geometric mean of the weight distribution. Geometric mean was chosen as we hypothesized that this would result in the minimum RSE of CLpop, as discussed in the Introduction. The proportion of successful covariance steps was compared for a statistically significant difference between the different normalization strategies, using a two-sample test for equality of proportions with continuity correction (prop.test function in R). For datasets that yielded successful covariance steps in both model fits, we calculated the ratio of the RSEs for the estimate of CLpop of both model fits:
| 6 |
Results
Case Study
Table 1 shows that only the RSE of the estimate of CLpop varied with normalization weight. Normalizing to median weight (2.7 kg) resulted in a lower RSE of CLpop, compared with 1 kg and 70 kg normalization. RSEs were 10.6, 8.0, and 48.2% for 1, 2.7 and 70 kg, respectively (Table 1). These RSE values were obtained from NONMEM’s covariance step, but similar results were obtained using a bootstrap (1 kg: 10.5%; 2.7 kg: 8.1%; 70 kg: 48.8%). Additionally, there was a stronger correlation between the uncertainty of the parameter estimates of CLpop and EXPWT when normalizing to 1 kg or 70 kg (Table 1). Finally, increased collinearity between the parameters was observed for the 1 kg and 70 kg normalizations, as identified by a higher condition number (Table 1).
Table 1.
Parameter estimates and relative standard errors (%) from the NONMEM covariance step for the neonatal dataset using different normalization weights
| WTnorm | 1 kg | 2.7 kg (median) | 70 kg |
|---|---|---|---|
| OFV | 1091 | 1091 | 1091 |
| CLpop (L/h) | 0.00615 (10.6%) | 0.0119 (8.0%) | 0.104 (48.2%) |
| V (L) | 2.37 (4.4%) | 2.37 (4.4%) | 2.37 (4.4%) |
| EXPWT | 0.665 (20.3%) | 0.665 (20.3%) | 0.665 (20.3%) |
| Proportional error [%] | 2.89 (23.5%) | 2.89 (23.5%) | 2.89 (23.5%) |
| Condition number | 4.4 | 2.8 | 16.2 |
| CorrelationCLpop, EXPwta | − 0.840 | 0.545 | 0.988 |
The parameter estimates of CLpop varied with normalization weight (Table 1), which results from the fact that this parameter represents the typical CL of a subject whose weight is equal to the normalization weight. However, the same predicted CLTV (Eq. 1) is obtained for the three model fits with different normalization weights since
This is further illustrated by Fig. 1 where the results on predicted CLTV for the model with a normalization weight of 2.7 kg is shown, while the models with different normalization weights produced equivalent results. Additionally, Fig. 1a shows that the 95% bootstrap confidence interval of predicted CLTV broadens the further the weight moves away from the centre of the weight distribution of the patient population in the dataset. Figure 1b illustrates that the CLTV functions from different bootstrap samples are very similar within the range of the data, and diverge outside the weight range of the original dataset, explaining the broader confidence intervals seen in Fig. 1a.
Fig. 1.
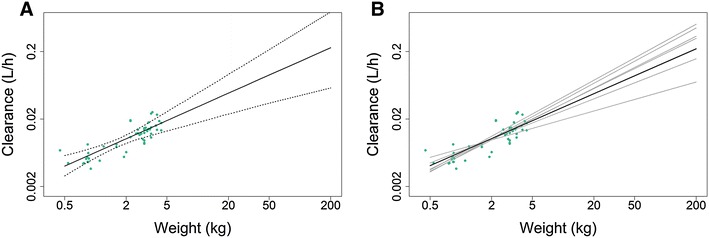
Clearance predictions versus weight (0.5–200 kg) in an example neonatal dataset. (a) Median (solid black line) and 95% confidence interval (dotted line) of 1000 functions of CLTV versus weight obtained from 1000 bootstrap runs; green dots represent the individual post hoc CLi estimates of the studied patients. (b) Estimated function of CLTV versus weight from the original dataset (solid black line) and illustrative set of functions of CLTV versus weight (grey solid lines) obtained in six (of 1000) separate bootstrap runs; green dots represent the individual post hoc CLi estimates of patients in the original dataset. Depicted results were obtained using a normalization weight of 2.7 kg. CLTV clearance for a typical individual, CLi clearance for individual i
Figure 2 shows the RSE of the predicted function of CLTV over weight, as well as the RSE of the estimate of the parameter CLpop. The RSE of the CLTV function was calculated using either bootstrap or the variance–covariance matrix (Eq. 5), with both methods resulting in similar results (Fig. 2). Using Eq. 5, the minimum RSE of CLTV (5.8%) was calculated at 1.9 kg, which is close to, but not equal to, measures of the central tendency of the weight distribution, such as the mean (2.4 kg, RSE = 6.6%), median (2.7 kg, RSE = 7.5%) or geometric mean (2.1 kg, RSE = 6.0%). The RSE of the estimate of the CLpop parameter reported by NONMEM for a model with a given normalization weight matches the RSE of the CLTV function at the normalization weight. The former results in 6.4% RSE with 1.9 kg normalization, and 8.0% RSE with normalization to the median.
Fig. 2.
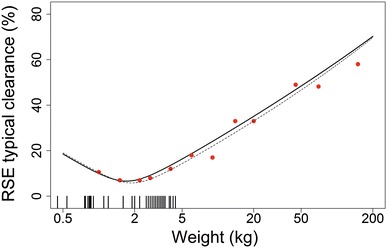
Relation between weight and the RSE of both CLTV and CLpop in an illustrative neonatal dataset. The solid line represents the RSE of CLTV predictions from 1000 bootstrap runs, the dotted line represents the RSE of CLTV predictions obtained from the variance–covariance matrix (Eq. 5), and the red dots represent the RSE of the estimated CLpop parameter using the corresponding normalization weight, obtained from the covariance step of a single NONMEM run. The vertical tick marks on the bottom of the graph depict the body weights of subjects in the dataset. RSE relative standard error, CLTV clearance for a typical individual, CLpop population clearance
Impact of 70 kg Normalization in Different Paediatric and Obese Populations
Evaluation of the impact of 70 kg normalization on the RSE of CLpop estimates for simulated paediatric and obese datasets with various weight distributions showed that, generally, 70 kg normalization resulted in a higher RSE of the CLpop estimate compared with normalization to geometric mean weight, resulting in an RSE ratio above 1 (Table 2, Fig. 3). The results show that the degree of impact of 70 kg normalization depends on the weight distribution in the dataset. The three paediatric weight distributions had a geometric mean weight of 20 kg, but different dispersion around the geometric mean. The RSE ratio was highest (median RSE ratio = 4.3) for the log-normal distribution with a standard deviation on the logarithmic scale of 0.25 (PEDIAT1).
Fig. 3.
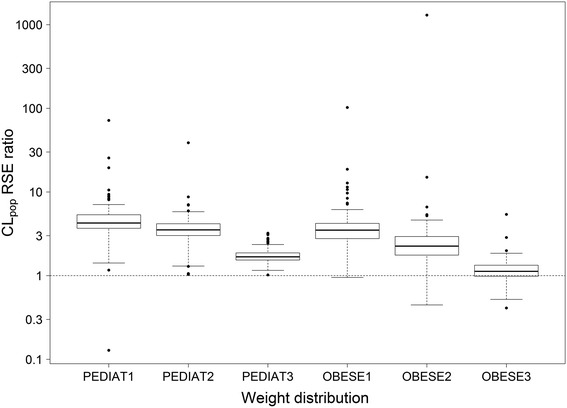
RSE ratio of CLpop (Eq. 6) when using 70 kg normalization compared with geometric mean weight normalization. For each weight distribution, 250 datasets were generated and refitted. Only results of datasets for which the covariance step was successful for both the 70 kg and geometric mean weight normalization were included in this graph (Table 2). RSE relative standard error, CLpop population clearance
The weight distributions of OBESE1 and OBESE2 have a similar standard deviation on a logarithmic scale, but different geometric means of the weight (162 and 118 kg, respectively). The results in Table 2 and Fig. 3 show that this results in a lower RSE ratio in OBESE2, compared with OBESE1 (median RSE ratio of 2.3 and 3.5, respectively). Normalizing to 70 kg has only marginal impact on RSE in OBESE3 (median RSE ratio = 1.1), with a geometric mean that is closer to 70 kg than OBESE2 (97 kg vs. 118 kg), as well as a higher standard deviation on a logarithmic scale (0.35 vs. 0.2).
Table 2 shows that for both paediatric and adult populations, the impact of 70 kg normalization on the RSE of the estimate of CLpop was highest for studies with a narrow weight range (low standard deviation on a logarithmic scale) and a median weight away from 70 kg.
The percentage of datasets for which a successful covariance step could be obtained in both model fits ranged from 28 to 84% in the different scenarios. The proportion of successful covariance steps was significantly higher using geometric mean normalization compared with 70 kg normalization in scenarios PEDIAT1 (p < 0.001) and PEDIAT2 (p < 0.001), but not in any of the other scenarios. The most common cause of the missing covariance steps was boundary issues due to the difficulty in estimating the variance for the interindividual variability of CL.
Discussion
This report illustrates the statistical principle that, when estimating the exponent in a body weight-based covariate submodel of a population pharmacokinetic model (Eq. 1), the use of a normalization weight outside the observed weight range can result in an inflated RSE of the estimate of CLpop. This holds true for 70 kg normalization, but also for 1 kg normalization (which is sometimes referred to as ‘no normalization’) [9].
The RSE of the CL parameter CLpop represents the RSE of the predicted typical CL (CLTV) at a particular normalization weight, and is therefore not a universal measure for the precision of the estimate of CL (Table 2). As we show, the RSE of the predicted CLTV is not constant, but dependent on the weight of the subject for whom CLTV is predicted (Figs. 1 and 2). This means that the RSE of CLpop represents how precisely CL can be estimated for a subject at the applied normalization weight. When estimating CLpop at a normalization weight outside the observed weight range, the RSE of CLpop will be inflated and cannot be used as a criterion for model selection. Our example with 70 kg normalization in a neonatal pharmacokinetic model showed, for instance, that the RSE of the CLpop estimate increased sixfold compared with the estimate obtained with normalization to the median.
In linear regression, the minimum RSE is obtained by normalizing to the mean value. Because the power function becomes a linear model in the log domain, one might expect the minimum RSE at the geometric mean weight as this is equivalent to normalizing to the mean value of log of weight. The results of the case study show that this is not necessarily the case for covariate models of non-linear mixed-effects models. In our case study, we found that the minimum RSE of CLpop was obtained by normalizing to 1.9 kg, rather than the mean, median or geometric mean weight. The normalization weight with minimum RSE of CLpop can be predicted by minimizing Eq. 5, although this does require that an initial model with a test normalization weight is run to obtain an estimate of the variance–covariance matrix. In our case study, we normalized to the median weight as this is the most commonly used normalization weight. This increased the RSE of CLpop to 8.0%, from the minimum RSE of 6.4% at a normalization weight of 1.9 kg. However, normalizing to the median weight will likely be fit-for-purpose in most cases.
Regardless of the normalization weight that is used, Eqs. 4 and 5 can be used to calculate RSE values for any given body weight based on a variance–covariance matrix. This can be useful as normalization to 70 kg as a ‘standardized individual’ is sometimes advocated to improve comparisons of results from different studies [5]. In these cases, normalization to 70 kg can applied and Eqs. 4 and 5 can be used to calculate the RSE values for a relevant body weight to enable its use as a model selection criterion. Alternatively, model estimation can be performed with median weight normalization, and both the estimated CLpop parameter for the median weight and the derived CLTV for a 70 kg individual are reported, in which case the calculated RSE of the latter is relevant when comparing results from different studies.
The expected increase in RSE of centring to 70 kg is dependent on both the variance of the weight distribution and the distance of the mean of the distribution from the chosen normalization weight (Fig. 3). The effect seems to be largest in cases of narrow distributions with a mean covariate value far away from the centre of the data. This especially holds true for (pre)term neonates, infants and young children and (morbidly) obese patients. In cases where the population mean is away from 70 kg but the range of weights includes 70 kg (such as scenario OBESE3), normalizing to 70 kg will likely result in an RSE increase of CLpop that will not affect its application in model selection (Fig. 3).
It is important to realize that weight normalization only impacts the precision of the CLpop parameter estimate, in case a covariate model according to Eq. 1 is chosen. If the exponent is fixed, the RSE of the estimate of CLpop will be unaffected by normalization weight. Similarly, in this situation, the RSE of the predicted CLTV will be independent of the subject’s weight. Whether or not the exponent should be estimated or fixed is an ongoing discussion that is outside the scope of this paper [5, 11, 12].
In the simulation study, the calculation of the RSE ratio required that both model fits resulted in a successful covariance step. This requirement introduces the potential for selection bias as the results from the excluded datasets might have had a different impact of normalization on the RSE of CLpop. However, performing a bootstrap on each of the 3000 model fits to obtain the RSE ratio independent of covariance step success was not feasible due to its computational demands.
Interestingly, the simulation study showed that in two of the six simulated scenarios, a higher percentage of runs with successful covariance steps was obtained when using geometric mean normalization instead of 70 kg normalization. Additionally, the paediatric case study showed that with normalization outside the observed weight range, collinearity between the CLpop and EXPWT parameter estimates is increased (Table 1). This suggests that normalization affects the stability of the parameter estimation process, which has been described for both linear models, as well as population pharmacokinetic models [15]. Although the increased stability was not observed in all simulated scenarios, it might be a reason to advocate median weight normalization over normalization outside the observed weight range.
Conclusions
Normalizing body weight-based covariate relationships in population pharmacokinetic models to 1 kg or 70 kg can inflate the RSE of the parameter estimate of CLpop in population pharmacokinetic models. The predictive performance of the models are unaffected by normalization. However, when normalizing with a weight outside the observed weight range, the CLpop RSE represents the precision of the CLTV at this extrapolated weight, and this value should therefore not be used for model selection. Instead, the precision of CLTV at a relevant weight value can be calculated from the covariance matrix.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the researchers who provided the data used for the case study in this paper, most notably Dr. Leo M. Stolk and Dr. Sinno H. P. Simons.
Funding
Catherijne A. J. Knibbe was supported by the Innovational Research Incentives Scheme of the Dutch Organization for Scientific Research (NWO, Vidi grant, May 2013).
Conflicts of interest
Sebastiaan C. Goulooze, Swantje Völler, Pyry A. J. Välitalo, Elisa A. M. Calvier, Leon Aarons, Elke H.J. Krekels, and Catherijne A.J. Knibbe have no conflicts of interest that are relevant to the content of this paper.
Footnotes
Sebastiaan Goulooze and Swantje Völler contributed equally to this work.
References
- 1.Mould DR, Upton RN. Basic concepts in population modeling, simulation, and model-based drug development. CPT Pharmacometrics Syst Pharmacol. 2012;1:e6. doi: 10.1038/psp.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mould DR, Upton RN. Basic concepts in population modeling, simulation, and model-based drug development—part 2: introduction to pharmacokinetic modeling methods. CPT Pharmacometrics Syst Pharmacol. 2013;2:e38. doi: 10.1038/psp.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krekels EHJ, van Hasselt JGC, van den Anker JN, Allegaert K, Tibboel D, Knibbe CAJ. Evidence-based drug treatment for special patient populations through model-based approaches. Eur J Pharm Sci. 2017;109S:S22–S26. doi: 10.1016/j.ejps.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Anderson BJ, Allegaert K, Holford NH. Population clinical pharmacology of children: modelling covariate effects. Eur J Pediatr. 2006;165(12):819–829. doi: 10.1007/s00431-006-0189-x. [DOI] [PubMed] [Google Scholar]
- 5.Anderson BJ, Holford NH. Mechanism-based concepts of size and maturity in pharmacokinetics. Annu Rev Pharmacol Toxicol. 2008;48:303–332. doi: 10.1146/annurev.pharmtox.48.113006.094708. [DOI] [PubMed] [Google Scholar]
- 6.Shi R, Derendorf H. Pediatric dosing and body size in biotherapeutics. Pharmaceutics. 2010;2(4):389–418. doi: 10.3390/pharmaceutics2040389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knibbe CA, Brill MJ, van Rongen A, Diepstraten J, van der Graaf PH, Danhof M. Drug disposition in obesity: toward evidence-based dosing. Annu Rev Pharmacol Toxicol. 2015;55:149–167. doi: 10.1146/annurev-pharmtox-010814-124354. [DOI] [PubMed] [Google Scholar]
- 8.Eleveld DJ, Proost JH, Absalom AR, Struys MM. Obesity and allometric scaling of pharmacokinetics. Clin Pharmacokinet. 2011;50(11):751–753. doi: 10.2165/11594080-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Mahmood I, Tegenge MA. Population pharmacokinetics: some observations in pediatric modeling for drug clearance. Clin Pharmacokinet. 2017;56(12):1567–1576. doi: 10.1007/s40262-017-0542-4. [DOI] [PubMed] [Google Scholar]
- 10.Holford NH. A size standard for pharmacokinetics. Clin Pharmacokinet. 1996;30(5):329–332. doi: 10.2165/00003088-199630050-00001. [DOI] [PubMed] [Google Scholar]
- 11.Fisher DM, Shafer SL. Allometry, Shallometry! Anesth Analg. 2016;122(5):1234–1238. doi: 10.1213/ANE.0000000000001257. [DOI] [PubMed] [Google Scholar]
- 12.Calvier EA, Krekels EH, Valitalo PA, Rostami-Hodjegan A, Tibboel D, Danhof M, et al. Allometric scaling of clearance in paediatric patients: when does the magic of 0.75 fade? Clin Pharmacokinet. 2017;56(3):273–285. doi: 10.1007/s40262-016-0436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Jadhav PR, Lala M, Gobburu JV. Clarification on precision criteria to derive sample size when designing pediatric pharmacokinetic studies. J Clin Pharmacol. 2012;52(10):1601–1606. doi: 10.1177/0091270011422812. [DOI] [PubMed] [Google Scholar]
- 14.Joerger M. Covariate pharmacokinetic model building in oncology and its potential clinical relevance. AAPS J. 2012;14(1):119–132. doi: 10.1208/s12248-012-9320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonate PL. Pharmacokinetic-pharmacodynamic modeling and simulation. 2. New York: Springer; 2011. [Google Scholar]
- 16.Snee RD, Marquardt DW. Comment: collinearity diagnostics depend on the domain of prediction, the model, and the data. Am Stat. 1984;38(2):83–87. [Google Scholar]
- 17.Belsley DA. Demeaning conditioning diagnostics through centering. Am Stat. 1984;38(2):73–77. [Google Scholar]
- 18.Voller S, Flint RB, Stolk LM, Degraeuwe PLJ, Simons SHP, Pokorna P, et al. Model-based clinical dose optimization for phenobarbital in neonates: an illustration of the importance of data sharing and external validation. Eur J Pharm Sci. 2017;109S:S90–S97. doi: 10.1016/j.ejps.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 19.Boeckmann AJ, Sheiner LB, Beal SL. NONMEM users guide—part VIII. Ellicott City: ICON Development Solutions; 2010. p. 220. [Google Scholar]
- 20.Keizer RJ, Karlsson MO, Hooker A. Modeling and simulation workbench for NONMEM: tutorial on Pirana, PsN, and Xpose. CPT Pharmacometrics Syst Pharmacol. 2013;2:e50. doi: 10.1038/psp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


