Abstract
The question whether and which nonhuman peptides or proteins are present in human milk was raised many decades ago. However, due to cross-reactivity or nonspecific antibody recognition, the accuracy of detection by immunochemical methods has been a concern. Additionally, the relative low-abundance of nonhuman peptides/proteins in the complex milk sample makes them a challenging target to detect. Here, by deep proteome profiling, we detected several nonhuman peptides, which could be grouped as nonhuman proteins. We next estimated their concentration in human milk by combining data-dependent shotgun proteomics and parallel reaction monitoring. First, we fractionated human milk at the protein level and were able to detect 1577 human proteins. Additionally, we identified 109 nonhuman peptides, of which 71 were grouped into 36 nonhuman proteins. In the next step, we targeted 37 nonhuman peptides and nine of them could be repeatedly quantified in human milk samples. Peptides/proteins originating from bovine milk products were the dominant nonhuman proteins observed, notably bovine caseins (α-S1-, α-S2-, β-, κ-caseins) and β-lactoglobulin. The method we present here can be expanded to investigate more about nonhuman peptides and proteins in human milk and give a better understanding of how human milk plays a role in allergy prevention.
Keywords: nonhuman peptides, nonhuman proteins, shotgun proteomics, parallel reaction monitoring, mass spectrometry, human milk
Introduction
At the molecular level, human milk is a complex mixture where the composition reflects the secretory activity of the mammary gland.1 Proteins in milk are either synthesized by mammary cells and released by exocytotic fusion or are transported to the mammary gland by transcytosis from blood plasma.2 Besides human proteins, the passage of some nonhuman proteins in human milk was reported many decades ago.3 Many nonhuman proteins were detected in human milk using immunoassays, especially proteins from cow’s milk,4−9 eggs,4,8,10−12 peanuts,13,14 and wheat,15 which were targeted due to their potential roles in food allergy and breastfeeding. Kilshaw and Cant found out by using solid-phase radioimmunoassays that the levels of β-lactoglobulin, ovalbumin, and ovomucoid in human milk and serum samples were increased after ingestion of eggs and cow’s milk.4 Several studies have used various enzyme-linked immunosorbent assays (ELISA) after consumption of food containing allergens to demonstrate that nonhuman proteins are detectable in human milk. For example, β-lactoglobulin was detected from 3 h to 7 days after consumption of 240 mL of cow milk with variation in each individual.9 In another study, within 6 h of eating one cooked egg, a dose–response correlation of ovalbumin content in human milk was noticed.10 Also, several peanut allergens were detectable after eating gram amounts of peanuts. For instance, Ara h1 and Ara h2 were detected in human milk after consumption of 50 g of dry roasted peanuts.13 Ara h6 was also found in human milk after eating 30 g of commercial roasted peanuts.14 Additionally, components of gluten such as nondegraded gliadins and their immune complexes with IgA were observed in human milk.15 This may indicate an interplay between food antigens and immune complexes which potentially then can play a role in the immune development of the infant. In summary, all these studies suggest that there is a relationship between nonhuman proteins in human milk and maternal diet, although individual differences are quite noticeable. Moreover, what this means for the infant’s developing immune system is not well understood and needs to be further investigated.
A major limitation of studies investigating nonhuman proteins in human milk is the accuracy of detection by immunochemical methods. These methods are of concern since cross-reactivity or nonspecific antibody recognition have been noticed in many studies. Such problems have been observed during testing of rabbit antibodies against bovine β-lactoglobulin and human lactoferrin16 or searching for specific IgEs binding to cow milk proteins versus human milk proteins.17 The cause for this extensive cross-reactivity is mainly due to the shared epitope between nonhuman proteins and their human equivalents.18−20 Therefore, there is a need for more sensitive and accurate assays allowing for a reliable distinction between human and nonhuman proteins in human milk.
The reported observations of nonhuman proteins in human milk has led to ongoing controversial debates, arising from mistrust in the methods commonly used for evidencing the existence of these nonhuman proteins. To overcome these inherent issues of immunochemical methods for determining nonhuman proteins in human milk, mass spectrometry (MS), especially shotgun proteomics methods, have been introduced.21−24 Bovine α-S1-casein was found in both term and preterm colostrum via two-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (2D-SDS PAGE) separation and subsequent LC-MS/MS analysis of the in-gel digested tryptic peptides observing bovine α-S1-casein sequence unique peptides.21,22 Additionally, LC-MS/MS analyses have identified unique peptides from bovine β-lactoglobulin and α-S1-casein from the milk of mothers receiving daily bovine milk, while those peptides were not detected in milk from mothers on a dairy-free diet. On the other hand, not all samples from mothers consuming daily bovine milk contained those peptides.23 The peanut allergen Ara h2 was detected via a combination of immunoaffinity enrichment and LC-MS/MS analysis and then quantified by a competitive inhibition ELISA after consumption of 100 g of dry roasted peanuts.24 These studies demonstrate that LC-MS/MS methods alone, or in combination with immunochemical methods, are suitable to assess nonhuman peptides/proteins in human milk. However, most of these methods remain semiquantitative at best and may not be reliable for detecting low concentrations of nonhuman proteins in human milk.
Shotgun proteomics is frequently applied for discovery experiments in a data-dependent acquisition (DDA) mode, which nowadays enables the identification of thousands of proteins in biological samples.25,26 In this setup, a number of precursor ions are automatically selected for MS/MS fragmentation based on their signal intensity in a preceding survey scan. Although the DDA mode is very powerful for indentifying a large number of proteins, low abundance analytes are usually under-represented, especially in complex biological samples.27 In contrast, targeted proteomics methods allow the consistent monitoring of peptides of interest with a high degree of specificity and sensitivity. In parallel reaction monitoring (PRM) mode, predefined precursors are sequentially measured, and all ions resulting from the fragmentation of a single, or several, precursor ions are measured simultaneously in one MS/MS scan. The trapping capabilities of a quadrupole-Orbitrap instrument significantly improve the analysis of low abundance peptides.28 The combination of shotgun proteomics and targeted proteomics allows comprehensive and accurate analysis in biological samples.
The aim of this study was to address the question whether (and which) low-abundant nonhuman peptides/proteins are present in human milk. By using in-gel separation to prefractionate the milk proteome and shotgun proteomics, we were able to cumulatively identify 1577 human proteins and 109 nonhuman peptides, of which 71 were grouped as 36 nonhuman proteins. We then developed PRM assays with spiked stable isotope-labeled standards of 37 nonhuman peptides and unambiguously detected and reproducibly quantified nine nonhuman peptides originating from six bovine/bovidae proteins. For all the nonhuman proteins found in human milk samples, proteins which are likely from bovine milk products were the dominant nonhuman proteins, such as bovine caseins (α-S1-, α-S2-, β-, κ-caseins) and β-lactoglobulin, for which we detected numerous unique peptides.
Materials and Methods
Chemicals and Materials
Unless otherwise specified, all chemicals and reagents were obtained from Sigma-Aldrich (Steinheim, Germany). Formic acid (FA) was from Merck (Darmstadt, Germany). Acetonitrile (ACN) was purchased from Biosolve (Valkenswaard, The Netherlands). Sequencing grade trypsin was obtained from Promega (Madison, WI). The 12% Criterion XT Bis-Tris Protein Gel and SDS-PAGE sample buffer were purchased from Bio-Rad (Hercules, CA, US). Pierce peptide retention time calibration mixture (PRTC), BenchMark ProteinLadder, and GelCold Blue Stain Reagent were from Thermo Fisher Scientific (Bremen, Germany). The Sep-Pak C18 vac cartridge was purchased from Waters (Massachusetts, USA). Milli-Q was produced by an in-house system (Millipore, Billerica, MA). The stable isotope-labeled peptides (crude SpikeTides_L) were synthesized by JPT Peptide Technologies (Berlin, Germany).
Human Milk Sample Collection
Human milk samples (n = 6) were obtained from milk collections forming part of the MediDiet study from healthy Italian mothers aged between 25 and 40 in a period between 5 and 7 weeks postpartum. All samples were collected from women delivering at term childbirth in the hospital center. Mothers were instructed to wash their hands carefully with soap and water before providing milk by breast pump into sterile hard plastic containers. The milk samples were stirred and aliquoted into smaller sterile plastic containers, overlaid with nitrogen gas, and tightly closed within 3 min. Samples were stored at −80 °C until analysis. This trial was registered in the Dutch Trial Register (www.trialregister.nl; NTR3468).
Skimmed Milk Separation and Proteolytic Digestion
For each individual, an aliquot of 300 μL of a thawed whole milk sample was centrifuged at 1000g for 1 h at 4 °C to separate the upper-fat layer and lower skimmed milk. The fat layer was discarded, and protein concentration in skimmed milk was measured by Bradford assays using bovine serum albumin (BSA) as the standard. For data-dependent shotgun analysis, a total of 25 μg of protein was reduced with 25 mM dithiothreitol (DTT), alkylated with 75 mM iodoacetamide (IAA) followed by SDS-PAGE fractionation on a 12% Criterion XT Bis-Tris protein gel at 70 mA with a BenchMark ProteinLadder. The gel was fixed with 40% ethanol/10% acetic acid for 30 min, stained with GelCold Blue Stain reagent for 1 h, and destained with 1% acetic acid overnight. A gel of each sample was excised in six bands in a similar manner and digested using trypsin (1:50, w/w skimmed milk proteins) for 16 h at 37 °C. After extraction with 10% FA and 100% ACN, the peptides were dried and stored at −20 °C until MS analysis. For PRM analysis, skimmed milk (20 μg protein) was added with 1% (w/v) sodium deoxycholate (SDC) in 100 mM Tris and then reduced with 5 mM tris(2-carboxyethyl)phosphine (TCEP) and alkylated with 30 mM 2-chloroacetamide (CAA) at room temperature. Trypsin (1:50, w/w skimmed milk proteins) was added, and samples were incubated for 16 h at 37 °C. After acidic precipitation of SDC with 0.5% TFA, peptides were cleaned via a Sep-Pak C18 vac cartridge, dried, and stored at −20 °C until MS analysis.
Data-Dependent Shotgun Analysis
All peptides were separated and analyzed using an Agilent 1290 Infinity HPLC system (Agilent Technologies, Waldbronn. Germany) coupled online to a Q-Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Reversed-phase separation was accomplished using a 100 μm inner diameter 2 cm trap column (in-house packed with ReproSil-Pur C18-AQ, 3 μm; Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) coupled to a 50 μm inner diameter 50 cm analytical column (in-house packed with Poroshell 120 EC-C18, 2.7 μm; Agilent Technologies, Amstelveen, The Netherlands). Mobile-phase solvent A consisted of 0.1% FA in water, and mobile-phase solvent B consisted of 0.1% FA in ACN. Trapping was performed at a flow rate of 5 μL/min for 5 min with 0% B, and peptides were eluted using a passively split flow of 300 nL/min for 80 min with 13% to 44% B over 65 min, 44% to 100% B over 3 min, 100% B for 1 min, 100% to 0% B over 1 min, and finally held at 0% B for 10 min. Mass spectra were collected in a data-dependent mode, automatically switching between MS and MS/MS. Full scans (from m/z 375 to 1600) were acquired at a resolution of 35 000 at 400 m/z and a target of 3e6 ions or a maximum injection time of 250 ms. Fragmentation was induced for the top 10 peaks using a 12 s dynamic exclusion. Target peaks were isolated in a 1.5 Da isolation window and subjected to high-energy collision dissociation (HCD) with a normalized collision energy value of 25%. MS/MS spectra (from m/z 200 to 2000) were acquired with a resolution of 17 500 at 400 m/z using an AGC setting of 5e4 ions or a maximum injection time of 120 ms. Charge state screening was enabled, and precursors with unknown charge state or a charge state of 1 were excluded.
Analysis of Shotgun LC-MS/MS data
Raw shotgun LC-MS/MS data were searched with Proteome Discoverer (version 1.4, Thermo Scientific) using the Mascot search engine (version 2.5). Since a search space for nonhuman peptides can become enormous, we conducted a two-step database search strategy. Generated peak lists were first searched against the Swiss-Prot database: Homo sapiens (canonical and isoform) and allergens (September 2016, 42 910 entries) with fixed Cys carbamidomethylation and variable Met oxidation of peptides. Trypsin was chosen for cleavage specificity with a maximum of two missed cleavages allowed. The searches were performed using a precursor mass tolerance of 50 ppm and a fragment mass tolerance of 0.05 Da, followed by data filtering using Percolator, resulting in a 1% false discovery rate (FDR). Only ranked 1 PSMs with Mascot scores > 20 were accepted. The second search was conducted against the Swiss-Prot database: Homo sapiens (canonical and isoform) and organisms with identified allergens (September 2016, 56 329 entries) using the same parameters as in the first database search. Identified nonhuman peptides via the second search were refined to exclude those which could have originated from a human protein by manually checking all matches to the spectrum using the Mascot server. Since we did not search the whole proteome, all refined nonhuman peptides were next entered into the NCBI protein BLAST search engine (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) and searched against UniProtKB/Swiss-Prot(SwissProt) using the blastp algorithm to further check uniqueness of the identified nonhuman peptides. Some peptides were identified as belonging to multiple nonhuman proteins when applied to the blastp algorithm. These peptides may potentially have originated from several nonhuman organisms, so we devised a specific and nonspecific classification scheme. If a peptide belonged to several proteins associated with multiple organisms under more than one tribe, no direct peptide to protein match could be made, and peptides were generally classified as nonhuman. If specific tribe to organism associations could be made, then the common organism nomenclature was used to represent the peptide to protein relationship. For example, “Bos taurus” is the representative of tribe Bovini, which includes Bison, Bos, Bubalus, Syncerus, and Pseudoryx. Amino acid sequence alignments were conducted by putting identifiers of nonhuman proteins and human homologues using the Clustal Omega program implemented in Uniprot (http://www.uniprot.org/align/).
Estimated Abundance of Proteins in Human Milk
Mascot.msf files were opened with Proteome Discoverer (version 2.2, Thermo Scientific) using the consensus step to enable retention time (RT) alignment with a maximum RT shift of 10 min in order to match the precursor between runs. Label-free quantification29 of peptides was performed using the intensities of the extracted ion chromatograms (XICs). Protein intensity was determined by the average intensity of the three (or fewer, if fewer peptides were detected) peptides with the highest intensity, using the so-called Top3MS method.30 Peptides found in different protein isoforms were combined to generate one protein intensity. A protein quantitation index (PQI) was calculated using the derived Top3 numbers assuming direct proportionality between PQI and protein abundance as demonstrated previously.31 Briefly, the total protein concentration in each sample was measured via Bradford assays, and then the estimated abundance of each protein was simply calculated by taking the proportion of the Top3 intensity for each protein to the total Top3 intensity.
Sequence Alignment and Visualization of Nonhuman Peptides
Nonhuman peptides were aligned in protein sequence by an in-house python script, which is available at github. In brief, an input file in fasta format containing the same entries as used in the second database search was required as a sequence database to be mapped on. Another input file in Excel format containing the identified nonhuman peptide sequence in the individual sample and related protein IDs from the UniProt database was used to locate the peptides in the protein sequences. Identified nonhuman peptides were shuffled in the sequence database, and unique peptides were matched with protein sequences, from which we then generated peptide alignment maps.
Targeted PRM Analysis
For PRM analysis, several nonhuman peptides were selected based on general rules for targeted proteomics.32 In brief, we attempted to avoid too short (less than six amino acids) or too long (more than 25 amino acids) peptides as well as peptides containing Met residues. Additionally, the cleavage of a tryptic peptide should be complete. For the selected peptides, stable isotope-labeled standards terminated with C-terminal heavy Arg/Lys were purchased. The heavy labeled peptides were mixed equally together, and a PRTC mixture was spiked in to monitor and calibrate the scheduled RT window.
Around 70 fmol of stable isotope-labeled peptides and 1 pmol of PRTC mixture were analyzed by LC-MS/MS, as described above for shotgun proteomics, except that the LC separation used a 35 min gradient of 13–40% buffer B and the mass analyzer was a Q-Exactive High Field X quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) to enable dynamic calibration for retention time prediction. MS/MS data were searched in Mascot. Skyline33 (Skyline-daily, version 4.0.9.11664) was used to build a spectral library and assign integrated fragment peaks. The Skyline document initially scanned for all b and y fragment ions and was then automatically refined to include only the top 10 ions (375 to 1600 m/z) for each peptide. Each peak was manually assessed; y1 ions and product ions with interference were removed. Increasing concentrations of stable isotope-labeled peptide mixture were spiked into pooled skimmed milk in-solution tryptic digest, generating a standard curve range from approximately 7 fmol to 1120 fmol and analyzed by PRM mode. Detection was done with a resolution of 60 000 using an AGC setting of 2e5, maximum IT of 128 ms, one microscan, a 1.4 m/z isolation window, 27% normalized collision energy, and 5 min retention time window by the same LC setting and mass spectrometer as above. Response curves of the sum of the extracted peak areas of all coeluted fragment ions and the amount of each stable isotope-labeled peptide were generated for the linear regression r2 and for the calculation of the coefficient of variation (CV). An equal amount of stable isotope-labeled peptide mixture and PRTC peptides was spiked into three replicates of an in-solution tryptic digest of six samples. Nonhuman peptides in samples were detected in PRM mode with the same parameter as above except that the LC separation used a 65 min gradient of 13–40% buffer B. A neat mixture of stable isotope-labeled peptides and PRTC peptides was analyzed for both light and heavy targets to assess the purity of synthetic, stable isotope-labeled peptides. Summing extracted peak areas of the top 5 (or fewer) ranked noninterfered fragment ions per each precursor ion were used for calculating the ratio of light to heavy in the pair of a nonhuman peptide and its stable isotope standard. Nonhuman peptide amount was generated by the ratio of light to heavy multiplying amounts of stable isotope-labeled peptide.
Results
Study Workflow
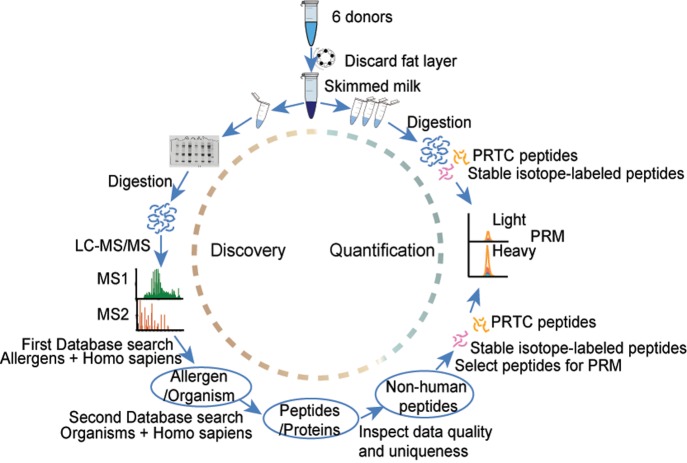
In this study, we analyzed skimmed milk samples from six individual donors. The overall workflow of the analysis is described in Figure 1. After the total protein concentration was measured using a Bradford assay, each skimmed milk sample was divided into four aliquots. One aliquot containing 25 μg of total protein was taken into the discovery part where human and nonhuman peptides and proteins were identified by in-gel digestion, data-dependent shotgun LC-MS/MS, and then in the two-step database search described in the Materials and Methods section. Three aliquots containing 20 μg of total protein each were used for quantification of nonhuman peptides by PRM after in-solution digestion and the addition of the spiked stable isotope heavy peptides and the PRTC peptides (see Materials and Methods for more detailed description).
Figure 1.
Schematic overview of the discovery and quantification workflow for the human milk proteome and nonhuman peptides present in human milk. Milk samples from six donors were assessed as skimmed milk; each sample was divided into four aliquots. From here, the workflow is composed of two parts: In the discovery part, accounting for one aliquot per donor, proteins were first prefractionated by 1D gel electrophoresis, then digested into peptides, and then analyzed and identified by data-dependent shotgun LC-MS/MS. Human and nonhuman proteins/peptides were identified by a two-step database search (see Materials and Methods section). In the quantification part, from the remaining three aliquots per donor, individual nonhuman peptides were quantified using parallel reaction monitoring (PRM) implementing stable isotope heavy peptide standards and PRTC peptides; peptide retention time calibration mixture.
Discovery of Nonhuman Peptides
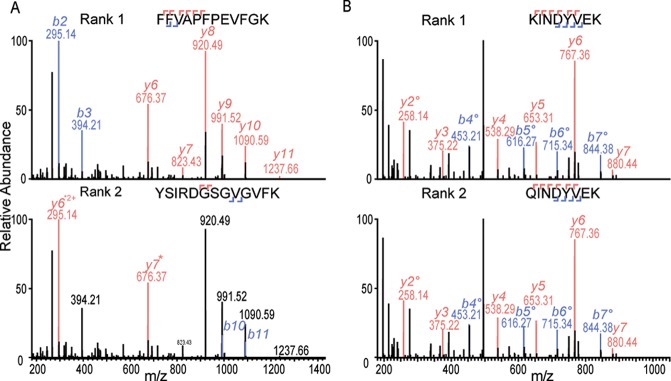
In a typical shotgun LC-MS/MS experiment, MS/MS data are used in an automated search of a protein sequence database to find the peptide that most closely matches each observed fragmentation spectrum. A score representing the degree of correlation is then calculated for each peptide. A major problem associated with automated search algorithms is the appearance of false positive hits caused by random matching between the experimental and theoretical data.34,35 We used a common Mascot searching algorithm equipped with Percolator that uses semisupervised machine learning to improve the discrimination between correct and incorrect spectrum identifications. Each peptide found by the Mascot software is reported with a certain probability represented by Percolator scores. The peptide list was further filtered by several parameters (see Materials and Methods) to obtain the lowest possible number of false matches. However, in our case, one simple search by using standard score filters would not be sufficient to obtain reliable peptide matches, due to the expected high sequence homology among human and nonhuman peptide/proteins presented in human milk. Therefore, we visually checked all the reported best nonhuman matches (rank 1) to eliminate them from consideration as positive peptide hits if the second match (rank 2) had a score greater than 20 and originated from humans.
In the first database search, we identified not only human proteins presented in the gel pieces containing milk proteins, but also eight allergens from seven foreign organisms (Bos taurus, Canis lupus familiaris, Equus caballus, Felis catus, Gallus gallus, and Oryza sativa subsp. japonica). These identified allergens defined a group of organisms, whose proteome databases we further used in the second step for searching more nonhuman peptides. The second database search was conducted with the same parameters as the first one and resulted in a discovery of 191 nonhuman peptides. To validate our list of nonhuman peptides, we performed a manual inspection of all the matched MS/MS spectra. First, we filtered out all common contaminants such as peptides from keratin proteins and trypsin. Since we use BSA as a daily quality control in our lab, it should also be considered as a contaminant in this study. We analyzed therefore separately our BSA, using the same parameters, and confirmed the purity of BSA, then we eliminated all BSA peptides found in human milk samples. In fact, we decided to remove from our nonhuman peptides list all serum albumins due to their high sequence homology (typically around 80%). Second, we deteled all matched peptides from variable immunoglobulin regions due to the high probability of false matches for those peptides in general. We can therefore not conclude that the milk samples contain endogeneous peptides from BSA peptides or immunoglobulins, as we disregarded them in our analysis.
For the remaining peptide hits, we further visually checked all matched MS/MS spectra. As mentioned before we accepted only the matches with Mascot scores >20. If any MS/MS spectrum was assigned with two or more possible peptides, Mascot ranks them according to their score value and sorts them from rank 1 to 10, where rank 1 is the best match. We did generate the distribution of scores of rank 1 peptides (human, nonhuman) and rank 2 peptides (only for the spectra matched with rank 1 nonhuman peptides) using a density plot (Figure S1). Rank 1 peptides (human, nonhuman) have a similar apex of ion scores, while rank 2 peptides show a different trend. However, some rank 2 peptides which have an ion score greater than 20 may be the true match instead of the rank 1 nonhuman peptide. Figure 2 shows two examples of how we checked matches to spectra for each nonhuman peptide and decided to keep or remove one hit in the list of nonhuman peptides. Figure S2 shows four more examples of MS/MS spectra of nonhuman peptides with nearly full peptide sequence coverage. Finally, we were skeptical about some peptides which may be semitryptic peptides or modified peptides from a human protein (Table S1). For instance, “QSPIDLQR” from dog carbonic anhydrase 6 (Q865C0) can possibly also be the deamidated version of “QSPINLQR” from human carbonic anhydrase 6 (P23280). Our final list of nonhuman peptides contained 109 carefully validated nonhuman peptides identified in the six human milk samples we analyzed (Table 1). Peptides originating from caseins, especially bovine caseins, were found to be the most dominant nonhuman peptides.
Figure 2.
Illustrative matches of nonhuman peptide sequences to the MS/MS spectrum. Examples of manual checking the matches to the MS/MS spectrum from the Mascot server. (A) The MS/MS spectrum was matched to the nonhuman peptide “FFVAPFPEVFGK” (ion score 46.5, rank 1, precursor mass 1383.7227), the rank 2 matched peptide “YSIRDGSGVGVFK” (ion score 0.2, precursor mass 1383.7147) from human protocadherin fat 1 was substantially less confident, verifying that the rank 1 sequence was correctly matched to this spectrum. (B) Nonhuman peptide “KINDYVEK” (ion score 32.2, rank 1, precursor mass 1007.5287) and human peptide “QINDYVEK” (ion score 32.2, rank 2, precursor mass 1007.4924) both matched equally well to one spectrum. Moreover, the fragmentation spectra did not allow us to distinguish one from another. We removed therefore the first sequence out of the list of nonhuman peptides.
Table 1. Overview of All Unique Nonhuman Peptides Observed in Human Milk.
| annotated sequence | # PSMs | protein names | accessions | organisms | ions score | found in 6 samples |
|---|---|---|---|---|---|---|
| [R].FFVAPFPEVFGKEK.[V] | 110 | Alpha-S1-casein | P02662 | Bos taurus | 53 | 6 |
| [K].HIQKEDVPSER.[Y] | 69 | Alpha-S1-casein | P02662 | Bos taurus | 67 | 6 |
| [K].HPIKHQGLPQEVLNENLLR.[F] | 41 | Alpha-S1-casein | P02662 | Bos taurus | 75 | 6 |
| [K].HQGLPQEVLNENLLR.[F] | 28 | Alpha-S1-casein | P02662 | Bos taurus | 62 | 6 |
| [K].EGIHAQQKEPMIGVNQELAYFYPELFR.[Q] | 26 | Alpha-S1-casein | P02662 | Bos taurus | 52 | 6 |
| [K].EKVNELSK.[D] | 19 | Alpha-S1-casein | P02662 | Bos taurus | 39 | 6 |
| [R].FFVAPFPEVFGK.[E] | 12 | Alpha-S1-casein | P02662 | Bos taurus | 47 | 6 |
| [R].LHSMKEGIHAQQK.[E] | 8 | Alpha-S1-casein | P02662 | Bos taurus | 55 | 6 |
| [K].EGIHAQQKEPMIGVNQELAYFYPELFR.[Q] | 6 | Alpha-S1-casein | P02662 | Bos taurus | 58 | 6 |
| [R].YLGYLEQLLR.[L] | 104 | Alpha-S1-casein | Bos taurus and Ovis aries | 66 | 6 | |
| [K].EDVPSER.[Y] | 19 | Alpha-S1-casein | Bos taurus and Ovis aries | 36 | 6 | |
| [K].TTMPLW.[-] | 19 | Alpha-S1-casein | Bos taurus and Ovis aries | 33 | 6 | |
| [K].TTMPLW.[-] | 8 | Alpha-S1-casein | Bos taurus and Ovis aries | 35 | 6 | |
| [K].FALPQYLK.[T] | 57 | Alpha-S2-casein | P02663 | Bos taurus | 36 | 6 |
| [R].NAVPITPTLNR.[E] | 29 | Alpha-S2-casein | P02663 | Bos taurus | 50 | 6 |
| [K].AMKPWIQPK.[T] | 20 | Alpha-S2-casein | P02663 | Bos taurus | 32 | 6 |
| [K].TKVIPYVR.[Y] | 16 | Alpha-S2-casein | P02663 | Bos taurus | 40 | 6 |
| [K].AMKPWIQPK.[T] | 2 | Alpha-S2-casein | P02663 | Bos taurus | 28 | 6 |
| [K].TKVIPYVRYL.[-] | 1 | Alpha-S2-casein | P02663 | Bos taurus | 30 | 5 |
| [K].ITVDDKHYQK.[A] | 18 | Alpha-S2-casein | Bos taurus and Ovis aries | 53 | 6 | |
| [K].ALNEINQFYQK.[F] | 16 | Alpha-S2-casein | Bos taurus and Ovis aries | 52 | 6 | |
| [K].LTEEEKNRLNFLK.[K] | 1 | Alpha-S2-casein | Bos taurus and Ovis aries and Sus scrofa | 21 | 6 | |
| [K].AVPYPQR.[D] | 24 | Beta-casein | P02666 | Bos taurus | 24 | 6 |
| [K].VKEAMAPK.[H] | 22 | Beta-casein | P02666 | Bos taurus | 45 | 6 |
| [R].GPFPIIV.[-] | 12 | Beta-casein | P02666 | Bos taurus | 31 | 6 |
| [K].VLPVPQKAVPYPQR.[D] | 4 | Beta-casein | P02666 | Bos taurus | 36 | 6 |
| [K].VLPVPQK.[A] | 164 | Beta-casein | Bos taurus and Ovis aries | 37 | 6 | |
| [K].HKEMPFPK.[Y] | 9 | Beta-casein | Bos taurus and Ovis aries | 36 | 6 | |
| [K].HKEMPFPK.[Y] | 3 | Beta-casein | Bos taurus and Ovis aries | 23 | 6 | |
| [R].SPAQILQWQVLSNTVPAK.[S] | 64 | Kappa-casein | P02668 | Bos taurus | 114 | 6 |
| [R].FFSDKIAK.[Y] | 21 | Kappa-casein | P02668 | Bos taurus | 45 | 6 |
| [R].YPSYGLNYYQQKPVALINNQFLPYPYYAKPAAVR.[S] | 1 | Kappa-casein | P02668 | Bos taurus | 30 | 5 |
| [K].YIPIQYVLSR.[Y] | 54 | Kappa-casein | Bos taurus and Ovis aries | 44 | 6 | |
| [K].AGLCQTFVYGGCR.[A] | 44 | Pancreatic trypsin inhibitor | P00974 | Bos taurus | 78 | 6 |
| [R].NNFKSAEDCMR.[T] | 8 | Pancreatic trypsin inhibitor | P00974 | Bos taurus | 39 | 6 |
| [R].IIRYFYNAK.[A] | 1 | Pancreatic trypsin inhibitor | P00974 | Bos taurus | 24 | 6 |
| [R].TPEVDDEALEKFDK.[A] | 19 | Beta-lactoglobulin | P02754 | Bos taurus | 81 | 6 |
| [R].VYVEELKPTPEGDLEILLQK.[W] | 4 | Beta-lactoglobulin | P02754 | Bos taurus | 47 | 6 |
| [R].TPEVDDEALEK.[F] | 1 | Beta-lactoglobulin | P02754 | Bos taurus | 58 | 5 |
| [K].ALPMHIR.[L] | 5 | Beta-lactoglobulin | Bos taurus and Ovis aries | 24 | 6 | |
| [K].VLVLDTDYKK.[Y] | 3 | Beta-lactoglobulin | Bos taurus and Ovis aries | 57 | 6 | |
| [K].TKIPAVFK.[I] | 2 | Beta-lactoglobulin | Bos taurus and Ovis aries | 25 | 6 | |
| [K].ALPMHIR.[L] | 1 | Beta-lactoglobulin | Bos taurus and Ovis aries | 21 | 6 | |
| [K].LGSVYTEGGFVEGVNKK.[L] | 14 | Bile salt-activated lipase (Fragment) | P30122 | Bos taurus | 55 | 6 |
| [K].RAISQSGVGLCPWAIQQDPLFWAK.[R] | 3 | Bile salt-activated lipase (Fragment) | P30122 | Bos taurus | 27 | 6 |
| [R].CMLDRNEDMLITGGRHPFLAR.[Y] | 12 | Xanthine dehydrogenase/oxidase | P80457 | Bos taurus | 34 | 6 |
| [R].NQPEPTVEEIEDAFQGNLCR.[C] | 4 | Xanthine dehydrogenase/oxidase | P80457 | Bos taurus | 44 | 6 |
| [R].CMLDRNEDMLITGGRHPFLAR.[Y] | 3 | Xanthine dehydrogenase/oxidase | P80457 | Bos taurus | 27 | 6 |
| [R].CMLDRNEDMLITGGRHPFLAR.[Y] | 3 | Xanthine dehydrogenase/oxidase | P80457 | Bos taurus | 31 | 6 |
| [K].LGCGEGGCGACTVMLSKYDRLQDK.[I] | 1 | Xanthine dehydrogenase/oxidase | P80457 | Bos taurus | 31 | 6 |
| [R].VFVQKEILDQFTEEVVKQTQR.[I] | 11 | 4-trimethylaminobutyraldehyde dehydrogenase | Q2KJH9 | Bos taurus | 47 | 6 |
| [R].VFVQKEILDQFTEEVVK.[Q] | 3 | 4-trimethylaminobutyraldehyde dehydrogenase | Q2KJH9 | Bos taurus | 42 | 6 |
| [K].EILDQFTEEVVK.[Q] | 1 | 4-trimethylaminobutyraldehyde dehydrogenase | Q2KJH9 | Bos taurus | 44 | 6 |
| [R].VIATFTCSGEKEVNLAVQDAK.[A] | 1 | 4-trimethylaminobutyraldehyde dehydrogenase | Q2KJH9 | Bos taurus | 20 | 1 |
| [K].ALGGEDVR.[V] | 11 | Alpha-2-HS-glycoprotein | P12763 | Bos taurus | 53 | 6 |
| [K].TPIVGQPSIPGGPVR.[L] | 1 | Alpha-2-HS-glycoprotein | P12763 | Bos taurus | 45 | 1 |
| [K].HTLNQIDSVK.[V] | 2 | Alpha-2-HS-glycoprotein | Bos taurus and Ovis aries | 34 | 3 | |
| [R].GYKHTLNQIDSVK.[V] | 2 | Alpha-2-HS-glycoprotein | Bos taurus and Ovis aries | 68 | 6 | |
| [K].SPPFFEDLTLDLQPPK.[S] | 4 | Cytoplasmic aconitate hydratase | Q0VCU1 | Bos taurus | 47 | 6 |
| [R].ADSLKKNQDLEFER.[N] | 4 | Cytoplasmic aconitate hydratase | Q0VCU1 | Bos taurus | 61 | 6 |
| [K].TVDNFVALATGEKGFGYKDSK.[F] | 3 | Peptidyl-prolyl cis–trans isomerase B | P80311 | Bos taurus | 44 | 5 |
| [K].GFGYKDSK.[F] | 1 | Peptidyl-prolyl cis–trans isomerase B | P80311 | Bos taurus | 27 | 6 |
| [R].VYVVDVATEPR.[A] | 2 | Selenium-binding protein 1 | Q2KJ32 | Bos taurus | 43 | 6 |
| [R].LVGQIFLGGSIVK.[G] | 1 | Selenium-binding protein 1 | Q2KJ32 | Bos taurus | 52 | 4 |
| [K].LSISETYDLK.[S] | 1 | Alpha-1-antiproteinase | P34955 | Bos taurus | 41 | 4 |
| [K].AALTIDEK.[G] | 1 | Alpha-1-antiproteinase | Bos taurus and Ovis aries | 28 | 6 | |
| [K].SVLGDVGITEVFSDR.[A] | 1 | Alpha-1-antiproteinase | Bos taurus and Ovis aries | 28 | 5 | |
| [K].QIPLTCIVDK.[R] | 6 | Metalloendopeptidase OMA1, mitochondrial | Q3SZN3 | Bos taurus | 24 | 6 |
| [K].VTISCSGGR.[S] | 5 | Complement component C7 | Q29RQ1 | Bos taurus | 48 | 6 |
| [R].FLEDYFDGNLKR.[Y] | 3 | Protein disulfide-isomerase A3 | P38657 | Bos taurus | 32 | 6 |
| [R].AIQAAFFYLEPR.[H] | 2 | Alpha-1-acid glycoprotein | Q3SZR3 | Bos taurus | 80 | 6 |
| [K].YVRPGGGFTPNFQLFEKGDVNGEKEQK.[F] | 2 | Glutathione peroxidase 3 | P37141 | Bos taurus | 39 | 6 |
| [K].YHALYINALQK.[L] | 2 | Diacylglycerol O-acyltransferase 2-like protein 6 | A6QP72 | Bos taurus | 31 | 6 |
| [K].TPTLEKQGK.[K] | 2 | Synaptonemal complex protein 3 | Q3T0E2 | Bos taurus | 29 | 6 |
| [K].LAVPIILR.[V] | 1 | Alpha-fetoprotein | Q3SZ57 | Bos taurus | 35 | 1 |
| [K].VLSGSIEKAK.[Q] | 1 | DNA excision repair protein ERCC-6-like 2 | A3KMX0 | Bos taurus | 27 | 6 |
| [R].TVSISPTK.[K] | 1 | Fibrous sheath CABYR-binding protein | Q2T9N0 | Bos taurus | 32 | 6 |
| [K].LVEFPLVAAWYQR.[I] | 1 | Glutathione S-transferase C-terminal domain-containing protein | A5PKL6 | Bos taurus | 27 | 2 |
| [K].LQHFFIGNR.[K] | 1 | Inositol 1,4,5-trisphosphate receptor-interacting protein | A7MB64 | Bos taurus | 21 | 6 |
| [R].EVSNKIVGYLDEEGVLDQNR.[S] | 1 | Lactoperoxidase | P80025 | Bos taurus | 53 | 6 |
| [R].ATDLVPR.[I] | 1 | Peroxisomal membrane protein 11A | Q0VCP2 | Bos taurus | 32 | 6 |
| [R].FEILPTR.[S] | 1 | Protein-glutamine gamma-glutamyltransferase E | A6QP57 | Bos taurus | 36 | 5 |
| [R].GSPAANVGVK.[V] | 2 | Transthyretin | Bos taurus and Ovis aries | 50 | 6 | |
| [K].SSELVSANR.[L] | 1 | Antithrombin-III | Bos taurus and Ovis aries | 48 | 5 | |
| [K].LLSTLCSADVCQCAEGK.[C] | 1 | Complement C4 | Bos taurus and Rattus norvegicus | 20 | 1 | |
| [K].ECHLAQVPSHAVVAR.[S] | 1 | Lactotransferrin | Bos taurus and Ovis aries | 25 | 6 | |
| [K].NVNESLLELHK.[L] | 2 | Ferritin heavy chain | Q8MIP0 | Equus caballus | 53 | 2 |
| [K].ADAVTLDGGLVYEAGLHPYK.[L] | 2 | Lactotransferrin (Fragment) | O77811 | Equus caballus | 24 | 2 |
| [R].ITGGRDFIDIESK.[F] | 1 | Lipoprotein lipase | P55031 | Felis catus | 24 | 6 |
| [R].WCAVAGGRLDSGK.[Q] | 2 | 17 kDa alpha-amylase/trypsin inhibitor 2 | Q7 × 8H9 | Oryza sativa subsp. japonica | 21 | 1 |
| [K].LFNIIEPDVAVFGK.[K] | 1 | Pantoate--beta-alanine ligase | O24210 | Oryza sativa subsp. japonica | 37 | 1 |
| [K].TIEVDNTDAEGR.[L] | 4 | Proline aminopeptidase 1/Leucyl aminopeptidase | Oryza sativa subsp. Japonica and Gloeobacter violaceus | 60 | 6 | |
| [K].QIKTLIEQTNEERK.[S] | 8 | Clusterin | P25473 | Canis lupus familiaris | 75 | 6 |
| [K].DFAEEGKK.[D] | 2 | S-arrestin | Q28281 | Canis lupus familiaris | 26 | 6 |
| [R].HPGWQGTLK.[A] | 17 | various nonhuman organisms | 39 | 6 | ||
| [K].GLTSLLR.[S] | 9 | various nonhuman organisms | 38 | 6 | ||
| [K].AMIAYWTNFAR.[T] | 4 | various nonhuman organisms | 26 | 6 | ||
| [K].AMIAYWTNFAR.[T] | 3 | various nonhuman organisms | 43 | 6 | ||
| [K].EVVLEEGTIAFKNWVK.[T] | 2 | various nonhuman organisms | 45 | 3 | ||
| [R].LGGAEIAR.[T] | 2 | various nonhuman organisms | 25 | 6 | ||
| [-].KIFERCELAR.[T] | 1 | various nonhuman organisms | 28 | 6 | ||
| [K].FESNFNTQATNR.[N] | 1 | various nonhuman organisms | 50 | 3 | ||
| [K].KEVVLEEGTIAFK.[N] | 1 | various nonhuman organisms | 27 | 1 | ||
| [R].ADLSGITK.[E] | 1 | various nonhuman organisms | 29 | 6 | ||
| [R].IEYDPNR.[S] | 1 | various nonhuman organisms | 23 | 5 | ||
| [R].LDSPATPERIR.[N] | 1 | various nonhuman organisms | 37 | 4 | ||
| [R].NTDGSTDYGILQINSR.[W] | 1 | various nonhuman organisms | 137 | 4 | ||
| [R].SALFAQINQGESITHALK.[H] | 1 | various nonhuman organisms | 45 | 6 | ||
| [R].YEVPLETPR.[V] | 1 | various nonhuman organisms | 24 | 5 |
Characterization of Human and Nonhuman Proteins in Human Milk
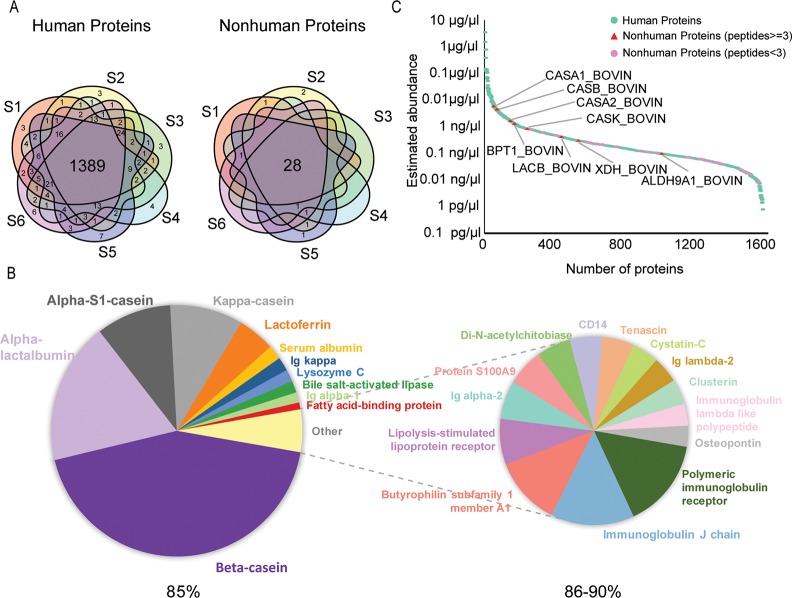
Cumulatively, 1577 human proteins and 36 nonhuman proteins were identified in the human milk samples of the six donors. The majority, 88%, of the total number of human proteins (1389 human proteins) and 78% of the total number of nonhuman proteins (28 nonhuman proteins), were detected in all six samples (Figure 3A). From our MS data, estimated protein abundances were averaged over all six samples. Similarly to human plasma,36 the 11 most abundant human proteins represent approximately 85% of the total protein mass in the skimmed milk proteome. The 25 most abundant human proteins then represent approximately 90% of the total protein mass in human milk (Figure 3B). The protein abundances converted into concentration (μg/μL to pg/μL) show an extremely high dynamic range, which makes milk proteins and especially low abundance nonhuman proteins challenging to detect and identify (Figure 3C, Table S2). Compared with human milk proteins, nonhuman proteins were less abundant, except for several proteins likely originating from bovine milk products, such as bovine caseins (α-S1-, α-S2-, β-, and κ-caseins) and β-lactoglobulin.37 Nevertheless, even the most abundant nonhuman protein, bovine α-S1-casein, with a concentration estimated around 0.006 μg/μL, is approximately 600× less abundant than the most abundant human milk protein β-casein (3.60 μg/μL).
Figure 3.
Overview of the human milk proteome and nonhuman proteins identified in human milk. (A) Our prefractionation approach enabled us to overcome the dominance of a dozen highly abundant human milk proteins, leading to the identification of 1577 human proteins (left) and 36 nonhuman proteins (right) across six human milk samples, of which about 88/78% were detected in all individuals, respectively. (B) Estimated contribution of individual proteins to the total protein mass of the most abundant proteins in human milk. About 11 proteins contribute about 85% of the total protein mass, and in total 25 proteins make up 90%. (C) Estimated abundance of all human and nonhuman proteins in the analyzed human milk samples. The majority of nonhuman proteins are relatively low in abundance with less than three identified peptides, except for several proteins, likely originating from bovine milk.
Sequence Alignment Map of Nonhuman Peptides
To provide our shotgun proteomics data as a future resource for researchers, we made Supporting Information Table S3, in which, for each individual protein, the following information is displayed: UniProt ID, gene name, protein name, organism, protein existence (numerical value describing the evidence for the existence of the protein), sequence version (version number of the sequence), sequence, and identified peptides in the different samples.
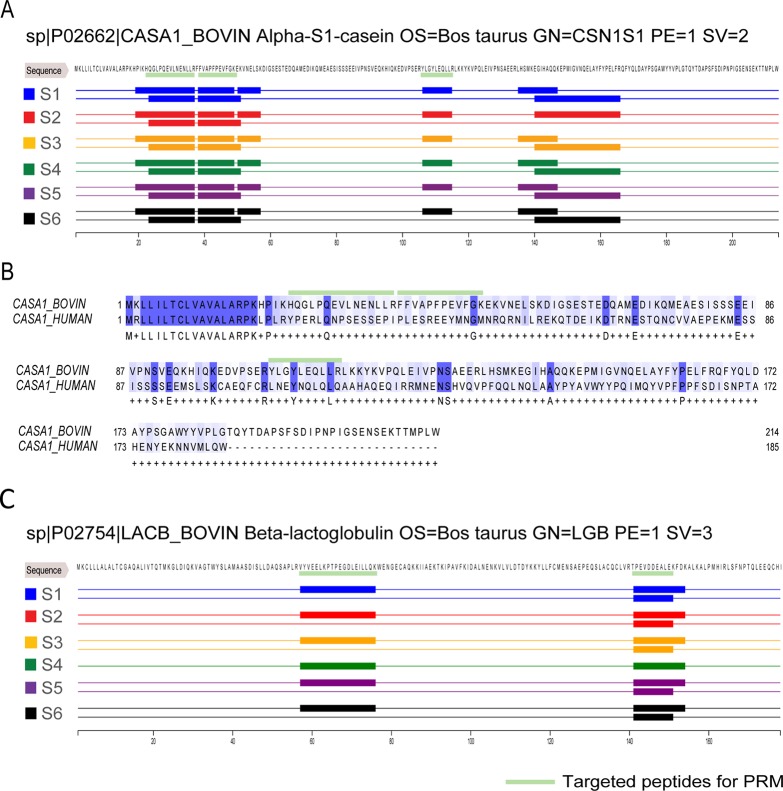
Figure 4 provides example summaries of information gathered on two more abundant nonhuman proteins, bovine α-S1-casein and bovine β-lactoglobulin. Bovine α-S1-casein was found in all six samples with nearly identical peptide sequences (Figure 4A). This remarkable reproducibility and distribution of the identified peptides provides credence that this protein likely is present intact in human milk. Moreover, the amino acid sequence alignment between bovine α-S1-casein and its human homologue showed a significant difference with only 31% of sequence homology (Figure 4B), which further supports that this nonhuman protein was unambigeously present in human milk. Similar sequence mapping was found for other nonhuman proteins, such as bovine β-lactoglobulin (Figure 4D). Selected peptides for followed PRM analysis marked in Figure 4 showed they were both reproducible across individuals.
Figure 4.
The same nonhuman peptides identified in all six donors. (A) Sequence alignment of the identified peptides of bovine alpha-S1 casein. Identified peptides are depicted as bars under the sequence colored by donor (blue (S1), red (S2), orange (S3), green (S4), purple (S5), and black (S6)). Observed sequence overlap is due to missed cleavages in some identified peptides. (B) Sequence alignment between bovine alpha-S1-casein and its human homologue. Identical amino acids are depicted by dark blue blocks and written below; biochemically similar amino acids are depicted by light blue blocks. “+” is displayed when the amino acids are not identical. Bovine and human alpha-S1-casein display about 30% of sequence homology. Selected peptides for PRM analysis are marked by light green bars. (C) As in A, albeit now for beta-lactoglobulin. Again, the same nonhuman peptides are detected in all six donors.
Parallel Reaction Monitoring Assays; Evaluation of Linearity and Reproducibility
Thirty-seven nonhuman peptides observed in our discovery mode experiments were next selected for further PRM analysis, and corresponding stable isotope standards were purchased. Through building a spectral library in Skyline, we generated expected PRM fragment ions and their relative intensity to assign integrated fragment peaks (Figure 5A). We first build a PRM assay for the 37 stable isotope-labeled peptides in a 55 min run with a 5 min-RT scheduling window, resulting in a maximum of 13 concurrent precursors (Figure S3A). We evaluated this PRM assay for linearity and dynamic range using concentration curves. PRM concentration curves were acquired for this set of 37 heavy peptides by in-solution digestion of pooled skimmed milk samples. After MS acquisition and peak extraction, the linear regression r2 and coefficient of variation (CV) were calculated. The r2 distribution of all calculated concentration curves showed more than 50% of the heavy peptides having an r2 higher than 0.95 (Figure S3B). This represents a good linearity in the setup PRM assay. CV values calculated at different concentrations showed the majority had a CV of less than 20%; however, the CV values were bigger when the peak area became smaller (Figure S3C).
Figure 5.
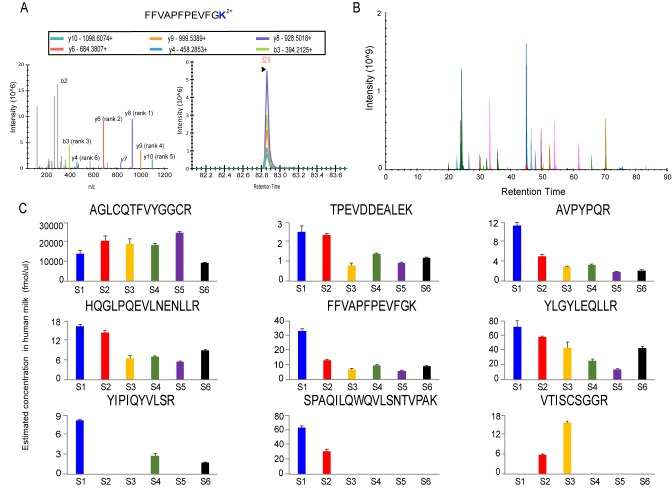
Quantitation of nonhuman peptides in human milk by parallel reaction monitoring (PRM). (A) Illustrative example of MS/MS peptide fragmentation spectra and extracted ion chromatogram of the bovin alpha-S1-casein peptide FFVAPFPEVFGK[+8]. (B) LC-MS chromatogram of all parallel monitored nonhuman peptides, representing a mixture of endogeneous and stable isotope standards. (C) Detected estimated concentrations (fmol/ul) of nonhuman peptides in human milk across the six donors. Error bars indicate standard deviation of n = 2–3 replicates.
Quantification of the Detected Nonhuman Peptides in Human Milk
We next applied the PRM assay to detect and quantify the nonhuman peptides in each of the six milk samples using a longer gradient to gain enough data points for each precursor. With a mass resolution of 60 000 using an AGC setting of 2e5, maximum IT of 128 ms, and 5 min retention time window, the maximum concurrent precursor was 13, resulting in around a maximum 1.6 s cycle time and minimum 10 data points for precursors. We noticed the retention time of some hydrophobic peptides were less stable between runs, which led us to increase the retention time window to 10 min for these selected peptides. The increased retention time window did not affect the maximum cycle time and minimum number of data points (Figure S4A). The targeted PRM inclusion list as given in Table S4 and LC-MS chromatograms of monitoring these targets are shown in Figure 5B. In the PRM analysis, we did not use the 1D gel based prefractionation step we used in the discovery phase, which makes the analysis less time-consuming, albeit likely at the expense of proteome depth.
Nine nonhuman peptides could be reproducibly quantified in at least two replicates in one of the six samples (Figure S5). For these peptides, we also evaluated the CV values of the ratio of light to heavy within replicates in different samples (Figure S4B). The majority CV values were within 20%. We calculated the concentration of nonhuman peptides across the six donors (Figure 5C). The most abundant peptide was “AGLCQTFVYGGCR” from a bovine pancreatic trypsin inhibitor (P00974), which was around 9–25 μM. Three peptides from α-S1-casein were quantified, in which two (“HQGLPQEVLNENLLR” and “FFVAPFPEVFGK”) were unique in bovine and one (“YLGYLEQLLR”) was shared within bovidae family members, with concentrations ranging from 5 to 16 nM, 6–33 nM, and 14–72 nM, respectively. Additionally, peptides from bovine or bovidae families’ β-casein, κ-casein, β-lactoglobulin, and complement component C7 were quantified in the nanomolar range. The majority of these proteins are considered milk proteins. Although we did not have two to three unique peptides for each protein, the detection of two unique peptides in α-S1-casein and their similar trend across six donors provide further validity that intact bovine α-S1-casein can be present in human milk.
Discussion
To our knowledge, this study provides the most comprehensive proteome analysis of nonhuman peptides and proteins in human milk to date. To overcome the challenging database issue of finding nonhuman proteins in human samples, we used a two-step database search strategy to reduce the database size followed by a manual universal blast of identified peptides to define their uniqueness. We further manually verified the data output from our database search deleting contaminants, possible modified versions of human peptides, and peptides with low quality MS/MS spectra. To increase the depth of our analysis, we applied in-gel separation, which allowed us to boost the number of protein identifications to 1577 human proteins and 36 nonhuman proteins. We next estimated protein abundances of human proteins and nonhuman proteins in human milk using standard methods used in label-free quantification. To increase quantitation accuracy for nonhuman proteins, we next applied PRM using stable isotope-labeled peptides. In this analysis we were then able to robustly quantify nine nonhuman peptides in human milk, without any prefractionation. Moreover, the usage of stable isotope synthetic standards provided further evidence of the existence of these detected nonhuman peptides in human milk. Our study provides strong evidence for the presence of intact nonhuman proteins in human milk, from mothers not exposed to any dietary intervention. Notably, there was an earlier study23 wherein they identiified the YVEELKPTPEGDL peptide from bovine Beta-lactoglobulin in human milk. We additionally searched raw files of previously published human milk proteomics studies deposited in PRIDE38 (PXD00365 and PXD00296) using our search parameters and identified quite a few nonhuman tryptic peptides, mostly originating from bovine milk proteins (e.g., “HIQKEDVPSER” and “FFVAPFPEVFGK” from bovine Alpha-S1-casein, “YLGYLEQLLR” from bovine or sheep Alpha-S1-casein, “AVPYPQR” from bovine Beta-casein, “VLPVPQK” from bovine or sheep Beta-casein, “YIPIQYVLSR” from bovine or sheep Kappa-casein) that were also identified in the present study. This reanalysis of earlier data sets further supports our observation of nonhuman proteins/peptides in human milk.
In this study, across all donors, we found that the nonhuman proteins originated mostly from bovine origin. It is still not clear how intact dietary proteins are transferred from the gut to the mammary gland and thus introduced in the human milk. Additionally, it is elusive as to why we found mainly bovine milk proteins as the most common nonhuman proteins in human milk over other nonhuman proteins that could have been introduce via the maternal diet (e.g., originating from meat, eggs, grains, and/or rice). Previous studies have shown that dietary proteins can escape from gastrointestinal digestion, at least in part, and be taken up across the intestinal epithelium and circulate in human body fluids as proteins or peptides.39 A recent study investigated the apical-to-basal transepithelial transport of bovine caseins and other food proteins using human intestinal Caco-2 cell monolayers and found that caseins appeared to survive to some extent lysosomal proteolysis and remained much more intact in vesicles around the basolateral side of Caco-2 cells, when compared to other food proteins, such as ovalbumin and β-lactoglobulin.40 A possible reason might be that caseins have a unique property of coagulation at acidic pH which may affect their susceptibility to proteolysis. This incomplete digestion of bovine caseins, may allow them to cross the intestinal epithelium and transfer into various body fluids, including milk when they further cross over into mammary epithelial cells. The study of bovine-derived proteins in human milk is of importance as the first onset of food allergy in infants is predominantly due to cow’s milk protein allergy. The mechanisms underlying this allergy are not well understood. Recently a study aimed to evaluate WHO guidelines for breastfeeding in two birth cohorts and showed heterogeneous effects between exclusive and nonexclusive breastfeeding and infant allergy outcomes up to 10 years of age.41 Therefore, a better understanding of which nonhuman proteins can be transported into human milk is of importance.
Allergic symptoms occurring in exclusively breastfed infants42 being documented even at the first exposure13 have steered interest into the possible occurrence of nonhuman proteins in human milk. However, avoidance of allergenic food in lactating mothers’ diets and delayed introduction of risky allergens in children have so far failed.43,44 Alternatively, the early (from 3 to 4 months of age) and sustained exposure of a food antigen may reduce risk for developing food allergy to that antigen.45 The immunological mechanisms underlying these effects are still largely unclear. Previous work aimed at detecting nonhuman proteins in human milk has predominantly relied on detection by various ELISA and other immunochemical methods. These methods require specific antibodies for specific protein detection, hampering an unbiased search of all potential peptides and proteins. This in turn could lead to a biased analysis of nonhuman peptides and proteins in human milk. Therefore, the method presented allows for high throughput detection of a large assortment of nonhuman peptides and proteins in human milk in the micromolar to nanomolar range. This broad dynamic range of peptide and protein detection at low levels of concentration better reflects the amount and diversity consumed by the breastfed infant. Additionally, recent study in mice showed feeding a diet enriched with a synbiotic mixture of short- and long-chain fructo-oligosaccharides (scGOS/lcFOS 9:1) and Bifidobacterium breve M-16V could promote the efficacy of oral administration of β-lactoglobulin-derived peptides as protecting the intestinal Th1/Th2 balance and reducing the allergic response to whole whey protein,46 emphasizing the importance of a proper immune environment during antigen presentation and oral tolerance induction in early life. As human milk contains various oligosaccharides47 and microbiota,48 the early and sustained exposure of a low amount of nonhuman peptides and proteins in human milk may be an ideal way to introduce possible antigens to breastfed infants, which may have health benefits of oral tolerance induction. Knowing the identity and origin of the nonhuman peptides and proteins allows the setup of targeted proteomics approaches for robust and sensitive detection of these products over larger cohorts, as demonstrated as a proof of concept in our work. Such analysis may then next provide more information about which foreign peptides and proteins may transfer into human milk and a better understanding on how human milk may help to support oral tolerance development via early introduction of those peptides and proteins, thus reducing the risk of developing an allergy.49−51
Acknowledgments
We thank the AIBLUD association (Italian Association of Human Milk Banks) for providing the human milk samples. We would like to acknowledge Marko Mank (Danone Nutricia Research) and Lucrece Matheron (Utrecht University) for their contributions of optimizing the protocol of in-gel separation and Vojtech Franc and Kelly A. Dingess (Utrecht University) for their suggestions and proofreading of the manuscript. A.J.R.H acknowledges support from The Netherlands Organization for Scientific Research (NWO) funding the large-scale proteomics facility Proteins@Work (project 184.032.201) embedded in The Netherlands Proteomics Centre and the TOP-Punt Grant 718.015.003. This project has received additional funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement 668036 (RELENT) and 686547 (MSMed). J.Z. acknowledges support from the Chinese Scholarship Council (CSC).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00550.
Density plot of Mascot ion scores, illustrative spectra of nonhuman peptides peptides, PRM assay on stable heavy isotopic peptides, PRM analysis on a mixture of nonhuman peptides and stable heavy isotopic standards, spectral library and fragment ion XICs from nonhuman peptides and stable heavy isotopic standards (PDF)
Identified peptides in six samples (XLSX)
Identified proteins and estimated abundances (XLSX)
Nonhuman peptide sequence alignment (PDF)
Targeted PRM inclusion list (XLSX)
The authors declare no competing financial interest.
Notes
The mass spectrometry data of shotgun proteomics have been deposited to the ProteomeXchange Consortium via the PRIDE38 partner repository with the data set identifier PXD011599. PRM data have been deposited to Panorama,52 which is a freely available, open-source repository server application for targeted mass spectrometry assays. Script of peptide alignment maps is available at the following link: https://github.com/juer120/Protein-sequence-alignment/tree/master.
Supplementary Material
References
- McManaman J. L.; Neville M. C. Mammary physiology and milk secretion. Adv. Drug Delivery Rev. 2003, 55 (5), 629–41. 10.1016/S0169-409X(03)00033-4. [DOI] [PubMed] [Google Scholar]
- Burgoyne R. D.; Duncan J. S. Secretion of milk proteins. J. Mammary Gland Biol. Neoplasia 1998, 3 (3), 275–86. 10.1023/A:1018763427108. [DOI] [PubMed] [Google Scholar]
- Donnally H. H. The question of the elimination of foreign protein (egg-white) in woman’s milk. J. Immunol 1930, 19 (1), 15–40. [Google Scholar]
- Kilshaw P. J.; Cant A. J. The Passage of Maternal Dietary Proteins into Human-Breast Milk. Int. Arch. Allergy Immunol. 2004, 75 (1), 8–15. 10.1159/000233582. [DOI] [PubMed] [Google Scholar]
- Jakobsson I.; Lindberg T.; Benediktsson B.; Hansson B. G. Dietary Bovine Beta-Lactoglobulin Is Transferred to Human-Milk. Acta Paediatr. 1985, 74 (3), 342–345. 10.1111/j.1651-2227.1985.tb10981.x. [DOI] [PubMed] [Google Scholar]
- Host A.; Husby S.; Hansen L. G.; Osterballe O. Bovine beta-lactoglobulin in human milk from atopic and non-atopic mothers. Relationship to maternal intake of homogenized and unhomogenized milk. Clin. Exp. Allergy 1990, 20 (4), 383–7. 10.1111/j.1365-2222.1990.tb02798.x. [DOI] [PubMed] [Google Scholar]
- Sorva R.; Makinen-Kiljunen S.; Juntunen-Backman K. Beta-lactoglobulin secretion in human milk varies widely after cow’s milk ingestion in mothers of infants with cow’s milk allergy. J. Allergy Clin. Immunol. 1994, 93 (4), 787–92. 10.1016/0091-6749(94)90259-3. [DOI] [PubMed] [Google Scholar]
- Fukushima Y.; Kawata Y.; Onda T.; Kitagawa M. Consumption of cow milk and egg by lactating women and the presence of beta-lactoglobulin and ovalbumin in breast milk. Am. J. Clin. Nutr. 1997, 65 (1), 30–5. 10.1093/ajcn/65.1.30. [DOI] [PubMed] [Google Scholar]
- Matangkasombut P.; Padungpak S.; Thaloengsok S.; Kamchaisatian W.; Sasisakulporn C.; Jotikasthira W.; Benjaponpitak S.; Manuyakorn W. Detection of beta-lactoglobulin in human breast-milk 7 days after cow milk ingestion. Paediatr Int. Child Health 2017, 37 (3), 199–203. 10.1080/20469047.2017.1289310. [DOI] [PubMed] [Google Scholar]
- Palmer D. J.; Gold M. S.; Makrides M. Effect of cooked and raw egg consumption on ovalbumin content of human milk: a randomized, double-blind, cross-over trial. Clin. Exp. Allergy 2005, 35 (2), 173–8. 10.1111/j.1365-2222.2005.02170.x. [DOI] [PubMed] [Google Scholar]
- Palmer D. J.; Gold M. S.; Makrides M. Effect of maternal egg consumption on breast milk ovalbumin concentration. Clin. Exp. Allergy 2008, 38 (7), 1186–91. 10.1111/j.1365-2222.2008.03014.x. [DOI] [PubMed] [Google Scholar]
- Metcalfe J. R.; Marsh J. A.; D’Vaz N.; Geddes D. T.; Lai C. T.; Prescott S. L.; Palmer D. J. Effects of maternal dietary egg intake during early lactation on human milk ovalbumin concentration: a randomized controlled trial. Clin. Exp. Allergy 2016, 46 (12), 1605–1613. 10.1111/cea.12806. [DOI] [PubMed] [Google Scholar]
- Vadas P.; Wai Y.; Burks W.; Perelman B. Detection of peanut allergens in breast milk of lactating women. JAMA 2001, 285 (13), 1746–1748. 10.1001/jama.285.13.1746. [DOI] [PubMed] [Google Scholar]
- Bernard H.; Ah-Leung S.; Drumare M. F.; Feraudet-Tarisse C.; Verhasselt V.; Wal J. M.; Creminon C.; Adel-Patient K. Peanut allergens are rapidly transferred in human breast milk and can prevent sensitization in mice. Allergy 2014, 69 (7), 888–897. 10.1111/all.12411. [DOI] [PubMed] [Google Scholar]
- Chirdo F. G.; Rumbo M.; Anon M. C.; Fossati C. A. Presence of high levels of non-degraded gliadin in breast milk from healthy mothers. Scand. J. Gastroenterol. 1998, 33 (11), 1186–1192. 10.1080/00365529850172557. [DOI] [PubMed] [Google Scholar]
- Monti J. C.; Mermoud A. F.; Jolles P. Anti-bovine beta-lactoglobulin antibodies react with a human lactoferrin fragment and bovine beta-lactoglobulin present in human milk. Experientia 1989, 45 (2), 178–80. 10.1007/BF01954867. [DOI] [PubMed] [Google Scholar]
- Cantisani A.; Giuffrida M. G.; Fabris C.; Bertino E.; Coscia A.; Oggero R.; Monti G.; Stroppiana P.; Conti A. Detection of specific IgE to human milk proteins in sera of atopic infants. FEBS Lett. 1997, 412 (3), 515–7. 10.1016/S0014-5793(97)00828-4. [DOI] [PubMed] [Google Scholar]
- Bernard H.; Negroni L.; Chatel J. M.; Clement G.; Adel-Patient K.; Peltre G.; Creminon C.; Wal J. M. Molecular basis of IgE cross-reactivity between human beta-casein and bovine beta-casein, a major allergen of milk. Mol. Immunol. 2000, 37 (3–4), 161–167. 10.1016/S0161-5890(00)00029-8. [DOI] [PubMed] [Google Scholar]
- Han N.; Jarvinen K. M.; Cocco R. R.; Busse P. J.; Sampson H. A.; Beyer K. Identification of amino acids critical for IgE-binding to sequential epitopes of bovine kappa-casein and the similarity of these epitopes to the corresponding human kappa-casein sequence. Allergy 2008, 63 (2), 198–204. 10.1111/j.1398-9995.2007.01539.x. [DOI] [PubMed] [Google Scholar]
- Villa C.; Costa J.; Oliveira M. B. P.; Mafra I. Bovine Milk Allergens: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17 (1), 137–164. 10.1111/1541-4337.12318. [DOI] [PubMed] [Google Scholar]
- Coscia A.; Orru S.; Di Nicola P.; Giuliani F.; Varalda A.; Peila C.; Fabris C.; Conti A.; Bertino E. Detection of cow’s milk proteins and minor components in human milk using proteomics techniques. J. Matern.-Fetal Neonat. Med. 2012, 25 (Suppl 4), 49. 10.3109/14767058.2012.715015. [DOI] [PubMed] [Google Scholar]
- Orru S.; Di Nicola P.; Giuliani F.; Fabris C.; Conti A.; Coscia A.; Bertino E. Detection of bovine alpha-S1-casein in term and preterm human colostrum with proteomic techniques. Int. J. Immunopathol. Pharmacol. 2013, 26 (2), 435–44. 10.1177/039463201302600216. [DOI] [PubMed] [Google Scholar]
- Picariello G.; Addeo F.; Ferranti P.; Nocerino R.; Paparo L.; Passariello A.; Dallas D. C.; Robinson R. C.; Barile D.; Canani R. B. Antibody-independent identification of bovine milk-derived peptides in breast-milk. Food Funct. 2016, 7 (8), 3402–9. 10.1039/C6FO00731G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schocker F.; Baumert J.; Kull S.; Petersen A.; Becker W. M.; Jappe U. Prospective investigation on the transfer of Ara h 2, the most potent peanut allergen, in human breast milk. Pediat Allerg Imm-Uk 2016, 27 (4), 348–355. 10.1111/pai.12533. [DOI] [PubMed] [Google Scholar]
- Frese C. K.; Mikhaylova M.; Stucchi R.; Gautier V.; Liu Q.; Mohammed S.; Heck A. J. R.; Altelaar A. F. M.; Hoogenraad C. C. Quantitative Map of Proteome Dynamics during Neuronal Differentiation. Cell Rep. 2017, 18 (6), 1527–1542. 10.1016/j.celrep.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelstrup C. D.; Jersie-Christensen R. R.; Batth T. S.; Arrey T. N.; Kuehn A.; Kellmann M.; Olsen J. V. Rapid and deep proteomes by faster sequencing on a benchtop quadrupole ultra-high-field Orbitrap mass spectrometer. J. Proteome Res. 2014, 13 (12), 6187–95. 10.1021/pr500985w. [DOI] [PubMed] [Google Scholar]
- Domon B.; Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010, 28 (7), 710–721. 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- Bourmaud A.; Gallien S.; Domon B. Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: Principle and applications. Proteomics 2016, 16 (15–16), 2146–2159. 10.1002/pmic.201500543. [DOI] [PubMed] [Google Scholar]
- Bantscheff M.; Schirle M.; Sweetman G.; Rick J.; Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007, 389 (4), 1017–1031. 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Silva J. C.; Gorenstein M. V.; Li G. Z.; Vissers J. P.; Geromanos S. J. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 2006, 5 (1), 144–56. 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- Ahrne E.; Molzahn L.; Glatter T.; Schmidt A. Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics 2013, 13 (17), 2567–78. 10.1002/pmic.201300135. [DOI] [PubMed] [Google Scholar]
- Lange V.; Picotti P.; Domon B.; Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2008, 4, 222. 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B.; Tomazela D. M.; Shulman N.; Chambers M.; Finney G. L.; Frewen B.; Kern R.; Tabb D. L.; Liebler D. C.; MacCoss M. J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26 (7), 966–968. 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J.; Chait B. T.; Fenyo D. A statistical basis for testing the significance of mass spectrometric protein identification results. Anal. Chem. 2000, 72 (5), 999–1005. 10.1021/ac990792j. [DOI] [PubMed] [Google Scholar]
- Eriksson J.; Fenyo D. A model of random mass-matching and its use for automated significance testing in mass spectrometric proteome analysis. Proteomics 2002, 2 (3), 262–70. . [DOI] [PubMed] [Google Scholar]
- Tirumalai R. S.; Chan K. C.; Prieto D. A.; Issaq H. J.; Conrads T. P.; Veenstra T. D. Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteomics 2003, 2 (10), 1096–103. 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- Le T. T.; Deeth H. C.; Larsen L. B. Proteomics of major bovine milk proteins: Novel insights. Int. Dairy J. 2017, 67, 2–15. 10.1016/j.idairyj.2016.11.016. [DOI] [Google Scholar]
- Vizcaino J. A.; Csordas A.; Del-Toro N.; Dianes J. A.; Griss J.; Lavidas I.; Mayer G.; Perez-Riverol Y.; Reisinger F.; Ternent T.; Xu Q. W.; Wang R.; Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44 (22), 11033. 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenepoel P.; Claus D.; Geypens B.; Hiele M.; Geboes K.; Rutgeerts P.; Ghoos Y. Amount and fate of egg protein escaping assimilation in the small intestine of humans. Am. J. Physiol. 1999, 277 (5 Pt 1), G935–G943. 10.1152/ajpgi.1999.277.5.G935. [DOI] [PubMed] [Google Scholar]
- Sakurai N.; Nishio S.; Akiyama Y.; Miyata S.; Oshima K.; Nadano D.; Matsuda T., Apical-to-basolateral transepithelial transport of cow’s milk caseins by intestinal Caco-2 cell monolayers: MS-based quantitation of cellularly degraded alpha- and beta-casein fragments. J. Biochem. 2018.164113. 10.1093/jb/mvy034 [DOI] [PubMed] [Google Scholar]
- Bion V.; Lockett G. A.; Soto-Ramirez N.; Zhang H.; Venter C.; Karmaus W.; Holloway J. W.; Arshad S. H. Evaluating the efficacy of breastfeeding guidelines on long-term outcomes for allergic disease. Allergy 2016, 71 (5), 661–70. 10.1111/all.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant A.; Marsden R. A.; Kilshaw P. J. Egg and Cows Milk Hypersensitivity in Exclusively Breast Fed Infants with Eczema, and Detection of Egg Protein in Breast-Milk. Brit Med. J. 1985, 291 (6500), 932–935. 10.1136/bmj.291.6500.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroudi A.; Xinias I. Dietary interventions for primary allergy prevention in infants. Hippokratia 2011, 15 (3), 216–22. [PMC free article] [PubMed] [Google Scholar]
- Recto M. S. T.; Genuino M. L. G.; Castor M. A. R.; Casis-Hao R. J.; Tamondong-Lachica D. R.; Sales M. I. V.; Tan M. G.; Mondonedo K. S.; Dionisio-Capulong R. C. Psaai; the, P., Dietary primary prevention of allergic diseases in children: the Philippine guidelines. Asia Pac Allergy 2017, 7 (2), 102–114. 10.5415/apallergy.2017.7.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ierodiakonou D.; Garcia-Larsen V.; Logan A.; Groome A.; Cunha S.; Chivinge J.; Robinson Z.; Geoghegan N.; Jarrold K.; Reeves T.; Tagiyeva-Milne N.; Nurmatov U.; Trivella M.; Leonardi-Bee J.; Boyle R. J. Timing of Allergenic Food Introduction to the Infant Diet and Risk of Allergic or Autoimmune Disease: A Systematic Review and Meta-analysis. JAMA 2016, 316 (11), 1181–1192. 10.1001/jama.2016.12623. [DOI] [PubMed] [Google Scholar]
- Kostadinova A. I.; Meulenbroek L. A.; van Esch B. C.; Hofman G. A.; Garssen J.; Willemsen L. E.; Knippels L. M. A Specific Mixture of Fructo-Oligosaccharides and Bifidobacterium breve M-16V Facilitates Partial Non-Responsiveness to Whey Protein in Mice Orally Exposed to beta-Lactoglobulin-Derived Peptides. Front. Immunol. 2017, 7, 673. 10.3389/fimmu.2016.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurl S.; Munzert M.; Boehm G.; Matthews C.; Stahl B. Systematic review of the concentrations of oligosaccharides in human milk. Nutr. Rev. 2017, 75 (11), 920–933. 10.1093/nutrit/nux044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L.; Langa S.; Martin V.; Maldonado A.; Jimenez E.; Martin R.; Rodriguez J. M. The human milk microbiota: origin and potential roles in health and disease. Pharmacol. Res. 2013, 69 (1), 1–10. 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Matheson M. C.; Allen K. J.; Tang M. L. Understanding the evidence for and against the role of breastfeeding in allergy prevention. Clin. Exp. Allergy 2012, 42 (6), 827–51. 10.1111/j.1365-2222.2011.03925.x. [DOI] [PubMed] [Google Scholar]
- Azad M. B.; Becker A. B.; Guttman D. S.; Sears M. R.; Scott J. A.; Kozyrskyj A. L. Canadian Healthy Infant Longitudinal Development Study, I., Gut microbiota diversity and atopic disease: does breast-feeding play a role?. J. Allergy Clin. Immunol. 2013, 131 (1), 247–8. 10.1016/j.jaci.2012.10.044. [DOI] [PubMed] [Google Scholar]
- Perkin M. R.; Logan K.; Tseng A.; Raji B.; Ayis S.; Peacock J.; Brough H.; Marrs T.; Radulovic S.; Craven J.; Flohr C.; Lack G. Randomized Trial of Introduction of Allergenic Foods in Breast-Fed Infants. N. Engl. J. Med. 2016, 374 (18), 1733–43. 10.1056/NEJMoa1514210. [DOI] [PubMed] [Google Scholar]
- Sharma V.; Eckels J.; Taylor G. K.; Shulman N. J.; Stergachis A. B.; Joyner S. A.; Yan P.; Whiteaker J. R.; Halusa G. N.; Schilling B.; Gibson B. W.; Colangelo C. M.; Paulovich A. G.; Carr S. A.; Jaffe J. D.; MacCoss M. J.; MacLean B. Panorama: a targeted proteomics knowledge base. J. Proteome Res. 2014, 13 (9), 4205–10. 10.1021/pr5006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.