Abstract
APOBEC3G is a potent inhibitor of HIV-1 replication, and act by deaminating cytidines in uracil on the negative strand of the viral cDNA. In this case-control study, APOBEC3G expression in subjects’ naïve to HAART infected by HIV-1 and the effect of APOBEC3G polymorphism on its expression were evaluated. The results show that the HIV-1 infected carriers of the G minor alleles of the variant rs8177832 had a higher expression of APOBEC3G mRNA than the controls carriers of the G minor allele. APOBEC3G polymorphisms could play an important role in the modulation of the HIV-1 dissemination.
Key words: HIV-1, APOBEC3G mRNA, rs8177832, rs35228531, rs6001417, Burkina Faso
Introduction
HIV-1 during its replication cycle interacts with the host restrictions factors of which APOBEC3G. The apolipoprotein B mRNA editing-enzyme catalytic polypeptide- like editing complex 3G (APOBEC3G), belongs to the cytidine deaminase family which comprise 10 proteins. 1 It is a potent inhibitor of HIV-1 replication, and act by deaminating cytidines in uracil on the negative strand of the viral cDNA and the virion infectivity factor Vif of HIV can hinder APOBECG action by targeting it to degradation in the proteasomes.2,3 APOBEC3G expression has been reported to be elevated in seronegative compared to HIV subtype B and C infected individuals.4,5 Although the latter findings were different, when the individuals were infected by circulating recombinant forms of HIV. A gene expression can be reduced by mutations. For this case-control study the aims were to evaluate the level of APOBEC3G expression in subjects’ naïve to HAART infected by HIV-1 and the effect of APOBEC3G polymorphisms on its expression.
Materials and Methods
Study population
All experiments were performed in compliance with relevant laws and institutional guidelines and in accordance with the ethical standards of the Declaration of Helsinki. Approval for the study was obtained from the Ethics Committee for Health Research of Burkina Faso. All study participants or guardians gave their free written and informed consent.
The study group consisted of 38 participants who were newly detected HIV+ naïve to treatment and 40 HIV negative individuals as controls. Both groups were recruited from the Saint Camille Hospital and the Pietro Annigoni Biomolecular Research Center (CERBA) cohorts and outpatients. Blood samples obtained by venipuncture were tested for HIV infection, using alere determine HIV-1/2 test, and the SD Bioline HIV1/2 test was used to differentiate between HIV-1 and HIV-2. CD4 cells were enumerated using the BD FACSCount CD4 Reagent kit on a BD FACS COUNT (Becton Dickinson, San Jose). Viral load was determined using the Abbott HIV-1 Real Time Quantitative kit (Promega, USA) on the Abbott m2000rt (Abbot Laboratories, Illinois) according to the manufacturer’s protocol.
DNA extraction and APOBEC3G expression
Genomic DNA was extracted from leucocytes using the DNA Rapid Salting-Out technique as described by Miller et al.6 APOBEC3G variants, rs8177832 (H186R), rs35228531 and rs6001417 were genotyped as previously described.7
Total RNA was extracted from frozen Plasma using the purelink pro kit (Invitrogen).
APOBEC3G mRNA expression was quantified in 38 HIV positive subjects and in 40 HIV negative subjects. Amplification of APOBEC3G was performed using a reverse transcription and target specific primers which are respectively: 5’-GCGGCCTTCAAGGAAACC- 3’, forward: 5’- CTGCTGAACCAGCGCAGG-3’ reverse: 5’-GCGGCCTTCAAGGAAACC-3’ and probe: 5’-CTTTCTATGCAACCAGGCTCCACATAAAC- 3’. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene to normalize for differences in cell counts or differences in RNA extraction.
Each 20 μL optimized PCR reaction mix contained: 10 μL of Express superscript Qpcr SuperMix Universal, 2 μL of each 50 μM primer, or GAPDH, 3 μL of probe mix (10 pmol/μL), 0.04 μL of ROX reference Dye, 2 μL of EXPRESS SUPERSCRIPT MIX for One step qPCR and 5 μL of RNA. The TaqMan quantitative real time RT-PCR was performed using the Applied Biosystems Prism 7500 Sequence Detection System, (Foster City, CA) according to the following Cycling conditions: 1 cycle at 50˚C for 15minute hold (cDNA synthesis), 1 cycle at 92˚C for 20 seconds and 45 cycles of 92˚C for 15 seconds and 60˚C for 1 minute. Values are expressed as calculated as fold change using the delta-Ct method.
Statistical analysis
The software SPSS version 20.0 and GRAPHPAD were used for data analysis. Power-Marker software Version 3.25 was used for the determination of the Hardy- Weinberg equilibrium and the calculation of allele and genotype frequencies. Differences between the groups were assessed using the Mann-Whitney U test for non-parametric analyses. Any value was considered statistically significant for P<0.05.
Ethical consideration
Approval for the study was obtained from the National Ethics Committee for Health Research of Burkina Faso. (Deliberation No. 2014-7-086). All study participants or guardians gave their free written and informed consent according to the Helsinki Declarations.
Results
Differences in mRNA expression
APOBEC3G mRNA was analyzed for all subjects in the study by Real-time qPCR. Higher expression levels of APOBEC3G were observed in HIV positive-untreated compared to controls (HIV-1 seronegative) but the difference was not significant (P=0.5457).
Among HIV positive participants, APOBEC3G expression level was higher in those with a CD4 count over 500 compared to the participants having a T CD4 counts less 500. This difference was significant (P=0.0411).
When comparing the mRNA expression among HIV positive individuals who had a viral load over 5000 copies/ml and those who had a viral load lower than 5000; the ladder had a higher expression of APOBEC3G. This difference was not statistically significant.
In this study, the population of interest was genotyped for 3 APOBEC3G polymorphisms rs6001417, rs8177832 and rs3522835. The expression level of APOBEC3G was compared among HIV positive and HIV negative participants, in relation to their different genotypes (Figures 1 and 2). For rs6001417, APOBEC3G expression level seems to be higher in HIV positive carriers of G alleles compared to controls, but the difference was not significant. When comparing APOBEC3G expression in relations to the genotypes of rs8177832 in cases and controls; the carriers of the G alleles also had a higher expression of APOBEC3G mRNA (P=0.0469). For rs352283, the carriers of the T allele in the HIV positive group have roughly a high expression level of APOBEC3G mRNA compared to HIV negative carrier of the T allele.
Figure 1.
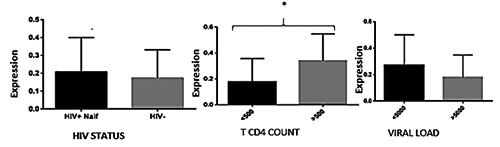
Comparison of APOBEC3G variants between case and control in function of its expression.
Figure 2.
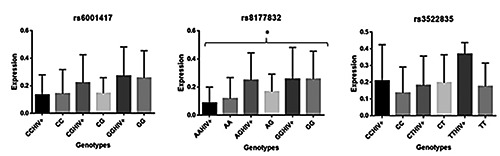
Comparison of APOBEC3G variants between case and control in function of its genotypes.
Discussion
In this study, the investigation was whether the expression level of APOBEC3G would be higher in the seronegative participants (control group) compared to the HIV positive participants. The results show that the APOBEC3G expression level is higher in the HIV positive group compared to the control group, which implies that APOBEC3G expression levels are upregulated upon HIV-1 infection. Although this finding is very different from many studies which found that APOBEC3G expression level was downregulated upon HIV-1 infection4,5,8,9; these results are similar to that of studies with samples from Senegal,10,11 and could be explained by the fact that in both Senegal and Burkina Faso located in West Africa where HIV-1 CRFs are predominant.12-14
When correlating the CD4 T count and the viral load to APOBEC3G expression level among HIV positive participants; individuals with a high CD4 count had a higher expression of APOBEC3G than the individuals with a low CD4 count, while participants with a low viral load seem to have a higher APOBEC3G expression level compared to the ones that have a high viral load. These results could be explained by the fact, that individuals with a high CD4 count express more APOBEC3G and thus control HIV replication thus a low viral load.11
The APOBEC3G mRNA expression in relations to the genotypes of rs8177832 in cases and controls shows that the HIV-1 infected carriers of the G minor alleles had a higher expression of APOBEC3G mRNA than the controls on the coding exon 4 and that a nucleotide change could modulate the expression of the putative gene. This study has some limits that are due on one hand by the small sample size and on the other hand by the limited number of the tested mutations which could influence APOBEC3G expression among the seropositive and seronegative patients.
In fact, different mutations have been associated either with the augmentation or the reduction of APOBEC3G gene and localized on the gene promoter have not been studied in the present work.15
In conclusion, this study supports previous findings on samples from West Africa that APOBEC3G expression level is higher in the HIV positive group compared to the control group, and that individuals with a high CD4 count had a higher expression of APOBEC3G than the individuals with a low CD4 count, while participants with a low viral load seem to have a higher APOBEC3G expression level compared to the ones with a high viral load.
Conclusions
The highlight of the study is that the HIV-1 infected carriers of the G minor alleles of the variant rs8177832 had a higher expression of APOBEC3G mRNA than the controls carriers of the G minor allele. Further investigations are needed to understand if the differences are due to HIV-1 subtypes or populations genetic variations.
Acknowledgements
The authors thank all the individuals who made this study possible. Their appreciation also goes to the whole team of the CERBA/LABIOGENE, and the Medical Center Saint Camille of Ouagadougou.
Funding Statement
Funding: The Permanent Secretary for the fight against AIDS and STIs (CNLS/IST) of Burkina Faso; the WAEMU/PACER2 and the Pietro Annigoni Association.
References
- 1.Jarmuz A, Chester A, Bayliss J, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 2002;79:285-96. [DOI] [PubMed] [Google Scholar]
- 2.Mehle A, Strack B, Ancuta P, et al. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem 2004;279:7792-8. [DOI] [PubMed] [Google Scholar]
- 3.Farrow MA, Sheehy AM. Vif and Apobec3G in the innate immune response to HIV: a tale of two proteins. Future Microbiol 2008;3:145-54. [DOI] [PubMed] [Google Scholar]
- 4.Jin X, Brooks A, Chen H, et al. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol 2005;79:11513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy K, Winkler CA, Werner L, et al. APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load. AIDS 2010;24:195-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Compaore TR, Soubeiga ST, Ouattara AK, et al. APOBEC3G Variants and Protection against HIV-1 Infection in Burkina Faso. PLoS One 2016;11: e0146386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandhi SK, Siliciano JD, Bailey JR, et al. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol 2008;82:3125-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho SJ, Drechsler H, Burke RC, et al. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol 2006;80:2069-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mous K, Jennes W, Camara M, et al. Expression analysis of LEDGF/p75, APOBEC3G, TRIM5alpha, and tetherin in a Senegalese cohort of HIV-1- exposed seronegative individuals. PLoS One 2012;7:e33934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulenga NK, Sarr AD, Thakore-Meloni S, et al. Relationship between human immunodeficiency type 1 infection and expression of human APOBEC3G and APOBEC3F. J Infect Dis 2008;198: 486-92. [DOI] [PubMed] [Google Scholar]
- 12.Nadembega WM, Giannella S, Simpore J, et al. Characterization of drug-resistance mutations in HIV-1 isolates from non-HAART and HAART treated patients in Burkina Faso. J Med Virol 2006;78:1385-91. [DOI] [PubMed] [Google Scholar]
- 13.Hamel DJ, Sankale JL, Eisen G, et al. Twenty years of prospective molecular epidemiology in Senegal: changes in HIV diversity. AIDS Res Hum Retroviruses 2007;23:1189-96. [DOI] [PubMed] [Google Scholar]
- 14.Kagone TS, Hien H, Meda N, et al. Characterization of HIV-1 genotypes and antiretroviral drug-resistance mutations among patients in Burkina Faso. Pak J Biol Sci 2011;14:392-8. [DOI] [PubMed] [Google Scholar]
- 15.Langlois MA, Beale RC, Conticello SG, et al. Mutational comparison of the single-domained APOBEC3C and double- domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res 2005;33: 1913-23. [DOI] [PMC free article] [PubMed] [Google Scholar]


