Abstract
Charcot-Marie-Tooth (CMT) disease type 1 is an inherited peripheral neuropathy characterized by demyelination and reduced nerve conduction velocities. We present a multi-generational family with peripheral neuropathy in whom clinical CMT panel testing failed to conclude a molecular diagnosis. We found a PMP2 pathogenic variant c.155T>C, p.(Ile52Thr) that segregates with disease suggesting that PMP2 variants should be considered in patients with neuropathy and that it may be prudent to include in clinical CMT gene panels.
Keywords: Charcot-Marie-Tooth disease, peripheral myelin protein 2, myelin P2 protein, peripheral neuropathy, CMT, PMP2
1. INTRODUCTION
Charcot-Marie-Tooth (CMT) disease, or hereditary motor and sensory neuropathy (HMSN), is an inherited peripheral neuropathy with an estimated prevalence of ~1 in 2500 [1,2]. CMT types are categorized based on clinical, electrophysiological, and histopathological features observed, i.e. demyelinating (type 1) or axonal (type 2) neuropathy [3,4]. CMT shows considerable genetic heterogeneity. To date, variants in more than 80 genes have been shown to cause CMT [5]. CMT type 1 is characterized by decreased motor nerve conduction velocities (< 38 m/s), sensory loss, progressive muscle weakness, distal limb atrophy and myelin defects [6]. Most patients with CMT have a duplication or missense variant in the PMP22 gene that encodes peripheral myelin protein of 22 kDa [7,8].
Recent studies have reported PMP2, encoding peripheral myelin protein 2, a myelin structural protein of 15 kDa, as a novel CMT1 disease gene in subjects from four families with autosomal dominant peripheral neuropathy [9,10,11]. PMP2 is present in Schwann cells in the peripheral nervous system (PNS) [12] and is a part of the highly conserved fatty acid-binding proteins (FABPs) family. PMP2 is involved in lipid homeostasis of myelin and may play a role in remyelination of the injured PNS [13,14]. Homozygous knockout Pmp2(−/−) mice showed a temporary reduction in motor nerve conduction velocities but did not show any overt myelin defects [13]. We describe an additional family with CMT1 with a rare variant in PMP2 c.155T>C, p.(Ile52Thr) identified by whole exome sequencing (WES) that segregates with disease; clinical gene panel testing was unable to conclude a molecular diagnosis
2. CASE PRESENTATION
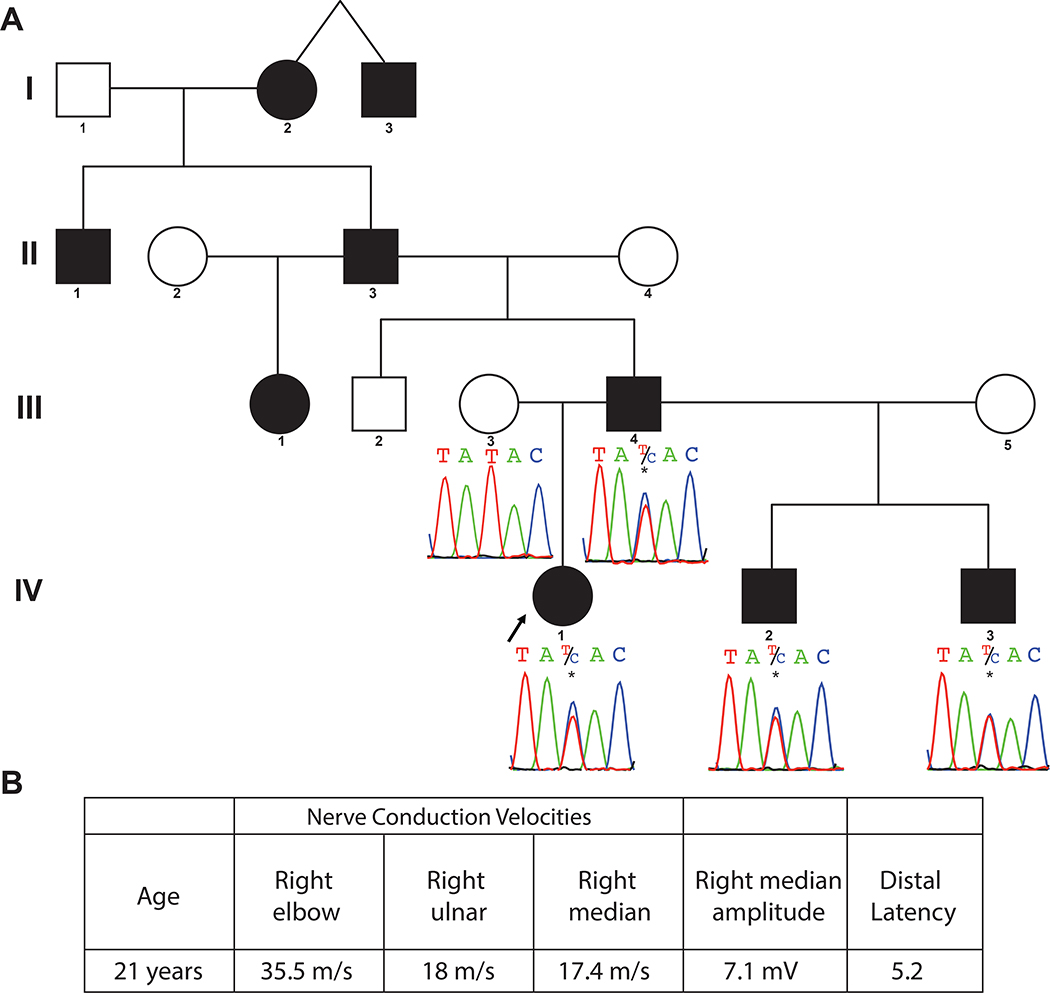
The proband is a 30 year old female with a clinical diagnosis of hereditary demyelinating neuropathy classified as CMT. Symptom onset was at 5 years of age, with a gradual increase in lower extremity weakness. Bilateral pes cavus was noted at 12 years, progressing to pes planus by 20 years; she has undergone at least two right foot orthopedic procedures. Examination at 21 years demonstrated lower extremity weakness, particularly of dorsiflexion at the ankles, absence of deep tendon reflexes, and an abnormal steppage gait. Four generations of similarly affected relatives were noted, with male to male transmission of the disease trait, most consistent with an autosomal dominant condition (Figure 1A). Her nerve conduction studies were consistent with a demyelinating neuropathy (Figure 1B). Prior to WES, the proband underwent extensive genetic testing including deletion and duplication testing for PMP22, as well as clinical CMT gene panel testing through Athena diagnostics (11 genes) and Fulgent Genetics (49 genes); the list of genes tested in these panels is included in Supplementary material (Table S1).
Figure 1.
A. Pedigree and Sanger variants. Four-generation pedigree shows the affected proband and 8 additional paternal relatives (filled circles and squares). Sanger sequencing of the identified PMP2 c.155T>C, p.(Ile52Thr) variant demonstrates segregation of the variant with the phenotype in the proband, father, and two paternal half-brothers, and absence of the variant in the unaffected mother.
B. Nerve conduction studies of proband. Markedly reduced motor nerve conduction velocities of proband at 21 years were consistent with CMT1 (<38 m/s) demyelinating neuropathy.
3. METHODS
Informed consent was obtained from the proband and available family members in accordance with the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) research protocol (Baylor College of Medicine IRB protocol number: H-29697). Whole exome sequencing (WES) was performed on the proband, and affected paternal half-brothers. Sequencing and variant prioritization workflow was performed as previously described [15]. Sanger sequencing was used for confirmation and segregation of the potential disease-causing variant.
4. RESULTS
WES analysis revealed a heterozygous rare variant c.155T>C, p.(Ile52Thr) in PMP2 (NM_002677) in the proband and affected paternal half-brothers. Sanger sequencing confirmed the variant and segregation according to Mendelian expectations for an autosomal dominant trait; i.e. multigenerational vertical transmission and evidence for male-to-male transmission (Figure 1). This variant has been reported as disease-causing in a family with CMT1 [11], is predicted likely damaging by multiple bioinformatic algorithms [16,17,18 19], is conserved (PhyloP) [20], and has a likely pathogenic CADD [21] score of 28.3.
5. DISCUSSION
We first reported PMP2 as a candidate CMT gene in a family with demyelinating neuropathy [9]. The putative pathogenic variant in PMP2 (p.Ile43Asn) segregated with disease. Further functional evidence of PMP2 pathogenicity was shown by morpholino knockdown of PMP2 orthologues in zebrafish leading to a motor neuron phenotype; the disease phenotype could be rescued by wild-type human PMP2 mRNA. However, overexpression of wild-type human mRNA also resulted in a motor neuron phenotype suggesting dosage sensitivity of the PMP2 transcript. Hong et al [10] described an autosomal dominant family with demyelinating CMT neuropathy with the same PMP2 (p.Ile43Asn) variant. They assessed PMP2 pathogenicity using transgenic mouse models to show that overexpression of wild-type as well as mutant PMP2 caused abnormal motor function resembling the CMT1 phenotype. Motley et al [11] described two families with demyelinating neuropathy with potential de novo dominant pathogenic variants in PMP2 (p.Thr51Pro, p.Ile52Thr) causing disease. Ruskamo et al [22] studied the molecular basis of known PMP2 disease variants by X-ray crystallography and determined that the specific variants did not alter overall folding of the protein but altered its biophysical properties and functional dynamics.
The identification of antibodies to PMP2 in animal models of Guillain-Barre syndrome [23,24], provided an intriguing molecular link between inherited and autoimmune-mediated neuropathies. As PMP2 plays a role in lipid homeostasis of myelin and may bind to cholesterol [25,26], potentially considering future therapeutic interventions with a cholesterol rich diet may be warranted [27]. Additionally, patients being treated with statins should be carefully monitored for exacerbation of neuromuscular symptoms.
Our report presents additional evidence classifying PMP2 as an established neuropathy disease gene, and supports the inclusion of PMP2 in routine clinical testing for distal symmetric polyneuropathy [28]. Moreover, we document limitations of disease gene panel testing for molecular diagnosis as newly described disease genes may not be part of the clinical testing panel. The continued elucidation of the molecular etiology of CMT informs understanding of the pathogenesis of inherited neuropathy, improves molecular testing, further empowers genetic counseling, enables more robust prognostic information, and guides future development of targeted therapies for this chronic, progressive condition [29].
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the family for their participation.
FUNDING SOURCES
This work was supported in part by the National Institutes of Health, National Institute of Neurologic Disorders and Stroke [R35 NS105078] and the Muscular Dystrophy Association [#512848] to JRL; and a jointly funded National Human Genome Research Institute (NHGRI), and National Heart, Lung, and Blood Institute (NHLBI) grant to the Baylor-Hopkins Center for Mendelian Genomics [UM1 HG006542]. JEP is supported by NHGRI [K08 HG008986].
Abbreviations
- CMT
Charcot-Marie-Tooth disease
- NCV
Nerve conduction velocity
- UL
Upper limb
- LL
Lower limb
- CADD
Combined annotation dependent depletion
Footnotes
CONFLICTS OF INTEREST
J.R.L. has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. Other authors have no potential conflicts to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Skre H Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet. 1974;6(2):98–118. [DOI] [PubMed] [Google Scholar]
- [2].Braathen GJ. Genetic epidemiology of Charcot-Marie-Tooth disease. Acta Neurol Scand Suppl. 2012;(193):iv–22. [DOI] [PubMed] [Google Scholar]
- [3].Murakami T, Garcia CA, Reiter LT, Lupski JR.Charcot-Marie-Tooth disease and related inherited neuropathies. Medicine (Baltimore). 1996;75(5):233–50. [DOI] [PubMed] [Google Scholar]
- [4].Saporta AS, Sottile SL, Miller LJ, Feely SM, Siskind CE, Shy ME. Charcot-Marie-Tooth disease subtypes and genetic testing strategies. Ann Neurol. 2011;69(1):22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vaeth S, Christensen R, Dunø M, Lildballe DL, Thorsen K, Vissing J, et al. Genetic analysis of Charcot-Marie-Tooth disease in Denmark and the implementation of a next generation sequencing platform. Eur J Med Genet. 2018; pii: S1769–7212(17)30717–6. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- [6].Harding AE, Thomas PK. The clinical features of hereditary motor and sensory neuropathy types I and II. Brain. 1980;103(2):259–80. [DOI] [PubMed] [Google Scholar]
- [7].DiVincenzo C, Elzinga CD, Medeiros AC, Karbassi I, Jones JR, Evans MC, et al. The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol Genet Genomic Med. 2014;2(6):522–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rossor AM, Tomaselli PJ, Reilly MM. Recent advances in the genetic neuropathies. Curr Opin Neurol. 2016;29(5):537–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015;12(7):1169–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hong YB, Joo J, Hyun YS, Kwak G, Choi YR, Yeo HK, et al. A Mutation in PMP2 Causes Dominant Demyelinating Charcot-Marie-Tooth Neuropathy. PLoS Genet. 2016;12(2):e1005829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Motley WW, Palaima P, Yum SW, Gonzalez MA, Tao F, Wanschitz JV, et al. De novo PMP2 mutations in families with type 1 Charcot-Marie-Tooth disease. Brain. 2016;139(Pt 6):1649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Trapp BD, Dubois-Dalcq M, Quarles RH. Ultrastructural localization of P2 protein in actively myelinating rat Schwann cells. J. Neurochem 1984;43:944–948. [DOI] [PubMed] [Google Scholar]
- [13].Zenker J, Stettner M, Ruskamo S, Domènech-Estévez E, Baloui H, Médard JJ, et al. A role of peripheral myelin protein 2 in lipid homeostasis of myelinating Schwann cells. Glia. 2014;62(9):1502–12. [DOI] [PubMed] [Google Scholar]
- [14].Stettner M, Zenker J, Klingler F, Szepanowski F, Hartung HP, Mausberg AK, et al. The Role of Peripheral Myelin Protein 2 in Remyelination. Cell Mol Neurobiol. 2018;38(2):487496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].White JJ, Mazzeu JF, Coban-Akdemir Z, Bayram Y, Bahrambeigi V, Hoischen A, et al. WNT Signaling Perturbations Underlie the Genetic Heterogeneity of Robinow Syndrome. Am J Hum Genet. 2018;102(1):27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–2. [DOI] [PubMed] [Google Scholar]
- [19].Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. [DOI] [PubMed] [Google Scholar]
- [20].Cooper GM, Shendure J. Needles in stacks of needles: finding disease causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. [DOI] [PubMed] [Google Scholar]
- [21].Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ruskamo S, Nieminen T, Kristiansen CK, Vatne GH, Baumann A, Hallin EI, et al. Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2. Sci Rep. 2017;7(1):6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ishaque A, Szymanska I, Ramwani J, Eylar EH. Allergic neuritis: phospholipid requirement for the disease-inducing conformation of the P2 protein. Biochim Biophys Acta. 1981;669(1):28–32. [DOI] [PubMed] [Google Scholar]
- [24].Ishaque A, Hofmann T, Eylar EH. The complete amino acid sequence of the rabbit P2 protein. J Biol Chem. 1982;257(2):592–5. [PubMed] [Google Scholar]
- [25].Majava V, Polverini E, Mazzini A, Nanekar R, Knoll W, Peters J, et al. Structural and Functional Characterization of Human Peripheral Nervous System Myelin Protein P2. PLoS One. 2010; 5(4): e10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sedzik J, Jastrzebski JP. High-resolution structural model of porcine P2 myelin membrane protein with associated fatty acid ligand: fact or artifact? J Neurosci Res. 2011;89(6):909–20. [DOI] [PubMed] [Google Scholar]
- [27].Saher G, Rudolphi F, Corthals K, Ruhwedel T, Schmidt KF, Löwel S, et al. Therapy of Pelizaeus-Merzbacher disease in mice by feeding a cholesterol-enriched diet. Nat Med. 2012;18(7):1130–5. [DOI] [PubMed] [Google Scholar]
- [28].England JD, Gronseth GS, Franklin G, Carter GT, Kinsella LJ, Cohen JA, et al. Practice Parameter: evaluation of distal symmetric polyneuropathy: role of laboratory and genetic testing (an evidence-based review). Report of the American Academy of Neurology, American Association of Neuromuscular and Electrodiagnostic Medicine, and American Academy of Physical Medicine and Rehabilitation. Neurology. 2009;72(2):185–92. [DOI] [PubMed] [Google Scholar]
- [29].Harel T, Lupski JR. Charcot-Marie-Tooth disease and pathways to molecular based therapies. Clin Genet. 2014;86(5):422–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.