Abstract
The development of multiscale models of infectious disease systems is a scientific endeavour whose progress depends on advances on three main frontiers: (a) the conceptual framework frontier, (b) the mathematical technology or technical frontier, and (c) the scientific applications frontier. The objective of this primer is to introduce foundational concepts in multiscale modelling of infectious disease systems focused on these three main frontiers. On the conceptual framework frontier we propose a three-level hierarchical framework as a foundational idea which enables the discussion of the structure of multiscale models of infectious disease systems in a general way. On the scientific applications frontier we suggest ways in which the different structures of multiscale models can serve as infrastructure to provide new knowledge on the control, elimination and even eradication of infectious disease systems, while on the mathematical technology or technical frontier we present some challenges that modelers face in developing appropriate multiscale models of infectious disease systems. We anticipate that the foundational concepts presented in this primer will be central in articulating an integrated and more refined disease control theory based on multiscale modelling - the all-encompassing quantitative representation of an infectious disease system.
Keywords: Multiscale models of infectious diseases, Immuno-epidemiological models, Linking individual/lower/micro and population/upper/macro scales, Comparative effectiveness research
1. Introduction
Infectious diseases continue to pose a major threat to human health. Although advances in medicine and public health have helped control many endemic diseases, World Health Organization (WHO) study on the global burden of diseases indicates that by 2002, infectious diseases were the cause for more than one quarter of approximately 57 million deaths worldwide (World Health Organization (WHO), 2004). In addition, approximately two thirds of all deaths in developing countries among children younger than 5 years of age are due to infectious diseases (World Health Organization (WHO), 2005). In order to respond effectively to the growing health threats of infectious diseases such as HIV/AIDS, tuberculosis, malaria and the transfer of these health risks we need to identify and implement high-impact health interventions. Therefore, there is need to develop appropriate mathematical modelling techniques to evaluate the effectiveness and value of health interventions in the control, elimination and eradication of infectious diseases to complement clinical trial studies, systematic reviews, innovative research strategies, and clinical registries. However, there is limited use of appropriate mathematical modelling techniques to achieve this. Since disease dynamics is strongly scale-dependent, mathematical models of mechanisms underlying health interventions, especially multiscale models that synthesize information from at least two scales describing variation in disease mechanisms in the patient population, are useful tools for health intervention research.
The past few years have witnessed a dramatic increase in our awareness and the need to understand the multiscale nature of infectious disease dynamics (see (Cen, Feng, & Zhao, 2014; Feng et al., 2013; Feng et al., 2015; Garira et al., 2014; Netshikweta and Garira, 2017) and references therein). Despite the recognition of the importance of multiscale analysis of infectious disease dynamics, a large number of published papers focused on modelling the dynamics of infectious diseases to date still do not address the multi-spatial and/or the multi-temporal scales of these disease systems. This may be largely attributed to the absence of a powerful foundational knowledge to support the development of multiscale modelling of infectious disease systems on three main frontiers: (a) the conceptual framework frontier, (b) the scientific applications frontier, and (c) the mathematical technology frontier. The objective of this primer is to introduce foundational concepts in multiscale modelling of infectious disease systems focusing on these three main frontiers. The conceptual framework frontier is introduced by proposing a three-level hierarchical framework of an infectious disease system. This is achieved by conceptualizing an infectious disease system as being organized into three main fundamental hierarchical levels which are: (i) the cell level, (ii) the tissue level, and (iii) the host level which serve as the units of multiscale analysis. Each of these three levels can be resolved into two adjacent scales which are the individual/lower/micro scale and population/upper/macro scale. For the cell level we have the within-cell scale and the between-cell scale, for the tissue level we have the within-tissue scale and the between-tissue scale, and for the host level we have the within-host scale and the between-host scale. At each of these three hierarchical levels (the cell level, the tissue level, and the host level) five different categories of multiscale models of an infectious disease system can be developed that integrate the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) and population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) (Garira, 2017). In what follows, we briefly describe each of these five categories of multiscale models of an infectious disease system.
Category I - individual based multiscale models (IMSMs)
In this category of multiscale models of infectious disease systems the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) submodel is used to describe the entire infectious disease system across both the individual/lower/micro scale and population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale). There is no information flow from the population/upper/macro scale submodel to the individual/lower/micro scale submodel. The population/upper/macro scale is observed as emergent behaviour of the individual/lower/micro scale entities. The four main classes of multiscale models in this category are (Garira, 2017):
-
•
Class 1: Network modelling individual-based multiscale models (NETW-IMSMs): These are multiscale models which are developed using graph theoretic or network modelling techniques.
-
•
Class 2: Empirical data modelling individual-based multiscale models (EMPI-IMSMs): These are multiscale models of infectious disease systems which are developed using statistical modelling techniques to model hierarchical empirical data. For example those which use regression-based approaches where the assumption that the outcomes of infection at individual/lower/micro scale for the units of analysis (i.e. the individual hosts) are independent is violated because they share the same population/upper/macro scale characteristics and are therefore influenced by the same measured population/upper/macro scale factors (e.g. access to health services, demographic factors, environmental factors, economic factors, etc.) or unmeasured factors (e.g. cultural factors, religious factors, behavioural factors, etc.).
-
•
Class 3: Simulation modelling individual-based multiscale models (SIMU-IMSMs): No mathematical equations are used to model the infectious disease system in this class of multiscale models. Instead, infectious disease systems are modelled using computational algorithms such computational algorithm-based models which include agent-based models (ABM), cellular automata (CA) and petri-nets (PN). If mathematical equations appear in this class, they are only used to describe specific entities with a particular scale, rather than to describe the dynamics of a whole scale.
-
•
Class 4: Hybrid individual-based multiscale models (BRID-IMSMs): These are individual-based multiscale models where the individual entities within a single scale are represented using different formalisms or mathematical representations. For example, some entities in an agent-based model (ABM) my be described by ODEs while others are described by PDEs. The use of hybrid petri nets will also result in BRID-MSMs.
Category II - nested multiscale models (NMSMs)
These are multiscale models of infectious diseases in which there is only unidirectional flow of information (only from individual/lower/micro scale submodel to the population/upper/macro submodel). Therefore, in this category of multiscale models the individual/lower/micro scale dynamics is independent of the population/upper/macro scale. The individual/lower/micro scale submodel and the population/upper/macro scale submodel must be described by the same formalism or mathematical representation. The three main classes of multiscale models in this category are (Garira, 2017):
-
•
Class 1: Transformation based nested multiscale models (TRAN-NMSMs): Here the individual/lower/micro scale submodel is formally transformed into a population/upper/macro scale model. They are formulated through developing physiologically structured population/upper/macro scale submodels. This task is accomplished by subdividing the entire host population into various sub-classes corresponding to the different levels of immune protection: naive or completely susceptible, completely or partially immune, vaccinated, immune compromised or protected from infection due to certain genetic factors.
-
•
Class 2: Unidirectional coupling based nested multiscale models (UNID-NMSMs): The nature of the multiscale model in this class is such that there is strictly one-way inter-scale information flow among the two submodels (from the individual/lower/micro scale submodel to the population/upper/macro scale submodel).
-
•
Class 3: Simplification based nested multiscale models (SIMP-NMSMs): These are multiscale models of infectious disease systems which are formulated by simplifying or reducing the order/dimensions of UNID-MSMs in class 2 of this category. The simplification or reduction of order is sometimes achieved by using methods such as slow and fast time scale analysis (Feng et al., 2015) or dynamical systems based methods such as centre manifold theory (Carr, 2012).
Category III - embedded multiscale models (EMSMs)
These are MSMs of infectious disease systems where both the individual/lower/micro scale submodel and the population/upper/macro scale submodel influence each other in a reciprocal way. The individual/lower/micro scale submodel and the population/upper/macro scale submodel must be described by the same formalism or mathematical representation. The two main classes of multiscale models in this category are (Garira, 2017):
-
•
Class 1: Bidirectionally Coupled Embedded Multiscale Models (BIDI-EMSMs): These are multiscale models of infectious disease systems which are such that there is strictly two-way inter-scale information flow (the individual/lower/micro scale submodel and the population/upper/macro scale submodel are bidirectionally coupled).
-
•
Class 2: Simplification based embedded multiscale models (SIMP-EMSMs): These are multiscale models of infectious disease systems which are formulated by simplifying or reducing the order/dimensions of BIDI-MSMs in class 1 of this category. The simplification or reduction of order is sometimes achieved by identifying assumptions that lead to the decoupling of the submodels. Although the two submodels will then be independent and can be analysed independently, their synergy gives an added value to the overall multiscale description of the infectious disease system.
Category IV: hybrid multiscale models (HMSMs)
These are multiscale models of infectious disease systems in which the individual/lower/micro scale submodel and population/upper/macro scale submodel are described by different mathematical representations. Examples of such paired formalisms are deterministic/stochastic, discrete time/continuous time, mechanistic/phenomenological, ODE/PDE, ODE/ABM, ODE/CA, etc. The three main classes of multiscale models in this category are (Garira, 2017):
-
•
Class 1: Unidirectionally coupled hybrid multiscale models (UNID-HMSMs): These are multiscale models of infectious disease systems in which there is strictly one-way inter-scale information flow from the individual/lower/micro scale to the population/upper/macro scale. They are similar to UNID-NMSMs of class 2 in category II except that in this case the submodels are described by different formalisms.
-
•
Class 2: Bidirectionally Coupled Hybrid Multiscale Models (BIDI-HMSMs): These are MSMs of infectious disease systems which are such that there is strictly two-way inter-scale information flow between the individual/lower/micro scale to the population/upper/macro scale. They are similar to BIDI-EMSMs in class 1 of category III except that in this case the individual/lower/micro scale and the population/upper/macro scale submodels are described by different mathematical representations.
-
•
Class 3: Simplification based hybrid multiscale models (SIMP-HMSMs): This class of hybrid multiscale is established by finding simplified versions of the UNID-HMSM of class 1 or BIDI-HMSMs of class 2 in this category.
Category V - coupled multiscale models (CMSMs)
These are multiscale models of infectious disease systems which consider multi-strain infections, multi-pathogen infections, multi-group infections, multi-host infections, multi-level infections, multi-geographical environments infections, multi-biological environments infections. They are not like categories I, II, II, and IV which focus on specific one-host-one-pathogen relationships in multiscale modelling of infectious disease systems. The five main classes of multiscale models in this category are (Garira, 2017):
-
•
Class 1: Single-host multi-pathogen coupled multiscale models (SHMP-CMSMs): These are multiscale models of infectious disease systems which take into account the diversity of pathogens implicated in the transmission of an infectious disease system (either multi-pathogen infections or multi-strain infections or multi-group infections) so that the individual/lower/micro scale submodel and the population/upper/macro scale submodel for each pathogen are developed and then integrated into a single multiscale model.
-
•
Class 2: Multi-host single-pathogen coupled multiscale models (MHSP-CMSMs): These are multiscale models of infectious disease systems which take into account the diversity of hosts implicated in the transmission of an infectious disease system (i.e. infectious disease systems arising from infection of multiple host species by a single pathogen).
-
•
Class 3: Multiple level single-pathogen coupled multiscale models (MLSP-CMSMs): These are multiscale models of infectious disease systems which take into account the diversity of organizational levels implicated in the transmission of an infectious disease system (the cell level, the tissue level, the host level).
-
•
Class 4: Multiple geographical environments/biological environments single-pathogen coupled multiscale models (MGSP/MBSP-CMSMs): These are HL-MSMs which take into account the diversity of geographical environments/biological environments implicated in the transmission of an infectious disease system.
-
•
Class 5: Multi-host, multi-pathogen, multiple-levels coupled multiscale models (MHMP-CMSMs): These are MSMs of infectious disease systems which take into account the diversity of organizational levels and/or diversity of geographical environments/biological environments and/or diversity of hosts and/or diversity of pathogens implicated in the transmission of an infectious disease system.
For details of examples of multiscale models of infectious disease systems in each of the classes within each of the five categories see (Garira, 2017). However, in addition to presenting foundational concepts on the conceptual framework frontier of multiscale modelling of infectious disease systems based on a three-level hierarchical framework, this primer also builds on the conceptual framework and use it to introduce foundational concepts on the mathematical technology frontier and the scientific applications frontier. The foundational concepts on the scientific applications frontier are introduced by highlighting the applications of the different structures of multiscale models in the control, elimination and eradication of infectious disease systems, while the foundational concepts on the mathematical technology or technical frontier are introduced by way of a comprehensive discussion on the challenges of multiscale modelling of infectious disease systems. In this primer we first present the three-level hierarchical of an infectious disease system and the different structures of multiscale models that can be developed at each of the three hierarchical levels in section 2. The causality for linkage across scales in the different structures of multiscale models is explained in section 3 in terms of key disease processes. In section 4 we propose scientific applications of multiscale models in the control, elimination and eradication of infectious disease systems. Section 5 considers the mathematical technology frontier and explain the main challenges in multiscale modelling of infectious disease systems. The paper concludes with section 6 which explains the usefulness of the foundational concepts of multiscale modelling of infectious disease systems presented in this primer.
2. The three-level hierarchical framework of an infectious disease system and the structure of multiscale models
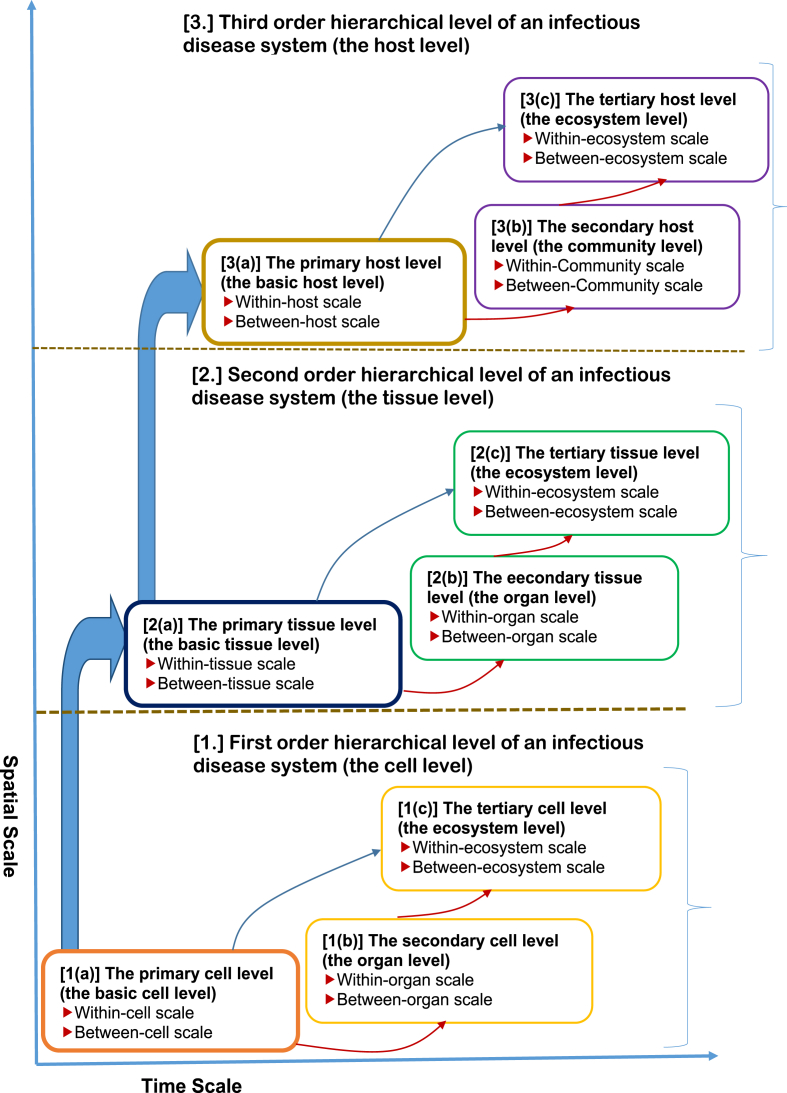
When developing multiscale models of disease dynamics, it is important to define the perspective that is used to inform the development of such models. The perspective may be different for infectious diseases and non-infectious diseases. Such a perspective needs to specify the causality for linkage across scales. In this study the multiscale nature of infectious disease dynamics is informed by a pathogen centred-perspective. Within this perspective an infectious disease system is conceptualized to be composed of three main orders of hierarchical levels which are (i) the cell level, (ii) the tissue level and (iii) the organism/host level. Each of these three main orders of hierarchical levels of organization of an infectious disease system constitute a different pathogen habitat or environment in which the pathogen can survive, grow, shed/excreted, replicate, and be transmitted to different scales of an infectious disease system. In the context of this perspective, each of these three orders of hierarchical levels of organization of an infectious disease system (cell level, tissue level and organism/host level) can, depending on the causative agent of the infectious disease system, constitute a different unit of multiscale analysis of an infectious disease system defined by its pathogen burden which is determined by (a) the intensity of its replication at one scale within a level, (b) pathogen transmission at another scale within the same level, and (c) disease interventions related to control, elimination and even eradication of the pathogen which are implemented and operating at different scales of infection. Central to this perspective is the idea that the ideal minimum requirements for describing the dynamics of an infectious disease system is two scales and that infectious disease dynamics can be analysed in successive scales of two adjacent scales at a time by introducing the concept of hierarchical levels (Garira, 2017) in multiscale analysis: (i) a lower scale (within-cell scale, within-tissue scale, within-host scale) in which pathogen replication often occurs and (ii) an upper scale (between-cell scale, between-tissue scale, between-host scale) in which pathogen transmission often occurs. The three main orders of hierarchical levels of an infectious disease system are perhaps best illustrated by a space-time diagram, which portrays the hierarchical nature of these levels and the positive correlation in spatial and temporal scales of varying disease processes. Fig. 1 is a conceptual representation of the three main orders of hierarchical level of an infectious disease system and the associated two limiting scales of infection for each level.
Fig. 1.
A conceptual diagram of the three-level hierarchical framework of an infectious disease system and the associated two limiting scales of infection for each level.
In what follows, we briefly describe each of the three main orders of hierarchical levels of organization of an infectious disease system.
-
1.First order hierarchical level of an infectious disease system (the cell level). At this level the cell is the basic unit of multiscale analysis. Multiscale models developed at this order of hierarchical level of an infectious disease system are called cell level immuno-epidemiological models (CL-IEMs) or cytoimmuno-epidemiological models (Garira, 2017) or simply cell-level multiscale models (CL-MSMs). Within this hierarchical level of an infectious disease system there are three sub-levels which are (a) the primary cell level which we also alternatively refer to as the basic cell level, (b) the secondary cell level which we also refer to as the organ level and (c) the tertiary cell level which we alternatively call the ecosystem level. Infectious disease systems modelled at this level of organization of an infectious disease system are those in which the pathogen infects specific cells and cause damage to these cells and the associated tissues and organs of the host. Examples of infectious disease systems which can be modelled at this hierarchical level of organization are some bacterial infections such as paratuberculosis and viral infections. We briefly describe each of the sub-levels and the structure of cell level multiscale models (CL-MSMs) of infectious disease systems that can be developed at each of these sub-levels as follows.
-
1(a)The primary cell level (the basic cell level): At this sub-level of the first order hierarchical level of organization of an infectious disease system multiscale models are developed that integrate within-cell scale and between-cell scale in the context of one-pathogen and one-target cell species scenario. In addition such CL-IEMs are developed to describe disease dynamics associated with a single anatomical compartment/organ of the host. We refer to the CL-IEMs developed to describe this sub-level of the first order hierarchical level of an infectious disease system as having a primary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these CL-IEMs with a primary structure fall into categories I, II, III and IV of multiscale models which are individual based multiscale models (IMSMs), nested multiscale models (NMSMs) and hybrid multiscale models (HMSMs) and embedded multiscale models (EMSMs).
-
1(b)The secondary cell level (the organ level): At this sub-level of the first order hierarchical level of an infectious disease system multiscale models are developed that also integrate within-cell scale and between-cell scale in the context of one-target cell species or multiple-target cell species and one-pathogen species scenario. However, unlike at the primary cell level, multiscale models here are developed to describe disease dynamics in more than one anatomical compartment/organ of the host. Although such multiscale models still integrate within-cell scale and between-cell scale, they are developed to incorporate more than one anatomical compartment/organ of the whole body/organism. Thus CL-IEMs developed at this sub-level of the first order hierarchical level of an infectious disease system describe disease dynamics in which infected cell populations are distributed over distinctly different anatomical compartments/organs of the host body (e.g. the liver and the blood in the case of malaria) with the cell as the unit of multiscale analysis. We refer to the CL-IEMs developed to describe this sub-level of the first order hierarchical level of an infectious disease system as having a secondary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these CL-IEMs with a secondary structure fall into class 4 (multiple biological environments single-pathogen coupled multiscale models (MBSP-CMSMs)) of category V which takes into account the different infected anatomical compartments/organs of the host by a single pathogen in multiscale modelling of infectious disease systems.
-
1(c)The tertiary cell level (the ecosystem level): At this sub-level of the first order hierarchical level of an infectious disease system, multiscale models are still developed that integrate the within-cell scale and the between-cell scale. However, unlike the primary cell level and the secondary cell level, the CL-IEMs developed at this sub-level of the first order hierarchical level of an infectious disease system consider the cell level as an ecosystem. Thus the CL-IEMs developed at this sub-level of an infectious disease system incorporate more than one anatomical compartment/organ of the whole body/organism in the context of multi-target cell species and/or multi-group target cell species and/or multi-pathogen species and/or multi-pathogen strain species in the development of the multiscale models with the cell as the unit of multiscale analysis. We refer to the CL-IEMs developed to describe this sub-level of the first order hierarchical level of an infectious disease system as having a tertiary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these CL-IEMs with a tertiary structure fall into classes 1, 2, 3 and 5 of category V of multiscale models of infectious disease systems.
-
1(a)
Overall, multiscale models developed at this hierarchical level of an infectious disease system describe whole body/organism infection using the cell as the unit of multiscale analysis.
-
2.Second order hierarchical level of an infectious disease system (the tissue level). At this level the tissue is the basic unit of multiscale analysis. Examples of tissues considered in the multiscale modelling effort are the granulomas (see (Garira, 2017) and references therein) or the microabscess (Pigozzo et al., 2012). Multiscale models developed at this order of hierarchical level of an infectious disease system are called tissue level immuno-epidemiological models (TL-IEMs) or histoimmuno-epidemiological models (Garira, 2017). Within this hierarchical level of organization of an infectious disease system there are three sub-levels which are (a) the primary tissue level which we alternatively call the basic tissue level, (b) the secondary tissue level which we also alternatively call the organ level and (c) the tertiary tissue level which we sometimes refer to as the ecosystem level. Infectious disease systems modelled at this level of organization of an infectious disease system are those in which the pathogen does infect specific tissues such as granulomas for some bacteria, fungi, protozoa and helminthes infections (James and Zumla, 1999; Permi et al., 2012; Zumla and James, 1996) and microabscess (Pigozzo et al., 2012), and cause damage to the host organs. In what follows, we briefly describe each of the three sub-levels and the structure of tissue level multiscale models (TL-MSMs) of infectious disease systems that can be developed at each of these sub-levels.
-
2(a)The primary tissue level (the basic tissue level): At this sub-level of the second order hierarchical level of an infectious disease system multiscale models are developed that integrate within-tissue scale and between-tissue scale in the context of one-pathogen and one-target tissue species scenario. In addition such multiscale models are developed to describe disease dynamics at a single anatomical compartment/organ of the host. We refer to TL-IEMs developed to describe this sub-level of the second order hierarchical level of an infectious disease system as having a primary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these TL-IEMs with a primary structure fall into categories I, II, III and IV of multiscale models which are individual based multiscale models (IMSMs), nested multiscale models (NMSMs) and hybrid multiscale models (HMSMs) and embedded multiscale models (EMSMs).
-
2(b)The secondary tissue level (the organ level): At this sub-level of the second order hierarchical level of an infectious disease system multiscale models are developed that also integrate within-tissue scale and between-tissue scale in the context of one-target tissue or multiple-target tissue species and one-pathogen species scenario. However, unlike the primary tissue level, multiscale models here are developed incorporating more than one anatomical compartment/organ of the host. Although such multiscale models still integrate within-tissue scale and between-tissue scale, they are developed to incorporate more than one anatomical compartment/organ of the whole body/organism. Thus TL-IEMs developed at this sub-level of the second order hierarchical level of an infectious disease system describe disease dynamics in which infected tissue populations are distributed over distinctly different anatomical compartments/organs of the whole body with the tissue as the unit of multiscale analysis. We refer to TL-IEMs developed to describe this sub-level of the second order hierarchical level of an infectious disease system as having a secondary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these TL-IEMs with a secondary structure fall into class 4 (multiple biological environments single-pathogen coupled multiscale models (MBSP-CMSMs)) of category V which takes into account the different infected anatomical compartments/organs of the host by a single pathogen in multiscale modelling of infectious disease systems.
-
2(c)The tertiary tissue level (the ecosystem level): At this sub-level of the second order hierarchical level of an infectious disease system, multiscale models of an infectious disease system are developed that still integrate the within-tissue scale and the between-tissue scale. However, unlike the primary tissue level and the secondary tissue level the TL-IEMs developed at this sub-level of the second order hierarchical level of an infectious disease system consider the tissue level as an ecosystem. Thus the multiscale models at this sub-level incorporate more than one anatomical compartment/organ of the whole body/organism in the context of multi-target tissue species and/or multi-group target tissue species and/or multi-pathogen species and/or multi-pathogen strain species in the development of the multiscale models with the tissue as the unit of multiscale analysis. At this sub-level of the second order hierarchical level of an infectious disease system the TL-IEMs are referred to as having a tertiary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these TL-IEMs with a tertiary structure fall into classes 1, 2, 3 and 5 of category V of multiscale models of infectious disease systems.
-
2(a)
Multiscale models developed at this hierarchical level of an infectious disease system also describe whole body/organism infection using the tissue (granuloma, microabscess, hydatid cyst, etc.) as the unit of multiscale analysis.
-
3.Third order hierarchical level of an infectious disease system (the host level). At this order of hierarchical level of an infectious disease system, the host is the basic unit of multiscale analysis. Multiscale models developed at this order of hierarchical level of an infectious disease system are called host level immuno-epidemiological models (HL-IEMs) (Garira, 2017). At this hierarchical level of an infectious disease system there are also three sub-levels which are (a) the primary host level which we also refer to as the basic host level, (b) the secondary host level which we alternatively call the community level, and (c) the tertiary host level which we also call the ecosystem level. Infectious disease systems modelled at this level of organization of an infectious disease system are those in which the pathogen does not infect specific cells and/or tissues, but still cause damage to cells and tissues. Examples of infectious disease systems which can be modelled at this hierarchical level of organization are some bacterial infections such as vibrio cholera and some helminthes infections. We briefly describe each of the three sub-levels and the structure of host level multiscale models (HL-MSMs) of infectious disease systems that can be developed at each of these sub-levels as follows.
-
3(a)The primary host level (the basic host level): At this sub-level of the third order hierarchical level of organization of an infectious disease system multiscale models are developed that integrate within-host scale and between-host scale in the context of one-pathogen and one-host species scenario. In addition such multiscale models are developed to describe disease dynamics in a single geographical area/country/region of the world. Multiscale models of an infectious disease system (HL-IEMs) developed to describe this sub-level of the third order hierarchical level of an infectious disease system are said to have a primary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these HL-IEMs with a primary structure fall into category I, II, III and IV of multiscale models which are individual based multiscale models (IMSMs), nested multiscale models (NMSMs) and hybrid multiscale models (HMSMs) and embedded multiscale models (EMSMs).
-
3(b)The secondary host level (the community level): At this sub-level of the third order hierarchical level of an infectious disease system multiscale models are developed that also integrate within-host scale and between-host scale in the context of single-host or multiple-host species and one-pathogen species scenario. However, unlike the primary host level, multiscale models here are developed to describe disease dynamics in more than one geographical area/country/region of the world. Although such multiscale models still integrate within-host scale and between-host scale, they are developed to incorporate different geographical areas/countries/regions of the world. Thus, the HL-IEMs developed at this sub-level of the third order hierarchical level of an infectious disease system describe disease dynamics in which infected host populations (humans or plants or animals or vectors) are distributed over distinctly different geographical areas/countries/regions or geographically separated areas/countries/regions of the world with the host as the unit of multiscale analysis. Within this sub-level of the third order hierarchical level of an infectious disease system the HL-IEMs are said to have a secondary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these HL-IEMs with a secondary structure fall into class 4 (multiple geographical environments single-pathogen coupled multiscale models (MGSP-CMSMs)) of category V which takes into account the different geographical areas/countries/regions of the world infected by a single pathogen in multiscale modelling of infectious disease systems.
-
3(c)The tertiary host level (the ecosystem level): At this sub-level of the first order hierarchical level of organization of an infectious disease system, multiscale models are still developed that integrate the within-host scale and the between-host scale. However, unlike the primary host level and the secondary host level, the multiscale models at this sub-level consider the host level as an ecosystem (Lofgren et al., 2016; Rynkiewicz et al., 2015; Sicard et al., 2014). Thus the multiscale models developed at this sub-level incorporate more than one geographical area/country/region of the world in the dynamics of the infectious disease system in the context of multi-host species and/or multi-group host species and/or multi-pathogen species and/or multi-pathogen strain species in the development of the multiscale models with the host as the unit of multiscale analysis. Within this sub-level of the third order hierarchical level of an infectious disease system the HL-IEMs are said to have a tertiary structure. Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017), these HL-IEMs with a tertiary structure fall into classes 1, 2, 3 and 5 of category V of multiscale models of infectious disease systems.
-
3(a)
Therefore, multiscale models developed at this level can be used to describe whole world disease dynamics with the host as the unit of multiscale analysis.
3. Integration of scales via disease process relationships in multiscale modelling of infectious disease systems
Pathogen specific disease processes are the causality of the linkage across scales at the three hierarchical levels of organization of infectious diseases. In particular four pathogen specific disease processes are responsible for the linkage across scales which are: (i) infection/superinfection by pathogen, (ii) pathogen replication, (iii) pathogen shedding/excretion and (iv) pathogen transmission. At each of the three hierarchical levels of an infectious disease system where the cell, the tissue and the host are the basic units of multiscale analysis, disease dynamics begins with infection/superinfection of the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) by the pathogen, that is, pathogen invasion of the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale). This disease process involves movement of pathogen from the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) and its entry inside a cell or tissue or host. Thus, infection/superinfection links the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) to the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale). From a mathematical point of view infection links the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) to the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) through initial conditions while superinfection links the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) to the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) through down-scaled population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) variables. For more information on the general problem of scaling (down-scaling and up-scaling) in the development multiscale models of infectious disease systems see challenge 1 in section 5. Once infection/superinfection of the individual/lower scale (within-cell scale, within-tissue scale, within-host scale) has successfully occurred, pathogen replication at the individual/lower scale (within-cell scale, within-tissue scale, within-host scale) follows. The process of pathogen replication at the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) is followed by pathogen shedding/excretion to the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale). Therefore, pathogen shedding/excretion links the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) to the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale). Shedding/excretion of the pathogen is then followed by pathogen transmission at the population/upper scale (between-cell scale, between-tissue scale, between-host scale). This implies that for each of the three hierarchical levels of an infectious disease system, the characteristic scale at which pathogen replication and pathogen transmission occur often do not match. Therefore, at each of the three orders of hierarchical level of an infectious disease system, infection/superinfection and shedding/excretion of the pathogen introduce a cycle of influence between pathogen replication at the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) and pathogen transmission at the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale). The limitation of single scale submodels alone associated with the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) or those associated with the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) in describing disease dynamics at each of the three orders of hierarchical levels of an infectious disease system is that they restrict themselves to one part of the replication-transmission cycle, with each single scale model making ‘black box’ or rudimentary assumptions about the other half – when the other half is considered at all. For the full understanding of disease dynamics at each of the three hierarchical levels of an infectious disease system one has to close the replication-transmission loop and consider the full feedback between the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) pathogen replication processes and the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) transmission processes. This is largely because at each of the three hierarchical levels of an infectious disease system, the intensity of pathogen replication at the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) determines how much pathogen load the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) will further shed/excrete into the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale), which will eventually affect the transmission of the pathogen at the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) and may eventually in turn feed back to the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) through infection/superinfection.
In general, we know from (Garira, 2017) that any multiscale model (CL-IEM or TL-IEM or HL-IEM) in which the individual/lower scale (within-cell, within-tissue, within-host) and the population/upper scale (between-cell, between-tissue, between-host) influence each other in a reciprocal way is called an embedded multiscale model and that any multiscale model (CL-IEM or TL-IEM or HL-IEM) in which the individual/lower/micro scale (within-cell, within-tissue, within-host) influences he population/upper scale (between-cell, between-tissue, between-host) without any reciprocal feedback is called a nested multiscale model. An interesting question is: when is it more appropriate to develop a nested multiscale instead of an embedded multiscale model and vice versa to describe infectious disease dynamics at each of the three hierarchical levels of an infectious disease system? Based on the conceptual framework of the organization of an infectious disease system presented is this primer (in which movement of pathogen across scales constitutes the flow of information across scales, we propose that for any of the three main hierarchical levels of an infectious disease system (cell level, tissue level, host level) if infection of the individual/lower/micro scale (within-cell, within-tissue, within-host) happens as a one-off event and infection at this individual/lower/micro scale is further sustained by pathogen replication only, then a nested multiscale model is the most appropriate model to describe disease dynamics at this hierarchical level of an infectious disease system. Thus nested multiscale models are usually more appropriate in modelling infectious disease systems in which there is mismatch between the scale at which pathogen replication occurs - which is usually the individual/lower/micro scale (within-cell, within-tissue, within-host) and the scale at which pathogen transmission occurs - which is usually the population/upper/macro scale (between-cell, between-tissue, between-host). At the host level, this usually happens for all directly transmitted infectious disease systems (in which pathogen replication only happens at the within-host scale while pathogen transmission happens at the between-host scale). At the cell level this also usually happens for all infectious disease systems caused by obligatory intra-cellular pathogens such as viruses where pathogen replication occurs at within-cell scale and pathogen transmission happens at between-cell scale. Further, for each of the three main hierarchical levels of an infectious disease system (cell level, tissue level, host level) if infection of the individual/lower/micro scale (within-cell, within-tissue, within-host) happens repeatedly (superinfection) and there is no pathogen replication at this individual/lower scale so that infection at the individual/lower scale (within-cell, within-tissue, within-host) is sustained by superinfection, then an embedded multiscale model is the most appropriate multiscale model to describe infection dynamics at that hierarchical level of an infectious disease system. Thus embedded multiscale models are usually more appropriate in modelling infectious disease systems in which there is no mismatch between the scale at which pathogen replication occurs and the scale at which pathogen transmission occurs. At the host level, this usually happens for some environmentally (indirectly) transmitted infectious disease systems in which both pathogen replication and pathogen transmission happen at the between-host scale. Typically examples for such embedded multiscale models at the host level are for schistosomiasis (Garira et al., 2014) and Guinea worm disease (Netshikweta and Garira, 2017).
4. The scientific applications frontier for multiscale modelling of infectious disease systems
Multiscale modelling is central to characterizing the epidemiology of infectious disease systems, developing new strategies for their control, informing disease management, identifying targets for new drugs and vaccines, and in turn evaluating the impact of these medical interventions. For the control, elimination and even eradication of an infectious disease system at each of three main orders of hierarchical levels of an infectious disease system conceptually represented in Fig. 1, we can broadly identify two types of health interventions: (i) those which operate at the individual/lower scale (within-cell, within-tissue, within-host), which usually interfere with the pathogen's replication processes and (ii) those which operate at the population/upper scale (between-cell scale, between-tissue scale, between-host scale) which normally interfere with the transmission processes of the pathogen. Thus health interventions either target the replication/multiplication/growth processes of the pathogen or they target transmission processes of the pathogen. At each of these three orders of hierarchical level of an infectious system, the words control, elimination and eradication of an infectious disease system may mean different things and multiscale models with different structures (primary structure, secondary structure or tertiary structure) are more appropriate for either the evaluation of control or the evaluation elimination or the evaluation of eradication of an infectious disease system. In this study, where we use a pathogen-centred perspective to conceptualize an infectious disease system, ‘baseline pathogen load’ is a phrase used to define control of infectious disease systems at each of the three hierarchical levels of an infectious disease system. We explicitly define baseline pathogen load as ‘the pathogen load/burden that would exist at a specific hierarchical level (cell level or tissue level or host level) of an infectious disease system if no interventions are implemented targeted at that level’.
-
a.
Control, elimination and eradication of infectious disease systems at the cell and tissue levels: At these two hierarchical levels of an infectious disease system, whole body/organism disease dynamics is described using either the cell level or the tissue level as the unit of multiscale analysis. Therefore, a common definition for control, elimination and even eradication of an infectious disease system applies at these two levels. Based on the understanding of the proposed three-level hierarchical framework of infectious disease systems (see Fig. 1), we define control, elimination and eradication of infectious disease systems at these two levels (the cell level and the tissue level) as follows. At the cell and tissue levels, control of an infectious disease system means ‘reduction of pathogen load/burden to below baseline levels in a specific anatomical compartment/organ of the whole body/organism through implementing health interventions that reduce cell level or tissue level pathogen transmission and/or pathogen replication’. Control of infection at any of these two levels does not result in healing or cure of infection at the whole body/organism level because reduction of pathogen load to below baseline levels in a specific anatomical compartment/organ of the whole body/organism means that the infectious agent persists in all anatomical compartments/organs of the whole body. The CL-IEMs (for the level) and the TL-IEMs (for the tissue level) with primary structure are appropriate for evaluation of control of infection at these levels. At these two levels (the cell level or the tissue level), elimination of an infectious disease system means ‘reduction of pathogen load/burden to zero or to below detection levels in a specific anatomical compartment/organ of the whole body/organism through implementing health interventions that stop cell level or tissue level pathogen transmission and/or pathogen replication’. Elimination does not also result in cure of the host infection because reduction of the pathogen load to zero or to below detection levels is only restricted to specific anatomical compartments/organs of the whole body. For example, at the cell level, HIV provirus can be eliminated in specific anatomical compartments/organs of the human body such as the blood by using antiviral drugs while corresponding elimination is not achieved in other anatomical compartments/organs of the human body such as the brain or the central nervous system. As a result, HIV-infected individuals must remain indefinitely on anti-retroviral therapy. The CL-IEMs (for the cell level) and TL-IEMs (for the tissue level) with a secondary structure are more appropriate for evaluating the elimination of infection at these hierarchical levels. Finally, at these two levels (the cell level or the tissue level), eradication of an infectious disease system means ‘reduction of pathogen load/burden to zero in all anatomical compartments/organs of the whole body/organism through implementing health interventions that stop cell level or tissue level pathogen transmission and/or pathogen replication’. Therefore, eradication of an infectious disease system means complete absence of the pathogen from all anatomical compartments/organs of the whole body/organism and there is complete recovery of the host from disease (cure) from illness. The CL-IEMs (for the cell level) and TL-IEMs (for the tissue level) with a tertiary structure are appropriate for evaluating eradication of an infectious disease system at these two levels.
-
b.
Control, elimination and eradication of infectious disease systems at the host level: At this level of an infectious disease system whole world infectious disease dynamics is described using the host level as the unit of multiscale analysis. Based on the understanding of the proposed three-level hierarchical framework of infectious disease systems (see Fig. 1), we define control, elimination and eradication of infectious disease systems at the host level as follows. Control of an infectious disease system means ‘reduction of pathogen load/burden to below baseline levels in a specific geographical area/country/region of the world through implementing health interventions that reduce host level pathogen transmission and/or pathogen replication’. Control of an infectious disease system at this level does not end the epidemic in the whole world because reduction of pathogen load to below baseline levels in specific geographical areas/countries/regions of the world implies that the infectious agent persists in the world. The HL-IEMs with a primary structure are more appropriate for evaluation of control of an infectious disease system at this level. Elimination of an infectious disease system means ‘reduction of pathogen load/burden to zero in a specific geographical area/country/region of the world through implementing health interventions that stop host level pathogen transmission and/or pathogen replication’. Elimination does not also end an epidemic in the whole world because reduction of the pathogen load to zero is only restricted to specific geographical areas/countries/regions of the world. The HL-IEMs with a secondary structure are more appropriate for evaluating the elimination of infection at this hierarchical level. Eradication of an infectious disease system means ‘reduction of pathogen load to zero in all geographical areas/countries/regions of the world through implementing health interventions that stop host level pathogen transmission and/or pathogen replication’. Eradication of an infectious disease system does end the epidemic in the whole world because reduction of the pathogen load to zero is attained in all geographical areas/countries/regions of the world. The HL-IEMs with a tertiary structure are more appropriate for evaluating the eradication of an infectious disease system at this hierarchical level.
The use of pathogen load as a common metric for (i) infectiousness and (ii) disease transmission potential and as (iii) an indicator of the effectiveness of health interventions in the above definitions for control, elimination and eradication at all the three hierarchical levels of an infectious disease system has two advantages over the use of incidence and prevalence which are often used in single scale framework at between-host scale (Dowdle, 1998; Heymann, 2006; Molyneux et al., 2004). First, it provides a common metric for disease dynamics across different scales. Second, it also provides a common metric for disease dynamics across different disease transmission mechanisms (i.e. directly transmitted and environmentally transmitted infectious disease systems). The current understanding of the words ‘control’, ‘elimination’ and ‘eradication’ of infectious disease systems based on prevalence and incidence as in (Dowdle, 1998; Heymann, 2006; Molyneux et al., 2004) does not take into account the multiscale nature of infectious disease systems because prevalence and incidence cannot be used a common metrics of disease dynamics across scales other than the between-host scale only.
In general, at each of these three hierarchical levels of an infectious diseases, there is always a reciprocal influence health interventions implemented at the individual/lower/micro scale (within-cell, within-tissue, within-host) intended to control, eliminate or even eradicate an infectious disease system by reducing/stopping pathogen replication on one hand, and health interventions implemented at the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) intended to control, eliminate or even eradicate an infectious disease system by reducing/stopping pathogen transmission on the other hand. For example, at the lower scale (within-host scale), one of the latest preventive health intervention that has been adopted for HIV/AIDS control, elimination and even eradication is Treatment as Prevention (TasP) which is based on the understanding that the single most important risk factor in determining the likelihood of HIV transmission at between-host scale is an HIV infected individual's plasma viral load at the within-host scale. With effective treatment targeting the within-host scale, the whole body/host plasma viral load can be greatly reduced for HIV infected individuals thereby significantly preventing severe disease, death and onward transmission (both heterosexual transmission and mother-to-child transmission) of HIV at between-host scale. Encouraged by results from TasP in HIV/AIDS (Cohen et al., 2012; Das, Chu & Santos, 2004; Delva et al., 2012; Fang et al., 2004; Granich et al., 2009; Montaner et al., 2010; Smith et al., 2012; Wilson, 2012), researchers are now also considering TasP as an approach for trying to deal with many other infectious diseases such as malaria (Chaccour et al., 2013; El-Sayed et al., 2007; Gerardin et al., 2015; Greenwood, 2006; John, 2016; Johnston et al., 2014; Kiszewski, 2010; Maude et al., 2012; McMorrow et al., 2011; Tanner et al., 2015; Tseroni et al., 2015; White, 2008), neglected tropical diseases (Bockarie, Kelly-Hope, Rebollo, & Molyneux, 2013; Keenan et al., 2013; Mbah et al., 2013; Smits, 2009), tuberculosis (Griffith et al., 2007; Centers for Disease Control and Prevention CDC, 2000; Halsey et al., 1998) and many other infectious diseases (Webster et al., 2014; Yamshchikov et al., 2009). In all of these infectious diseases, the use of TasP as a preventive health intervention at between-host scale is based on the fact that the transmission of an infectious disease system at between-host scale can be prevented by implementing treatment at within-host scale of infected individuals so that they become less likely to transmit the infection to others at between-host scale. Since treatment is administered at within-host scale while other health interventions such as male condom use, male circumcision (Mukandavire et al., 2007; Mukandavire and Garira, 2007) for HIV/AIDS, indoor residual spraying (IRS) and insecticide-treated nets (ITNs) (Griffin et al., 2010) for malaria and water supply, water quality, sanitation, and hygiene (WASH) intervention systems (Fewtrell et al., 2005; Fung, 2014) for diarrhea and cholera are implemented at between-host scale, mathematical models that link the within-host and between-host scales of infection can better inform public health policies on the comparative effectiveness of several health interventions which are implemented at different scale domains for the same infectious disease system. Further, the performance of health interventions at the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) normally referred to as efficacy determined using empirical studies (observational, culture, experimental systems, clinical trials, etc.) which usually target the replication of the pathogen does not reflect their performance at the corresponding population/upper/macro scale (between-cells scale, between-tissues scale, between-host scale) normally referred to as effectiveness which usually target the transmission of the pathogen. Therefore, within each of the three hierarchical levels of an infectious disease system, the characteristic scale of efficacy and effectiveness of an intervention often do not match. This establishes an efficacy-effectiveness gap (Eichler et al., 2011; Nordon et al., 2016; Panayidou et al., 2016). One of the most important scientific applications proposed in this study for multiscale models of infectious disease systems is to bridge the efficacy-effectiveness gap in comparative effectiveness research.
5. The mathematical technology frontier and challenges for multiscale modelling of infectious disease systems
Efforts to develop multiscale models of infectious disease dynamics at the different hierarchical levels (the cell level, the tissue level, the host level) of an infectious disease system have recently been underpinned by a categorization framework that helps to identify the different categories of multiscale models that can be developed (Garira, 2017) at each of these three levels. Given this foundation which is further refined in this study, the frontiers of multiscale modelling of infectious disease dynamics lie more on the mathematical technology/technical frontier - to find methods for developing multiscale models of infectious disease systems that belong to the different categories (with either primary structure or secondary structure or tertiary structure). However, the mathematical technology frontier for multiscale modelling of infectious disease systems is still full of roadblocks and potholes associated with the following ten main challenges.
- Challenge 1: Development of methods for linking/coupling/integration of the submodels: There is a critical need to develop mathematical methods for linking/coupling submodels across scales in the development of multiscale models of infectious disease systems. Linking the submodels, also referred to as coupling the submodels or simply integration of submodels works by feeding the outputs (variables and parameters) of one submodel as input to another submodel. To date the development models of infectious disease dynamics that integrate submodels across scales has been informed by different types of coupling principles for linking submodels across scales (Garira et al., 2014). We can roughly demarcate these coupling principles into three categories:i.
-
iDirect unidirectional coupling: In this type of linking submodels in a multiscale model, the lower scale is unidirectionally coupled to the upper scale. That is, the lower scale influences the upper scale and not the other way round. Nested multiscale models (category II of multiscale models) and some hybrid multiscale models (category IV of multiscale models) are developed using this coupling method.
-
ii.Direct bidirectional coupling: In this type of linking submodels in a multiscale model, there is direct bidirectional coupling between the upper scale and the lower scale. In this case the two scales (lower and upper scale) influence each other in a reciprocal way. Embedded multiscale models (category III of multiscale models) and some hybrid multiscale models (category IV of multiscale models) are developed using this coupling method.
-
iii.Indirect unidirectional coupling: In this type of linking submodels in a multiscale model, there is no direct coupling between the lower scale and the upper scale. However, there is direct coupling between the entities of the lower scale and the upper scale is only observed as emergent behaviour (a property of complex systems) of the lower scale entities. Individual based multiscale models (category I of multiscale models) are developed using this coupling method.
-
i
However, coupled multiscale models (category V of multiscale models), that is, multiscale models of infectious diseases that integrate several scales of infection, are developed using a combination of these coupling methods (direct unidirectional coupling, direct bidirectional coupling, and indirect unidirectional coupling). In general linking/coupling/integration of submodels across scales involves up-scaling or down-scaling variables associated with some disease processes. This is because for an infectious disease system, the hierarchical levels and their associated scales (see Fig. 1) indicate/represent shifts in disease processes (e.g pathogen replication at the lower scale and pathogen transmission at the upper scale). However, the greatest challenge is in methodological difficulties on specific implementation approaches for scaling (down-scaling and up-scaling) in space and time and in converting dimensions across spatial and temporal scales when coupling the submodels of a multiscale model. The lack of rigorous frameworks for scaling (down-scaling and up-scaling) undermines progress in the development of multiscale models of infectious disease systems. Furthermore, down-scaling to lower scale (within-cell, within-tissue, within-host, etc.) or up-scaling to upper scale (between-cell scale, between-tissue scale, between-host scale, etc.) involves developing conceptual models that adequately capture the dominant disease processes at these scales. In particular, we inevitably need to know more details when down-scaling while we also need to ignore some fine details when up-scaling. But another challenge is that when either down-scaling or up-scaling, we do not know in general how scale transition occurs. In particular when down-scaling, we do not know what lower scale (within-cell, within-tissue, within-host, etc.) disease processes and parameters are essential in representing the upper scale (between-cell scale, between-tissue scale, between-host scale, etc.) processes and vice-versa when up-scaling. For classic examples of up-scaling and down-scaling when developing embedded multiscale models of infectious disease system see (Garira et al., 2014; Netshikweta and Garira, 2017).
Challenge 2: Development of methods for validating multiscale models: Validation of multiscale models of infectious disease systems need to be conducted accurately to account for uncertainties in data (clinical, experimental, demographic, environmental, epidemiological, immunological, etc.) at various spatial and temporal scales. However, there are no simple rules to follow when validating multiscale models of infectious disease systems.
Challenge 3: Obtaining all relevant and accurate data for development of multiscale models: Obtaining all relevant and accurate data for development of multiscale models of infectious disease systems is critical for reliability of a given multiscale model particularly as the submodels in a multiscale model are bridged across scales. Minor errors introduced in data collection at one scale can introduce significant errors as the errors propagate across scales in a multiscale model. More readily available relevant and accurate infectious disease data (clinical, experimental, demographic, environmental, epidemiological, immunological, etc.) across scales. would facilitate wider adoption of multiscale modelling approaches in infectious disease study.
Challenge 4: Development of methods for reduced order multiscale models: There are many methods for analysis of multiscale infectious disease systems, enabling reduction of their size, complexity, and/or dimensionality. Reduction of order of multiscale models of infectious disease systems is an essential tool for reducing the degrees of freedom (e.g., the number of variables in a multiscale model) so as to increase computational efficiency and make high dimensional multiscale models more tractable. However, there are still a number of barriers to development of reduced order multiscale models of infectious disease systems. Dimensional reduction of a multiscale model of an infectious disease system is important for three main reasons (Garira, 2017): (i) to reduce the computational needs for simulating the multiscale model, (ii) to identify the most fundamental components with respect to the drivers of the multiscale model behaviour, and (iii) to simplify the process of analysis of the multiscale model. To date, several dimensional reduction methods have been developed which include (a) slow and fast time scale analysis (Cen et al., 2014; Feng et al., 2013, 2015), (b) response surface modelling (Myers et al., 2016), (c) statistical methods such as principal component analysis (Kambhatla and Leen, 2006), (d) response and statistical surrogate modelling (Frangos et al., 2010) as well as (e) dynamical systems based methods such as centremanifold theory (Carr, 2012) are examples of a reduced order approaches that can be applied to multiscale modelling of infectious disease systems. However, more work is needed in this area because reduced order multiscale modelling of infectious disease systems still presents some challenges.
Challenge 5: Development of more efficient computational tools for multiscale models: Currently, the use of computational tools to solve multiscale models of infectious disease systems face limitations of computing power. This is because numerically solving some multiscale models of infectious disease systems use numerical methods that are computationally intensive. This limits the practical use of computational methods in multiscale modelling of infectious diseases. Further, the computational expense presents some challenges when scaling up from the lower scale to the upper scale of an infectious disease system and vise-versa because of time scale mismatch between the lower scales and the upper scales. Thus a key challenge to multiscale modelling of infectious disease systems is that there is a timescale mismatch over different length scales. More efficient computational tools need to be developed in order to increase available computing power for solving multiscale models of infectious disease systems. In addition, numerical methods used to solve multiscale models of infectious disease systems using discretization methods where specific discretization methods in terms of length and/or time are employed. A major challenge to employing such methods at multiple scales is that the different scales require different grid densities and time steps, creating difficulties in passing information across different scales.
Challenge 6: The inability to account for emergent phenomena in multiscale models: A fundamental limitation to multiscale modelling of infectious disease systems is the inability to account for emergent behaviour. “Emergent phenomena” in this context refers to behaviour that emerges at the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale), often unexpectedly or unexplainably, from interactions at an individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale)- a fundamental property of complex systems. In particular, upper scale (between-cell, between-tissue, between-host) submodels are often not tailored to effectively capture emergent phenomena, that is, behaviours realized at upper scales resulting from interactions of entities at lower scales that can be critical contributing factors to the property of evolution of infectious disease systems. Because of emergent effects some disease processes might show different patterns when studied at different scales. Therefore, if we model a disease process at inappropriate scale we may not be able to detect its actual dynamics at that scale, but may instead identify behaviour patterns that are emergent effects of its actual dynamics.
Challenge 7: Development of methods for uncertainty quantification for multiscale models: Uncertainties exist in specification of the full detailed multiscale model, identification of reduced order multiscale models, integration of submodels into a single multiscale model, data assimilation, calibration and validation of multiscale models. The need to adequately address uncertainty quantification and propagation is important but is still not adequately addressed in multiscale modelling of infectious disease systems. Uncertainty quantification has been addressed for some multiscale modelling approaches at individual scales, but propagating the uncertainty across scales is challenging. This is the case because uncertainty quantification is usually handled differently at different length or time scales. In addition, in situations where it is feasible to pass uncertain values up from a lower scale to a higher scale, it is not always clear how to understand these values or what practical insights can be made from them. More dialogue between researchers with disparate skillsets in modelling infectious disease systems at different scales of infection is needed to address this challenge.
Challenge 8: Missing underlying biological mechanisms in the development of multiscale models: One of the challenges which also presents opportunities for growth lies in the fact that although some large strides have been made in the development of multiscale modelling approaches of infectious diseases in recent years, there are still significant limitations and gaps in the individual single scale modelling capabilities that serve as the building blocks for modelling across length and time scales. These limitations in multiscale modelling of infectious disease systems are often due to failures of the model to properly incorporate the underlying biological mechanisms that drive dynamics of infectious disease systems.
Challenge 9: Development of multiscale models to account for evolution of pathogen across scales: Evolution of pathogens drives many infectious disease processes ranging from immune escape to changes in virulence and drug resistance. Pathogen evolution plays out at scales ranging from within-host to between-host and beyond. The challenge here is to develop tractable multiscale models of infectious disease to address evolutionary questions and to understand how multiple scales of selection influence the evolutionary emergence of novel pathogens.
Challenge 10: Development of multiscale models to account for co-evolution of host and pathogen across scales: The main difference between evolution and co-evolution is the reciprocity of selective pressure and change occurring in both the pathogen population and the host population. The dynamics of an infectious disease systems involves the reciprocal evolution between both the host and the pathogen whereby both the host and the pathogen impose selection on the other. In this context, co-evolution of host and pathogen is a dynamic process of ongoing reciprocal change where a pathogen population imposes a selective influence on a host population which responds to the selection, in turn imposing a selective influence on the pathogen population, with this cycle potentially repeated over and over. The outcome of this arms race may involve traits like parasite infectivity, host resistance, parasite host-finding ability and parasite avoidance behaviour by the host. The challenge here is to develop appropriate multiscale models of infectious disease that account for this tug-of-war between pathogens and hosts as antagonistic co-evolving populations to improve our ability to forecast the trajectory of infectious disease systems in various situations.
Research to address these challenges will have substantial potential impact on progress in multiscale modelling of infectious diseases systems.
6. Discussion and conclusions
In this primer, we presented some foundational concepts on multiscale models of infectious disease systems focused on three main frontiers which are the conceptual framework frontier, the mathematical technology or technical frontier and the scientific applications frontier. We anticipate these foundational concepts to be central in articulating an integrated, more refined infectious disease control theory of the reciprocal influence between health interventions that target pathogen replication at the individual/lower/micro scale (within-cell scale, within-tissue scale, within-host scale) on one hand and health interventions that target pathogen transmission at the population/upper/macro scale (between-cell scale, between-tissue scale, between-host scale) on the other hand. In particular, we identified three different structures of multiscale models at each of the three different hierarchical levels of an infectious disease system with each of them being more appropriate for either the control, or the elimination, or the eradication of an infectious disease system which are:
-
a.
Multiscale models with a primary structure: Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017) only categories I, II, III, and IV of multiscale models have such a structure: IMSMs, NMSMs, EMSMs and HMSMs. They can be either CL-IEMs (for the cell level) or TL-IEMs (for the tissue level) or HL-IEMs (for the host level). Such multiscale models are more relevant for evaluating the control of infection at specific hierarchical levels of an infectious disease system.
-
b.
Multiscale models with a secondary structure: Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017) only class 4 of category V of multiscale models have such a structure. They can be either CL-IEMs (for the cell level) or TL-IEMs (for the tissue level) or HL-IEMs (for the host level). Such multiscale models are more relevant for evaluating the elimination of infection at specific hierarchical levels of an infectious disease system.
-
c.
Multiscale models with a tertiary structure: Based on the categorization of multiscale models of infectious disease systems given in (Garira, 2017) only classes 1, 2, 3 and 5 of category V of multiscale models have such a structure. They can be either CL-IEMs (for the cell level) or TL-IEMs (for the tissue level) or HL-IEMs (for the host level). Such multiscale models are more relevant for evaluating the eradication of infection at all hierarchical levels of an infectious disease system.
Given this foundation, the frontiers of multiscale modelling of infectious disease systems lie more on technical or mathematical technology frontier.
Acknowledgments
The author acknowledges with thanks financial support from NRF, South Africa Grant No. IPRR (UID 81235).
Handling editor: J. Wu
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.idm.2018.09.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bockarie M.J., Kelly-Hope L.A., Rebollo M., Molyneux D.H. Preventive chemotherapy as a strategy for elimination of neglected tropical parasitic diseases: Endgame challenges. Philosophical Transactions of the Royal Society of London B Biological Sciences. 2013;368(1623):20120144. doi: 10.1098/rstb.2012.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr J. Vol. 35. Springer Science & Business Media; 2012. (Applications of centre manifold theory). [Google Scholar]
- Cen X., Feng Z., Zhao Y. Emerging disease dynamics in a model coupling within-host and between-host systems. Journal of Theoretical Biology. 2014;361:141–151. doi: 10.1016/j.jtbi.2014.07.030. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention CDC Updated guidelines for the use of rifabutin or rifampin for the treatment and prevention of tuberculosis among HIV-infected patients taking protease inhibitors or nonnucleoside reverse transcriptase inhibitors. MMWR. Morbidity and Mortality Weekly Report. 2000;49(9):185. [PubMed] [Google Scholar]
- Chaccour C.J., Kobylinski K.C., Bassat Q., Bousema T., Drakeley C., Alonso P. Ivermectin to reduce malaria transmission: A research agenda for a promising new tool for elimination. Malaria Journal. 2013;12(153) doi: 10.1186/1475-2875-12-153. 10-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M.S., Dye C., Fraser C., Miller W.C., Powers K.A., Williams B.G. HIV treatment as prevention: Debate and commentary—will early infection compromise treatment-as-prevention strategies? PLoS Medicine. 2012;9(7) doi: 10.1371/journal.pmed.1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M., Chu P., Santos G.M. Proceedings of the 18th Conference on Retroviruses and Opportunistic Infections. 2011, February. Success of test and treat in San Francisco? Reduced time to virologic suppression, decreased community viral load, and fewer new HIV infections, 2004 to 2009. [Google Scholar]
- Delva W., Eaton J.W., Meng F., Fraser C., White R.G., Vickerman P. HIV treatment as prevention: Optimising the impact of expanded HIV treatment programmes. PLoS Medicine. 2012;9(7) doi: 10.1371/journal.pmed.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle W.R. The principles of disease elimination and eradication. Bulletin of the World Health Organization. 1998;76(Suppl 2):22. [PMC free article] [PubMed] [Google Scholar]
- Eichler H.G., Abadie E., Breckenridge A., Flamion B., Gustafsson L.L., Leufkens H. Bridging the efficacy–effectiveness gap: A regulator's perspective on addressing variability of drug response. Nature Reviews Drug Discovery. 2011;10(7):495. doi: 10.1038/nrd3501. [DOI] [PubMed] [Google Scholar]
- El-Sayed B., El-Zaki S.E., Babiker H., Gadalla N., Ageep T., Mansour F. A randomized open-label trial of artesunate-sulfadoxine-pyrimethamine with or without primaquine for elimination of sub-microscopic P. falciparum parasitaemia and gametocyte carriage in eastern Sudan. PLoS One. 2007;2(12):e1311. doi: 10.1371/journal.pone.0001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C.T., Hsu H.M., Twu S.J., Chen M.Y., Chang Y.Y., Hwang J.S. Decreased HIV transmission after a policy of providing free access to highly active antiretroviral therapy in Taiwan. Journal of Infectious Diseases. 2004;190(5):879–885. doi: 10.1086/422601. [DOI] [PubMed] [Google Scholar]
- Feng Z., Cen X., Zhao Y., Velasco-Hernandez J.X. Coupled within-host and between-host dynamics and evolution of virulence. Mathematical Biosciences. 2015;270:204–212. doi: 10.1016/j.mbs.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Feng Z., Velasco-Hernandez J., Tapia-Santos B. A mathematical model for coupling within-host and between-host dynamics in an environmentally-driven infectious disease. Mathematical Biosciences. 2013;241(1):49–55. doi: 10.1016/j.mbs.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Fewtrell L., Kaufmann R.B., Kay D., Enanoria W., Haller L., Colford J.M. Water, sanitation, and hygiene interventions to reduce diarrhoea in less developed countries: A systematic review and meta-analysis. The Lancet Infectious Diseases. 2005;5(1):42–52. doi: 10.1016/S1473-3099(04)01253-8. [DOI] [PubMed] [Google Scholar]
- Frangos M., Marzouk Y., Willcox K., van Bloemen Waanders B. Large-scale inverse problems and quantification of uncertainty. 2010. Surrogate and reduced-order modeling: A comparison of approaches for large-scale statistical inverse problems; pp. 123–149. [Google Scholar]
- Fung I.C.-H. Cholera transmission dynamic models for public health practitioners. Emerging Themes in Epidemiology. 2014;11(1):1. doi: 10.1186/1742-7622-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garira W. A complete categorization of multiscale models of infectious disease systems. Journal of Biological Dynamics. 2017;11(1):378–435. doi: 10.1080/17513758.2017.1367849. [DOI] [PubMed] [Google Scholar]
- Garira W., Mathebula D., Netshikweta R. A mathematical modelling framework for linked within-host and between-host dynamics for infections with free-living pathogens in the environment. Mathematical Biosciences. 2014;256:58–78. doi: 10.1016/j.mbs.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Gerardin J., Eckhoff P., Wenger E.A. Mass campaigns with antimalarial drugs: A modelling comparison of artemether-lumefantrine and DHA-piperaquine with and without primaquine as tools for malaria control and elimination. BMC Infectious Diseases. 2015;15(1):144. doi: 10.1186/s12879-015-0887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granich R.M., Gilks C.F., Dye C., De Cock K.M., Williams B.G. Universal voluntary HIV testing with immediate antiretroviral therapy as a strategy for elimination of HIV transmission: A mathematical model. The Lancet. 2009;373(9657):48–57. doi: 10.1016/S0140-6736(08)61697-9. [DOI] [PubMed] [Google Scholar]
- Greenwood B. Review: Intermittent preventive treatment–a new approach to the prevention of malaria in children in areas with seasonal malaria transmission. Tropical Medicine and International Health. 2006;11(7):983–991. doi: 10.1111/j.1365-3156.2006.01657.x. [DOI] [PubMed] [Google Scholar]
- Griffin J.T., Hollingsworth T.D., Okell L.C., Churcher T.S., White M., Hinsley W. Reducing plasmodium falciparum malaria transmission in Africa: A model-based evaluation of intervention strategies. PLoS Medicine. 2010;7(8) doi: 10.1371/journal.pmed.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith D.E., Aksamit T., Brown-Elliott B.A., Catanzaro A., Daley C., Gordin F. An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. American Journal of Respiratory and Critical Care Medicine. 2007;175(4):367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- Halsey N.A., Coberly J.S., Desormeaux J., Losikoff P., Atkinson J., Moulton L.H. Randomised trial of isoniazid versus rifampicin and pyrazinamide for prevention of tuberculosis in HIV-1 infection. The Lancet. 1998;351(9105):786–792. doi: 10.1016/S0140-6736(97)06532-X. [DOI] [PubMed] [Google Scholar]
- Heymann D.L. Control, elimination, eradication and re-emergence of infectious diseases: Getting the message right. Bulletin of the World Health Organization. 2006;84(2) doi: 10.2471/blt.05.029512. 82-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D.G., Zumla A. Cambridge University Press; 1999. The granulomatous disorders. [Google Scholar]
- John C.C. Primaquine plus artemisinin combination therapy for reduction of malaria transmission: Promise and risk. BMC Medicine. 2016;14:1. doi: 10.1186/s12916-016-0611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G.L., Gething P.W., Hay S.I., Smith D.L., Fidock D.A. Modeling within-host effects of drugs on Plasmodium falciparum transmission and prospects for malaria elimination. PLoS Computational Biology. 2014;10(1) doi: 10.1371/journal.pcbi.1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambhatla N., Leen T.K. Dimension reduction by local principal component analysis. Dimension. 2006;9(7) [Google Scholar]
- Keenan J.D., Hotez P.J., Amza A., Stoller N.E., Gaynor B.D., Porco T.C. Elimination and eradication of neglected tropical diseases with mass drug administrations: A survey of experts. PLoS Neglected Tropical Diseases. 2013;7(12) doi: 10.1371/journal.pntd.0002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiszewski A.E. Blocking plasmodium falciparum malaria transmission with drugs: The gametocytocidal and sporontocidal properties of current and prospective antimalarials. Pharmaceuticals. 2010;4(1):44–68. [Google Scholar]
- Lofgren E.T., Egizi A.M., Fefferman N.H. Patients as patches: Ecology and epidemiology in healthcare environments. Infection Control and Hospital Epidemiology. 2016:1–6. doi: 10.1017/ice.2016.224. [DOI] [PubMed] [Google Scholar]
- Maude R.J., Socheat D., Nguon C., Saroth P., Dara P., Li G. Optimising strategies for plasmodium falciparum malaria elimination in Cambodia: Primaquine, mass drug administration and artemisinin resistance. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0037166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbah M.L.N., Poolman E.M., Atkins K.E., Orenstein E.W., Meyers L.A., Townsend J.P. Potential cost-effectiveness of schistosomiasis treatment for reducing HIV transmission in Africa–the case of Zimbabwean women. PLoS Neglected Tropical Diseases. 2013;7(8) doi: 10.1371/journal.pntd.0002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMorrow M.L., Aidoo M., Kachur S.P. Malaria rapid diagnostic tests in elimination settings—can they find the last parasite? Clinical Microbiology and Infections. 2011;17(11):1624–1631. doi: 10.1111/j.1469-0691.2011.03639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux D.H., Hopkins D.R., Zagaria N. Disease eradication, elimination and control: The need for accurate and consistent usage. Trends in Parasitology. 2004;20(8):347–351. doi: 10.1016/j.pt.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Montaner J.S., Lima V.D., Barrios R., Yip B., Wood E., Kerr T. Association of highly active antiretroviral therapy coverage, population viral load, and yearly new HIV diagnoses in British Columbia, Canada: a population-based study. The Lancet. 2010;376(9740):532–539. doi: 10.1016/S0140-6736(10)60936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukandavire Z., Bowa K., Garira W. Modelling circumcision and condom use as HIV/AIDS preventive control strategies. Mathematical and Computer Modelling. 2007;46(11):1353–1372. [Google Scholar]
- Mukandavire Z., Garira W. Effects of public health educational campaigns and the role of sex workers on the spread of HIV/AIDS among heterosexuals. Theoretical Population Biology. 2007;72(3):346–365. doi: 10.1016/j.tpb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Myers R.H., Montgomery D.C., Anderson-Cook C.M. John Wiley & Sons; New Jersey: 2016. Response surface methodology: Process and product optimization using designed experiments. [Google Scholar]
- Netshikweta R., Garira W. A multiscale model for the world's first parasitic disease targeted for eradication: Guinea worm disease. Computational and Mathematical Methods in Medicine. 2017 doi: 10.1155/2017/1473287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordon C., Karcher H., Groenwold R.H., Ankarfeldt M.Z., Pichler F., Chevrou-Severac H. The efficacy-effectiveness gap: Historical background and current Conceptualization. Value in Health. 2016;19(1):75–81. doi: 10.1016/j.jval.2015.09.2938. [DOI] [PubMed] [Google Scholar]
- Panayidou K., Gsteiger S., Egger M., Kilcher G., Carreras M., Efthimiou O. Get real in mathematical modelling: A review of studies predicting drug effectiveness in the real world. Research Synthesis Methods. 2016;7(3):264–277. doi: 10.1002/jrsm.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permi H.S., Shetty J., Padma S.K., Teerthanath S., Mathias M., Kumar S. A histopathological study of granulomatous inflammation. Nitte University Journal of Health Science. 2012;2:15–19. [Google Scholar]
- Pigozzo A.B., Macedo G.C., Weber dos Santos R., Lobosco M. Computational modeling of microabscess formation. Computational and Mathematical Methods in Medicine. 2012;2012 doi: 10.1155/2012/736394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynkiewicz E.C., Pedersen A.B., Fenton A. An ecosystem approach to understanding and managing within-host parasite community dynamics. Trends in Parasitology. 2015;31(5):212–221. doi: 10.1016/j.pt.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Sicard M., Dittmer J., Greve P., Bouchon D., Braqquart-Varnier C. A host as an ecosystem: Wolbachia coping with environmental constrains. Environmental Microbiology. 2014 doi: 10.1111/1462-2920.12573. [DOI] [PubMed] [Google Scholar]
- Smith M.K., Powers K.A., Muessig K.E., Miller W.C., Cohen M.S. HIV treatment as prevention: The utility and limitations of ecological observation. PLoS Medicine. 2012;9(7) doi: 10.1371/journal.pmed.1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits H.L. Prospects for the control of neglected tropical diseases by mass drug administration. Expert Review of Anti-infective Therapy. 2009;7(1):37–56. doi: 10.1586/14787210.7.1.37. [DOI] [PubMed] [Google Scholar]
- Tanner M., Greenwood B., Whitty C.J., Ansah E.K., Price R.N., Dondorp A.M. Malaria eradication and elimination: Views on how to translate a vision into reality. BMC Medicine. 2015;13(1):1. doi: 10.1186/s12916-015-0384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseroni M., Baka A., Kapizioni C., Snounou G., Tsiodras S., Charvalakou M. Prevention of Malaria Resurgence in Greece through the Association of Mass Drug Administration (MDA) to Immigrants from Malaria-Endemic Regions and Standard Control Measures. PLoS Neglected Tropical Diseases. 2015;9(11) doi: 10.1371/journal.pntd.0004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster J.P., Molyneux D.H., Hotez P.J., Fenwick A. The contribution of mass drug administration to global health: Past, present and future. Philosophical Transactions of the Royal Society of London B Biological Sciences. 2014;369(1645):20130434. doi: 10.1098/rstb.2013.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N.J. The role of anti-malarial drugs in eliminating malaria. Malaria Journal. 2008;7(1):1. doi: 10.1186/1475-2875-7-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.P. HIV treatment as prevention: Natural experiments highlight limits of antiretroviral treatment as HIV prevention. PLoS Medicine. 2012;9(7) doi: 10.1371/journal.pmed.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO). reportThe world health report: 2004 — Changing history; http://www.who.int/whr/2004/en/.
- World Health Organization (WHO). reportThe world health report: 2005 — Make every mother and child count; http://www.who.int/whr/2005/en/index.html.
- Yamshchikov A., Desai N., Blumberg H., Ziegler T., Tangpricha V. Vitamin D for treatment and prevention of infectious diseases: A systematic review of randomized controlled trials. Endocrine Practice. 2009;15(5):438–449. doi: 10.4158/EP09101.ORR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., James D.G. Granulomatous infections: Etiology and classification. Clinical Infectious Diseases. 1996;23(1):146–158. doi: 10.1093/clinids/23.1.146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.