Abstract
While influenza has been simulated extensively to better understand its behavior and predict future outbreaks, most other respiratory viruses have seldom been simulated. In this study, we provide an overview of four common respiratory viral infections: respiratory syncytial virus (RSV), respiratory adenovirus, rhinovirus and parainfluenza, present specimen data collected 2004–2014, and simulate outbreaks in 19 overlapping regions in the United States. Pairing a compartmental model and data assimilation methods, we infer key epidemiological parameters governing transmission: the basic reproductive number R0 and length of infection D. RSV had been previously simulated, and our mean estimate of D and R0 of 5.2 days and 2.8, respectively, are within published clinical and modeling estimates. Among the four virus groupings, mean estimates of R0 range from 2.3 to 3.0, with a lower and upper quartile range of 2.0–2.8 and 2.6–3.2, respectively. As rapid PCR testing becomes more common, estimates of the observed virulence and duration of infection for these viruses could inform decision making by clinicians and officials for managing patient treatment and response.
1. Introduction
Respiratory viruses target particular host cells from the sinus cavity to the alveolar ducts and cause a variety of ailments. These ailments, including influenza-like illness (ILI), defined by the symptoms of fever, cough, and a runny nose, can only be reliably distinguished through laboratory testing (Ebell & Afonso, 2011). In addition to being clinically indistinguishable, co-infection with multiple viruses is not uncommon; one study found 23% of patients with ILI were co-infected with at least two pathogens (Pavia, 2011). In the U.S., acute lower respiratory infection accounts for 14% of mortality among children under 5 years old (Black et al., 2010), and about half of children visits to a primary care doctor or hospital (Schluger, 2010). Globally, acute respiratory infections are a leading cause of death in children (Lozano et al., 2012; Mayor, 2010; Williams, Gouws, Boschi-Pinto, Bryce, & Dye, 2002), in part due to indoor air pollution (Mishra, 2003; Smith, Samet, Romieu, Bruce) and under-nutrition (Black et al., 2010). Excluding influenza, other respiratory viruses (ORVs) can produce severe illness in children and the elderly; even in adults, ORVs can produce more hospitalizations than influenza (Gilca et al., 2014). In this paper, we simulate and estimate key epidemiological characteristics of four common respiratory viruses: respiratory syncytial virus, respiratory adenovirus, rhinovirus and parainfluenza. Table 1 summarizes the estimated contribution of each of these viruses to ILI.
Table 1.
Contribution of ORVs to ILI.
| Respiratory Virus | Estimated percent of ILI |
|---|---|
| Respiratory syncytial virus | 3–29% (Pavia, 2011); 16.6% (Li et al., 2013); 19.6% in adults (Bellei et al., 2008); 31% among ≤6 yrs (Jansen et al., 2011) |
| Respiratory adenovirus | 0.4–4% (Williams et al., 2002); 4.7 (Aberle et al., 2003); 7–8% (Brandt et al., 1969; Munoz, Piedra, & Demmler, 1998; Osiowy, 1998); 7–12% (Pavia, 2011); 14.11% (Li et al., 2013) |
| Rhinovirus | 3–45% any season (Pavia, 2011); 25% among ≤6 yrs (Jansen et al., 2011) |
| Parainfluenza virus | PIV 8–13% (Pavia, 2011); PIV 1–3: 5% (Miller et al., 2013); PIV 1: 2.6–18.5% (Henrickson, 2003); PIV 1–2: 3.6–21% (Henrickson, 2003) PIV 1: 56% (Hall, Geiman, Breese, & Douglas, 1977); PIV 1–3: 9.25% (Li et al., 2013) |
1.1. Clinical description of the four respiratory viruses
Respiratory syncytial virus (RSV) infects the bronchiole tubes in the lungs, causing bronchiolitis. RSV is the most common respiratory ailment in infants under one year of age (Goldstein, Greene, Olson, Hanage, & Lipsitch, 2015; Hall, Douglas, & Geiman, 1976; Handforth, Friedland, & Sharland, 2000; Leader & Kohlhase, 2003), and is also prevalent in older ages (Falsey, 1998; Falsey, Hennessey, Formica, Cox, & Walsh, 2005). RSV has been found to infect up to 29% of children with respiratory symptoms (Juvén et al., 2000). It has a very low asymptomatic rate among the very young (Jansen et al., 2011), but can manifest as mild ILI or be asymptomatic among adults (Munywoki et al., 2015). Table 2 provides estimates of key epidemiological characteristics for RSV, including duration of illness, D, and the basic reproductive number, R0, which were derived from clinical and challenge studies of healthy individuals. RSV is symptomatic for roughly a week. Modeling work has estimated the force of infection or R0 within a range of 1.7–8.2, in part depending on the model structure and whether age stratification was included (Pitzer et al., 2015; Poletti et al., 2015; Reis & Shaman, 2016; Velasco-Hernández, Núñez-López, Comas-García, Cherpitel, & Ocampo, 2015; Weber, Weber, & Milligan, 2001; White et al., 2007).
Table 2.
Summary of clinical studies of each respiratory virus and estimates of the epidemiological parameters D and R0. We were unable to find an estimate of R0 for respiratory adenovirus. Where data were available, we show the mean estimate from each paper.
| D (days) | R0 | |
|---|---|---|
| Respiratory syncytial virus (RSV) |
Mean: 7.3; 5–10 (Hall et al., 2009); 11.2 shedding detected by PCR (Munywoki et al., 2015); 4.5 by antibody detection (Okiro et al., 2010); 6 inferred by modeling (Reis & Shaman, 2016) Assumed in prior modeling studies: 10.14 (Weber et al., 2001); 5–10 with progressive immunity (Pitzer et al., 2015); 9 (White et al., 2007) |
Mean: 4.5; 1.1–2.1 SIRS model (Weber et al., 2001) 8.9–9.2 SIRS model with age structures (Pitzer et al., 2015) 2.3–8.9 SIR model (Velasco-Hernández et al., 2015) 3.0 (Reis & Shaman, 2016) 2.18 (Poletti et al., 2015) 1.2–9.5 (White et al., 2007) depending on model structure 1.6 (Levy, Iv, & Yom-Tov, 2018) |
| Respiratory adenovirus (rAD) |
Mean: 7.7; With treatment: 5.5 (Larrañaga, Kajon, Villagra, & Avendaño, 1988–1996); 10.6 (Hong et al., 2001); 7 (Aberle et al., 2003) |
Cross-infection with adenovirus 7 h type: 55% (Palomino, Larrañaga, & Avendaño, 2000) |
| Rhinovirus |
Mean: 7.7; 3 (Corne et al., 2002); 7.4 (Gwaltney, Hendley, Simon, & Jordan, 1967) 10 (Pappas, Hendley, Hayden, & Winther, 2008); ill 7–9, shedding > 10 (Douglas, Cate, Gerone, & Couch, 1966) |
1.5 family members infected (Peltola et al., 2008a); 36% volunteers infected (small sample) (Hendley, Gwaltney, & Jordan, 1969); 95% infected, 74% developed a cold (Gwaltney, 2002); 74–90% volunteers developed a cold (Cohen, Tyrrell, & Smith, 1991); 1.2 from simulation (Levy et al., 2018) |
| Parainfluenza virus (PIV) |
Mean: 9.8; All subtypes: 3–17 d (Henrickson, 2003); PIV1: 4 d PIV2, PIV4: 6–13 d (Henrickson, 2003); PIV3: 9–10 d (Frank et al., 1981); 16 d to zero shedding after given vaccine (Belshe et al., 2004) |
2.7 from simulation (Levy et al., 2018) |
Respiratory adenovirus (rAD) can cause bronchiolitis, croup, and pneumonia, and infects 70%–80% of children by age 5 (Aberle et al., 2003). Children have been found exposed to an average of 3.4 rAD serotypes by age two, with the highest incidence in infants aged 6–12 months (Pacini, Collier, & Henderson, 1987). Up to 14% of patients with ILI were rAD positive (Li et al., 2013). As listed in Table 2, untreated, rAD patients are infected for more than a week. A highly contagious strain, 7 h, can infect 55% of contacts (Palomino et al., 2000).
Rhinovirus, an enterovirus, is most associated with a mild upper respiratory tract infection, especially a runny nose (Garcia et al., 2013). Nearly half of acute respiratory infections in infants have rhinovirus (Kusel et al., 2007). Rhinovirus has a high asymptomatic rate, estimated between 15% and 30% in children (Jartti, Lehtinen, Vuorinen, Koskenvuo, & Ruuskanen, 2004; Johnston et al., 1993; van Benten et al., 2003; van Gageldonk-Lafeber et al., 2005), and as much as 50% in adults (Peltola et al., 2008b). (Jansen et al., 2011). In temperate climates, rhinovirus outbreaks have been observed to peak in the fall and early spring (Gwaltney et al., 1967; Jacobs, Lamson, St George, & Walsh , 2013). All three serotypes have been found in each season, albeit sometimes on an alternating basis (Jacobs et al., 2013; Kaida et al., 2011; Miller et al., 2011; Savolainen-Kopra et al., 2009). Investigations continue to tease apart the contribution of each serotype to the seasonal pattern of rhinovirus (Kaida et al., 2011). Rhinovirus infections can last up to two weeks, and are easily spread among family members (Table 2).
Parainfluenza virus (PIV) serotypes 1 and 2 cause croup symptoms (seal barking cough caused by infection of vocal cords) and infect most children by age five. PIV types 1 and 2 are responsible for an estimated 250,000 hospital visits and 70,000 hospitalizations (Henrickson, Kuhn, & Savatski, 1994). PIV type 3 is the second most common cause of acute respiratory infection after RSV, and also causes bronchiolitis and pneumonia; most children get PIV-3 by age two (Glezen, Frank, Taber, & Kasel, 1984). PIV type 4 is detected less frequently in sick patients (Fry et al., 2006), although serological data show that PIV-4 circulates widely (Henrickson, 2003). PIV also infects older generations and those with a weakened immune system (Fry et al., 2006). PIV-3 infections tend to last longer than a week, whereas PIV 1–2 infections usually clear within a week (Table 2). We were unable to find estimates of R0 for any PIV subtypes.
1.2. Modeling work and aims
Of the four respiratory viruses to be examined here (Table 1), most prior studies have simulated RSV, using either a compartmental structure or more complicated model form (Pitzer et al., 2015; Reis & Shaman, 2016; Weber et al., 2001; White et al., 2007). Most likely, the work on RSV has been driven by its severity in infants and seasonal regularity. Recently Levy et al. (Levy et al., 2018) used a compartmental model in conjunction with twitter data, viral specimen data, and a matching pursuit decomposition approach to simulate nine respiratory pathogens producing ILI signal in England. In this paper, we use a compartmental model, data assimilation methods, and specimen data to simulate RSV, rAD, rhinovirus, and PIV subtypes and estimate epidemiological transmission parameters for each of these four viruses. Among the four viruses, outbreaks range from those that occur with regular, highly seasonal epidemic curves (e.g. RSV) to those with highly unstructured chains of transmission (e.g. rAD), where patterns only emerge at coarser temporal or geographical resolution.
2. Methods
2.1. Data
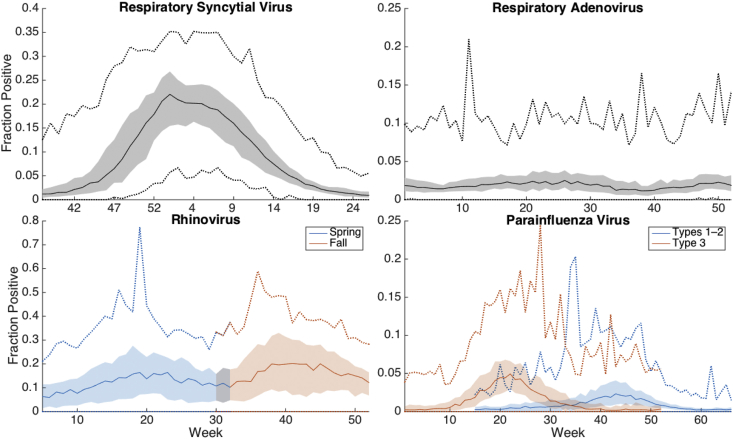
Specimen data, sampled using antigen detection, viral culture, and PCR testing, were provided by the Centers for Disease Control and Prevention (CDC), as reported through the National Respiratory and Enteric Virus Surveillance System (NREVSS). Data were available from 2004 to 2014 at the Census Division and Health and Human Services geographic regions in the United States. Rhinovirus samples were mainly tested using PCR-assay, and were only available in adequate volume since 2009 (i.e. at least ten tests per week). In this paper, we study positive specimen tests divided by the total number of tests performed each week in each region. The age structure of those tested was not available. Fig. 1 shows the mean outbreak for all each regional groupings and years. The shaded region indicates fraction positive data within 25–75% percentiles.
Fig. 1.
The mean outbreak across each Census Division and HHS region and year from 2004 to 2015 (rhinovirus dates: 2009–2015). The shaded region shows the data quartiles, and the dotted lines show the extremes.
For each virus type or subtype, we simulated each individual seasonal outbreak; thus, we modeled 19 overlapping geographical areas and 10 outbreak seasons for RSV, rAD, and PIV, and five seasons for rhinovirus. We simulated outbreaks of rhinovirus beginning in the fall and winter separately. We also grouped PIV types 1 and 2, and simulated PIV type 3 separately, due to PIV's marked seasonal variation by subtype; we did not include PIV type 4 due to a low number of positive tests (S1 Fig). Table 3 shows the Julian calendar weeks used to define the period of simulation for each virus. As shown in Fig. 1, mean outbreak structure differs among the viruses, ranging from regular outbreaks of RSV to highly irregular incidence of rAD. By separately simulating these four virus types, we can estimate the epidemiological characteristics of each, including the force of transmission.
Table 3.
Julian calendar weeks simulated for each virus.
| Respiratory Virus | RSV | Adenovirus | Rhinovirus winter | Rhinovirus fall | PIV1-2 | PIV3 |
|---|---|---|---|---|---|---|
| Weeks simulated | 37–26 | 1–52 | 5–32 | 30–52 | 15–14 | 1–52 |
2.2. Compartmental model
We used a susceptible-infected-recovered (SIR) compartmental model to represent transmission dynamics. As previously described for simulation of RSV outbreaks (Reis & Shaman, 2016), the SIR model structure is as follows:
| (1) |
| (2) |
where S is the susceptible population, I is the number of infected, is the basic reproductive number, D is the mean infection period, and N is the size of the population. N was held constant at an arbitrary size of 500,000 people.
2.3. Mapping fraction positive to number infected in the population
To estimate the incidence in a population of arbitrary size N, the positive fraction of specimen tests were multiplied by both N and a scalar γ. Generally, γ scales N to the actual population sampled in NREVSS regional ‘catchments’, including the number of reporting NREVSS laboratories and the population served by each laboratory. We found γ ranging from 0.02 to 0.05 produced posterior outbreaks with lowest RMSE for the six outbreak types, and γ = 0.02 produced the lowest RMSE summed across all six viruses.
2.4. Inference of parameter and state variables
We used the ensemble adjustment Kalman filter (EAKF) to infer parameter and state variables during model integration (Anderson, 2001; Karspeck & Anderson, 2007a, 2007b; Reis & Shaman, 2016; Shaman & Karspeck, 2012; Shaman, Karspeck, Yang, Tamerius, & Lipsitch, 2013). The EAKF generates posterior distributions by taking a weighted average of the prior distribution and observations, weighting the two estimates based on the variance of the observed model variable (here incidence), estimated directly from the ensemble of simulations, and the error variance of the observational data stream (OEV), estimated as:
| (3) |
The initial OEV (OEV0) was assumed to be 105 and the constant a was set to 50, values selected during prior work developing an SIR-EAKF model for RSV (Reis & Shaman, 2016). The observed infections are taken as a running mean of the prior 3 weeks (), allowing OEV to rise and fall with observed incidence.
To utilize the EAKF, an ensemble of model realizations is required. Here we used 300 ensemble members for each outbreak simulation, where each ensemble member was initialized independently from a wide range of initial state variable and parameter conditions (Table 4), as previously described (Reis & Shaman, 2016). We further ran each 300-member simulation five times to account for any stochastic effects during initialization. As our principal aim was to estimate D and R0, we ran a series of estimation tests using the SIR-EAKF and ‘synthetic’ (i.e. model-generated) data that resembled different viral outbreaks (see SI). These synthetic tests demonstrated the ability of our SIR-EAKF to adequately converge to the parameters used to generate the synthetic data.
Table 4.
The range of initial state variables for the SIR model for each virus.
| Variable or parameter | Initialized Range | Distribution |
|---|---|---|
| Susceptible, S | 1.1 × 105 –3.9 × 105 people; μ = 23.6 × 104; σ = 5.7 × 104 |
Normal |
| Infectious, I | One to 1.5 × 103 people; μ = 14.8 | Exponential |
| Initial D ± range | 3–8 days | Uniform |
| Initial R0 range | 1.3–4 | Uniform |
3. Results
3.1. Simulation
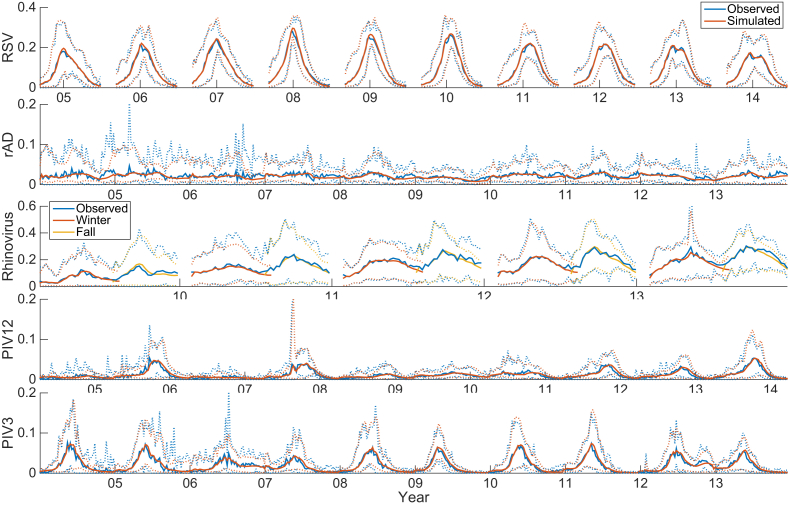
The observed and simulated outbreaks, taken as an average over each of the 19 regions, again reveal differences among the epidemic curves of the different respiratory viruses (Fig. 2). RSV is the most regular and rAD is the least regular of the simulated viruses. Simulation of highly regular outbreaks such as RSV had a high correlation with observed data (r2 = 0.98), whereas simulation of irregular outbreaks, such as rAD, had low correlations (r2 = 0.30, S1 Table). The fraction of observations that falls within the 90% and 50% credible intervals of all posterior ensemble ranges between 0.70–0.86 and 0.55–0.66, respectively, depending on the virus (see S1 Table). These ranges indicate that the ensemble-estimated credible intervals are slightly too narrow.
Fig. 2.
Observed outbreaks of each virus shown as a mean across the 19 overlapping U.S. regional data. Each season was simulated independently. Rhinovirus winter and fall outbreaks were each simulated independently. The dotted lines show the 5th and 95th percentile across all regions.
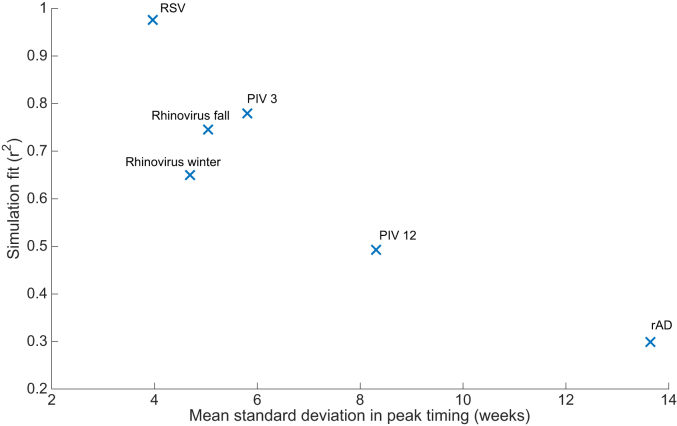
We quantified the variability of four epidemic characteristics: value of peak intensity, timing of peak intensity, mean cases (percentage of positive tests per week), and onset of the epidemic. The CDC defines an onset threshold for RSV as 10% positivity, but does not specify an onset threshold for the other viruses we simulate. Here, we define onset threshold as the mean level for each virus over all locations and epidemic weeks simulated, e.g. the RSV data on average were 9.4% positive. Based on quantification of these four characteristics, RSV outbreaks are the least variable, whereas the other viruses show considerably more variability (S2 Fig). Of the four epidemic characteristics, the strongest predictor of simulation fit was variability in peak timing. Fig. 3 shows that model fit, as measured by r2, has a strong negative correlation with the observed variability of the timing of peak intensity. Fig. 3 suggests that a compartmental model will yield an r2 ≥ 0.5 with a mean observed standard deviation of peak timing of eight weeks or fewer.
Fig. 3.
The model fit (r2) is negatively correlated with observed standard deviation of observed peak timing (weeks).
3.2. Parameter estimation
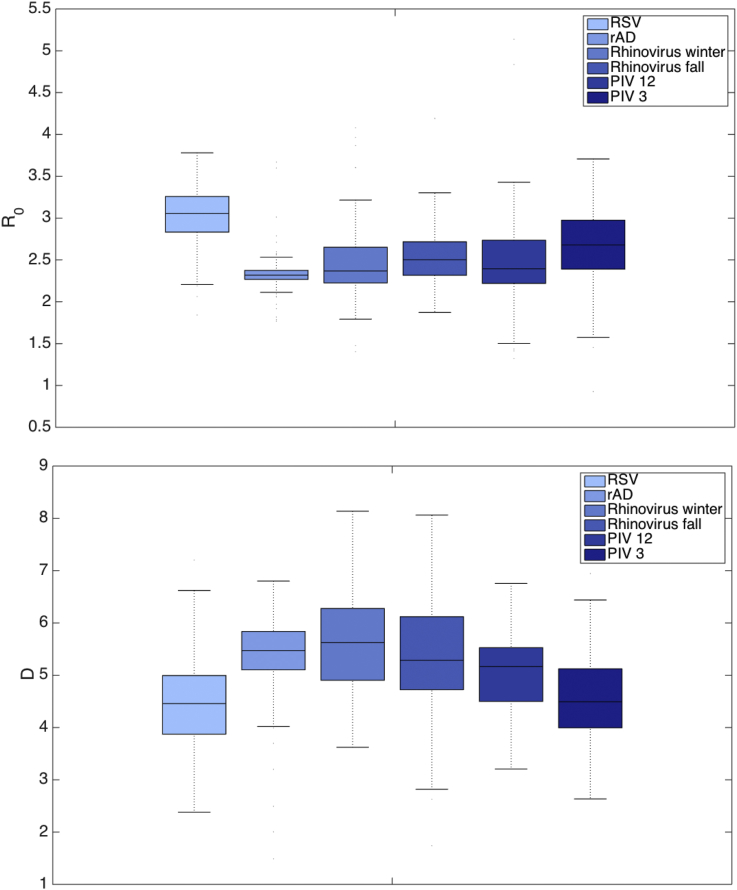
Fig. 4 presents boxplots of the basic reproductive number, R0, and the infection period, D, estimated on the last week of simulation for each outbreak. Across the six different viral groupings, the mean estimate of R0 range is 2.3–3.0, with a lower and upper quartile range of 2.0–2.8 and 2.6–3.2, respectively. Synthetic tests show that estimation R0 but not RE depend on the initialization of S, and median estimates of RE are shown in S5 Fig. The mean length of infection D varies 4.4–5.6 days, with the upper and lower interquartile ranges varying 3.7–4.9 and 5.1–6.3, respectively. As discussed, rAD has a relatively irregular seasonality and is unstructured, and this irregular shape may account for its relatively low R0 and high D. Table 5 lists the mean estimates of the parameters at the week of outbreak peak intensity and on the last week of simulation. RSV has the highest R0 (2.8 at peak) and the lowest length of infection, D, (5.2 days). Conversely, rAD and PIV types 1–2 had the lowest R0 (2.3 each at peak timing) and greatest value of D (5.5 each).
Fig. 4.
Estimates of R0 (top plot) and D (bottom plot) for each virus shown as a boxplot. Estimates were obtained on the last week of outbreak simulation.
Table 5.
Mean estimates of parameters R0 and D at the week of peak intensity and on the last week of outbreak simulation.
| Respiratory Virus | RSV | rAD | Rhinovirus winter | Rhinovirus fall | PIV1-2 | PIV3 |
|---|---|---|---|---|---|---|
| R0 at peak timing | 2.82 | 2.34 | 2.60 | 2.71 | 2.31 | 2.53 |
| D at peak timing | 5.16 | 5.49 | 5.24 | 5.24 | 5.53 | 5.29 |
| R0 on last week | 3.02 | 2.33 | 2.46 | 2.52 | 2.49 | 2.69 |
| D on last week | 4.44 | 5.39 | 5.64 | 5.35 | 4.99 | 4.58 |
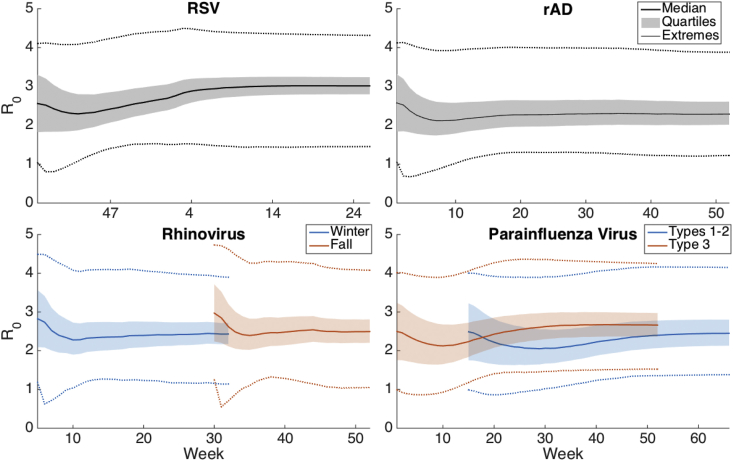
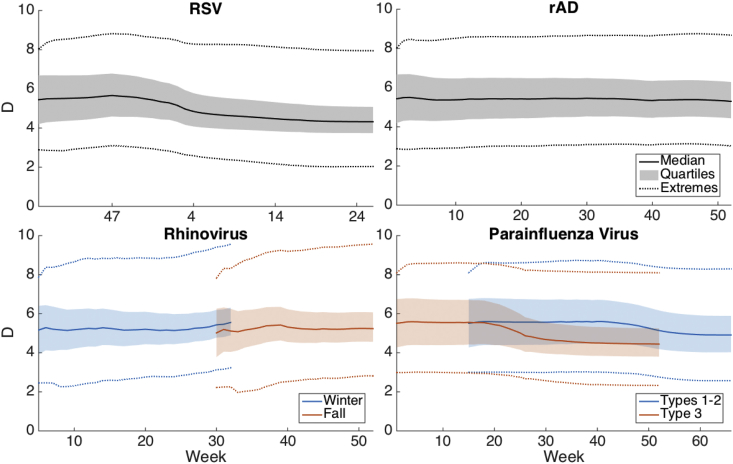
Fig. 5, Fig. 6 show the median parameter estimates of R0 and D as a time series throughout the simulation over each region. The shaded region shows the 25–75% ensemble result and the dotted lines show the minimum and maximum. We performed sensitivity analysis for the above estimates by shifting the initial prior range of D and repeating the simulation and parameter estimation. S3 and S4 Figures show R0 and D, respectively, over the simulation period when D is initialized with an initial prior range shifted ±4 days above or below the values listed in Table 4 (for convenience this simulation will be referred to as the baseline). As shown, the values of D are sensitive to the initial prior. The value of R0 converges to the baseline value for rAD and rhinovirus, but the change in initialization has a greater impact on the final value of R0 for RSV and PIV, although these values vary by less than one, suggesting that R0 is not overly sensitive to changes in the simulation initialization. The effective reproductive number, RE, moreover, converges within the simulation period (S5 Fig.). We also provide estimates of RE and β (R0/D) for the baseline simulation (S6 Fig.).
Fig. 5.
Estimates of R0 as an average time series for each virus. The shaded region shows the 25–75% interval. Doted lines show the minimum and maximum ensemble values.
Fig. 6.
Estimates of D as an average time series for each virus. The shaded region shows the 25–75% interval, with dotted lines indicating the maximum and minimum.
3.3. Geographical variation of parameter estimates
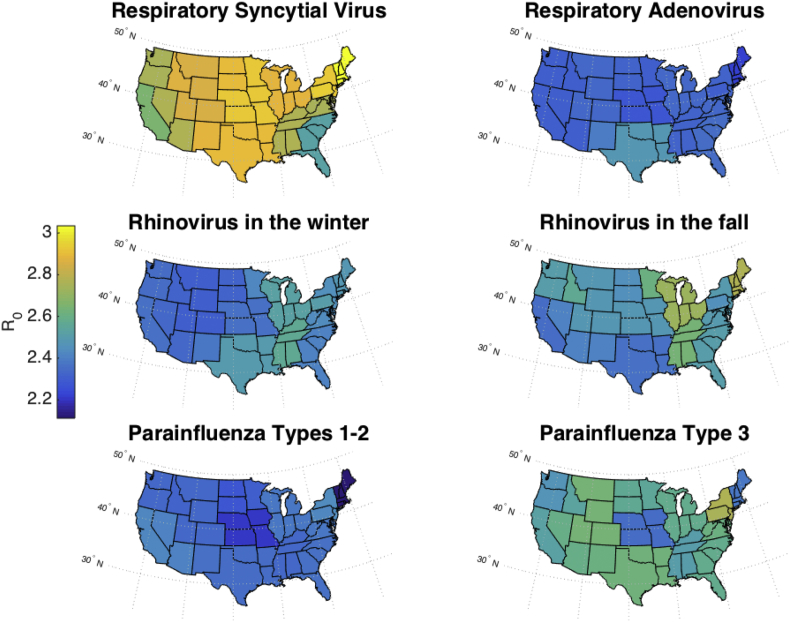
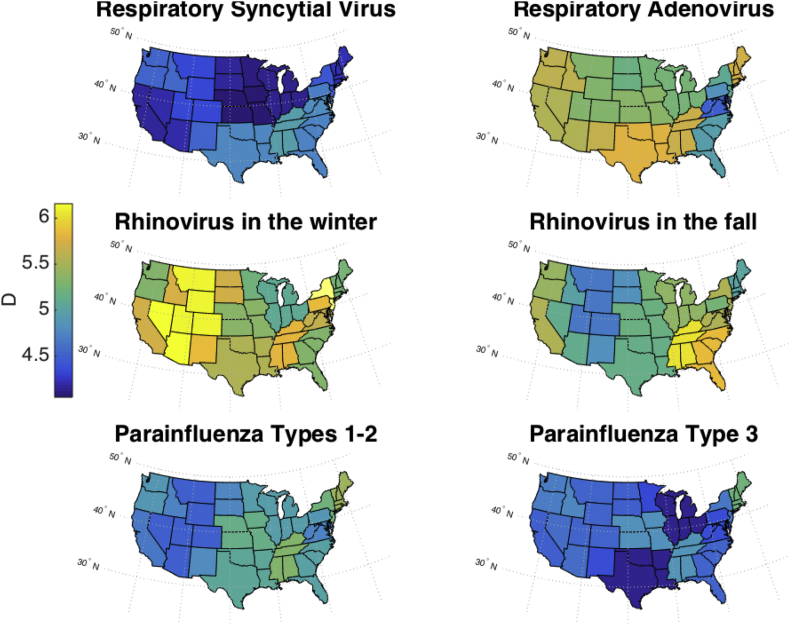
Fig. 7, Fig. 8 map the values of R0 and D, respectively, in the continental U.S., showing the mean parameter values, from weeks 7 to the end of the simulation, in each state as an average of the corresponding CD and HHS regions. RSV has a lower R0 in Florida and the southeast than in other states, which may be a reflection of the type of data simulated here, percent positive, versus the number of hospitalizations that has been used in previous work (Pitzer et al., 2015). Overall, R0 was the highest for RSV outbreaks and the lowest for outbreaks of parainfluenza virus types 1–2 in northern states. These maps show a greater spatial variation in D than in R0.
Fig. 7.
Estimates of mean R0 plotted in the continental U.S. Because Census Divisions and HHS regions overlap, the mean estimate from both regional groupings was taken for each state.
Fig. 8.
Estimates of mean D plotted in the continental U.S. Because Census Divisions and HHS regions overlap, the mean estimate from both regional groupings was taken for each state.
4. Discussion
In this paper, we simulated outbreaks of four respiratory viruses that often present as ILI, including simulations of respiratory adenovirus, rhinovirus, and parainfluenza virus. Prior estimates of epidemiological parameters for these three viruses were derived mainly from clinical work (Table 2), but here we provide estimates of the reproductive number, R0, and the length of illness, D, using broad epidemiological surveillance data.
Model simulations were well correlated with observed infection rates for rhinovirus, RSV, and PIV. The simulations were less strongly correlated with rAD observations, whose irregularity may reflect co-circulation of multiple serotypes or sparse reporting due to high asymptomatic infection rates or more limited testing. Should more active surveillance and testing for rAd occur in the future, outbreaks may be better resolved and simulated. Alternatively, other model forms might be tested in the future and might improve simulation. For the other viruses, the high correlation of the modeled data to the observations suggests that the SIR compartmental model is a reasonable form for representing transmission dynamics. We deliberately chose this simple model form so that it could be applied to a wide variety of respiratory viral outbreaks.
For RSV, our mean estimate of D and R0 of 5.2 days and 2.8, respectively, are within published clinical and modeling estimates (Table 2). Our estimate of the infection period, D, for rAD conforms to clinical observations, while our estimate of R0 of 2.3 is lower than the attack rate of 55% observed for the virulent strain 7 h, which seems to be more prevalent in low-income countries (Hatherill et al., 2004; Palomino et al., 2000; Rodríguez-Martínez, Rodríguez, & Nino, 2015; Straliotto, Siqueira, Machado, & Maia, 2004) but has also been found in the US and Canada (Erdman et al., 2002). The great diversity of rAD serotypes and their characteristics (Gray et al., 2007; Morris, Cooper, Barr, & Bailey, 1996) precludes identifying a single R0 for all rAD species. Fall and winter outbreaks of rhinovirus, which we simulated separately, may be caused by different strains; however, the mean R0 value of rhinovirus in the fall (2.7, September to December) was only slightly higher than it was for outbreaks beginning in the winter (2.6, February through July), but with a greater upper range (Fig. 8). Overall the R0 we inferred was at the high end of the range estimated from observations of household viral spread (roughly 1.5–2.5) and higher than a previous modeling estimate (Levy et al., 2018). Even so, it is possible that the R0 of rhinovirus is in fact still greater than here estimated due to the relatively mild symptoms expressed with rhinovirus infection that typically do not require medical attention. Clinical estimates of the infection period, D, for PIV subtypes vary widely from 3 days to two weeks (Belshe et al., 2004; Frank et al., 1981; Henrickson, 2003), and our estimates fall in the middle at slightly less than one week (5.5 and 5.3 days for PIV 1–2 and PIV 3, respectively), though our sensitivity analyses indicate that this parameter is not strongly constrained. Although a prior estimate found a higher estimate of R0 for PIV (Levy et al., 2018), our inferred values (Table 5) are among the smallest of the four viruses simulated, suggesting a less contagious pattern of transmission than RSV or rhinovirus.
Here we inferred epidemiological parameters using the EAKF, which provides a probability distribution for each estimate. In this study, we initialized the model susceptible and infectious state variables and D and R0 parameters from a random uniform range. To evaluate the sensitivity of the estimates, we varied the initialization of parameter D ± 4 days from the values derived from clinical work (Table 4). S3 –S5 Figs show that the impact on the estimate of R0 and RE was minor while the estimate of D was sensitive to initialization.
In the future, these four respiratory viruses could be further interrogated to identify factors affecting outbreak timing and intensity, such as events where vulnerable populations mix with other critical subpopulations, or geospatial or climatic associations with intensification of viral outbreaks (Pitzer et al., 2015; Shaman, Pitzer, Viboud, Grenfell, & Lipsitch, 2010). These viral outbreaks could be simulated to then explore scenarios that foster intense transmission, which could alert public health authorities to the most dangerous combination of risk factors. Similarly, computational experiments could be used to help evaluate potential interventions to limit infection, such as reduced contact with infected groups when infection rates are high or greater preparedness in hospitals and clinics for high volume of respiratory illnesses.
Handling Editor: Jianhong Wu
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.idm.2018.03.006.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Aberle S.W., Aberle J.H., Steininger C., Matthes-Martin S., Pracher E., Popow-Kraupp T. Adenovirus DNA in serum of children hospitalized due to an acute respiratory adenovirus infection. The Journal of Infectious Diseases. 2003;187:311–314. doi: 10.1086/367808. Springer, Berlin. [DOI] [PubMed] [Google Scholar]
- Anderson J.L. An ensemble adjustment kalman filter for data assimilation. Monthly Weather Review. 2001;129:2884–2903. [Google Scholar]
- Bellei N., Carraro E., Perosa A., Watanabe A., Arruda E., Granato C. Acute respiratory infection and influenza-like illness viral etiologies in Brazilian adults. Journal of Medical Virology. 2008;80:1824–1827. doi: 10.1002/jmv.21295. Wiley Subscription Services, Inc., A Wiley Company. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshe R.B., Newman F.K., Anderson E.L., Wright P.F., Karron R.A., Tollefson S. Evaluation of combined live, attenuated respiratory syncytial virus and parainfluenza 3 virus vaccines in infants and young children. The Journal of Infectious Disease. 2004;190:2096–2103. doi: 10.1086/425981. [DOI] [PubMed] [Google Scholar]
- van Benten I., Koopman L., Niesters B., Hop W., van Middelkoop B., de Waal L. Predominance of rhinovirus in the nose of symptomatic and asymptomatic infants. Pediatric Allergy & Immunology. 2003;14:363–370. doi: 10.1034/j.1399-3038.2003.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black R.E., Cousens S., Johnson H.L., Lawn J.E., Rudan I., Bassani D.G. Global, regional, and national causes of child mortality in 2008: A systematic analysis. Lancet. 2010;375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- Brandt C.D., Kim H.W., Vargosko A.J., Jeffries B.C., Arrobio J.O., Rindge B. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. I. Adenovirus pathogenicity in relation to serologic type and illness syndrome. American Journal of Epidemiology. 1969;90:484–500. doi: 10.1093/oxfordjournals.aje.a121094. [DOI] [PubMed] [Google Scholar]
- Cohen S., Tyrrell D.A.J., Smith A.P. Psychological stress and susceptibility to the common cold. New England Journal of Medicine. 1991;325:606–612. doi: 10.1056/NEJM199108293250903. [DOI] [PubMed] [Google Scholar]
- Corne J.M., Marshall C., Smith S., Schreiber J., Sanderson G., Holgate S.T. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: A longitudinal cohort study. Lancet. 2002;359:831–834. doi: 10.1016/S0140-6736(02)07953-9. [DOI] [PubMed] [Google Scholar]
- Douglas R.G., Cate T.R., Gerone P.J., Couch R.B. Quantitative rhinovirus shedding patterns in volunteers. American Review of Respiratory Disease. 1966;94:159–167. doi: 10.1164/arrd.1966.94.2.159. [DOI] [PubMed] [Google Scholar]
- Ebell M.H., Afonso A. A systematic review of clinical decision rules for the diagnosis of influenza. Ann Fam Med. American Academy of Family Physicians. 2011;9:69–77. doi: 10.1370/afm.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman D.D., Xu W., Gerber S.I., Gray G.C., Schnurr D., Kajon A.E. Molecular epidemiology of adenovirus type 7 in the United States, 1966-2000. Emerging Infectious Disease. 2002;8:269–277. doi: 10.3201/eid0803.010190. Centers for Disease Control and Prevention. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A. Respiratory syncytial virus infection in older persons. Vaccine. 1998;16:1775–1778. doi: 10.1016/s0264-410x(98)00142-x. [DOI] [PubMed] [Google Scholar]
- Falsey A.R., Hennessey P.A., Formica M.A., Cox C., Walsh E.E. Respiratory syncytial virus infection in elderly and high-risk adults. New England Journal of Medicine. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- Frank A.L., Taber L.H., Wells C.R., Wells J.M., Glezen W.P., Paredes A. Patterns of shedding of myxoviruses and paramyxoviruses in children. The Journal of Infectious Disease. 1981;144:433–441. doi: 10.1093/infdis/144.5.433. [DOI] [PubMed] [Google Scholar]
- Fry A.M., Curns A.T., Harbour K., Hutwagner L., Holman R.C., Anderson L.J. Seasonal trends of human parainfluenza viral infections: United States, 1990–2004. Clinical Infectious Disease. 2006;43:1016–1022. doi: 10.1086/507638. American Society for Microbiology, Washington, DC. [DOI] [PubMed] [Google Scholar]
- van Gageldonk-Lafeber A.B., Heijnen M.-L.A., Bartelds A.I.M., Peters M.F., van der Plas S.M., Wilbrink B. A case-control study of acute respiratory tract infection in general practice patients in The Netherlands. Clinical Infectious Disease. 2005;41:490–497. doi: 10.1086/431982. Blackwell Science, Oxford. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J., Espejo V., Nelson M., Sovero M., Villaran M.V., Gomez J. Human rhinoviruses and enteroviruses in influenza-like illness in Latin America. Virology Journal. 2013;10:305. doi: 10.1186/1743-422X-10-305. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilca R., Amini R., Douville-Fradet M., Charest H., Dubuque J., Boulianne N. Other respiratory viruses are important contributors to adult respiratory hospitalizations and mortality even during peak weeks of the influenza season. Open Forum Infectious Diseases. 2014:1. doi: 10.1093/ofid/ofu086. ofu086-ofu086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glezen W.P., Frank A.L., Taber L.H., Kasel J.A. Parainfluenza virus type 3: Seasonality and risk of infection and reinfection in young children. The Journal of Infectious Diseases. 1984;150:851–857. doi: 10.1093/infdis/150.6.851. [DOI] [PubMed] [Google Scholar]
- Goldstein E., Greene S.K., Olson D.R., Hanage W.P., Lipsitch M. Estimating the hospitalization burden associated with influenza and respiratory syncytial virus in New York City, 2003-2011. Influenza and Other Respiratory Viruses. 2015;9:225–233. doi: 10.1111/irv.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G.C., McCarthy T., Lebeck M.G., Schnurr D.P., Russell K.L., Kajon A.E. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004-2006. Clinical Infectious Diseases. 2007;45:1120–1131. doi: 10.1086/522188. Churchill Livingstone, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwaltney J.M. Clinical significance and pathogenesis of viral respiratory infections. Americas Journal of Medicine. 2002;112(Suppl. 6A):13S–18S. doi: 10.1016/s0002-9343(01)01059-2. [DOI] [PubMed] [Google Scholar]
- Gwaltney J.M., Hendley J.O., Simon G., Jordan W.S. Rhinovirus infections in an industrial population. II. Characteristics of illness and antibody response. Journal of the American Medical Association. 1967;202:494–500. [PubMed] [Google Scholar]
- Hall C.B., Douglas R.G., Geiman J.M. Respiratory syncytial virus infections in infants: Quantitation and duration of shedding. The Journal of Pediatrics. 1976;89:11–15. doi: 10.1016/s0022-3476(76)80918-3. [DOI] [PubMed] [Google Scholar]
- Hall C.B., Geiman J.M., Breese B.B., Douglas R.G. Parainfluenza viral infections in children: Correlation of shedding with clinical manifestations. The Journal of Pediatrics. 1977;91:194–198. doi: 10.1016/s0022-3476(77)80811-1. [DOI] [PubMed] [Google Scholar]
- Hall C.B., Weinberg G.A., Iwane M.K., Blumkin A.K., Edwards K.M., Staat M.A. The burden of respiratory syncytial virus infection in young children. New England Journal of Medicine. 2009;360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handforth J., Friedland J.S., Sharland M. Basic epidemiology and immunopathology of RSV in children. Paediatric Respiratory Reviews. 2000;1:210–214. doi: 10.1053/prrv.2000.0050. [DOI] [PubMed] [Google Scholar]
- Hatherill M., Levin M., Lawrenson J., Hsiao N.-Y., Reynolds L., Argent A. Evolution of an adenovirus outbreak in a multidisciplinary children's hospital. Journal of Paediatrics and Child Health. 2004;40:449–454. doi: 10.1111/j.1440-1754.2004.00426.x. Blackwell Science Pty. [DOI] [PubMed] [Google Scholar]
- Hendley J.O., Gwaltney J.M., Jordan W.S. Rhinovirus infections in an industrial population. IV. Infections within families of employees during two fall peaks of respiratory illness. American Journal of Epidemiology. 1969;89:184–196. doi: 10.1093/oxfordjournals.aje.a120928. Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Henrickson K.J. Parainfluenza viruses. Clinical Microbiology Reviews. 2003;16:242–264. doi: 10.1128/CMR.16.2.242-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson K.J., Kuhn S.M., Savatski L.L. Epidemiology and cost of infection with human parainfluenza virus types 1 and 2 in young children. Clinical Infectious Disease. 1994;18:770–779. doi: 10.1093/clinids/18.5.770. Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Hong J.-Y., Lee H.-J., Piedra P.A., Choi E.-H., Park K.-H., Koh Y.-Y. Lower respiratory tract infections due to adenovirus in hospitalized Korean Children: Epidemiology, clinical features, and prognosis. Clinical Infectious Disease. 2001;32:1423–1429. doi: 10.1086/320146. Churchill Livingstone, New York. [DOI] [PubMed] [Google Scholar]
- Jacobs S.E., Lamson D.M., St George K., Walsh T.J. Human rhinoviruses. Clinical Microbiology Reviews. 2013;26:135–162. doi: 10.1128/CMR.00077-12. American Society for Microbiology (ASM) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.R., Wieringa J., Koekkoek S.M., Visser C.E., Pajkrt D., Molenkamp R. Frequent detection of respiratory viruses without Symptoms: Toward defining clinically relevant cutoff values. Journal of Clinical Microbiology. 2011;49:2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti T., Lehtinen P., Vuorinen T., Koskenvuo M., Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. Journal of Medical Virology. 2004;72:695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- Johnston S.L., Sanderson G., Pattemore P.K., Smith S., Bardin P.G., Bruce C.B. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. Journal of Clinical Microbiology. 1993;31:111–117. doi: 10.1128/jcm.31.1.111-117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvén T., Mertsola J., Waris M., Leinonen M., Meurman O., Roivainen M. Etiology of community-acquired pneumonia in 254 hospitalized children. The Pediatric Infectious Disease Journal. 2000;19:293–298. doi: 10.1097/00006454-200004000-00006. [DOI] [PubMed] [Google Scholar]
- Kaida A., Kubo H., Takakura K., Togawa M., Shiomi M., Kohdera U. Molecular epidemiology of human rhinovirus C in patients with acute respiratory tract infections in Osaka City, Japan. Japanese Journal of Infectious Diseases. 2011;64:488–492. [PubMed] [Google Scholar]
- Karspeck A.R., Anderson J.L. Experimental implementation of an ensemble adjustment filter for an intermediate ENSO model. Journal of Climate. 2007;20:4638–4658. [Google Scholar]
- Karspeck A.R., Anderson J.L. Experimental implementation of an ensemble adjustment filter for an intermediate ENSO model. Journal of Climate. 2007;20:4638–4658. [Google Scholar]
- Kusel M.M.H., de Klerk N.H., Kebadze T., Vohma V., Holt P.G., Johnston S.L. Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. The Journal of Allergy and Clinical Immunology. 2007;119:1105–1110. doi: 10.1016/j.jaci.2006.12.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrañaga C., Kajon A., Villagra E., Avendaño L.F. Adenovirus surveillance on children hospitalized for acute lower respiratory infections in Chile. Journal of Medical Virology. 1988-1996;2000(60):342–346. [PubMed] [Google Scholar]
- Leader S., Kohlhase K. Recent trends in severe respiratory syncytial virus (RSV) among US infants, 1997 to 2000. The Journal of Pediatrics. 2003;143:127–132. doi: 10.1067/s0022-3476(03)00510-9. [DOI] [PubMed] [Google Scholar]
- Levy N., Iv M., Yom-Tov E. Modeling influenza-like illnesses through composite compartmental models. Physica A: Statistical Mechanics and its Applications. 2018;494:288–293. [Google Scholar]
- Li H., Wei Q., Tan A., Wang L., Denny F., Loda F. Epidemiological analysis of respiratory viral etiology for influenza-like illness during 2010 in Zhuhai, China. Virology Journal. 2013;10:143. doi: 10.1186/1743-422X-10-143. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the global burden of disease study 2010. Lancet (London, England) 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. World Health Organization, Geneva. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S. Acute respiratory infections are world's third leading cause of death. BMJ. 2010:341. [Google Scholar]
- Miller E.K., Gebretsadik T., Carroll K.N., Dupont W.D., Mohamed Y.A., Morin L.-L. Viral etiologies of infant bronchiolitis, croup and upper respiratory illness during 4 consecutive years. The Pediatric Infectious Disease Journal. 2013;32:950–955. doi: 10.1097/INF.0b013e31829b7e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.K., Williams J.V., Gebretsadik T., Carroll K.N., Dupont W.D., Mohamed Y.A. Host and viral factors associated with severity of human rhinovirus–associated infant respiratory tract illness. The Journal of Allergy and Clinical Immunology. 2011;127:883–891. doi: 10.1016/j.jaci.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V. Indoor air pollution from biomass combustion and acute respiratory illness in preschool age children in Zimbabwe. International Journal of Epidemiology. 2003;32:847–853. doi: 10.1093/ije/dyg240. Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Morris D.J., Cooper R.J., Barr T., Bailey A.S. Polymerase chain reaction for rapid diagnosis of respiratory adenovirus infection. Journal of Infection. 1996;32:113–117. doi: 10.1016/s0163-4453(96)91250-5. [DOI] [PubMed] [Google Scholar]
- Munoz F.M., Piedra P.A., Demmler G.J. Disseminated adenovirus disease in immunocompromised and immunocompetent children. Clinical Infectious Disease. 1998;27:1194–1200. doi: 10.1086/514978. [DOI] [PubMed] [Google Scholar]
- Munywoki P.K., Koech D.C., Agoti C.N., Kibirige N., Kipkoech J., Cane P.A. Influence of age, severity of infection, and co-infection on the duration of respiratory syncytial virus (RSV) shedding. Epidemiology and Infection. 2015;143:804–812. doi: 10.1017/S0950268814001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiro E.A., White L.J., Ngama M., Cane P.A., Medley G.F., Nokes D.J. Duration of shedding of respiratory syncytial virus in a community study of Kenyan children. BMC Infectious Diseases. 2010;10:15. doi: 10.1186/1471-2334-10-15. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiowy C. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. Journal of Clinical Microbiology. 1998;36:3149–3154. doi: 10.1128/jcm.36.11.3149-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini D.L., Collier A.M., Henderson F.W. Adenovirus infections and respiratory illnesses in children in group day care. The Journal of Infectious Disease. 1987;156:920–927. doi: 10.1093/infdis/156.6.920. Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Palomino M.A., Larrañaga C., Avendaño L.F. Hospital-acquired adenovirus 7h infantile respiratory infection in Chile. The Pediatric Infectious Disease Journal. 2000;19:527–531. doi: 10.1097/00006454-200006000-00007. [DOI] [PubMed] [Google Scholar]
- Pappas D.E., Hendley J.O., Hayden F.G., Winther B. Symptom profile of common colds in school-aged children. The Pediatric Infectious Disease Journal. 2008;27:8–11. doi: 10.1097/INF.0b013e31814847d9. [DOI] [PubMed] [Google Scholar]
- Pavia A.T. Viral infections of the lower respiratory Tract: Old viruses, new viruses, and the role of diagnosis. Clinical Infectious Disease. 2011;52:S284–S289. doi: 10.1093/cid/cir043. Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola V., Waris M., Osterback R., Susi P., Ruuskanen O., Hyypiä T. Rhinovirus transmission within families with children: Incidence of symptomatic and asymptomatic infections. The Journal of Infectious Disease. 2008;197:382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- Peltola V., Waris M., Österback R., Susi P., Ruuskanen O., Hyypiä T. Rhinovirus transmission within families with Children: Incidence of symptomatic and asymptomatic infections. The Journal of Infectious Disease. 2008;197:382–389. doi: 10.1086/525542. Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Pitzer V.E., Viboud C., Alonso W.J., Wilcox T., Metcalf C.J., Steiner C.a. Environmental drivers of the spatiotemporal dynamics of respiratory syncytial virus in the United States. PLoS Pathogens. 2015;11:e1004591. doi: 10.1371/journal.ppat.1004591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poletti P., Merler S., Ajelli M., Manfredi P., Munywoki P.K., Nokes J. 2015. Evaluating vaccination strategies for reducing infant respiratory syncytial virus infection in low-income settings; p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis J., Shaman J. Retrospective parameter estimation and forecast of respiratory syncytial virus in the United States. PLoS Computational Biology. 2016;12:e1005133. doi: 10.1371/journal.pcbi.1005133. Wilke CO, editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Martínez C.E., Rodríguez D.A., Nino G. Respiratory syncytial virus, adenoviruses, and mixed acute lower respiratory infections in children in a developing country. Journal of Medical Virology. 2015;87:774–781. doi: 10.1002/jmv.24139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen-Kopra C., Blomqvist S., Kaijalainen S., Jounio U., Juvonen R., Peitso A. All known human rhinovirus species are present in sputum specimens of military recruits during respiratory infection. Viruses. 2009;1:1178–1189. doi: 10.3390/v1031178. Multidisciplinary Digital Publishing Institute (MDPI) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluger N.W. World Lung Foundation; 2010. World Lung Foundation (New York NY. Acute respiratory infections atlas. [Google Scholar]
- Shaman J., Karspeck A. Forecasting seasonal outbreaks of influenza. Proceedings of the National Academy of Sciences. 2012;109:20425–20430. doi: 10.1073/pnas.1208772109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman J., Karspeck A., Yang W., Tamerius J., Lipsitch M. Real-time influenza forecasts during the 2012-2013 season. Nature Communications. 2013;4:2837. doi: 10.1038/ncomms3837. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman J., Pitzer V.E., Viboud C., Grenfell B.T., Lipsitch M. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biology. 2010:8. doi: 10.1371/journal.pbio.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. R., Samet J. M., Romieu I., Bruce N.. Indoor air pollution in developing countries and acute lower respiratory infections in children. [DOI] [PMC free article] [PubMed]
- Straliotto S.M., Siqueira M.M., Machado V., Maia T.M. Respiratory viruses in the pediatric intensive care unit: Prevalence and clinical aspects. Clin Stud Drug Resist. Fundação Oswaldo Cruz. 2004;99:883–887. doi: 10.1590/s0074-02762004000800017. [DOI] [PubMed] [Google Scholar]
- Velasco-Hernández J.X., Núñez-López M., Comas-García A., Cherpitel D.E.N., Ocampo M.C. Superinfection between influenza and RSV alternating patterns in San Luis potosí state, México. PLoS One. 2015;10:e0115674. doi: 10.1371/journal.pone.0115674. Tang JW, editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A., Weber M., Milligan P. Modeling epidemics caused by respiratory syncytial virus ( RSV ) Mathematical Biosciences. 2001;172:95–113. doi: 10.1016/s0025-5564(01)00066-9. [DOI] [PubMed] [Google Scholar]
- White L.J., Mandl J.N., Gomes M.G.M., Bodley-Tickell A.T., Cane P.A., Perez-Brena P. Understanding the transmission dynamics of respiratory syncytial virus using multiple time series and nested models. Mathematical Biosciences. 2007;209:222–239. doi: 10.1016/j.mbs.2006.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.G., Gouws E., Boschi-Pinto C., Bryce J., Dye C. Estimates of world-wide distribution of child deaths from acute respiratory infections. The Lancet Infectious Disease. 2002;2:25–32. doi: 10.1016/s1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.