Abstract
Long non-coding RNAs (lncRNAs) have crucial roles via tethering with DNA, RNA or protein in diverse biological processes. These lncRNA-mediated interactions enhance gene regulatory networks and modulate a wide range of downstream genes. It has been demonstrated that several lncRNAs act as key regulators in hematopoiesis. This review highlights the roles of lncRNAs in normal hematopoietic development and discusses how lncRNA dysregulation correlates with disease prognoses and phenotypes.
Keywords: lncRNAs, Hematopoiesis, Leukemia
1. Introduction
The human genome is pervasively transcribed, with a percentage of almost 75% DNA sequence transcribed into RNAs. Nevertheless, only less than 2% of genome encodes proteins.1 Previous research showed that, in general, the more complex the eukaryotes are, the larger amounts of non-coding RNAs (ncRNAs) they possess.2 This suggests that ncRNAs play pivotal roles in the evolution of eukaryotic biological complexity and are indispensable components of transcriptomes.3, 4 According to the length, ncRNAs are categorized into two main types: small ncRNAs and long non-coding RNAs (lncRNAs). Small ncRNAs include microRNAs (miRNAs), PIWI-interacting RNAs (pi-RNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and other classes of small regulatory RNAs.5 LncRNAs are currently defined as transcripts greater than 200 nucleotides with no apparent open reading frames (ORFs).6
As research continues, the number of lncRNAs increases constantly due to deeper and more sensitive RNA sequencing technologies.7, 8 This exploding class of transcripts increasingly attracts attention of researchers. Although there are more and more documented lncRNAs,9 details about lncRNAs are still uncovered. Studies have revealed that several annotated lncRNAs turn to encode for small peptides. For instance, a micropeptide named myoregulin, which is essential for muscle performance, is encoded by a putative lncRNA.10 Also, lncRNA LINC00961 encodes for SPAR polypeptide, which is involved in mTORC1 pathway regulation and muscle regeneration.11 Thus, with the help of these hidden peptides, lncRNAs could more finely regulate specific biological process.
Although lncRNAs are expressed at lower level than protein-coding genes, they tend to be more cell-type specific.1, 3, 8 Studies have shown that lncRNAs are restricted to be transcribed in precise developmental and cellular context,12, 13, 14, 15 indicating they might act as ‘fine-tuners’ of cell fate. Moreover, it has been reported that lncRNAs participate in multiple important biological processes, including genomic imprinting, dosage compensation, pluripotency,15, 16 cell differentiation and other pathological processes.17, 18, 19, 20 For example, lncRNA Xist in X chromosome inactivation,21, 22, 23 Kcnq1ot1 in genomic imprinting,24, 25 Evf2 in neural cell fate determination,26 linc-MD1 in muscle differentiation,27, 28 MALAT1 in diabetes and cancers.29, 30
While most well-characterized lncRNAs are shaped with 5′ cap structure and 3′ poly(A) tails, studies have revealed new formats: such non-polyadenylated lncRNAs are transcribed by RNA polymerase III, or processed by RNase P cleavage to generate a mature 3′ end, or capped by snoRNP complexes at both ends, or by forming circular structures.31, 32 Due to their structural diversity, ways of lncRNAs involved in diverse regulation are plentiful.33 Evidence so far indicates that lncRNAs act mainly through interacting with RNA, DNA or protein.34 In cytoplasm, lncRNAs can bind to protein or RNA: they can act as “miRNA sponge” to decoy specific miRNA and lead to miRNA inactivation; also, they could function as scaffold, tethering protein to form complex, due to their structural flexibility.34, 35 In nucleus, lncRNAs cooperate with protein to regulate chromatin state in cis or in trans,36, 37 they also guide protein to particular genomic loci via RNA-DNA interactions.7 Therefore, lncRNAs could modulate multiple cellular contexts, including chromatin remodeling, transcription, pre-mRNA splicing and RNA stability.35, 38, 39
In this review, we discuss with a particular focus on how lncRNAs contribute to normal hematopoiesis, and how dysregulation of lncRNAs takes responsibility for malignant hematopoiesis.
2. LncRNAs in normal hematopoiesis
Hematopoiesis is a dynamic process of formation of all kinds of blood cells. In vertebrate, there are multiple waves of hematopoiesis during development.40 The first primitive wave produces unipotent blood cell types. It is transient and replaced by the definitive wave, which generates multipotent hematopoietic progenitors.40, 41 The definitive wave produces long-term hematopoietic stem cells (LT-HSCs) which have robust multilineage repopulating hematopoietic activity. The blood cell development is intricate and well-regulated and the hematopoietic hierarchy serves as a good paradigm for research. Lots of factors and pathways are involved in this process, including lncRNAs.40, 42, 43, 44 (See Fig. 1).
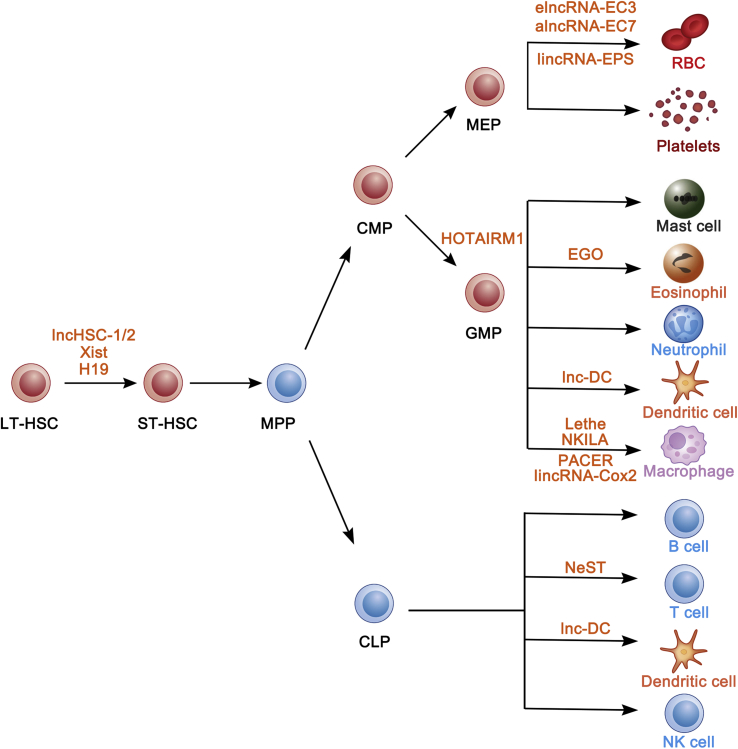
Fig. 1.
LncRNAs in normal hematopoiesis. Several lncRNAs are involved in the blood cell development. LncHSC-1/2, Xist and H19 regulate HSC quiescence and self-renewal. LinRNA-EPS, alncRNA-EC7 and elncRNA-EC3 modulate red blood cell formation. HOTAIRM1 functions in granulocytic differentiation. EGO promotes eosinophil differentiation. LincRNA-Cox2, NKILA, PACER, and Lethe participate in the innate immune response. NeST correlates to T cell formation. Lnc-DC is essential for dendritic cell differentiation.
2.1. Hematopoietic stem cells (HSCs)
Mature blood cells derive from the hematopoietic stem cells (HSCs), which are able to self-renew and have the capacity to give rise to multipotent progenitors, then yielding red blood cells, megakaryocytes, monocytes and lymphocytes via unilineage differentiation. In general, HSCs have the potential to rebuild the entire blood system.40 Observations suggest that several lncRNAs function as key regulators of HSC maintenance and differentiation.
To assess the functional lncRNAs in HSC, Goodell and her colleagues comprehensively identified lncRNAs specific to HSC.45 They first identified and annotated two functional lncRNAs, named lncHSC-1 and lncHSC-2. LncHSC-1 is an HSC-specific lncRNA, whereas lncHSC-2 occurs in both HSCs and other progenitors. Knockdown of lncHSC-1 results in increased myeloid differentiation. However, knockdown of lncHSC-2 promotes T cell differentiation and affects HSC self-renewal to some extent.45 The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting mitochondrial metabolism. Qian and colleagues described unique fingerprint lncRNAs in 17 Hematopoietic cell types and revealed that lncRNAs from the Dlk1-Gtl2 Locus predominantly enriched in CD49blo LT-HSCs. These lncRNAs might also preserve LT-HSC function.46 Furthermore, another study indicated that the imprinted lncRNA gene H19 is highly expressed in long-term HSCs (LT-HSCs), regulating HSC self-renewal.47 Conditional deletion of the maternal H19 leads to LT-HSCs decrease and short-term HSCs (ST-HSCs) increase. H19-deficient HSCs lose their quiescence and begin to proliferate, and finally are exhausted. Interestingly, this phenomenon is due to the de-repression of Igf2 and Igf1r. In turn, H19-deficient phenotype can be partially rescued by inactivation of Igf1r.47 Taken together, lncRNA H19 modulates HSC quiescence and self-renewal via the Igf2–Igfr1 pathway. Similarly, lncRNA Xist deficiency in HSCs causes aberrant maturation and age-dependent-loss.48 The metastasis-associated lung adenocarcinoma transcript 1, MALAT1, is a marker for lung cancer metastasis.49 LncRNA Malat1 is regulated by p53 and plays a significant role in maintaining the proliferation potential of early-stage hematopoietic cells.50
2.2. Erythroid, myeloid and lymphoid cells
With the help of next-generation sequencing of cells at key stages of erythropoiesis, erythroid specific lncRNA, LincRNA erythroid prosurvival (LincRNA-EPS), was annotated.51 Evidence suggests that, LincRNA-EPS promotes erythroid differentiation and inhibits apoptosis of mature erythrocytes. It is demonstrated that lincRNA-EPS represses many apoptotic gene, such as Pycard.51 In addition, lincRNA-EPS-deficient mice show enhanced inflammation, because lincRNA-EPS acts as a transcriptional brake to repress immune response genes expression by interacting with hnRNPL.52 Another lncRNA, lncRNA-EC7, derived from an enhancer region, could act in cis to activate its neighbor gene BAND3, which is a major anion transporter on erythrocyte membranes.43 Also, enhancer derived (elncRNAs) elncRNA-EC3 functions as cis-regulator to promote KIF2A expression during erythropoiesis.43 Shlnc-EC6 is highly expressed in late stage of erythroid cells and regulates murine erythroid enucleation via Rac1/PIP5K signal pathway.53 Long noncoding monocytic RNA (lnc-MC) promotes monocyte/macrophage differentiation by soaking up miR-199a-5p and releasing ACVR1B expression.54 Moreover, Eosinophil Granule Ontogeny (EGO) is an lncRNA involved in eosinophil development.55 EGO is highly expressed in mature eosinophils and diminished EGO leads to decreased basic protein expression and eosinophil derived neurotoxins, which are essential components of eosinophil development.55 HOXA transcript antisense RNA myeloid-specific 1 (HOTAIRM1) is an lncRNA expressed from the human HOXA cluster.56 This lncRNA specifically occurs in myeloid lineage and implicates in the regulation of granulocytic differentiation.57 Knockdown of HOTAIRM1 in NB4 promyelocytic leukemia cells inhibits HOXA1 and HOXA4 expression, as well as CD11b and CD18, the hallmarks of mature granulocytes.58 Nonetheless, the exact mechanisms still await for clarification.
Lymphocyte development involves a complex series of tightly choreographed events. Cao and his colleagues identify lnc-DC and make elaborate explanation on its role in human dendritic cell differentiation.59 Lnc-DC is exclusively expressed in classical antigen-presenting DCs (cDCs). Knockdown of lnc-DC during human monocyte-derived DCs (Mo-DC) differentiation results in impaired DC differentiation along with down-regulated expression of numerous DC-function-related genes. Mechanistically, lnc-DC binds to STAT3 and enhances its phosphorylation, which is a novel mechanism of lncRNAs, involving in the posttranslational modification.59 BIC is a primary microRNA (pri-miR-155) and can be processed to miR-155. B cell lymphomas patients show accumulation of miR-155 and ncRNA BIC.60 High levels of B-cell receptor triggers BIC expression, indicating BIC might involve in the selection of B cells.61
Moreover, lncRNAs also implicate in the modulation of innate immune and inflammatory responses.34, 62, 63 Lethe, a mammalian pseudogene lncRNA, is usually induced by inflammatory cytokines via NF-κB.64 Conversely, Lethe could bind to RelA, a subunit of NF-κB, and functions as a NF-κB decoy molecule, forming a negative feedback loop.64 Similarly, the p50-associated COX-2 extragenic RNA (PACER) also acts as a decoy lncRNA for the NF-κB signaling pathway and interacts with NF-κB subunit p50 to sequester it away from PTGS2 promoter.65
Another NF-κB interacting lncRNA, NKILA, could suppress breast cancer metastasis via negative regulating NF-κB signaling.66 Additionally, lincRNA-Cox2 modulates the expression of inflammatory molecules by interacting with hnRNP-A/B and hnRNP-A2/B1 in innate immune response.67 LncRNA NeST, also known as Tmevpg1, occurs in CD8+ T and CD4+ T helper 1 (Th1) cells, could control microbial susceptibility through its control of IFN-γ gene.68, 69, 70, 71
3. LncRNAs in malignant hematopoiesis
As we described above, normally expressed lncRNAs ensure healthy hematopoietic development. Conversely, dysregulated lncRNAs trigger blood diseases.72, 73, 74, 75, 76, 77 (See Fig. 2).
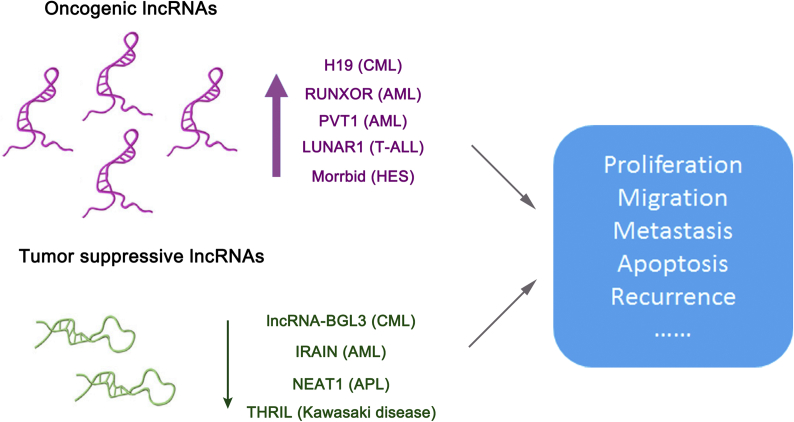
Fig. 2.
LncRNAs in malignant hematopoiesis. Some lncRNAs share close relationship with hematopoietic diseases. They act as oncogenic or tumor suppressive lncRNAs. H19 and lncRNA-BGL3 are associated with chronic myeloid leukemia (CML). RUNXOR, PVT1 and IRAIN are involved in acute myeloid leukemia (AML). LUNAR1 acts as an oncogene in T cell acute lymphoblastic leukemia (T-ALL). Morrbid is highly expressed in patients with hypereosinophilic syndrome (HES). NEAT1 expression is repressed in de novo acute promyelocytic leukemia (APL) patients. Similarly, Kawasaki disease patients with severe symptoms show decreased THRIL expression.
3.1. Chronic myelogenous leukemia (CML)
Chronic myelogenous leukemia (CML), also known as chronic myeloid leukemia, is characterized by unregulated growth of myeloid cells, especially immature granulocytes in bone marrow. The main cause of CML is Bcr–Abl translocation. It has been suggested that lncRNA-BGL3 acts as a tumor-suppressor in Bcr-Abl-mediated CML.78 BGL3 inhibits tumorigenesis by sensitizing Bcr-Abl-positive K562 leukemic cells to undergo imatinib-induced apoptosis. Mechanistically, BGL3 functions as a competitive endogenous RNA (ceRNA) for tumor suppressor gene Phosphatase and Tensin homolog (PTEN).78 Another lncRNA involved in the Bcr-Abl-induced hematopoietic malignancies is H19.79 In contrast to BGL3, H19 facilitates Bcr-Abl-mediated leukemogenesis.79 Loss of imprinting leads to H19 activation, which is found to be a frequent event in adult T-cell leukemia/lymphoma (ATL).80
3.2. Chronic lymphocytic leukemia (CLL)
B cell chronic lymphocytic leukemia (CLL), is a malignancy diagnosed by demonstration of an abnormal population of B lymphocytes in blood. Remarkably, deletion of 13q14.3 occurs in more than 50% patients.81, 82, 83, 84, 85, 86 Two lncRNAs, DLEU1 and DLEU2, are found to be transcribed from this genomic region and display hypermethylation in CLL patients. It has been demonstrated that these two lncRNAs could regulate NF-κB signaling pathway, affecting leukemia cell survival.83 Besides, deleting DLEU2 in mice results in CLL, and this phenotype is discovered to have a correlation with abnormal cell-cycle progression.87, 88
3.3. Acute myeloid leukemia (AML)
Acute myeloid leukemia (AML) progresses rapidly and patients often die within weeks if untreated. Surprisingly, more than 55% adult AML patients are found to be associated with recurrent genetic abnormalities.89, 90, 91 LncRNAs have been described to involve in AML and used as biomarkers for outcome.92 For instance, lncRNA IRAIN, an intragenic lncRNA transcribed from the antisense direction of Insulin-like Growth Factor type I Receptor gene (IGF1R), is implicated in AML cell growth.93 Moreover, the hypermethylation of lncRNA MEG3 promoter is often observed in AML patients, and might serve as a biomarker for poor prognosis.94, 95 lncRNA HOTAIRM1, an lncRNA located between HOXA1 and HOXA2, might provide novel insights into AML treatment, due to the well-known feature of dysregulated HOX genes in AML.96, 97, 98 The RUNX1 overlapping RNA (RUNXOR), an lncRNA transcribed from RUNX1 promoter and overlapping with RUNX1, could initiate leukemogenesis through interacting with RUNX1.99 Other lncRNAs correlate with AML include PVT1 and NEAT1.100, 101, 102, 103
3.4. Acute lymphoblastic leukemia (ALL)
Acute lymphoblastic leukemia (ALL) patients carry large number of immature lymphocytes. Several functional lncRNAs have been reported to contribute to ALL progression. For example, lncRNA LUNAR1 acts as an oncogenic gene in T cell acute lymphoblastic leukemia (T-ALL).104 LUNAR1 works by enhancing IGF1R mRNA expression and modulating Notch signaling pathway.104 Also, the lncRNA B-ALL-associated long RNAs-2 (BARL-2) is annotated to have correlation with corticosteroid treatment response in B-ALL.105
Intriguingly, it has been proposed that the famous lncRNA Xist is associated with hematological cancer.48 Actually, X chromosome aneuploidies have been documented to be with different human cancers, such as breast and ovarian cancers.106, 107 Given the significant role of Xist in this process,108, 109 it is reasonable that Xist might be linked to blood tumor. Another strong evidence is that Xist deletion in female mice results in myeloproliferative neoplasm and myelodysplastic syndrome (mixed MPN/MDS).48
3.5. Immunological diseases
Myeloid RNA regulator of Bim-induced death (Morrbid) is a conserved lncRNA, which implicates in the regulation of lifespan of short-lived myeloid cells, such as neutrophils and eosinophils.110 Interestingly, Morrbid is highly expressed in patients with hypereosinophilic syndrome (HES), a disease characterized by a persistently elevated eosinophil count in the blood. In vivo loss-of-function study shows that Morrbid deficiency results in short-lived myeloid cell reduction, and Morrbid-deficient mice are more vulnerable when facing bacterial infection. Mechanism behind this regulation is that Morrbid works in cis to repress its neighbor gene Bcl2l11, a pro-apoptotic molecular, by recruiting PRC2 to its promoter. Given the function of Morrbid during the myeloid cell regulation, it might serve as a potential therapeutic target in inflammatory diseases.110 Furthermore, TNFα and hnRNPL related immunoregulatory LincRNA (THRIL) also plays an important role in inflammatory immune responses.111 By way of interacting with hnRNPL and binding to TNFα promoter, THRIL promotes TNFα transcription and thus implicates in the innate immune response. In turn, high levels of TNFα decrease THRIL expression. Consistent with this discovery, Kawasaki disease patients with severe symptoms show decreased THRIL level, and this might be a potential biomarker for inflammatory diseases.111 Consistently, given the significant role of lncRNAs in inflammatory response, it is tantalizing to uncover new drug targets and effective therapeutic by intervene these crucial lncRNAs.64, 67
4. Conclusions
LncRNAs have gained increasing attention, as their physiological and pathological functions are being gradually understood. Among them, several lncRNAs are key regulators of normal hematopoiesis, and their dysregulation could lead to hematopoietic disorders. Therefore, the comprehensive characterization of lncRNAs in blood development might not only help us better understand hematopoiesis but provide a new set of opportunities for future clinical translation.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
We thank our laboratory members for the helpful discussions. This work was supported by the National Key Research and Development Program of China (2016YFA0100601 to J.Y), CAMS Innovation Fund for Medical Sciences (2017-I2M-3-009 to J.Y., 2016-I2M-3-002 to F.W.), the National Key Basic Research Program of China (2015CB943001 to J.Y.), the National Natural Science Foundation of China (81530007 and 31725013 to J.Y.; 31571523 and 31471227 to F.W.), the CAMS (2016GH310001 and 2017-I2M-B&R-04 to J.Y.), and Grant from Medical Epigenetics Research Center, CAMS (2017PT31035).
Footnotes
Peer review under responsibility of Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences.
Contributor Information
Fang Wang, Email: wo_wfang@hotmail.com.
Jia Yu, Email: j-yu@ibms.pumc.edu.cn.
References
- 1.Djebali S., Davis C.A., Merkel A. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taft R.J., Pheasant M., Mattick J.S. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays: News Rev Mol Cell Dev Biol. 2007;29(3):288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 3.Ravasi T., Suzuki H., Pang K.C. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Gen Res. 2006;16(1):11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 5.Mattick J.S., Makunin I.V. Non-coding RNA. Hum Mol Genet. 2006;15(1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 6.Clark M.B., Mercer T.R., Bussotti G. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat Methods. 2015;12(4):339–342. doi: 10.1038/nmeth.3321. [DOI] [PubMed] [Google Scholar]
- 7.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derrien T., Johnson R., Bussotti G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Gen Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Anderson D.M., Anderson K.M., Chang C.L. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto A., Pasut A., Matsumoto M. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541(7636):228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 12.Luo S., Lu J.Y., Liu L. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016;18(5):637–652. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 13.Amaral P.P., Mattick J.S. Noncoding RNA in development. Mamm Gen: Off J Int Mamm Gen Soc. 2008;19(7-8):454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 14.Perry R.B., Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development. 2016;143(21):3882–3894. doi: 10.1242/dev.140962. [DOI] [PubMed] [Google Scholar]
- 15.Yin Y., Yan P., Lu J. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015;16(5):504–516. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14(6):752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Matsui M., Corey D.R. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.T., Bartolomei M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science (New York, NY) 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C.K., Blanco M., Jackson C. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science (New York, NY) 2016;354(6311):468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 24.Mancini-Dinardo D., Steele S.J., Levorse J.M., Ingram R.S., Tilghman S.M. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20(10):1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley R.L., Kuroda M.I. Noncoding RNA genes in dosage compensation and imprinting. Cell. 2000;103(1):9–12. doi: 10.1016/s0092-8674(00)00099-4. [DOI] [PubMed] [Google Scholar]
- 26.Berghoff E.G., Clark M.F., Chen S., Cajigas I., Leib D.E., Kohtz J.D. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140(21):4407–4416. doi: 10.1242/dev.099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cesana M., Cacchiarelli D., Legnini I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell. 2014;53(3):506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y., Huang C., Meng X., Li J. Long noncoding RNA MALAT1: insights into its biogenesis and implications in human disease. Curr Pharmaceut Des. 2015;21(34):5017–5028. doi: 10.2174/1381612821666150724115625. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimoto R., Mayeda A., Yoshida M., Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859(1):192–199. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Yang L., Chen L.L. Life without A tail: new formats of long noncoding RNAs. Int J Biochem Cell Biol. 2014;54:338–349. doi: 10.1016/j.biocel.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Bhatia G., Goyal N., Sharma S., Upadhyay S.K., Singh K. Present scenario of long non-coding RNAs in plants. Noncoding RNA. 2017;3(2) doi: 10.3390/ncrna3020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paralkar V.R., Weiss M.J. Long noncoding RNAs in biology and hematopoiesis. Blood. 2013;121(24):4842–4846. doi: 10.1182/blood-2013-03-456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atianand M.K., Caffrey D.R., Fitzgerald K.A. Immunobiology of long noncoding RNAs. Annu Rev Immunol. 2017;35:177–198. doi: 10.1146/annurev-immunol-041015-055459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinn J.L., Kertesz M., Wang J.K. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toiber D., Leprivier G., Rotblat B. Long noncoding RNA: noncoding and not coded. Cell Death Discov. 2017;3:16104. doi: 10.1038/cddiscovery.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orkin S.H., Zon L.I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dzierzak E., Bigas A. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 2018;22(5):639–651. doi: 10.1016/j.stem.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Paralkar V.R., Mishra T., Luan J. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123(12):1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez-Dominguez J.R., Hu W., Yuan B. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123(4):570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jagannathan-Bogdan M., Zon L.I. Hematopoiesis. Development. 2013;140(12):2463–2467. doi: 10.1242/dev.083147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo M., Jeong M., Sun D. Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell. 2015;16(4):426–438. doi: 10.1016/j.stem.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian P., He X.C., Paulson A. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell. 2016;18(2):214–228. doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatraman A., He X.C., Thorvaldsen J.L. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500(7462):345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yildirim E., Kirby J.E., Brown D.E. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152(4):727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eissmann M., Gutschner T., Hammerle M. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma X.Y., Wang J.H., Wang J.L., Ma C.X., Wang X.C., Liu F.S. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genom. 2015;16:676. doi: 10.1186/s12864-015-1881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu W., Yuan B., Flygare J., Lodish H.F. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25(24):2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atianand M.K., Hu W., Satpathy A.T. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165(7):1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., Wu X., Shen F., Li Y., Zhang Y., Yu D. Shlnc-EC6 regulates murine erythroid enucleation by Rac1-PIP5K pathway. Dev Growth Differ. 2015;57(6):466–473. doi: 10.1111/dgd.12225. [DOI] [PubMed] [Google Scholar]
- 54.Chen M.T., Lin H.S., Shen C. PU.1-regulated long noncoding RNA lnc-MC controls human monocyte/macrophage differentiation through interaction with MicroRNA 199a-5p. Mol Cell Biol. 2015;35(18):3212–3224. doi: 10.1128/MCB.00429-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner L.A., Christensen C.J., Dunn D.M. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109(12):5191–5198. doi: 10.1182/blood-2006-06-027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X., Lian Z., Padden C. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Z.H., Wang W.T., Huang W. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24(2):212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X., Weissman S.M., Newburger P.E. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11(6):777–787. doi: 10.4161/rna.28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang P., Xue Y., Han Y. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science (New York, NY) 2014;344(6181):310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 60.Eis P.S., Tam W., Sun L. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Berg A., Kroesen B.J., Kooistra K. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37(1):20–28. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 62.Chen Y.G., Satpathy A.T., Chang H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18(9):962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Satpathy A.T., Chang H.Y. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42(5):792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 64.Rapicavoli N.A., Qu K., Zhang J., Mikhail M., Laberge R.M., Chang H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife. 2013;2 doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krawczyk M., Emerson B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. eLife. 2014;3 doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu B., Sun L., Liu Q. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 67.Carpenter S., Aiello D., Atianand M.K. A long noncoding RNA mediates both activation and repression of immune response genes. Science (New York, NY) 2013;341(6147):789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gomez J.A., Wapinski O.L., Yang Y.W. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vigneau S., Rohrlich P.S., Brahic M., Bureau J.F. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J Virol. 2003;77(10):5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Collier S.P., Collins P.L., Williams C.L., Boothby M.R., Aune T.M. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol (Baltimore Md: 1950) 2012;189(5):2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collier S.P., Henderson M.A., Tossberg J.T., Aune T.M. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by t-bet. J Immunol (Baltimore, Md: 1950) 2014;193(8):3959–3965. doi: 10.4049/jimmunol.1401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei P., Han B., Chen Y. Role of long non-coding RNAs in normal and malignant hematopoiesis. Sci China Life Sci. 2013;56(10):867–875. doi: 10.1007/s11427-013-4550-9. [DOI] [PubMed] [Google Scholar]
- 73.Garitano-Trojaola A., Agirre X., Prosper F., Fortes P. Long non-coding RNAs in haematological malignancies. Int J Mol Sci. 2013;14(8):15386–15422. doi: 10.3390/ijms140815386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alvarez-Dominguez J.R., Hu W., Gromatzky A.A., Lodish H.F. Long noncoding RNAs during normal and malignant hematopoiesis. Int J Hematol. 2014;99(5):531–541. doi: 10.1007/s12185-014-1552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morlando M., Ballarino M., Fatica A. Long non-coding RNAs: new players in hematopoiesis and leukemia. Front Med. 2015;2:23. doi: 10.3389/fmed.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei S., Wang K. Long noncoding RNAs: pivotal regulators in acute myeloid leukemia. Exp Hematol Oncol. 2015;5:30. doi: 10.1186/s40164-016-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alvarez-Dominguez J.R., Lodish H.F. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130(18):1965–1975. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo G., Kang Q., Zhu X. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34(14):1768–1779. doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
- 79.Guo G., Kang Q., Chen Q. High expression of long non-coding RNA H19 is required for efficient tumorigenesis induced by Bcr-Abl oncogene. FEBS Lett. 2014;588(9):1780–1786. doi: 10.1016/j.febslet.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 80.Takeuchi S., Hofmann W.K., Tsukasaki K. Loss of H19 imprinting in adult T-cell leukaemia/lymphoma. Br J Haematol. 2007;137(4):380–381. doi: 10.1111/j.1365-2141.2007.06581.x. [DOI] [PubMed] [Google Scholar]
- 81.Rosenwald A., Ott G., Krumdiek A.K. A biological role for deletions in chromosomal band 13q14 in mantle cell and peripheral t-cell lymphomas? Genes Chromosomes Cancer. 1999;26(3):210–214. doi: 10.1002/(sici)1098-2264(199911)26:3<210::aid-gcc4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 82.Mertens D., Stilgenbauer S. CLL and deletion 13q14: merely the miRs? Blood. 2012;119(13):2974–2975. doi: 10.1182/blood-2012-01-400747. [DOI] [PubMed] [Google Scholar]
- 83.Garding A., Bhattacharya N., Claus R. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the in Cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013;9(4) doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dohner H., Stilgenbauer S., Benner A. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 85.Rawstron A.C., Bennett F.L., O'Connor S.J. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359(6):575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y., Hermanson M., Grander D. 13q deletions in lymphoid malignancies. Blood. 1995;86(5):1911–1915. [PubMed] [Google Scholar]
- 87.Klein U., Lia M., Crespo M. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 88.He L., Thomson J.M., Hemann M.T. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyer S.C., Levine R.L. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 2014;15(9):e382–e394. doi: 10.1016/S1470-2045(14)70008-7. [DOI] [PubMed] [Google Scholar]
- 90.Shih A.H., Abdel-Wahab O., Patel J.P., Levine R.L. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 91.Woods B.A., Levine R.L. The role of mutations in epigenetic regulators in myeloid malignancies. Immunol Rev. 2015;263(1):22–35. doi: 10.1111/imr.12246. [DOI] [PubMed] [Google Scholar]
- 92.Garzon R., Volinia S., Papaioannou D. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci USA. 2014;111(52):18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun J., Li W., Sun Y. A novel antisense long noncoding RNA within the IGF1R gene locus is imprinted in hematopoietic malignancies. Nucleic Acids Res. 2014;42(15):9588–9601. doi: 10.1093/nar/gku549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khoury H., Suarez-Saiz F., Wu S., Minden M.D. An upstream insulator regulates DLK1 imprinting in AML. Blood. 2010;115(11):2260–2263. doi: 10.1182/blood-2009-03-212746. [DOI] [PubMed] [Google Scholar]
- 95.Benetatos L., Hatzimichael E., Dasoula A. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34(2):148–153. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 96.Eklund E.A. The role of HOX genes in myeloid leukemogenesis. Curr Opin Hematol. 2006;13(2):67–73. doi: 10.1097/01.moh.0000208467.63861.d6. [DOI] [PubMed] [Google Scholar]
- 97.Bei L., Lu Y., Bellis S.L., Zhou W., Horvath E., Eklund E.A. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding beta3 integrin during myeloid differentiation. J Biol Chem. 2007;282(23):16846–16859. doi: 10.1074/jbc.M609744200. [DOI] [PubMed] [Google Scholar]
- 98.Rice K.L., Licht J.D. HOX deregulation in acute myeloid leukemia. J Clin Investig. 2007;117(4):865–868. doi: 10.1172/JCI31861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang H., Li W., Guo R. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int J Cancer. 2014;135(12):2783–2794. doi: 10.1002/ijc.28922. [DOI] [PubMed] [Google Scholar]
- 100.Zeng C., Yu X., Lai J., Yang L., Chen S., Li Y. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol. 2015;8:126. doi: 10.1186/s13045-015-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeng C., Xu Y., Xu L. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cho S.W., Xu J., Sun R. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell. 2018;173(6):1398–1412. doi: 10.1016/j.cell.2018.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tseng Y.Y., Moriarity B.S., Gong W. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512(7512):82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Trimarchi T., Bilal E., Ntziachristos P. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158(3):593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fernando T.R., Rodriguez-Malave N.I., Waters E.V. LncRNA expression discriminates karyotype and predicts survival in B-lymphoblastic leukemia. Mol Cancer Res: MCR. 2015;13(5):839–851. doi: 10.1158/1541-7786.MCR-15-0006-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pageau G.J., Hall L.L., Ganesan S., Livingston D.M., Lawrence J.B. The disappearing Barr body in breast and ovarian cancers. Nat Rev Cancer. 2007;7(8):628–633. doi: 10.1038/nrc2172. [DOI] [PubMed] [Google Scholar]
- 107.Liao D.J., Du Q.Q., Yu B.W., Grignon D., Sarkar F.H. Novel perspective: focusing on the X chromosome in reproductive cancers. Cancer Investig. 2003;21(4):641–658. doi: 10.1081/cnv-120022385. [DOI] [PubMed] [Google Scholar]
- 108.McHugh C.A., Chen C.K., Chow A. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sarma K., Cifuentes-Rojas C., Ergun A. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159(4):869–883. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kotzin J.J., Spencer S.P., McCright S.J. The long non-coding RNA morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016;537(7619):239–243. doi: 10.1038/nature19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li Z., Chao T.C., Chang K.Y. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci USA. 2014;111(3):1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]