Summary
Neutrophils are short-lived blood cells that play a critical role in host defense against infections. To better comprehend neutrophil functions and their regulation, we provide a complete epigenetic overview, assessing important functional features of their differentiation stages from bone marrow-residing progenitors to mature circulating cells. Integration of chromatin modifications, methylation, and transcriptome dynamics reveals an enforced regulation of differentiation, for cellular functions such as release of proteases, respiratory burst, cell cycle regulation, and apoptosis. We observe an early establishment of the cytotoxic capability, while the signaling components that activate these antimicrobial mechanisms are transcribed at later stages, outside the bone marrow, thus preventing toxic effects in the bone marrow niche. Altogether, these data reveal how the developmental dynamics of the chromatin landscape orchestrate the daily production of a large number of neutrophils required for innate host defense and provide a comprehensive overview of differentiating human neutrophils.
Introduction
Human neutrophils, with a daily production rate of ~1011 cells (Strydom and Rankin, 2013), are the most abundant type of leukocytes and an essential element of the innate immune system. Patients with either acquired or inherited defects in neutrophil development, migration, or function show enhanced susceptibility to infection by opportunistic pathogens (Erlacher and Strahm, 2015; Fodil et al., 2016). Neutrophils have a variety of effector functions, such as phagocytosis of pathogens (Nauseef and Borregaard, 2014), intracellular killing in phagosomes with reactive oxygen species (ROS), and antimicrobial granule components (Darrah and Andrade, 2013; Garcia-Romo et al., 2011; Hakkim et al., 2010). They are produced in the bone marrow and reach the bloodstream across five main stages of differentiation. Myeloblasts (MBs) undergo 1 week of proliferation, followed by maturation (Bainton et al., 1971), then progressively move through promyelocytes and myelocytes (P/Ms), metamyelocytes (MMs), band neutrophils (BNs), and finally segmented neutrophils (SNs). During this time they produce different granules filled with antimicrobial compounds and develop the abilities of respiratory burst and chemotaxis. SNs represent a reserve pool from which cells are constantly released into the peripheral bloodstream, as non-dividing polymorphonuclear neutrophils (PMNs). The differentiation process exhibits a precise and tightly defined transcriptional program (Theilgaard-Mönch et al., 2005). In this study, we used total RNA sequencing (RNA-seq) to characterize and compare gene expression profiles of the various differentiation stages. Histone modifications, chromatin immunoprecipitation sequencing (ChIP-seq), and whole-genome bisulfite sequencing (WGBS) have been used to describe the epigenetic landscapes underlying these coordinated changes. For each stage we defined DNA methylation patterns and characterized four active histone marks—H3 lysine-4 trimethylation (H3K4me3), H3 lysine-27 acetylation (H3K27ac), H3 lysine-4 monomethylation (H3K4me1), H3 lysine-36 trimethylation (H3K36me3)—and two repressive marks—histone H3 lysine-9 trimethylation (H3K9me3) and histone H3 lysine-27 trimethylation (H3K27me3). The integration of these analyses reveals a high degree of coordination between the dynamics of histone modifications and transcriptional changes. The activation of PMNs triggers the host defense. This process is accomplished through some functional key responses as respiratory burst, cell migration, and degranulation. Integrating our functional assays with the epigenomic analysis gave us the opportunity to further characterize the timing of these important processes.
Results
Transcriptional Changes in Neutrophil Development
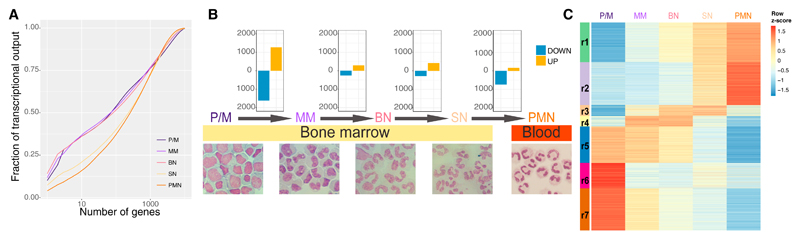
To better understand neutrophil functions and their regulation, we analyzed the transcriptomes of circulating neutrophils and their progenitor populations (Figure S1A). Our analysis revealed quantitative and qualitative differences in expression among PMNs, SNs, and the other differentiation stages. We used cumulative distribution of expression to explore the complexity of the transcriptome (Melé et al., 2015) and found that in PMNs and SNs, the genes with higher expression give a smaller contribution to the global transcriptional output than in other stages (Figure 1A). To deepen the transcriptome comparisons, we performed differential gene expression analysis between subsequent stages and between the initial and the final stage (Figure 1B; Table S1). Downregulated genes in the P/M-PMNs comparison enriched Gene Ontology (GO) categories related to cell functions, such as metabolism, translation, cell cycle, DNA replication, mitochondria, and cell divisions (Figures S1B and S1C). On the other hand, the upregulated genes enriched fewer categories, related mainly to immune system functions and vesicle transport processes (Figures S1B and S1C). The analysis of differentially expressed transcripts across all stages found 108 genes with significant changes in transcription starting site (TSS) use (Table S1). Only for 12 genes did this lead to changes in the open reading frame (Table S1). Cluster analysis and gap statistic (Tibshirani et al., 2001) summarized the differentially expressed genes in at least one of the five comparisons (P/M-MM, MM-BN, BN-SN, SN-PMN, or P/M-PMN) in seven main clusters (r1–r7; Figure 1C; Table S1). Genes in clusters r1 and r2 showed an increase in expression toward the latter stages of differentiation and enriched GO terms related to immune system and signal transduction (Table S1). Genes in clusters r5, r6, and r7 displayed a decreasing expression trend with differentiation progression and enriched GO terms related to translation, mitochondria, and cell cycle (Table S1). The remaining two clusters (r3 and r4) were made by genes whose expression increased during the intermediate stages of differentiation and then decreased at the PMN stage. These enriched GO terms related to vesicle transport and signal transduction (Table S1).
Figure 1. Transcriptome and Epigenome Dynamics of Neutrophil Differentiation.
(A) Cumulative distribution of the average fraction of total transcription contributed by protein-coding genes when sorted from most to least expressed in each differentiation stage.
(B) Top: bar plots reporting differentially expressed genes (posterior probability > 0.5 and absolute fold change > 2) in the comparisons P/M-MM, MM-BN, BN-SN, and SN-PMN. Bottom: May-Grünwald-Giemsa staining of the isolated cells.
(C) Heatmap displaying the expression patterns of the clusters (r1–r7) identified by the K-means analysis of the genes differentially expressed in at least one comparison.
Epigenetic Dynamics in Neutrophil Development
In order to define the chromatin dynamics underlying the changes in gene expression during neutrophil differentiation, we determined the epigenetic landscape in the five differentiation stages and integrated them with the results of the transcriptome characterization. Principal-component analysis (PCA) of the four active histone modifications (H3K4me3, H3K27ac, H3K4me1, and H3K36me3) revealed a good separation among the five neutrophil development stages (Figure S2A). In contrast, PCA for the two repressive marks (H3K9me3 and H3K27me3) revealed donor-specific, rather than stage-specific, clusters (Figure S2A). Moreover, PCA analysis of DNA methylation profiles did not reveal differences among the five differentiation stages (Figure S2B), and only a few differentially methylated regions (DMRs) were identified between subsequent stages of differentiation (Figure S2C; Table S2). Altogether, histone mark and DNA methylation results indicate a modest contribution of repressive epigenetic mechanisms in the described phases of neutrophil differentiation.
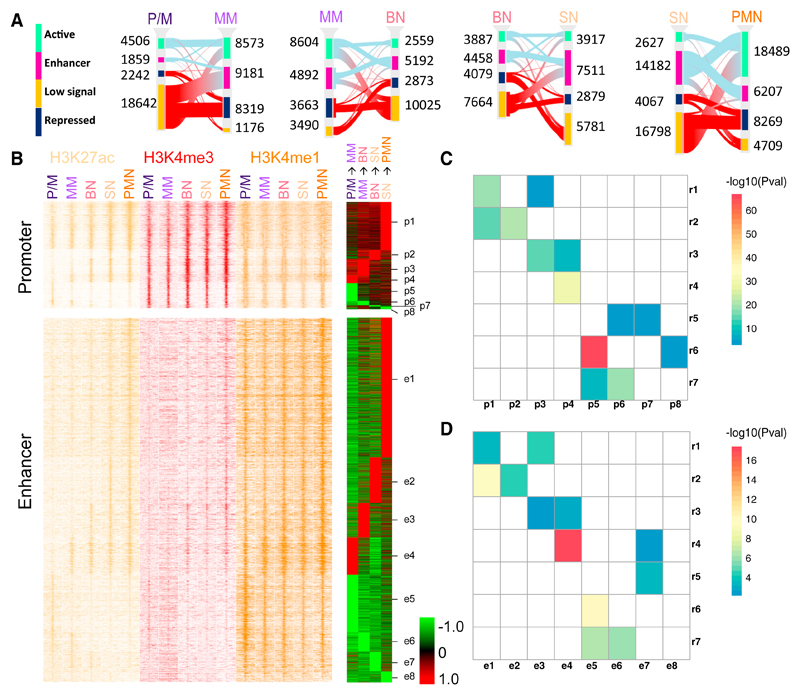
The six different histone modifications were also used to perform a genome segmentation analysis (Ernst and Kellis, 2012), which indicated 12 representative states (Figure S3A). These were collapsed into a smaller set for ease of interpretation (Carrillo-de-Santa-Pau et al., 2017) to obtain four main categories: active, enhancer, low signal, and repressed (Figure S3A). We then focused on the changes of chromatin states, identifying between 15,685 and 27,604 at each consecutive differentiation transition (Figure 2A; Table S3). In agreement with the differential gene expression analysis results, the consecutive transitions with most changes were P/M to MM and SN to PMN. Upregulated genes were enriched in changes toward active states (i.e., from low signal/repressed to active/enhancer), while downregulated genes were enriched in transitions toward inactive states (i.e., from active/enhancer to low signal/repressed) in the majority of differentiation stages (Figure S3B). H3K27ac, strongly associated with active and enhancer states, is among the most highly informative marks about gene regulation (Karlić et al., 2010; Rada-Iglesias et al., 2011). For this reason, we decided to focus our attention on the 9,100 genomic regions characterized by changes in H3K27ac levels during differentiation (see Experimental Procedures). Using co-occupancy with H3K4me3 to identify promoter regions and co-occupancy with H3K4me1 to identify enhancers, in addition to a supervised cluster of the H3K27ac log ratio RPKM (reads per kilobase of transcript per million mapped reads), we divided these regions in two categories each made of eight subgroups (Figure 2B; Table S4). The intersection of these regions with the chromatin states revealed an overlap with active (promoter) regions in the H3K4me3-enriched clusters and an overlap with enhancers in the H3K4me1-enriched clusters (Figure S3C), corroborating our partitioning. We refer to the clusters as promoter (p1–p8) and enhancer (e1–e8). In both promoter and enhancer clusters about two-thirds of dynamic regions displayed an increase in H3K27ac (i.e., increase in activity), while about one-third displayed a reduction in H3K27ac (i.e., reduction in activity). The contingency tables made by the intersection of the genes in the RNA-seq clusters gave significant Pearson’s chi-square statistics for both acetylation clusters (enhancer chi-square = 513, p = 0.0005; promoter chi-square = 1,364, p = 0.0005). We tested the association of each gene expression cluster with each dynamic acetylation cluster (Figures 2C and 2D). Interestingly we found that expression clusters r1 and r2, containing genes with increased expression, were significantly associated with promoter clusters p1 and p2 and enhancer clusters e1 and e2. Expression clusters r6 and r7, formed by genes with decreased expression, were significantly associated with promoter clusters p5 and p6 and enhancer clusters e5 and e6, all characterized by reduced histone acetylation levels. The expression cluster r4 included genes with an increased expression in the PM-to-MM transition, and the same trend was observed in the associated acetylation clusters e4 and p4.
Figure 2. H3K27 Acetylation Drives Consecutive Neutrophil Differentiation Stages.
(A) Sankey diagrams of consistent chromatin states for each of the neutrophil differentiation transitions. Different chromatin states are represented with specific color codes: green indicates active regions, purple enhancers, amber low signal regions, and dark blue heterochromatic regions. The red connecting lines indicate transitions toward inactive states (low signal and repressed); the light blue lines indicate transitions toward active states (active and enhancer).
(B) Left: stacking heatmaps of H3K27ac (yellow), H3K4me3 (red), and H3K4me1 (orange) ChIP-seq of promoters and enhancers (±5 kb) with dynamic acetylation during different stages of neutrophil differentiation. Right: differentially acetylated promoter and enhancer during two consecutive neutrophil progenitor stage transitions are clustered as acetylation clusters for promoters p1–p8 and for enhancers e1–e8, respectively. Acetylation gains are presented in red and acetylation losses in green.
(C and D) Heatmaps representing the association between the expression clusters identified in Figure 1C and the promoter (C) and enhancer (D) clusters identified in Figure 2B. Only significant corrected p values (<0.05) are reported in the figures.
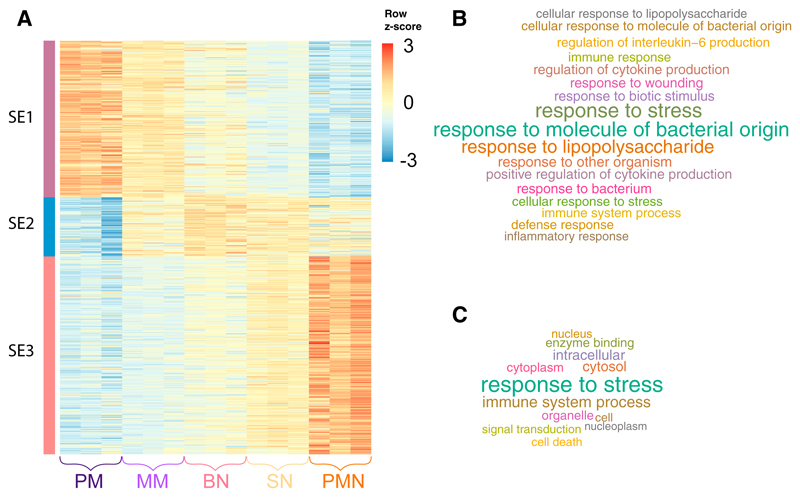
Dynamically acetylated enhancers of the same cluster had a genomic distance closer than expected by chance (permutation test Z = 21). This observation prompted us to investigate the presence of super-enhancers (SEs), regulatory elements made by clusters of enhancer that specify cell identity (Whyte et al., 2013; Witte et al., 2015). We made use of the established ROSE algorithm to identify 629 SEs, which could be clustered in three main groups (Figure 3A; Table S4). SE cluster 1 (SE1) was made by elements more acetylated in the P/M state, showing a decreasing trend of acetylation in the subsequent stages of differentiation; SE cluster 2 (SE2) had an opposite trend, with acetylation increasing in the first stages and then decreasing at the SN-PMN transition. Finally, SE cluster 3 (SE3) contained SEs activated at the end of differentiation process. Genes controlled by SEs belonging to cluster 3 were the only ones showing an enrichment for functional categories, all of them related to PMN’s activation (Figures 3B and 3C).
Figure 3. Identification of Super-enhancers Associated with Neutrophil Differentiation.
(A) Heatmap of H3K27ac density at superenhancers with dynamic acetylation during different stages of neutrophil differentiation. For each state tag density for the three replicates is included.
(B and C) GO Biological Processes (B) and GO Slim (C) categories enriched by genes associated with super-enhancers activated at the final stages of neutrophil differentiation (cluster SE3).
Transcription Factors and Release from Bone Marrow
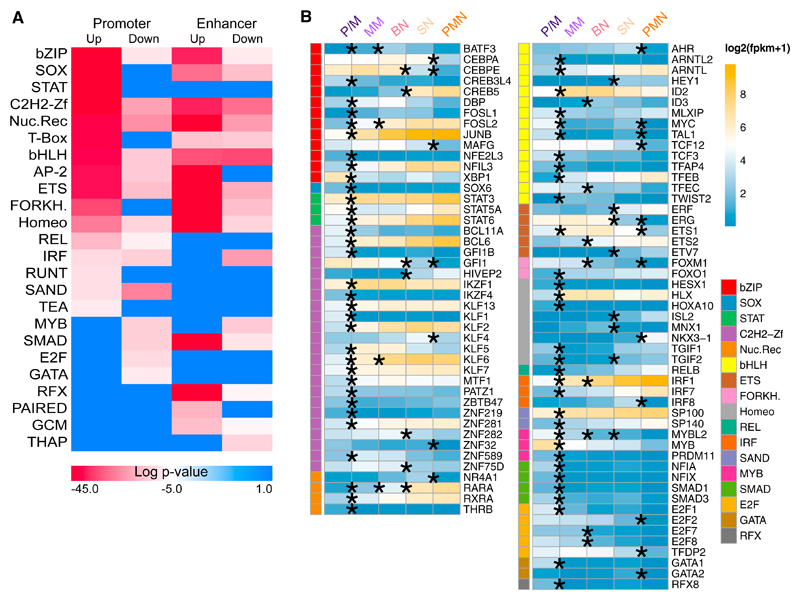
Transcription factors (TFs) bind regulatory elements (promoters and enhancers) and enforce transcriptional programs (Lambert et al., 2018; White et al., 2013). To identify the TFs that have an important role in granulopoiesis, we searched for TF family consensus binding sites, enriched in regions with increased acetylation (clusters p1–p4 and e1–e4) compared with regions with decreased acetylation (clusters p5–p8 and e5–e8) and vice versa. We found enrichments for basic leucine zippers (bZIP), SOX, zinc fingers (ZF), nuclear receptors (NR), basic helix-loop-helices (bHLH), NF-κB family members (REL), homeodomains, and ETS families, with stronger signal in regions with increased acetylation (Figure 4A). Regions with decreased H3K27ac signal showed enrichment for MYB- and GATA-binding motifs. We integrated these results with gene expression analysis results and identified for each enriched motif TF genes differentially expressed in at least one of the comparisons between differentiation stages (Figure 4B). The majority of these TFs (81 of 95) were differentially expressed in only one of the four comparisons. Twelve of the remaining 14 genes, differentially expressed at multiple transitions, were always either up- or downregulated. Only 2 genes, differentially expressed at multiple transitions, had a trend inversion: TGIF2 was downregulated in the P/M-MM transition and upregulated in the BN-SN transition, and ETS1 was upregulated in the P/M-MM transition and down-regulated in the SN-PMN transition.
Figure 4. Transcription Factors with Enriched Binding Sites in Dynamic Acetylated Regions.
(A) Transcription factor (TF) family motif enrichment in the dynamic acetylated regions in Figure 2B.
(B) RNA expression in log2(FPKM+1) of the TF family members in (A) differentially expressed in at least one of the comparison between differentiation stages; the asterisk denotes the transitions where the expression is significantly different.
The P/M-to-MM transition included the majority (65 of 95) of differentially expressed TFs, followed by the bone marrow-to-blood transition (SN to PMN) with 20 differentially expressed TFs (Figure 4B). Interestingly 19 of them were downregulated, and NKX3-1, the only upregulated one, had a reported transcriptional repressor activity (Simmons and Horowitz, 2006). The TF families MYB, GATA, and E2F were exclusively overrepresented in enhancers and promoters with decreased acetylation, reflecting their role in other lineages and cell cycle progression (Bartůnek et al., 2003; Ghazaryan et al., 2014). Accordingly, differentially expressed genes from these families were all down-regulated (Figure 4B).
Acquisition and Control of Cytotoxic Capabilities
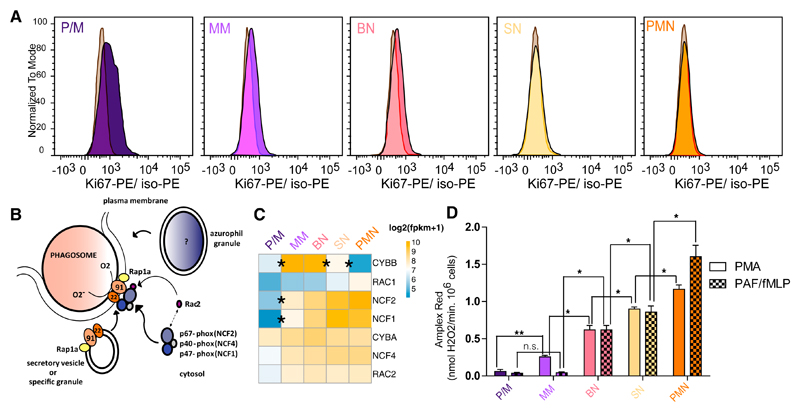
Cell proliferation is halted during neutrophil differentiation; our transcriptome and epigenome data indicated that this occurs in the bone marrow, as previously reported (Klausen et al., 2004; Theilgaard-M¦nch et al., 2005). Flow cytometry quantification of Ki67, a marker for proliferation (Gerdes, 1990), showed that the majority of the signal drops in the early stages of myeloid differentiation (P/M-to-MM transition) and a complete loss is observed at the SN stage (Figure 5A). Neutrophils are phagocytes, capable of ingesting and killing microorganisms by the release of ROS, highly toxic chemical species, potentially harmful for the bone marrow niche. The multicomponent nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex, consists of a membrane-associated heterodimer cytochrome b558 and a trio of cytosolic proteins (Nauseef and Borregaard, 2014). This complex confers the unique neutrophils’ ability to generate a family of reactive oxidizing agents (Weiss, 1989), and we focused on it to characterize the exact dynamic of this defense mechanism formation. Upon activation, the intact NADPH oxidase generates superoxide, the precursor to hydrogen peroxide and other ROS with microbial activity, at the plasma membrane and at the membranes of phagosomes where particles are ingested (Figure 5B). The oxidase complex subunits showed different expression dynamics during differentiation (Figure 5C). CYBA (p22-phox), NCF4 (p40-phox), RAC2, and RAC1 had stable expression levels, whereas CYBB (gp91-phox) was maximally expressed between the MM and BN stages, and the cytoplasmic components NCF1 (p47-phox) and NCF2 (p67-phox) were upregulated in the P/M-to-MM transition and then remained stable in the subsequent stages. In order to verify when the complex is functional, we used an Amplex Red hydrogen peroxide assay to detect the NADPH oxidase activity. Stimulation of PAF-primed neutrophils with the bacterial-derived tripeptide formyl-MLP (PAF/fMLP) (Drewniak et al., 2013; Kuijpers et al., 2005) was used to ensure the maximal stimulation via the natural receptor. At P/M the NAPDH oxidase activity was very low. It became readily detectable at the next stage of differentiation (MM), as shown by the activation with the cell-permeable phorbol ester PMA, which directly activates intracellular protein kinase C (Moore et al., 1991). In contrast, stimulation via the cell-impermeable PAF/fMLP revealed a delay in ROS formation, detectable from the BN stage onward (Figure 5D). In agreement with the increase in transcription level and functional activity of NADPH oxidase, we found that cytochrome b558, the heterodimer of gp91-phox and p22-phox, and fMLP-receptor FPR1 levels (both determined by flow cytometry) increased during differentiation (Figures S4A and S4B). Additionally we also noted that CXCR1, CXCR2, CXCR4, FPR1, FPR2, PTAFR, and C5AR1, activating G protein-coupled receptors for the major chemotactic factors in neutrophils, follow the same expression trend (Figure S4C), and are all in the expression cluster r1.
Figure 5. Cell Cycle and NADPH Oxidase Regulation in Neutrophil Differentiation.
(A) Flow cytometry analysis of Ki67 expression in bone marrow neutrophil progenitor fractions normalized to isotope control in gray for the three biological replicates.
(B) Schematic of the assembly of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase enzyme complex on the phagosomal or plasma membrane upon neutrophil activation.
(C) Heatmap representing the expression in log2(FPKM+1) of NADPH complex subunits during neutrophil differentiation.
(D) Respiratory burst in sorted neutrophil progenitors from bone marrow and mature PMN upon stimulation with PMA and PAF/fMLP. Results are expressed as maximal rates in nmol H2O2/min · 106 PMNs and as mean ± SEM from the three biological replicates. t test corrected p values of the consecutive stages are indicated in the figure: *p < 0.1 and **p < 0.01. ns, nonsignificant. The t test corrected p values of all comparisons are reported in Table S5.
Transcriptional and Epigenetic Control of Granule Proteins
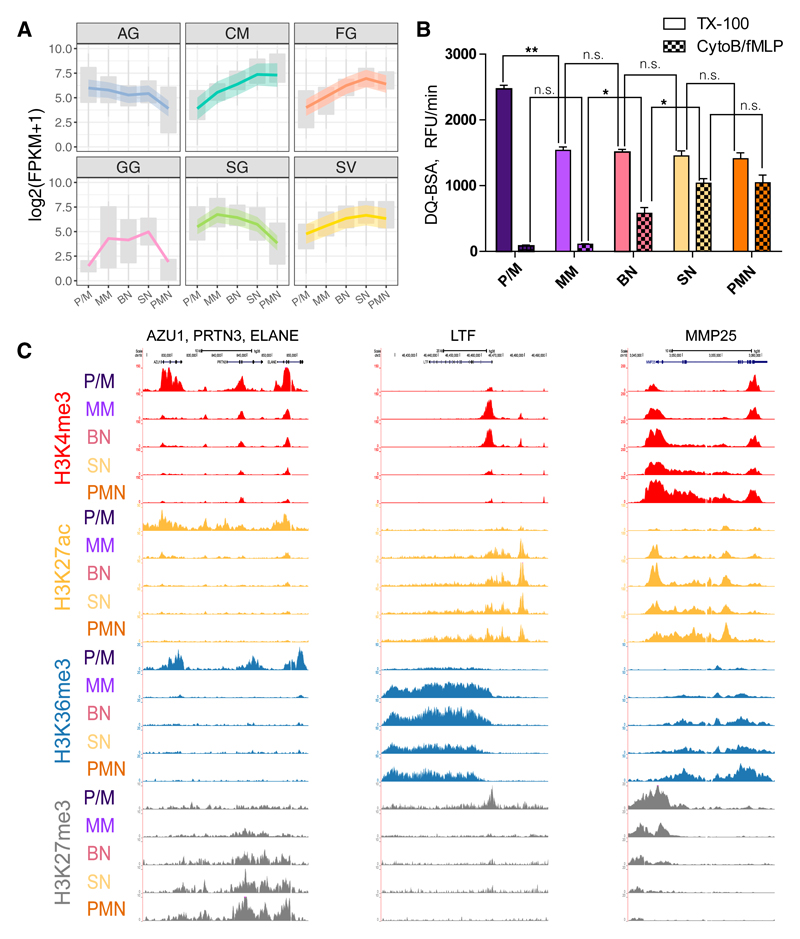
Another main function of the neutrophils is the degranulation consisting in the release of an assortment of proteins, stored in different granules. These granules are distinguishable by their contents: azurophilic granules (AG), specific granules (SG), gelatinase-containing granules (GG), ficolin-containing granules (FG), and secretory vesicles (SV) (Rørvig et al., 2013). To gain insight in the control of the biogenesis of these granules during differentiation, we examined available proteomic data (Rørvig et al., 2013) and selected the proteins whose granule assignments were supported by other existing literature (Table S6) and the granule proteins assigned to the cell membrane (CM) fraction. We found that genes encoding proteins belonging to various granules display different expression trends (Figure 6A), and in agreement with this, they showed a tendency to belong to different RNA-seq clusters (chi-square = 91.8062, p < 0.0005; Figure S5A). AG proteins were associated with clusters r6 and r7 (Fisher’s exact test corrected p < 0.05), while CM proteins were associated with cluster r1 (Fisher’s exact test corrected p < 0.005) and SG proteins with cluster r4 (Fisher’s exact test corrected p = 3.1 × 10−6).
Figure 6. Epigenetics and Transcriptomics of Granule Proteins.
(A) Granule gene expression across differentiation. Upper and lower boxplot margins indicate first and third quartiles. LOESS fitting of the data with relative confidence interval is represented by a colored line with a shadow area.
(B) Proteolytic activity of the different differentiation stages measured by DQ-BSA cleavage. Triton X-100 releases total proteolytic activity stored in the cell by lysis, while proteolytic activity by CytoB/fMLP is a receptor-mediated release of proteases. Results are shown as the maximal slopes in RFU/min and expressed as mean ± SEM from the three biological replicates. t test corrected p values of the consecutive stages are indicated in the figure: *p < 0.1 and **p < 0.01. ns, nonsignificant. The t test corrected p values of all comparisons are reported in Table S5.
(C) Representative views of epigenetic modifications dynamics at five loci encoding for granule proteins.
To determine the time of appearance of AG’s proteolytic capability, we measured their combined serine protease activity, in sorted neutrophil progenitors, upon CytoB/fMLP stimulation or Triton X-100 lysis. The results showed that although AG proteins have already been produced and stored at P/M stage, the protease release machinery is not in place before the BN stage. P/M cells were found to harbor the highest concentration of proteolytic activity, which was then diluted over to the MM daughter cells and remained stable in activity thereafter (Figure 6B). Genes of different granule proteins, similarly to what observed for gene expression, displayed different H3K27ac trends (Figure S5B). In agreement with this, genes encoding soluble AG proteins, such as AZU1, PRTN3, and ELANE, had a high level of the H3K27ac in the P/M stage and reduced in the MM stage (Figures 6C and S5B) and were enriched in clusters p5, p6, and e5 (Fisher’s test corrected p values: for p5, p = 6.64 × 10−3; for p6, p = 6.64 × 10−3; and for e5, p = 3.65 × 10−2). In contrast, SG-associated genes, such as LTF, with increased acetylation in the P/M-to-MM transition, were enriched mainly in clusters p4 and e4 (Fisher’s test corrected p values: for p4, p = 1.7 × 10−3; and for e4, p = 2.88 × 10−2) (Figures 6C and S5B). The SV-associated genes as MMP25 displayed increased acetylation at the final stages of differentiation (Figures 6C and S5B).
Discussion
Our study dissects human neutrophil differentiation through the analysis and integration of transcriptomes, epigenomes, and functional assays of five consecutive stages of maturation.
In the transcriptome of a cell, a relative small number of genes contribute to a large fraction of the expression (Melé et al., 2015). Interestingly, we found this trend to be less pronounced in the two latest stages of neutrophil maturation, SN and PMN, indicating a change in transcriptome features at the final stages of the differentiation.
The P/M-to-MN and SN-to-PMN were the consecutive transitions with the largest numbers of differentially expressed genes and changes in chromatin states. These transcriptomic and epigenomic differences reflect the important differences between these stages. Indeed, the P/M-to-MN transition denotes a passage from cells capable of dividing to cells with indented nuclei and unable to divide (Wahed and Dasgupta, 2015). Similarly, the SN-to-PMN transition denotes the passage from bone marrow to the bloodstream.
Different stages of maturation do not display relevant differences in DNA methylation. This suggests that the unique patterns of DNA methylation found in neutrophils (Schuyler et al., 2016) are established at earlier stages of differentiation, in agreement with a previous study reporting that the majority of changes in DNA methylation occur between the oligopotent common myeloid progenitor and the bipotent granulocyte monocyte progenitor, with only minor changes occurring in the P/M-to-PMN transition (Rönnerblad et al., 2014). Our study extends this result, with more differentiation steps during maturation of the granulocyte progenitors, thus providing a more detailed description of the dynamics of transcriptional enhancers’ commitment. Moreover, the use of WGBS allowed a 60-fold increase of the number of CpG analyzed residues.
In line with the DNA methylation results the two repressive histone marks analyzed cannot be used to discriminate among neutrophil differentiation stages. Previous studies demonstrated that repressive H3K9me3 and H3K27me3 marks and DNA methylation have a crucial role in cell commitment (Bock et al., 2012; Hawkins et al., 2010), and our results suggest that in neutrophils, they do not play any further roles once lineage commitment is achieved.
We focused our attention on promoter and enhancer regions subject to dynamic acetylation across differentiation. These regions, subdivided in clusters, showed significant overlaps of target genes with the clusters of differentially expressed genes, highlighting a strong correlation between epigenomic and transcriptomic profiles at the various differentiation stages. The study of dynamically acetylated regions led us to identify SEs differently modulated across differentiation; a consistent group of these is activated at the end of differentiation and have as potential targets the genes involved in immunological response and neutrophil activation.
To further characterize the dynamically acetylated regions, we looked for enriched TF binding sites and integrated them with the differential expression results. This approach aimed to highlight TFs playing a potential role in the differentiation process. CEBPA is a TF of the bZIP family with a major role in the neutrophil differentiation program, and it is known to be crucial for the development of acute myeloid leukemia (Avellino and Delwel, 2017). Our results show that CEBPA is highly expressed in the bone marrow and then significantly downregulated in the SN-to-PMN transition. CEBPE, another member of the bZIP family, is also highly expressed in the bone marrow and is downregulated in the SN-to-PMN transition but also in the preceding transition, BN to SN. Interestingly CEBPE modulates some of the lactoferrin-containing SG genes (Khanna-Gupta et al., 2007) and regulates the expression of GG and FG granule genes (Gombart et al., 2001), both characterized by a decreasing trend of expression at the PMN stage.
The transcriptome and epigenome characterizations depicted a precise differentiation program that, in agreement with previous findings (Theilgaard-Mönch et al., 2005), progressively activates immune functionalities, turning off essential cell processes. The study of five differentiation stages gave us the opportunity to detect with better resolution where the cell cycle inactivation takes place. We have shown that the P/M stage is the most proliferative, with Ki67 signal dimming in the following MM and BN stages and completely disappearing in the SN stage. These data, alongside with the expression of key cell cycle regulators, indicate that the cell cycle is arrested after the P/M stage.
The antimicrobial activity of neutrophils is achieved through ROS production by the NADPH oxidase enzyme complex. We showed that this enzyme complex activity reaches its peak only at the end of differentiation, although its expression is already detectable at the MM stage. The observed difference in ROS production during differentiation between PMA and PAF/fMLP kinetics highlights that G protein-coupled receptor and its signaling machinery are required to trigger the process. Indeed, we showed that when the surface expression of fMLP receptor FPR1 is low, at early stages of development, the response to PAF/fMLP is impaired.
Along the same line, we noticed that genes encoding for the proteins contained in the various granules displayed different transcriptional and epigenetic dynamics. Indeed, the proteases were released by cellular activation only at the mature stage, because the signaling cascade and surface expression of activating GPCR receptors were not present at earlier stages.
The results presented in this study shed light on the neutrophil differentiation processes and represent a useful resource for future studies on as yet uncharacterized primary immune disorders or neutrophil-related disorders such as congenital neutropenia syndromes, Chédiak-Higashi syndrome, and lazy leukocyte syndrome (Dinauer, 2014; Nunoi et al., 1999).
Experimental Procedures
Study Design
Bone marrow neutrophils were collected from three different healthy donors, and mature neutrophils were collected from other three other different healthy donors. ChiP-seq, RNA-seq, and WGBS experiments were done on samples from the same donors.
Cell Isolation and Library Preparation
The study was approved by the local ethics committee of Sanquin Blood Supply Organization and the Academic Medical Center (Amsterdam, the Netherlands). PMNs were enriched by gradient centrifugation and purified by negative selection with EasySep Human Neutrophil Enrichment Kit. Bone marrow neutrophil progenitors were purified by fluorescence-activated cell sorting (FACS) with CD11b and CD16 (see Supplemental Experimental Procedures for further details). ChIP-seq and total RNA-seq libraries were prepared according to the BLUEPRINT protocols (http://www.blueprint-epigenome.eu).
ChIP-Seq Analysis and Genome Segmentation
Sequenced reads were mapped against the GRCh38 genome assembly with BWA (Li and Durbin, 2009). ChromHMM software (version 1.10) (Ernst and Kellis, 2012) was then used to segment the genome in 200 bp intervals and assign epigenetic states. Chromatin states consistently identified in all three replicates were represented with a Sankey diagram at each stage of development using the “makeRiver” and “riverplot” functions included in the riverplot R package version 0.5.
RNA-Seq Analysis
Trim Galore (version 0.3.7) (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with parameters “-q 15 -s 3 –length 30 -e 0.05” was used to trim PCR and sequencing adapters. Trimmed reads were aligned to the Ensembl version 80 (Cunningham et al., 2015) human transcriptome with Bowtie 1.0.1 (Langmead et al., 2009) using the parameters “-a –best –strata -S -m 100 -X 500 –chunkmbs 256 –nofw –fr.” MMSEQ (version 1.0.8a) (Turro et al., 2014; Turro et al., 2011) was used with default parameters to quantify gene expression. Genes and transcripts with posterior probability > 0.5 (calculated by MMDiff), absolute fold change > 2, and FPKM (fragments per kilobase of transcript per million mapped reads) > 1 in at least one of the two compared cell types were considered differentially expressed. Differentially expressed genes in at least one of the comparisons P/M-MM, MM-BN, BN-SN, SN-PMN, and P/M-PMN (Figure S1) were clustered using the “kmeans” (Hartigan and Wong, 1979) clustering R function. Gene expression FPKMs were log2-transformed, and for each gene the Z score was calculated. We set the number of centers to seven according to the results of the gap statistic (Tibshirani et al., 2001) by using the “clusGap” function of the cluster R package.
SE Identification
SEs in each sample were predicted by the ROSE algorithm (Whyte et al., 2013) using H3K27ac as the surrogate mark. Briefly, all H3K27ac peaks within ±2.0 kb around TSSs were first excluded. The remaining peaks closer than default distance of 12.5 kb were stitched together and subsequently ranked by normalized H3K27ac level corrected by input background. Finally, SEs were separated from typical enhancers on the basis of the inflection point of H3K27ac signal curve.
Functional Enrichment Analysis
Functional enrichment analysis was done by using FIDEA (D’Andrea et al., 2013) with default settings.
Promoter/Enhancer/SE Gene Assignment
For each transcript annotated in Ensembl version 80 (Cunningham et al., 2015), we defined a promoter as the 2,000 bp (±1,000 bp) around its TSS. All the regions defined as promoter by the genome segmentation analysis and the H3K27ac dynamic analysis were assigned to the genes of the overlapping promoters. All the regions defined as enhancer by segmentation analysis and the H3K27ac analysis were assigned to the gene with the closest promoter (distant less than 10,000 bp) and to genes with promoters overlapping their interacting regions (CHiCAGO score ≥ 5 in neutrophils) derived from the study of Javierre et al. (2016). The “findOverlaps” and “distanceToNearest” functions of the GenomicRanges (Lawrence et al., 2013) R package were used to make region comparisons.
Motif analysis was performed with the HOMER (Heinz et al., 2010) program.
WGBS was carried out to minimum genome coverage of 303 with established protocols (Kulis et al., 2012).
Differential methylation was estimated for (1) individual CpG sites in pairwise comparisons by using methyl_diff (Raineri et al., 2014) and across regions in group-wise comparison by using replicates with metilene (Jühling et al., 2016).
Association between Acetylation and Expression Clusters
We built a contingency table with the number of genes for each acetylation cluster (rows) and gene expression cluster (columns). The Pearson’s chisquare test was used to verify the non-independence of the two categorical variables. The significance of the association of each gene expression cluster with each dynamic acetylation cluster was tested by a hyper-geometric test, and the derived p values were corrected using the Benjamini-Hochberg method (Hochberg and Benjamini, 1990).
NADPH-oxidase activity was assessed as the release of hydrogen peroxide determined with an Amplex Red kit (Drewniak et al., 2013; Kuijpers et al., 2005).
Protease release after degranulation was measured with DQ-green BSA (see Supplemental Experimental Procedures for further details).
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures, five figures, and six tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.08.018.
Highlights.
Dynamic acetylation enforces human neutrophil progenitor differentiation
Neutrophils’ cytotoxic capability is established early at the (pro)myelocyte stage
Coordinated signaling component expression prevents toxic effects to the bone marrow
Acknowledgments
The work presented here was supported by the Landsteiner Foundation for Blood Transfusion Research (LSBR-1121 to T.W.K.) and by the Dutch Cancer Foundation (KUN 2011-4937 to J.H.A.M.). The work was funded by a grant from the European Commission 7th Framework Program (FP7/2007-2013, grant 282510, BLUEPRINT). M.F. is supported by the British Heart Foundation (BHF) Cambridge Centre of Excellence (RE/13/6/30180). Additional support was provided by the Wellcome Trust (WT108749/Z/15/Z to P.F.) and by the European Molecular Biology Laboratory (E.L., A.D., L.C., P.F.). Research in the Ouwehand laboratory is also supported by grants from Bristol-Myers Squibb, the British Heart Foundation, the Medical Research Council, the National Institute for Health Research (NIHR) (W.H.O. is NIHR senior investigator), and NHS Blood and Transplant (NHSBT).
Footnotes
Data and Software Availability
The web site https://blueprint.haem.cam.ac.uk/neutrodiff collects the analyzed data presented in this article. All the genome-wide data generated in this study, as well as the sequencing details, can be accessed via http://dcc.blueprint-epigenome.eu/.
Author Contributions
F.P. and T.W.K. designed the study, performed and supervised research, and wrote the manuscript. L.G. analyzed the data and wrote the manuscript. S.U., A.M., F.W, E.C.d.S.P., G.Y., and J.P. analyzed the data. I.H.H., A.T.J.T., E.M., E.J.-M., K.B., H.K., E.H., M.G., M.L.Y., and M.L. performed experiments. E.L., A.D., L.C., and P.F. performed and supervised data management. M.V., D. Roos, T.K.v.d.B., S.H., D. Rico, M.K., I.G., A.V., W.H.O., and H.G.S. provided expert supervision. M.F. provided expert supervision and wrote the manuscript. J.H.A.M. designed the study, analyzed the data, supervised research, and wrote the manuscript. All authors read and approved the final version of the manuscript.
Declaration of Interests
P.F. is a member of the Scientific Advisory Boards of Fabric Genomics, Inc., and Eagle Genomics, Ltd.
References
- Avellino R, Delwel R. Expression and regulation of C/EBPα in normal myelopoiesis and in malignant transformation. Blood. 2017;129:2083–2091. doi: 10.1182/blood-2016-09-687822. [DOI] [PubMed] [Google Scholar]
- Bainton DF, Ullyot JL, Farquhar MG. The development of neutrophilic polymorphonuclear leukocytes in human bone marrow. J Exp Med. 1971;134:907–934. doi: 10.1084/jem.134.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartůnek P, Králová J, Blendinger G, Dvorák kM, Zenke M. GATA-1 and c-myb crosstalk during red blood cell differentiation through GATA-1 binding sites in the c-myb promoter. Oncogene. 2003;22:1927–1935. doi: 10.1038/sj.onc.1206281. [DOI] [PubMed] [Google Scholar]
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-de-Santa-Pau E, Juan D, Pancaldi V, Were F, Martin-Subero I, Rico D, Valencia A, BLUEPRINT Consortium Automatic identification of informative regions with epigenomic changes associated to hematopoiesis. Nucleic Acids Res. 2017;45:9244–9259. doi: 10.1093/nar/gkx618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea D, Grassi L, Mazzapioda M, Tramontano A. FIDEA: a server for the functional interpretation of differential expression analysis. Nucleic Acids Res. 2013;41:W84–W88. doi: 10.1093/nar/gkt516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah E, Andrade F. NETs: the missing link between cell death and systemic autoimmune diseases? Front Immunol. 2013;3:428. doi: 10.3389/fimmu.2012.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinauer MC. Disorders of neutrophil function: an overview. Methods Mol Biol. 2014;1124:501–515. doi: 10.1007/978-1-62703-845-4_30. [DOI] [PubMed] [Google Scholar]
- Drewniak A, Gazendam RP, Tool AT, van Houdt M, Jansen MH, van Hamme JL, van Leeuwen EM, Roos D, Scalais E, de Beaufort C, et al. Invasive fungal infection and impaired neutrophil killing in human CARD9 deficiency. Blood. 2013;121:2385–2392. doi: 10.1182/blood-2012-08-450551. [DOI] [PubMed] [Google Scholar]
- Erlacher M, Strahm B. Missing cells: pathophysiology, diagnosis, and management of (pan)cytopenia in childhood. Front Pediatr. 2015;3:64. doi: 10.3389/fped.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodil N, Langlais D, Gros P. Primary immunodeficiencies and inflammatory disease: a growing genetic intersection. Trends Immunol. 2016;37:126–140. doi: 10.1016/j.it.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Romo GS, Caielli S, Vega B, Connolly J, Allantaz F, Xu Z, Punaro M, Baisch J, Guiducci C, Coffman RL, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J. Ki-67 and other proliferation markers useful for immunohistological diagnostic and prognostic evaluations in human malignancies. Semin Cancer Biol. 1990;1:199–206. [PubMed] [Google Scholar]
- Ghazaryan S, Sy C, Hu T, An X, Mohandas N, Fu H, Aladjem MI, Chang VT, Opavsky R, Wu L. Inactivation of Rb and E2f8 synergizes to trigger stressed DNA replication during erythroid terminal differentiation. Mol Cell Biol. 2014;34:2833–2847. doi: 10.1128/MCB.01651-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombart AF, Shiohara M, Kwok SH, Agematsu K, Komiyama A, Koeffler HP. Neutrophil-specific granule deficiency: homozygous recessive inheritance of a frameshift mutation in the gene encoding transcription factor CCAAT/enhancer binding protein–epsilon. Blood. 2001;97:2561–2567. doi: 10.1182/blood.v97.9.2561. [DOI] [PubMed] [Google Scholar]
- Hakkim A, Fürnrohr BG, Amann K, Laube B, Abed UA, Brinkmann V, Herrmann M, Voll RE, Zychlinsky A. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci U S A. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. Algorithm AS 136: a k-means clustering algorithm. Appl Stat. 1979;28:100–108. [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Varnai C, Thiecke MJ, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühling F, Kretzmer H, Bernhart SH, Otto C, Stadler PF, Hoffmann S. metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;26:256–262. doi: 10.1101/gr.196394.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 2010;107:2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Gupta A, Sun H, Zibello T, Lee HM, Dahl R, Boxer LA, Berliner N. Growth factor independence-1 (Gfi-1) plays a role in mediating specific granule deficiency (SGD) in a patient lacking a gene-inactivating mutation in the C/EBPepsilon gene. Blood. 2007;109:4181–4190. doi: 10.1182/blood-2005-05-022004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen P, Bjerregaard MD, Borregaard N, Cowland JB. End-stage differentiation of neutrophil granulocytes in vivo is accompanied by up-regulation of p27kip1 and down-regulation of CDK2, CDK4, and CDK6. J Leukoc Biol. 2004;75:569–578. doi: 10.1189/jlb.1003474. [DOI] [PubMed] [Google Scholar]
- Kuijpers TW, Alders M, Tool AT, Mellink C, Roos D, Hennekam RC. Hematologic abnormalities in Shwachman Diamond syndrome: lack of genotype-phenotype relationship. Blood. 2005;106:356–361. doi: 10.1182/blood-2004-11-4371. [DOI] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queirós AC, Navarro A, Clot G, Martínez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JJ, Moore RM, Vander Kooy D. Protein kinase-C activation is required for oxytocin-induced prostaglandin production in human amnion cells. J Clin Endocrinol Metab. 1991;72:1073–1080. doi: 10.1210/jcem-72-5-1073. [DOI] [PubMed] [Google Scholar]
- Nauseef WM, Borregaard N. Neutrophils at work. Nat Immunol. 2014;15:602–611. doi: 10.1038/ni.2921. [DOI] [PubMed] [Google Scholar]
- Nunoi H, Yamazaki T, Tsuchiya H, Kato S, Malech HL, Matsuda I, Kanegasaki S. A heterozygous mutation of beta-actin associated with neutrophil dysfunction and recurrent infection. Proc Natl Acad Sci U S A. 1999;96:8693–8698. doi: 10.1073/pnas.96.15.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri E, Dabad M, Heath S. A note on exact differences between beta distributions in genomic (Methylation) studies. PLoS ONE. 2014;9:e97349. doi: 10.1371/journal.pone.0097349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnerblad M, Andersson R, Olofsson T, Douagi I, Karimi M, Lehmann S, Hoof I, de Hoon M, Itoh M, Nagao-Sato S, et al. FANTOM consortium Analysis of the DNA methylome and transcriptome in granulopoiesis reveals timed changes and dynamic enhancer methylation. Blood. 2014;123:e79–e89. doi: 10.1182/blood-2013-02-482893. [DOI] [PubMed] [Google Scholar]
- Rørvig S, Østergaard O, Heegaard NH, Borregaard N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J Leukoc Biol. 2013;94:711–721. doi: 10.1189/jlb.1212619. [DOI] [PubMed] [Google Scholar]
- Schuyler RP, Merkel A, Raineri E, Altucci L, Vellenga E, Martens JHA, Pourfarzad F, Kuijpers TW, Burden F, Farrow S, et al. Distinct trends of DNA methylation patterning in the innate and adaptive immune systems. Cell Rep. 2016;17:2101–2111. doi: 10.1016/j.celrep.2016.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SO, Horowitz JM. Nkx3.1 binds and negatively regulates the transcriptional activity of Sp-family members in prostate-derived cells. Biochem J. 2006;393:397–409. doi: 10.1042/BJ20051030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strydom N, Rankin SM. Regulation of circulating neutrophil numbers under homeostasis and in disease. J Innate Immun. 2013;5:304–314. doi: 10.1159/000350282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilgaard-Mönch K, Jacobsen LC, Borup R, Rasmussen T, Bjerregaard MD, Nielsen FC, Cowland JB, Borregaard N. The transcriptional program of terminal granulocytic differentiation. Blood. 2005;105:1785–1796. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Series B Stat Methodol. 2001;63:411–423. [Google Scholar]
- Turro E, Su SY, Gonçalves Â, Coin LJ, Richardson S, Lewin A. Haplotype and isoform specific expression estimation using multimapping RNA-seq reads. Genome Biol. 2011;12:R13. doi: 10.1186/gb-2011-12-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turro E, Astle WJ, Tavaré S. Flexible analysis of RNA-seq data using mixed effects models. Bioinformatics. 2014;30:180–188. doi: 10.1093/bioinformatics/btt624. [DOI] [PubMed] [Google Scholar]
- Wahed A, Dasgupta A. Hematology and Coagulation. San Diego: Elsevier; 2015. Chapter 2: Bone marrow examination and interpretation; pp. 15–29. [Google Scholar]
- Weiss SJ. Tissue destruction by neutrophils. N Engl J Med. 1989;320:365–376. doi: 10.1056/NEJM198902093200606. [DOI] [PubMed] [Google Scholar]
- White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci U S A. 2013;110:11952–11957. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte S, O’Shea JJ, Vahedi G. Super-enhancers: asset management in immune cell genomes. Trends Immunol. 2015;36:519–526. doi: 10.1016/j.it.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.