Abstract
Myelin is a specialized subcellular structure that evolved uniquely in vertebrates. A myelinated axon conducts action potentials many times faster than an unmyelinated axon of the same diameter; for the same conduction speed, the unmyelinated axon would need a much larger diameter and volume than its myelinated counterpart. Hence myelin speeds information transfer and saves space, allowing the evolution of a powerful yet portable brain. Myelination in the central nervous system (CNS) is controlled by a gene regulatory program that features a number of master transcriptional regulators including Olig1, Olig2 and Myrf. Olig family genes evolved from a single ancestral gene in non-chordates. Olig2, which executes multiple functions with regard to oligodendrocyte identity and development in vertebrates, might have evolved functional versatility through post-translational modification, especially phosphorylation, as illustrated by its evolutionarily conserved serine/ threonine phospho-acceptor sites and its accumulation of serine residues during more recent stages of vertebrate evolution. Olig1, derived from a duplicated copy of Olig2 in early bony fish, is involved in oligodendrocyte development and is critical to remyelination in bony vertebrates, but is lost in birds. The origin of Myrf orthologs might be the result of DNA integration between an invading phage or bacterium and an early protist, producing a fusion protein capable of self-cleavage and DNA binding. Myrf seems to have adopted new functions in early vertebrates – initiation of the CNS myelination program as well as the maintenance of mature oligodendrocyte identity and myelin structure - by developing new ways to interact with DNA motifs specific to myelin genes.
Keywords: myelin, oligodendrocyte, transcription factor, Olig1, Olig2, MyRF, evolution, phylogeny
Evolution of a genetic program specifying myelination of axons was a momentous event in the history of vertebrates. By allowing a dramatic increase in the speed and efficiency of action potential propagation, myelin overcame limits to the size of both brain and body as well as increasing greatly the potential processing power of the brain - opening the way to the evolution of tetrapods, for example. Several genes encoding myelin structural components are already present in the lancelet (a Cephalochordate) and/or lampreys (Agnatha) (Gould et al., 2005; Li and Richardson, 2008; Smith et al., 2013; Werner, 2013). Neither lampreys nor hagfish, the other extant member of the Agnatha, are myelinated in their central or peripheral nervous systems (CNS or PNS) (Bullock et al., 1984). This suggests that subsequent to the Agnatha, myelin evolved through a process of "pick and mix" whereby pre-existing genes encoding proteins with favourable physico-chemical properties were co-opted into a single myelin gene regulatory program that subsequently diverged into two closely-related programs in the central and peripheral nervous systems (in oligodendrocytes and Schwann cells, respectively). How this gene recruitment process occurred is a fascinating question, the answer to which is likely to illuminate evolutionary processes in general. A driving force for the evolution of gene regulatory networks is thought to be alterations in the properties of key transcription factors (TFs) and their sequence-specific binding to DNA (Cheatle Jarvela and Hinman, 2015). Attention has previously focused on genomic re-arrangements involving the insertion and deletion of cis-regulatory DNA sequence motifs but recent studies suggest that mutations in the coding regions of the TFs themselves - leading to altered protein expression, protein-protein interactions and/or post-translational modifications - could have figured in the evolution of gene regulatory networks more than previously imagined (Lynch et al., 2011; Reece-Hoyes et al., 2013; Voordeckers et al., 2015). In this review we focus on evolution of the myelin gene regulatory network in oligodendrocytes, which involves a considerable number of TFs such as Olig1/2, Sox10, Nkx2.2, Nkx6.2, Mash1, Myrf, Sip1, Tcf7l2 and so on (Li et al., 2009; Zuchero and Barres, 2013; Huang et al., 2013; Emery and Lu, 2015). Among these, Olig1/2, Sox10 and Myrf are considered the master regulators; here we concentrate on Olig1/2 and Myrf, since the role of Sox10 is reviewed separately in this issue (see chapter by Wegner).
Oligodendrocyte lineage genes 1 and 2 (Olig1 and Olig2)
Phylogeny of Olig genes
Olig genes, derived from a common ancestor, form a subgroup of the basic helix-loop-helix (bHLH) family, which includes Olig1, Olig2, Olig3, Olig4, Bhlhb4 and Bhlhb5. Olig1/2 genes were first identified in mouse almost simultaneously by three groups searching for genes that regulate oligodendrocyte development. Subsequently, Olig3, Olig4, Bhlhb4 and Bhlhb5 were also identified.
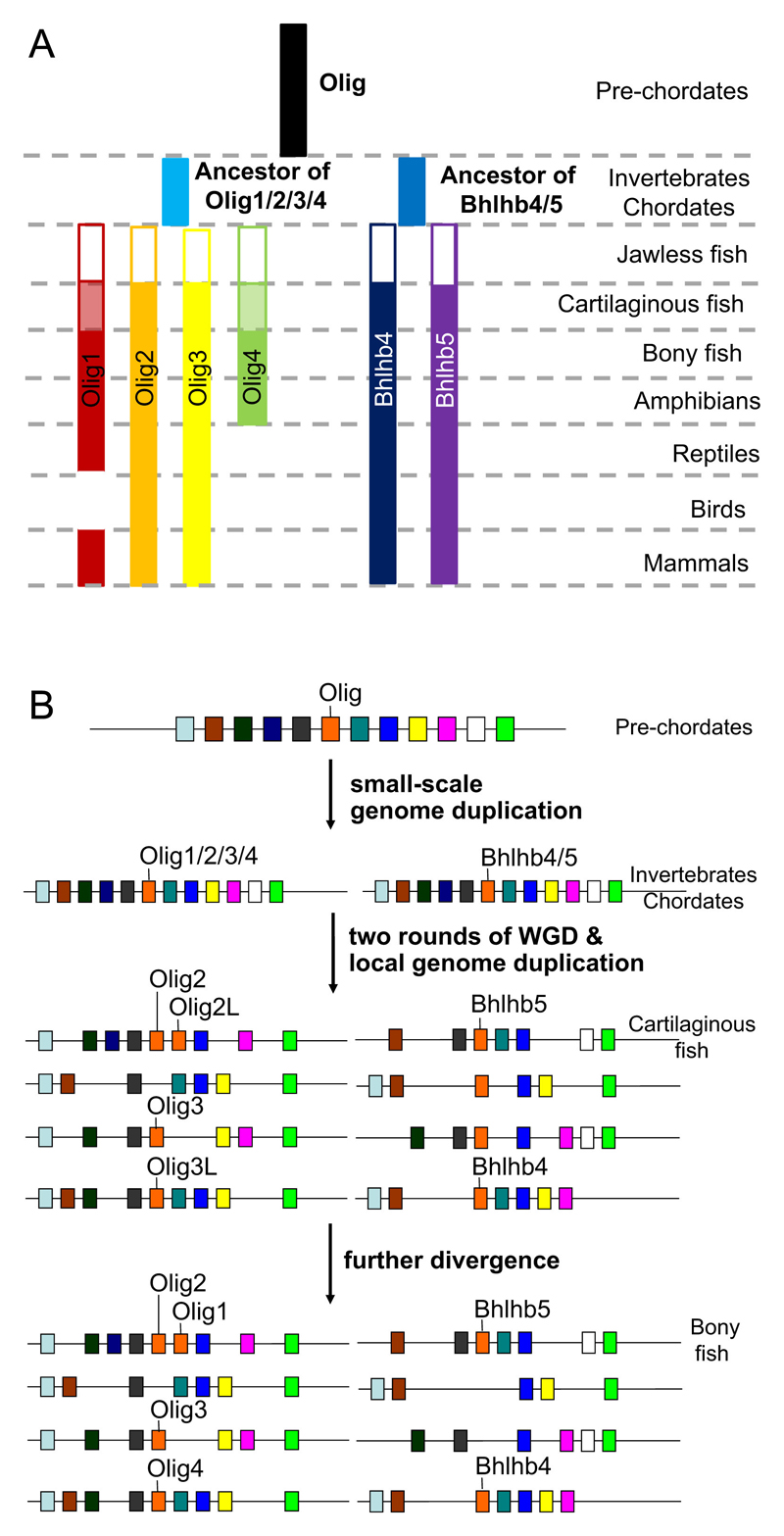
Evidence of a common ancestor for Olig genes can be found in arthropods, nematodes and platyhelminthes. In the nematode Caenorhabditis elegans, the Olig homolog Hlh-17 is expressed at all development stages in the cephalic sheath cells (considered to be glial cells) that ensheath four of the dopaminergic neurons (McMiller and Johnson, 2005) and plays a role in dopamine signaling (Felton and Johnson, 2011). The Drosophila Olig homolog, Oli, is not expressed in glial lineage cells but in certain ventral motor neuron subtypes; Oli is responsible for regulating larval and adult locomotion and its loss of function can be partially compensated for by over-expression of chick Olig2 (Oyallon et al., 2012). In the hemichordate Saccoglossus kowalevskii (acorn worm), the Olig homolog is first expressed in a cluster of dorsal cells of the prosome base right after gastrulation; later on, its expression expands across the entire proboscis ectoderm and to other dorsal cells along the dorsal midline (Lowe et al., 2006). As it is still early days for research on non-chordate Olig genes, we have yet to form a clear picture of their expression patterns, functions and evolutionary connections with their vertebrate descendants. Interestingly, in the genome of the cephalochordate Branchiostoma floridae (lancelet) (http://genome.jgi-psf.org/Brafl1/Brafl1.home.html), there are at least two copies of Olig, one the precursor of Olig1/2/3/4 and the other the precursor of Bhlhb4/5, hinting at a small scale genome duplication resulting in the initial divergence of Olig genes (Louis et al., 2012).
As jawless fish are the only vertebrates without compact myelin, they are important phylogenetic tools for studying Olig gene evolution. The recent sea lamprey (Petromyzon marinus) genome project has identified about 26,000 protein-coding genes including a group of genes for myelin components (Smith et al., 2013). However, we cannot find any Olig homolog in the current annotated gene set and performing a TBLASTN search with elephant shark (Callorhinchus milii) Olig2 protein sequence (GenBank:XM_007906736) against Petromyzon 7.0 genome sequence (www.ensembl.org/Petromyzon_marinus/) finds no match. The missing lamprey Olig might simply reflect still-incomplete genome sequencing but, given that adult liver tissue is the source of the sequenced DNA, it is also possible that the Olig locus in the lamprey somatic genome has been lost due to chromosome rearrangement at embryonic stages. It is known that during embryonic development the lamprey undergoes programmed genome rearrangement, which can lead to the removal of certain repetitive sequences, entire chromosomes or individual genes, resulting in deletion of~20% of germline DNA from somatic cells (Smith et al., 2009; Smith et al., 2012). Although the mechanism and effect of such genomic rearrangement are not understood, it seems to be a shared property of the jawless fish because hagfish (Myxine glutinosa) are believed to undergo similar large-scale rearrangements, based on the fact that the genome size of their somatic tissue is smaller than that of germline tissue (Kubota et al., 1997; Goto et al., 1998; Kubota et al., 2001). We have searched another relatively new genome database derived from testis of Japanese lamprey (Lethenteron japonicum) using TBLASTN with Olig2 protein sequence of the elephant shark against the draft genome assembly LetJap1.0 (http://jlampreygenome.imcb.a-star.edu.sg/blast/) and have identified DNA fragments encoding partial orthologs of Bhlhb4 and Bhlhb5 and two Olig2/3/4 orthologs – one full length and one a small portion. These data imply that at least 4 copies of Olig family genes have been deleted in lamprey somatic cells. On the other hand, lamprey genome analysis suggests that two-rounds of whole-genome duplication might have occurred in a common ancestor of jawless fish (Agnatha) and Gnathostomes, meaning that all Olig genes except for Olig1 would have already been in existence when lampreys first appeared. We cannot be certain about this until the lamprey germline genome project has been completed. It would also be of interest to sequence the hagfish genome.
Cartilaginous fish (family Chondrichthyes) are the most ancient living species to possess myelin. We are able to retrieve sequences of Olig2, Olig2-like, Olig3, Olig3-like and Bhlhb5 from the elephant shark genome database (http://esharkgenome.imcb.a-star.edu.sg) and Olig2, Olig3 and Bhlhb4 from the little skate (Leucoraja erinacea) database. It is interesting to note that the elephant shark has two Olig2 homologs just 28 kb apart in one scaffold (scaffold 170; Venkatesh et al., 2014), which is similar to the way that Olig1 and Olig2 are located in a synteny block in bony vertebrates, lending support to our previous hypothesis that Olig1 might have evolved from a local duplication at an ancestral Olig2 locus (Li and Richardson, 2008). Given that the elephant shark has two copies of Olig3 but no Olig4, it is likely that one of the Olig3 copies has diverged to become Olig4 in higher species. Therefore, since their divergence from the Chondrichthyes, jawed vertebrates have possessed all of the Olig family genes with two exceptions: the loss of Olig4 in amniotes (including birds, reptiles and mammals) and the curious absence of Olig1 from birds.
Taking all this into account we have updated our view of Olig gene evolution (Li and Richardson, 2008) (Fig. 1). We now hypothesize the following sequence of events: 1) a single founder Olig gene was present in non-chordates, 2) a small-scale genomic duplication in an invertebrate chordate generated two copies of Olig, 3) following two rounds of whole genome duplication and subsequent massive gene loss in a common ancestor of jawless fish and jawed vertebrates (Dehal and Boore, 2005), the two divergent Olig precursors gave rise to Olig2/3/4 and Bhlhb4/5 respectively, 4) a local genome duplication at the Olig2 locus formed a synteny block containing two copies of Olig2, 5) in early bony fish, one Olig2 locus recombined with a distant relative (another bHLH gene), undergoing domain insertion, rearrangement and/or reshuffling (Atchley and Fitch, 1997; Morgenstern and Atchley, 1999) to form Olig1, 6) subsequently Olig4 was lost from amniotes and Olig1 from birds.
Figure 1.
Phylogeny of Olig genes. (A) Olig genes were evolved from a common invertebrate ancestor gene which was duplicated in an early chordate to provide the ancestral genes of Olig1/2/3/4 and Bhlhb4/5. Although we are unsure how many Olig paralogs there are in jawless fish (Agnatha), pre-Olig1 (Olig2-like), Olig2, Olig3, pre-Olig4 (Olig3-like), Bhlhb4 and Bhlhb5 all exist in cartilaginous fish. Olig4 is subsequently lost from all Amniota and Olig1 from Aves. (B) Hypothetical model of Olig gene evolution. A small-scale genomic duplication in the early chordate resulted in the production of ancestors of Olig1/2/3/4 and Bhlhb4/5. After two rounds of whole genome duplication during early vertebrate evolution, accompanied by gene loss, the Olig1/2/3/4 gene ancestor gave rise to Olig2, Olig3 and Olig4 while the Bhlhb4/5 gene ancestor gave rise to Bhlhb4 and Bhlhb5. Meanwhile, a local duplication around the Olig2 locus produced a synteny block containing two Olig2 genes and one of these subsequently underwent recombination with another distantly related bHLH family gene to produce Olig1 (adapted and updated from Li and Richardson, 2009).
Despite having a common origin, Olig genes display diverse functions. Bhlhb4, which is expressed in the pancreas and brain (Bramblett et al., 2002), marks the diencephalic-mesencephalic boundary and is also required for the maturation of retinal rod bipolar cells (Bramblett et al., 2004). Bhlhb5, which can be detected in the CNS and in sensory organs such as the eye, hair follicle, cochlea of the developing inner ear and nasal epithelium (Brunelli et al., 2003), is critical for the specification of bipolar and amacrine neurons in the retina (Feng et al., 2006) and is necessary for the survival of a subset of inhibitory interneurons that control pruritic (itch) circuits in the dorsal horn (Ross et al., 2010). Olig3 is expressed in both dorsal and ventral precursor domains of the developing spinal cord but is only required for the patterning of dorsal spinal cord (Muller et al., 2005; Liu et al., 2014). Olig3 is also expressed in the dorsal hindbrain where it plays an essential role in the development of brainstem nuclei and rhombic-lip-derived neurons (Liu et al., 2008; Storm et al., 2009). The above roles of Olig genes in the development of neural progenitors and neurons presumably evolved prior to their relatively new functions in controlling myelination. To understand how myelination comes about, we need to start by looking into the two major regulators of myelination - Olig1 and Olig2.
Olig2 regulates oligodendrocyte development
The majority (≥80%) of oligodendrocyte lineage cells in the perinatal spinal cord are descended from a ventrally-located precursor domain of the embryonic ventricular zone, the pMN domain (for "progenitors of motor neurons") (Richardson et al., 2006; Tripathi et al., 2011). Prior to neural tube closure, Olig2 mRNA is expressed in the pMN domain as well as the neighbouring p3 and p2 domains that abut pMN on its ventral and dorsal sides, respectively (Lu et al., 2000; Takebayashi et al., 2000; Zhou et al., 2000). From about embryonic day 9.5 (E9.5) in the mouse, Olig2 expression is confined to pMN, possibly via a microRNA-mediated mRNA silencing mechanism in the adjacent domains (Chen et al., 2011). Induced by the gradient of a signaling molecule Shh (sonic hedgehog) secreted from the notochord and floor plate, Olig2 defines pMN by repressing Irx3 (for p2 identity) and Nkx2.2 (for p3 identity) (Marquardt and Pfaff, 2001; Novitch et al., 2001); in Olig2 knockout (KO) mice the pMN domain is lost and the p2 domain expands ventrally into what would normally be pMN territory (Lu et al., 2002; Takebayashi et al., 2002; Zhou and Anderson, 2002). Olig2 knockout (KO) mice therefore lack spinal motor neurons and the animals die at birth (Lu et al., 2002; Takebayashi et al., 2002; Zhou and Anderson, 2002). However, the role of Olig2 extends beyond MN specification because it continues to be expressed in the pMN domain after MN production is complete, preserving the progenitor cell properties of pMN for later production of oligodendrocyte precursors (OPs). Olig2 is down-regulated rapidly in migrating post-mitotic MN precursors and is absent from mature MNs; forced expression of Olig2 in MN precursors prevents their differentiation into functional MNs (Lee et al., 2005). At the end of MN production, pMN progenitors switch fate and start to generate migratory OPs that express platelet-derived growth factor receptors (alpha subunit, Pdgfra) and transcription factor Sox10. Olig2 is critical to OP specification in the spinal cord and, in Olig2-KO mice, Pdgfra/Sox10-positive cells are completely absent (Lu et al., 2002; Takebayashi et al., 2002; Zhou and Anderson, 2002). Olig2 also plays an important role in the later events of OP differentiation and oligodendrocyte maturation in the spinal cord, because conditional knockout (cKO) of Olig2 in OPs (using CNP-Cre) represses OP differentiation and results in hypomyelination, while cKO of Olig2 in immature oligodendrocytes (using PLP-CreER) facilitates oligodendrocyte maturation and accelerates myelination (Mei et al., 2013). In addition, Olig2’s function in oligodendrocyte development in the spinal cord of vertebrates is evolutionarily conserved. For instance, knockdown of Olig2 with morpholino antisense oligonucleotides in zebrafish spinal cord prevents development of both MNs and oligodendrocytes, suggesting that Olig2 is charged with the same task in teleost oligodendrocyte development as in mammals (Park et al., 2002).
It is unclear whether a similar neuron-to-oligodendrocyte fate switch takes place in the ventral VZ of the brain. In the developing forebrain, Olig2 starts to be expressed at E9.5, mainly in the thalamus (ventral diencephalon). Later on at E12.5, Olig2 is highly expressed in the medial ganglionic eminence (MGE) and the anterior entopeduncular area (AEP) and weakly expressed in the lateral ganglionic eminence (LGE) (Takebayashi et al., 2000; Woodruff et al., 2001). Olig2-positive precursor cells in the MGE and LGE can give rise to GABAergic interneurons, which migrate throughout the developing forebrain and into the cerebral cortex, and cholinergic projection neurons, which remain in the ventral telencephalon (Furusho et al., 2006; Miyoshi et al., 2007; Ono et al., 2008). Loss of Olig2 results in a 40% decrease in the number of cholinergic neurons in the basal forebrain while there is no change in the number of GABAergic interneurons (Furusho et al., 2006; Ono et al., 2008). In the forebrain, oligodendrocyte specification begins at E13.5 as indicated by the expression of OP marker Pdgfra at the ventral boundary of the MGE (Tekki-Kessaris et al., 2001). Unlike the spinal cord, Sox10/Pdgfra-expressing OPs are still present in the forebrain of Olig2 null mice although in dramatically reduced numbers compared to wild type controls (Lu et al., 2002). Conditionally deleting Olig2 in embryonic neural stem cells (NSCs), using a Cre transgene driven either by the human GFAP promoter (expressed in all NSCs in mice) or by the Emx1 promoter (restricted to cortical NSCs), has no obvious effect on OP specification but can inhibit further differentiation of OPs into oligodendrocytes, leading to decreased myelin gene expression and hypo-myelination (Yue et al., 2006). Another study found that when Olig2 is conditionally deleted in cortical OPs using an OP-specific NG2-Cre, they change fate and transform into GFAP+ astrocytes (Zhu et al., 2012).
Phosphorylation contributes to Olig2’s functional versatility
How can Olig2 play multiple roles in neural stem cell (NSC) specification and oligodendrocyte development? Unlike other neurogenic bHLH TFs such as Mash1 and Ngn2 that are transiently expressed and function at restricted times during development, Olig2 has sustained expression throughout development and into adulthood in all stages of oligodendrocyte lineage cells. Therefore, it is likely that post-translational modification is the key to Olig2’s functional versatility. Post-translational modification can greatly change the properties of the amino acids and the proteins that contain them in response to developmental need and evolutionary pressure, which might be essential to the evolution of organismal complexity (Prabakaran et al., 2012).
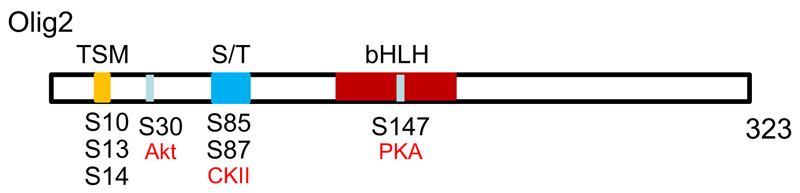
One common post-translational modification is protein phosphorylation on serine, threonine and tyrosine residues. We have found that TFs in general tend to possess more serine residues relative to other classes of protein, based on comparison of 542 TFs and 15,861 non-TFs in a mouse UniProtKB/Swiss-Prot database (Jiang, C. and Li, H., unpublished). Olig2 in particular is a serine-rich protein containing 50 serine residues as well as 14 threonines and 3 tyrosines out of 323 amino acids, showing great potential for phosphorylation. We chose GPS3.0 (Group-based Prediction System, version 3, http://gps.biocuckoo.org), a newly updated bioinformatics software program to predict potential protein phosphorylation sites given that the previous version GPS2.0 performed large-scale prediction on more than 13,000 mammalian protein phosphorylation sites with remarkable accuracy (Xue et al., 2008). By setting a high cut-off threshold (with a 2% false positive rate for serine/threonine kinases and a 4% false positive rate for tyrosine kinases), we called up all of the murine Olig2 serine/ threonine/ tyrosine sites that can be phosphorylated by at least one protein kinase. This computer prediction is backed up by our experimental data, which indicate that Olig2 over-expressed in COS-7 cells can be phosphorylated at multiple sites and that the phosphorylation can be reversed by alkaline phosphatase treatment (Li et al., 2011). Mass spectrometry has identified phosphorylation on serine 10 (S10), S13, S14, S81 and S263 and threonine 43 (T43) of mouse Olig2 in neural stem cells or COS-7 cells (Sun et al., 2011) although, due to limitations to the sensitivity of mass spectrometry, it is impossible to identify all phosphorylation sites on Olig2 in a few experiments.
Murine Olig2 contains a cluster of 12 contiguous serine/threonine residues from S77 to S88, i.e. an S/T box, which is a substrate of casein kinase II (CK2) (Huillard et al., 2010). Data obtained from neurosphere culture suggest that S/T box phosphorylation by CK2 serves to regulate neural progenitor proliferation and oligodendrocyte production (Huillard et al., 2010). Apart from the S/T box, mouse Olig2 has a triple serine motif comprising S10, S13 and S14, and the phosphorylation state of this triple serine motif changes during development, affecting the proliferation of neural progenitors. The triple serine motif is phosphorylated during OP proliferation in embryonic spinal cord but dephosphorylated in postnatal spinal white matter; phosphorylation of the triple serine motif is required for neural progenitor proliferation according to neurosphere culture but is not needed for OP specification and oligodendrocyte differentiation (Sun et al., 2011). The triple serine motif of Olig2 is also phosphorylated in p53-positive human gliomas, repressing the p53-mediated apoptotic pathway (Sun et al., 2011; Meijer et al., 2014). In addition, Olig2 with phosphorylated triple serine motif binds to a transcriptionally active chromatin domain, regulating target gene expression (Meijer et al., 2014). We have provided evidence that Olig2 controls the MN-OP fate switch by reversible phosphorylation on S147, a conserved protein kinase A (PKA) target site; Olig2-S147 is phosphorylated during ventral spinal cord patterning and MN generation but de-phosphorylated at the onset of OL specification (Li et al., 2011). In vivo evidence acquired from Olig2-S147A mutant mice indicates that phosphorylation at this site is required for the ventral patterning and MN generation, while data from in ovo electroporation of chick embryos and P19 embryonal carcinoma cells (which resemble NSCs) show that de-phosphorylation at S147 favours OP fate specification. Furthermore, S147 phosphorylation causes Olig2 to switch its preferred binding partner from itself (or Olig1) to Ngn2, and this regulated exchange of co-factors is required for and triggers the MN-OP fate switch (Li et al., 2011). In addition, S30 of Olig2 has been found to be a target of protein kinase B (e.g. Akt) and S30 phosphorylation triggers translocation of Olig2 from the nucleus to the cytoplasm (Setoguchi and Kondo, 2004). In vitro NSC culture shows that Akt, possibly by phosphorylating S30, facilitates Olig2’s exclusion from the nucleus and stimulates NSCs to differentiate into astrocytes in the presence of ciliary neuronotrophic factor (CNTF) (Setoguchi and Kondo, 2004).
Seen from an evolutionary perspective, of all these known Olig2 phosphorylation sites, the triple serine motif, S/T box, S30 and S147, only S147 is conserved among vertebrates and their invertebrate forerunners, which is not surprising considering the fact that Olig2 S147 phosphorylation is indispensible to motor neuron generation in mouse spinal cord. By analogy, Oli might also be required for motor neuron generation in the ventral nerve cord of Drosophila and phosphorylation might also be involved (Oyallon et al., 2012). However, flies have no oligodendrocytes or compact myelin and presumably do not need the dephosphorylated form of Olig2-S147 to direct NSCs to a glial fate. Nonetheless, it would be interesting to find out if Oli is phosphorylated/dephosphorylated during Drosophila nerve cord development. The triple serine motif of Olig2 is conserved in bony vertebrates but not in elephant sharks, suggesting that cartilaginous fish do not need extensive OP proliferation to generate a large number of oligodendrocytes as in mammals. In contrast, the S/T box and S30 are only present in mammals, although phosphorylation at those two sites has yet to be confirmed in vivo, leaving us wondering if their phosphorylation could lead to some Olig2 activities that are specific to mammals. Close examination of mouse Olig2 protein reveals that S/T142 and T151 of the bHLH domain are conserved in ancestral Olig genes and S320 at the Olig2 C-terminus is conserved in jawed vertebrates. These conserved phosphorylation sites might be pointers to key mechanisms of CNS development and would be worth exploring. In addition, the number of serine residues in Olig2 orthologs increases gradually along the evolutionary path, from 28 in elephant shark to 50 in mouse, raising the possibility that by increasing phosphorylation sites, Olig2 might have gained functional versatility to provide a genetic basis for species to adapt under selective pressure.
Olig1 contributes to myelination and remyelination
In the spinal cord, Olig1 mRNA is first detected in the p3 and pMN ventral progenitor domains and becomes confined to the pMN domain by E10.5 (Chen et al., 2011). However, Olig1 protein can only be detected after E18.5 (Fu et al., 2009). Loss of Olig1 does not impair MN development in the spinal cord, but leads to an about 30% increase in the number of adult cortical interneurons (Silbereis et al., 2014). Evidence from three independently generated Olig1 KO mouse lines (Lu et al., 2002; Paes de Faria et al., 2014) suggests that Olig1 has no effect on oligodendrocyte development in the spinal cord, apart from causing a slight delay in OP differentiation. In stark contrast to this are data from another Olig1 KO line (Xin et al., 2005), in which myelination in the spinal cord is completely blocked and the animals die around the third postnatal week. The Olig1 KO of Xin et al. (2005) was derived from the Olig1 KO of Lu et al. (2002) by removing the Pgk-neo cassette from the Olig1 null allele in the latter. Xin et al. (2005) invoked the presence of the Pgk-neo cassette in the Lu et al. (2002) line to explain the difference between the two KO lines, speculating that the Pgk promoter might up-regulate the adjacent Olig2 gene, thereby compensating for the loss of Olig1. However, a later study (Samanta et al. 2007) demonstrated that Olig2 gene expression is not altered in the Olig1 KO of Lu et al. (2002). In respect of oligodendrocyte development in the brain, our own Olig1 KO models (Paes de Faria et al., 2014) exhibit no abnormal phenotypes, while the Olig1 KO of Lu et al. (2002) displays a subtle delay in oligodendrocyte maturation in the spinal cord and impaired OP specification and oligodendrocyte differentiation in the corpus callosum (Dai et al., 2015). Such differences in the phenotypes of Olig1 null lines might be attributed to different genetic backgrounds, different nutritional and/or environmental conditions or unintended genetic alterations at the Olig1/2 locus during removal of the Pgk-neo cassette. Notwithstanding these uncertainties, Olig1 is believed to play a role in myelination in some contexts. For instance, Olig1 can physically interact with Sox10 to promote myelin basic protein (Mbp) transcription via evolutionarily conserved DNA motifs in the Mbp promoter (Li et al., 2007). Moreover, Olig1 is critical to remyelination following lysolecithin- or cuprizone-induced demyelination (Arnett et al., 2004) and is necessary for transplanted neural progenitor cells to differentiate and remyelinate in virus-induced demyelination (Whitman et al., 2012). In addition, at two weeks postnatal, Olig1 translocates from the cell nucleus to the cytoplasm, but returns to the nucleus during early remyelination, suggesting that the subcellular location of Olig1 might have an impact on myelination and remyelination (Arnett et al., 2004).
Traces of Olig1 can be found in elephant sharks. However, all avian genome databases (including chicken, turkey, budgerigar, medium ground finch and zebra finch genomes) lack any Olig1 homolog. Moreover, no bird Olig1 gene sequences can be found in NCBI Genbank, contrasting with 21 listed bird Olig2 gene sequences. Taken together, we infer that Aves do not have Olig1. Olig1 is likely to be one of many genes lost in birds. In fact, there are genomic clues pointing to a massive gene loss in birds: the genome size of Aves is smaller than that of other tetrapod classes (Szarski, 1976; Tiersch and Wachtel, 1991), the number of protein coding genes (paralogs) in a Gallus gene family is normally lower than that in its mammalian or reptile counterpart (Hughes and Friedman, 2008; Zhang et al., 2 and the gene number of avian genomes is ~70% of that of the human genome (Zhang et al., 2014). This large-scale loss of DNA segments might have happened after birds split from other reptiles around 100 million years ago and, as a result, birds might have taken a minimalist approach to adaptation by undergoing functional compensation and innovation in paralogous gene copies (Zhang et al., 2014). Given that Olig1 is involved in oligodendrocyte function in teleosts and mammals, it would be intriguing to find out how birds tackle myelination and remyelination without a contribution from Olig1.
Myelin regulatory factor
Myelin regulatory factor (Myrf or Mrf) is also known as GM98 (gene mode 98) in mouse and C11ORF9 (chromosome 11 open reading frame 9) in human. Unlike other master regulators of oligodendrocyte development such as Olig1/2 and Sox10, which are expressed at all stages of the oligodendrocyte lineage, Myrf is exclusively expressed in differentiated oligodendrocytes in the mouse CNS. Myrf does not start to be expressed until OP differentiation has been initiated. Myrf is critical to oligodendrocyte differentiation and is necessary to drive the CNS myelin transcriptional program during development (Emery et al., 2009). Moreover, Myrf is necessary for oligodendrocyte generation in the adult CNS and conditional deletion of Myrf in adult OPs can prevent new oligodendrocytes from being generated, leading to defects in motor learning (McKenzie et al., 2014). Myrf is also required for maintaining the identity of mature oligodendrocytes and myelin structure; cKO of Myrf in mature oligodendrocytes (e.g. using Plp-CreER or Sox10-CreER) causes dramatic down-regulation of myelin protein expression and the breakdown of myelin sheaths (Koenning et al., 2012; McKenzie et al., 2014). In addition, Myrf can interact physically with Sox10 to control myelin gene expression directly (Hornig et al., 2013). Current data on Myrf in non-mammals is scanty, but we have detected Myrf expression in oligodendrocytes of adult zebrafish (Li and Richardson, data not shown).
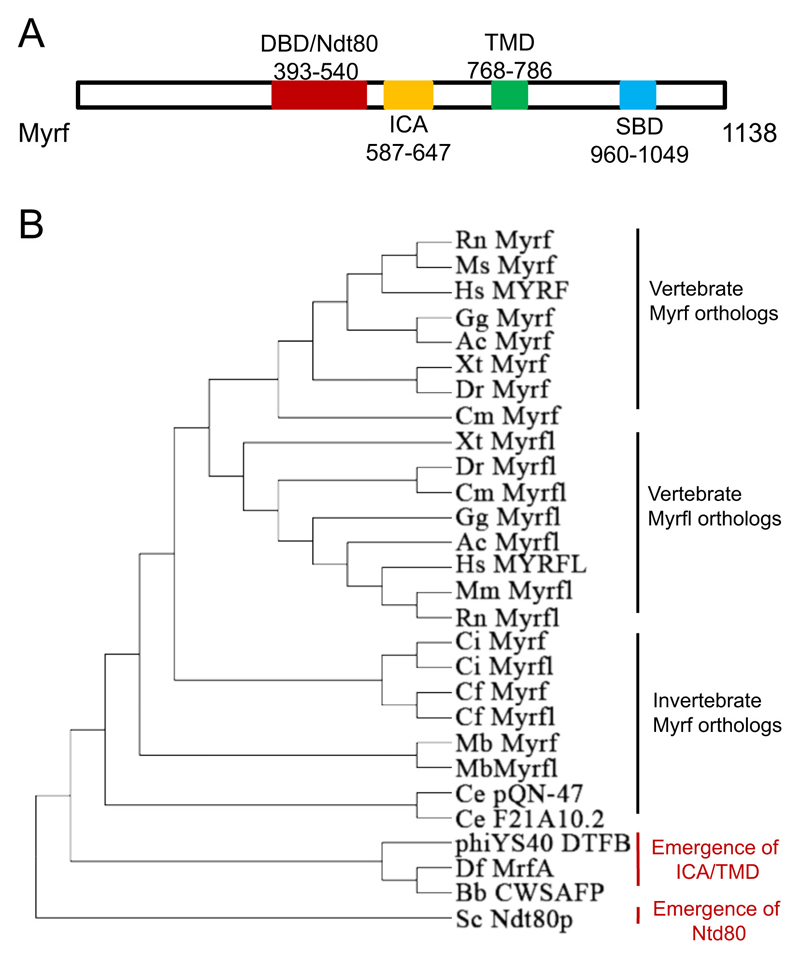
Myrf contains a DNA-binding domain, an intramolecular chaperone auto-processing (ICA) domain and a transmembrane domain (Bujalka et al., 2013; Li et al., 2013) (Fig. 2A) and belongs to a class of membrane-bound TFs that are translated as transmembrane proteins but subsequently undergo proteolytic processing, releasing a soluble TF that translocates to the nucleus to participate in transcriptional regulation.
Figure 2.
Phosphorylation sites of of murine Olig2 (GenBank: NP_058663). Functionally confirmed phosphorylation regions and sites in Olig2 are shown. Akt, CKII and PKA are confirmed kinases that can phosphorylate Olig2 at specific sites. bHLH: basic helix-loop-helix domain; TSM: tripleserine motif; S/T: serine and threonine rich domain.
The DNA binding domain of Myrf is homologous to the Ndt80 (non-dityrosine 80) of yeast. This Ndt80 DNA binding domain is conserved with a similar crystal structure among a range of eukaryotic organisms from protists and fungi to mammals (Montano et al., 2002; Xu et al., 1995). Yeast Ndt80 can bind to a so-called midsporulation element (MSE) with the consensus sequence CRCAA [where R = A or G; (Chu et al., 1998)]. However, it is not possible for a chimeric protein comprising the human Myrf DNA binding domain to repair defects in yeast caused by Ndt80 inactivation (Fingerman et al., 2004). A different view argues that Myrf and its orthologs are wrongly annotated as homologous to yeast Ndt80 because they have no genuine nuclear localization signal (NLS), illustrated by the fact that its ortholog in C. elegans, pqn-47 (prion-like glutamine/asparagine rich protein 47), is not localized in the nucleus at any developmental stage (Russel et al., 2011). Nevertheless, a DNA motif (CTGGYAC, Y = C or T) that is different from the MSE sequence has been identified and proven to be able to bind to murine Myrf, leading to activation of myelin gene expression (Bujalka et al., 2013). In addition, MrfA, the Myrf ortholog in Dictyostelium (slime mould), can bind to a 39-mer CA-rich motif (Senoo et al., 2012).
Unlike other membrane-bound TFs that require the ubiquitin/proteasome-dependent system or regulated intramembrane proteolysis for cleavage (Hoppe et al., 2000; Wang et al., 1994), Myrf can self-process its proteolytic activation through the ICA domain. To date, only two vertebrate membrane-bound TFs - Myrf and its paralog Myrfl (myelin regulatory factor-like) – have proven to have self-cleavage ability. The ICA domain of Myrf harks back to endosialidases, the tailspike proteins of bacteriophages, which are essential for bacteriophages to infect bacteria (Stummeyer et al., 2005). The ICA domain causes the tailspike protein to form trimers, triggering auto-proteolytic cleavage and ending in the release of the mature protein (Schulz et al., 2010). During phage infection of bacteria, the tailspike trimers first attach to the surface of the host, and then undergo ICA domain-mediated self-cleavage, producing an active endosialidase to break down the host’s cell wall (Schwarzer et al., 2007). In the endoplasmic reticulum (ER) of mouse oligodendrocytes, membrane-bound Myrf also forms trimers via a leucine zipper in the ICA domain; after self-cleavage, the detached N-terminal trimer of Myrf containing the DNA-binding domain translocates from ER into the nucleus to execute transcriptional regulation (Bujalka et al., 2013; Li et al., 2013). Similarly, the MrfA of Dictyostelium is first affixed to the ER membrane in a trimer fashion via the C-terminal transmembrane and ICA domains, then undergoes constitutive self-cleavage, releasing the N-terminal fragment, which remains in the cytoplasm of growing cells but accumulates in the nuclei of anterior-like cells and pre-stalk cells (Senoo et al., 2012).
Apart from the activities of DNA binding and ICA domains, the C-terminus of Myrf is thought to be involved in cellular secretion, which is supported by the observation that all known Myrf orthologs are initially located in the ER membrane and by the finding that pqn-47 of Nematodes is required for regulated secretion of cuticle components or hormones during larval molting cycles (Russel et al., 2011). Taking into account the fact that, during myelination, oligodendrocytes transport huge amounts of myelin component proteins, cholesterol and membrane lipids through the secretory pathway (Anitei and Pfeiffer, 2006), it is possible that Myrf and pqn-47 might play a similar secretory role despite being phylogenetically distant. In vitro assays with oligodendrocyte cell lines show that Myrf’s N-terminal transcription factor fragment is not sufficient to promote oligodendrocyte maturation compared to full-length Myrf, suggesting that the C-terminus of Myrf is also important to oligodendrocyte maturation, possibly by controlling the ER secretory pathway (Li et al., 2013). Interestingly, Myrf can bind to Sox10, not through its transcriptional domain but through the membrane bound C-terminal domain (Hornig et al., 2013), although it is still unclear whether this phenomenon is peculiar to Myrf and Sox10 or whether it underlies a common mechanism for the secretion of myelin related proteins.
Given the fact that yeast Ndt80 has no ICA domain and that those ICA domain-containing proteins in phages and prokaryotes such as Bdellovibrio bacteriovorus have no Ndt80 domain, while protists including choanoflagellates and Dictyostelium have Myrf orthologs containing both an ICA domain and an Ndt80-like DNA binding domain, a likely scenario for the emergence of Myrf's ancestor during evolution is that an invasion event of a phage or bacterium into an early protist resulted in the phage/bacterial DNA integrating into the protist genome, producing a fusion protein composed of an ICA domain and a DNA binding domain (Roberts, 2013). In addition, most Metazoa possess two Myrf homologs, hinting at early genome duplication in eukaryotes. Our Myrf phylogenetic tree illustrates that two Myrf homologs evolved simultaneously in invertebrates and that subsequently, in vertebrates, the two Myrf homologs diverged to form two separate gene branches, Myrf and Myrfl (Fig. 2B). According to a mouse CNS RNA-seq database, Myrfl is not expressed in the CNS (Zhang et al., 2014). It is curious that no extra Myrf homologs have appeared in vertebrates - considering the two-round genome duplication – however, they might first have been generated then deleted in the following massive gene loss. As with Olig genes, one Myrf homolog can be found in the Japanese lamprey testis genome, but no Myrf homolog can be retrieved at the moment in the database of sea lamprey genome derived from somatic tissue. Myrf might have gained the function of regulating myelin gene expression in early vertebrates; it would be worth looking into how Myrf functions in fish oligodendrocytes and whether invertebrate or mammalian Myrfl orthologs (full-length, N-terminal, or C-terminal) can rescue a null mutation of Myrf in fish.
Overview
The invention of myelin was a major event that changed the course of evolution. Based on developmental clues, we had previously hypothesized that myelin forming cells evolved from so-called “motor glia” that had the same developmental origin as motor neurons (Richardson et al., 1997; Li and Richardson, 2008). A new gene regulatory network was required to shape the motor glia into today’s oligodendrocytes; in this regard, cis-regulatory elements were integrated into a group of genes encoding myelin-forming proteins, while transcription factors simultaneously adopted new functions by undergoing post-transcriptional modification to permit interactions with novel transcriptional co-factors and cis-regulatory elements. This would have led to the expression of myelin genes in a spatially and temporally controlled manner. As jawless fish and cartilaginous fish occupy key nodes in myelin evolution, future research on myelin evolution would benefit from studying the expression patterns and molecular functions of master myelin regulators Olig1/2, Sox10 and Myrf in the lamprey and hagfish as well as in cartilaginous fish such as dogfish, elephant shark or little skate.
Figure 3.
Phylogeny of Myrf. (A) Structure of murine Myrf protein (GenBank: XP_006526992.1). DBD: DNA binding domain; ICA, intramolecular chaperone auto-processing; TMD: transmembrane domain; SBD, Sox10 binding domain. (B) Myrf phylogenetic tree. Myrf homolog/ortholog sequences were downloaded from NCBI (www.ncbi.nlm.nih.gov) and Ensembl (www.ensembl.org). MEGA6/ClustalW was used to draw the rooted phylogenetic tree (http://www.megasoftware.net/mega.php). Myrf orthologs might result from an invasion event of a phage or bacterium into an early protist resulting in the phage/bacterial DNA (encoding ICA domain) integrating into the protist genome (encoding DBD/Ndt80p domain). In invertebrates, two Myrf homologs evolved simultaneously; in vertebrates, these two Myrf homologs diverged to form Myrf and Myrfl branches. Hs: Homo sapiens; Mm: Mus musculus; Rn: Rattus norvegicus (brown rat); Gg: Gallus gallus (chicken); Ac: Anolis carolinensis (Carolina anole, a lizard); Xt: Xenopus tropicalis (clawed frog); Dr: Danio rerio (zebrafish); Cm: Callorhinchus milii (elephant shark or chimera); Ci: Ciona intestinalis (sea squirt, an ascidian) Cf: Camponotus floridanus (Florida carpenter ant); Sc: Saccharomyces cerevisiae (brewers' yeast); Ce: Caenorhabditis elegans (a nematode worm); Df: Dictyostelium fasciculatum (slime mould); Mb: Monosiga brevicollis (a choanoflagellate, close relative of metazoans); Bb: Bdellovibrio bacteriovorus (a motile gram-negative bacterium that invades and parasitizes other bacteria); CWSAFP: cell wall surface anchor family protein; DTFP: distal tail fibre protein.
Acknowledgments
Work in the authors’ laboratories was supported by the UK Biotechnology and Biological Sciences Research Council (BB/J006602/1 and BB/L003236/1), the Medical Research Council (G0800575), the Wellcome Trust (WT100269MA) and the European Research Council (ERC “Ideas” Programme 293544).
References
- Anitei M, Pfeiffer SE. Myelin biogenesis: sorting out protein trafficking. Curr Biol. 2006;16:R418–R421. doi: 10.1016/j.cub.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramblett DE, Copeland NG, Jenkins NA, Tsai MJ. BHLHB4 is a bHLH transcriptional regulator in pancreas and brain that marks the dimesencephalic boundary. Genomics. 2002;79:402–412. doi: 10.1006/geno.2002.6708. [DOI] [PubMed] [Google Scholar]
- Bramblett DE, Pennesi ME, Wu SM, Tsai MJ. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004;43:779–793. doi: 10.1016/j.neuron.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Brunelli S, Innocenzi A, Cossu G. Bhlhb5 is expressed in the CNS and sensory organs during mouse embryonic development. Gene Expr Patterns. 2003;3:755–759. doi: 10.1016/s1567-133x(03)00135-2. [DOI] [PubMed] [Google Scholar]
- Bujalka H, Koenning M, Jackson S, Perreau VM, Pope B, Hay CM, Mitew S, Hill AF, Lu QR, Wegner M, Srinivasan R, et al. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol. 2013;11:e1001625. doi: 10.1371/journal.pbio.1001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock TH, Moore JK, Fields RD. Evolution of myelin sheaths: both lamprey and hagfish lack myelin. Neurosci Lett. 1984;48:145–8. doi: 10.1016/0304-3940(84)90010-7. [DOI] [PubMed] [Google Scholar]
- Cheatle Jarvela AM, Hinman VF. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evodevo. 2015;6:3. doi: 10.1186/2041-9139-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JA, Huang YP, Mazzoni EO, Tan GC, Zavadil J, Wichterle H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69:721–735. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Dai J, Bercury KK, Ahrendsen JT, Macklin WB. Olig1 function is required for oligodendrocyte differentiation in the mouse brain. J Neurosci. 2015;35:4386–4402. doi: 10.1523/JNEUROSCI.4962-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Lu QR. Transcriptional and Epigenetic Regulation of Oligodendrocyte Development and Myelination in the Central Nervous System. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton CM, Johnson CM. Modulation of dopamine-dependent behaviors by the Caenorhabditis elegans Olig homolog HLH-17. J Neurosci Res. 2011;89:1627–1636. doi: 10.1002/jnr.22694. [DOI] [PubMed] [Google Scholar]
- Feng L, Xie X, Joshi PS, Yang Z, Shibasaki K, Chow RL, Gan L. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006;133:4815–4825. doi: 10.1242/dev.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman IM, Sutphen K, Montano SP, Georgiadis MM, Vershon AK. Characterization of critical interactions between Ndt80 and MSE DNA defining a novel family of Ig-fold transcription factors. Nucleic Acids Res. 2004;32:2947–2956. doi: 10.1093/nar/gkh625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Cai J, Clevers H, Fast E, Gray S, Greenberg R, Jain MK, Ma Q, Qiu M, Rowitch DH, Taylor CM, et al. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J Neurosci. 2009;29:11399–11408. doi: 10.1523/JNEUROSCI.0160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusho M, Ono K, Takebayashi H, Masahira N, Kagawa T, Ikeda K, Ikenaka K. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev Biol. 2006;293:348–357. doi: 10.1016/j.ydbio.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Goto Y, Kubota S, Kohno S. Highly repetitive DNA sequences that are restricted to the germ line in the hagfish Eptatretus cirrhatus: a mosaic of eliminated elements. Chromosoma. 1998;107:17–32. doi: 10.1007/s004120050278. [DOI] [PubMed] [Google Scholar]
- Gould RM, Morrison HG, Gilland E, Campbell RK. Myelin tetraspan family proteins but no non-tetraspan family proteins are present in the ascidian (Ciona intestinalis) genome. Biol Bull. 2005;209:49–66. doi: 10.2307/3593141. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Hornig J, Frob F, Vogl MR, Hermans-Borgmeyer I, Tamm ER, Wegner M. The transcription factors Sox10 and Myrf define an essential regulatory network module in differentiating oligodendrocytes. PLoS Genet. 2013;9:e1003907. doi: 10.1371/journal.pgen.1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhao XF, Zheng K, Qiu M. Regulation of the timing of oligodendrocyte differentiation: mechanisms and perspectives. Neurosci Bull. 2013;29:155–64. doi: 10.1007/s12264-013-1314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Genome size reduction in the chicken has involved massive loss of ancestral protein-coding genes. Mol Biol Evol. 2008;25:2681–2688. doi: 10.1093/molbev/msn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huillard E, Ziercher L, Blond O, Wong M, Deloulme JC, Souchelnytskyi S, Baudier J, Cochet C, Buchou T. Disruption of CK2beta in embryonic neural stem cells compromises proliferation and oligodendrogenesis in the mouse telencephalon. Mol Cell Biol. 2010;30:2737–2749. doi: 10.1128/MCB.01566-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, Emery B. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci. 2012;32:12528–12542. doi: 10.1523/JNEUROSCI.1069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S, Ishibashi T, Kohno S. A germline restricted, highly repetitive DNA sequence in Paramyxine atami: an interspecifically conserved, but somatically eliminated, element. Mol Gen Genet. 1997;256:252–256. doi: 10.1007/s004380050567. [DOI] [PubMed] [Google Scholar]
- Kubota S, Takano J, Tsuneishi R, Kobayakawa S, Fujikawa N, Nabeyama M, Kohno S. Highly repetitive DNA families restricted to germ cells in a Japanese hagfish (Eptatretus burgeri): a hierarchical and mosaic structure in eliminated chromosomes. Genetica. 2001;111:319–328. doi: 10.1023/a:1013751600787. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Richardson WD. The evolution of Olig genes and their roles in myelination. Neuron Glia Biol. 2008;4:129–135. doi: 10.1017/S1740925X09990251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009;19:479–485. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, de Faria JP, Andrew P, Nitarska J, Richardson WD. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron. 2011;69:918–929. doi: 10.1016/j.neuron.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Park Y, Marcotte EM. A Bacteriophage tailspike domain promotes self-cleavage of a human membrane-bound transcription factor, the myelin regulatory factor MYRF. PLoS Biol. 2013;11:e1001624. doi: 10.1371/journal.pbio.1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li H, Hu X, Yu L, Liu H, Han R, Colella R, Mower GD, Chen Y, Qiu M. Control of precerebellar neuron development by Olig3 bHLH transcription factor. J Neurosci. 2008;28:10124–10133. doi: 10.1523/JNEUROSCI.3769-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hu X, Huang C, Zheng K, Takebayashi H, Cao C, Qiu M. Olig3 is not involved in the ventral patterning of spinal cord. PLoS One. 2014;9:e111076. doi: 10.1371/journal.pone.0111076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis A, Roest CH, Robinson-Rechavi M. How much does the amphioxus genome represent the ancestor of chordates? Brief Funct Genomics. 2012;11:89–95. doi: 10.1093/bfgp/els003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CJ, Terasaki M, Wu M, Freeman RM, Jr, Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C, Smith M, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog--regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, May G, Wagner GP. Regulatory evolution through divergence of a phosphoswitch in the transcription factor CEBPB. Nature. 2011;480:383–386. doi: 10.1038/nature10595. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Pfaff SL. Cracking the transcriptional code for cell specification in the neural tube. Cell. 2001;106:651–654. doi: 10.1016/s0092-8674(01)00499-8. [DOI] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li H, de Faria JP, Emery B, Tohyama K, Richardson WD. Motor skill learning requires active central myelination. Science. 2014;346:318–322. doi: 10.1126/science.1254960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMiller TL, Johnson CM. Molecular characterization of HLH-17, a C. elegans bHLH protein required for normal larval development. Gene. 2005;356:1–10. doi: 10.1016/j.gene.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Wang H, Liu S, Niu J, Wang L, He Y, Etxeberria A, Chan JR, Xiao L. Stage-specific deletion of Olig2 conveys opposing functions on differentiation and maturation of oligodendrocytes. J Neurosci. 2013;33:8454–8462. doi: 10.1523/JNEUROSCI.2453-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer DH, Sun Y, Liu T, Kane MF, Alberta JA, Adelmant G, Kupp R, Marto JA, Rowitch DH, Nakatani Y, Stiles CD, et al. An amino terminal phosphorylation motif regulates intranuclear compartmentalization of Olig2 in neural progenitor cells. J Neurosci. 2014;34:8507–8518. doi: 10.1523/JNEUROSCI.0309-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano SP, Cote ML, Fingerman I, Pierce M, Vershon AK, Georgiadis MM. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc Natl Acad Sci USA. 2002;99:14041–14046. doi: 10.1073/pnas.222312199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B, Atchley WR. Evolution of bHLH transcription factors: modular evolution by domain shuffling? Mol Biol Evol. 1999;16:1654–1663. doi: 10.1093/oxfordjournals.molbev.a026079. [DOI] [PubMed] [Google Scholar]
- Muller T, Anlag K, Wildner H, Britsch S, Treier M, Birchmeier C. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Dev. 2005;19:733–743. doi: 10.1101/gad.326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ono K, Takebayashi H, Ikeda K, Furusho M, Nishizawa T, Watanabe K, Ikenaka K. Regional- and temporal-dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Dev Biol. 2008;320:456–468. doi: 10.1016/j.ydbio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Oyallon J, Apitz H, Miguel-Aliaga I, Timofeev K, Ferreira L, Salecker I. Regulation of locomotion and motoneuron trajectory selection and targeting by the Drosophila homolog of Olig family transcription factors. Dev Biol. 2012;369:261–276. doi: 10.1016/j.ydbio.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paes de Faria J, Kessaris N, Andrew P, Richardson WD, Li H. New Olig1 null mice confirm a non-essential role for Olig1 in oligodendrocyte development. BMC Neurosci. 2014;15:12. doi: 10.1186/1471-2202-15-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: nature's escape from genetic imprisonment and the basis for dynamic information encoding. Interdiscip Rev Syst Biol Med. 2012;4:565–583. doi: 10.1002/wsbm.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Pons C, Diallo A, Mori A, Shrestha S, Kadreppa S, Nelson J, Diprima S, Dricot A, Lajoie BR, Ribeiro PS, et al. Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol Cell. 2013;51:116–127. doi: 10.1016/j.molcel.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Pringle NP, Yu WP, Hall AC. Origins of spinal cord oligodendrocytes: possible developmental and evolutionary relationships with motor neurons. Dev Neurosci. 1997;19:58–68. doi: 10.1159/000111186. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RG. Myelination borrows a trick from phage. PLoS Biol. 2013;11:e1001626. doi: 10.1371/journal.pbio.1001626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Mardinly AR, McCord AE, Zurawski J, Cohen S, Jung C, Hu L, Mok SI, Shah A, Savner EM, Tolias C, et al. Loss of inhibitory interneurons in the dorsal spinal cord and elevated itch in Bhlhb5 mutant mice. Neuron. 2010;65:886–898. doi: 10.1016/j.neuron.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel S, Frand AR, Ruvkun G. Regulation of the C. elegans molt by pqn-47. Dev Biol. 2011;360:297–309. doi: 10.1016/j.ydbio.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Burke GM, McGuire T, Pisarek AJ, Mukhopadhyay A, Mishina Y, Kessler JA. BMPR1a signaling determines numbers of oligodendrocytes and calbindin-expressing interneurons in the cortex. J Neurosci. 2007;27:7397–407. doi: 10.1523/JNEUROSCI.1434-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EC, Schwarzer D, Frank M, Stummeyer K, Muhlenhoff M, Dickmanns A, Gerardy-Schahn R, Ficner R. Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF. J Mol Biol. 2010;397:341–351. doi: 10.1016/j.jmb.2010.01.028. [DOI] [PubMed] [Google Scholar]
- Schwarzer D, Stummeyer K, Gerardy-Schahn R, Muhlenhoff M. Characterization of a novel intramolecular chaperone domain conserved in endosialidases and other bacteriophage tail spike and fiber proteins. J Biol Chem. 2007;282:2821–2831. doi: 10.1074/jbc.M609543200. [DOI] [PubMed] [Google Scholar]
- Senoo H, Wang HY, Araki T, Williams JG, Fukuzawa M. An orthologue of the Myelin-gene Regulatory Transcription Factor regulates Dictyostelium prestalk differentiation. Int J Dev Biol. 2012;56:325–332. doi: 10.1387/ijdb.120030jw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi T, Kondo T. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol. 2004;166:963–968. doi: 10.1083/jcb.200404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbereis JC, Nobuta H, Tsai HH, Heine VM, McKinsey GL, Meijer DH, Howard MA, Petryniak MA, Potter GB, Alberta JA, Baraban SC, et al. Olig1 function is required to repress dlx1/2 and interneuron production in Mammalian brain. Neuron. 2014;81:574–587. doi: 10.1016/j.neuron.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci USA. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Baker C, Eichler EE, Amemiya CT. Genetic consequences of programmed genome rearrangement. Curr Biol. 2012;22:1524–1529. doi: 10.1016/j.cub.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–422. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm R, Cholewa-Waclaw J, Reuter K, Brohl D, Sieber M, Treier M, Muller T, Birchmeier C. The bHLH transcription factor Olig3 marks the dorsal neuroepithelium of the hindbrain and is essential for the development of brainstem nuclei. Development. 2009;136:295–305. doi: 10.1242/dev.027193. [DOI] [PubMed] [Google Scholar]
- Stummeyer K, Dickmanns A, Muhlenhoff M, Gerardy-Schahn R, Ficner R. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat Struct Mol Biol. 2005;12:90–96. doi: 10.1038/nsmb874. [DOI] [PubMed] [Google Scholar]
- Sun Y, Meijer DH, Alberta JA, Mehta S, Kane MF, Tien AC, Fu H, Petryniak MA, Potter GB, Liu Z, Powers JF, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906–917. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarski H. Cell size and nuclear DNA content in vertebrates. Int Rev Cytol. 1976;44:93–111. doi: 10.1016/s0074-7696(08)61648-4. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Tekki-Kessaris N, Woodruff R, Hall AC, Gaffield W, Kimura S, Stiles CD, Rowitch DH, Richardson WD. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. doi: 10.1242/dev.128.13.2545. [DOI] [PubMed] [Google Scholar]
- Tiersch TR, Wachtel SS. On the evolution of genome size of birds. J Hered. 1991;82:363–368. doi: 10.1093/oxfordjournals.jhered.a111105. [DOI] [PubMed] [Google Scholar]
- Tripathi RB, Clarke LE, Burzomato V, KessRis N, Anderson PN, Attwell D, Richardson WD. Dorsally and ventally derived oligodendrocytes have similar electrical properties but myelinate preferred tracts. J Neurosci. 2011;31:6809–6819. doi: 10.1523/JNEUROSCI.6474-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Lee AP, Ravi V, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K, Pougach K, Verstrepen KJ. How do regulatory networks evolve and expand throughout evolution? Curr Opin Biotechnol. 2015;34:180–188. doi: 10.1016/j.copbio.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Werner HB. Do we have to reconsider the evolutionary emergence of myelin? Front Cell Neurosci. 2013;7:217. doi: 10.3389/fncel.2013.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman LM, Blanc CA, Schaumburg CS, Rowitch DH, Lane TE. Olig1 function is required for remyelination potential of transplanted neural progenitor cells in a model of viral-induced demyelination. Exp Neurol. 2012;235:380–387. doi: 10.1016/j.expneurol.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff RH, Tekki-Kessaris N, Stiles CD, Rowitch DH, Richardson WD. Oligodendrocyte development in the spinal cord and telencephalon: common themes and new perspectives. Int J Dev Neurosci. 2001;19:379–385. doi: 10.1016/s0736-5748(00)00083-6. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T, Xian K, Hurlock E, Xin M, Kernie SG, Parada LF, Lu QR. A critical role for dorsal progenitors in cortical myelination. J Neurosci. 2006;26:1275–1280. doi: 10.1523/JNEUROSCI.4717-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zuo H, Maher BJ, Serwanski DR, LoTurco JJ, Lu QR, Nishiyama A. Olig2-dependent developmental fate switch of NG2 cells. Development. 2012;139:2299–2307. doi: 10.1242/dev.078873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Barres BA. Intrinsic and extrinsic control of oligodendrocyte development. Curr Opin Neurobiol. 2013;23:914–920. doi: 10.1016/j.conb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]