Abstract
All aspects of transcription and its regulation involve dynamic events. The basal transcription machinery and regulatory components are dynamically recruited to their target genes, and dynamic interactions of transcription factors with chromatin—and with each other—play a key role in RNA polymerase assembly, initiation, and elongation. These short-term binding dynamics of transcription factors are superimposed by long-term cyclical behavior of chromatin opening and transcription factor-binding events. Its dynamic nature is not only a fundamental property of the transcription machinery, but it is emerging as an important modulator of physiological processes, particularly in differentiation and development.
Introduction
Transcription is the key step in the regulation of gene expression. The transcription process involves several distinct steps. The core promoter structure contains sequences that serve to anchor a set of protein complexes generally referred to as the general transcription factors (Smale and Kadonaga, 2003; Kornberg, 2005). These factors lead to the formation of a preinitiation complex, which can be quite stable when assembled in vitro. RNA polymerases recognize in turn these multimeric complexes and initiate the synthesis of RNA. The rate of initiation is generally considered the primary regulatory step of transcription, but alternative processes are now known to modulate the accumulation of transcripts. Enhancers are positioned at distant sites in chromatin and modify the rate of initiation complex formation by mechanisms that are still poorly understood (Roeder, 2005). The elongating polymerase complex can be transiently retarded, leading to a “paused” complex that may be subject to reactivation (Price, 2008). The elongation complex brings with it several activities that modulate its postinitiation activity (Shilatifard et al., 2003; Price, 2008). Furthermore, during RNA chain extension, the elongating complex must pass through nucleosome structures, and activities are specifically recruited that allow for their transient disassembly and reformation (Carrozza et al., 2005). Complex histone modifications also take place during elongation (Guenther et al., 2007), and appear to interact with the extending polymerase, affecting its progress. Lastly, the termination and polyadenylation of transcripts represents a final step in the generation of the primary transcript (Rosonina et al., 2006).
The process of transcription is intrinsically dynamic. Yet, most of what we know about how the transcription machinery is assembled, how it initiates, and how it elongates along a gene comes from mostly static biochemical investigations. While these methods have been invaluable in defining the key factors involved in the transcription process and their interactions, they are not ideally suited to gain insight into the real-time kinetics of transcription. The reliance on purification approaches and in vitro reconstitution also raises the question of how accurately findings using these methods reflect the complex environment in which transcription takes place in an intact living cell. The application of recently developed cell biological methods, mostly based on in vivo imaging, to the study of transcription in its natural context and in real-time has overcome some of these limitations (Misteli, 2001; Darzacq et al., 2009). These new methods are now providing first insights into how transcription occurs in a live cell nucleus.
How Transcription Factors Find Their Targets: 3D Genome Scanning
The basis of all transcriptional activity and regulation is the recruitment of transcription complexes to target genes. The basal transcription machinery associates with well-defined binding sites in promoter regions, and regulatory factors bind to specific sites in control elements in the vicinity and, at times, at long distances away, from target genes. Specific binding sites for both the basal machinery, as well as gene-specific regulators, are exceedingly sparse in the genome compared to the number of nonspecific binding sites with which a given transcription factor (TF) may interact. Conservatively, assuming an average mammalian core promoter size of ~150 nt, promoter regions make up less than 0.1% of the human genome, and many TFs have only a few specific binding sites in the genome. How then do TFs find, often rapidly and in response to tightly controlled physiological signaling cascades, their few specific binding sites in the vast sea of nontarget sites in the genome? The key to efficient recruitment of the transcription machinery to its target site are two fundamental dynamic properties of TFs: their ability to rapidly diffuse through the nucleus and their propensity to very transiently bind to chromatin.
Diffusion is the prime means by which TFs move through the nucleus (Misteli, 2001; Gorski et al., 2006). FRAP (fluorescence recovery after photobleaching) experiments in which the motion of a fluorescently tagged TF is traced in living cells have revealed that most TFs move rapidly within the nucleus (Phair et al., 2004; Sprague et al., 2004; Hoogstraten et al., 2002; Stenoien et al., 2001). TF motion is not directional and does not require energy. Measured diffusion coefficients for TFs range from ~0.5 to 5 μm2s‒1 depending on a molecule’s shape, size, and its interactions with chromatin (Gorski et al., 2006). To put this into perspective, this diffusion behavior allows a molecule to traverse the entire length of a typical mammalian nucleus in a few seconds and makes it possible for a single molecule of a TF to visit the volume of the nucleus in a matter of minutes. The high mobility of TFs is the basis for their ability to, rapidly and without the cell’s expenditure of energy, find their sparse specific binding sites in the genome.
The second property that critically contributes to efficient TF targeting is the highly transient and dynamic nature of their binding to chromatin in vivo. In photobleaching experiments, the apparent diffusion coefficients of most TFs have been found to be typically 10- to 100-fold lower than would be expected based on their size and shape alone (Sprague et al., 2004; Mueller et al., 2008). While some of this retarded mobility may be due to their integration into larger complexes, the major contributor to slowing down overall TF motion in the nucleus is their binding to chromatin. This concept is most clearly illustrated by the linker histone H1. A wild-type H1 molecule that has full DNA-binding activity has a diffusion coefficient of <0.1 μm2s-1; however, the same molecule containing several point mutations that eliminate its ability to bind DNA, has a diffusion coefficient of ~20 μm2s-1, similar to that of a protein which has no DNA-binding activity. The diffusion kinetics of a chromatin protein is now recognized as a direct measure of its binding properties to DNA in vivo (Sprague et al., 2004; Mueller et al., 2008). These quantitative in vivo approaches have indicated that the vast majority of chromatin proteins, including architectural proteins such as linker histones, heterochromatin protein 1, and HMG proteins, but particularly transcription cofactors and regulators, bind chromatin transiently with rapid turnover kinetics, typically on the order of seconds (Phair et al., 2004; Gorski et al., 2006).
The rapid motion of TFs through nuclear space and the very transient interactions of TF with chromatin along the way point toward a 3D scanning model for how TFs find their specific target-binding sites in the genome (Misteli, 2001). As TFs diffuse thought the nucleus, they frequently encounter and physically interact with chromatin fibers along the way (Figure 1). Given the scarcity of specific binding sites, most often these encounters will be with off-target sites and do not result in any functional interactions. After short immobilization, the TF dissociates and continue its random walk through the nucleus until it encounters another chromatin fiber, where it undergoes another interaction. Estimates for the time between such encounters are typically on the order of 50–250 ms. This 3D hopping will continue until the molecule finds a specific target site where it binds, most likely for a somewhat longer period of time, and elicits a functional response (Figure 1). An important implication and prediction from this model is that the vast majority of molecules of any given TF at any time are bound to chromatin, albeit at nontarget sites. This prediction is confirmed by analysis of FRAP studies demonstrating that for most TFs large fractions of molecules are chromatin bound (Phair et al., 2004). Furthermore, single-molecule observations on the lac-repressor in living bacteria indicate that a single lac repressor molecule spends most of its time bound to chromatin although not associated with its specific target site (Elf et al., 2007).
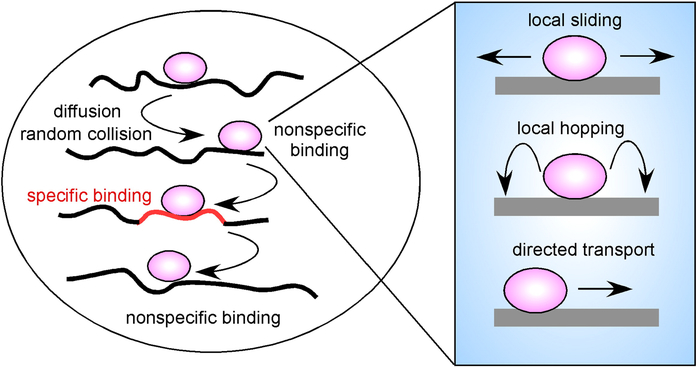
Figure 1. TFs Find Their Specific Binding Sites by Random Scanning of the Genome in 3D.
A TF (purple) diffuses through the nuclear space and by random collision associates with chromatin. Most encounters are at nontarget sites resulting in highly transient interactions. Occasionally, a specific binding site (orange) is encountered, and prolonged binding occurs. At each encounter a TF might undergo local motion on the chromatin fiber by either sliding along the DNA, hopping locally or by directed, motor driven motion.
An interesting corollary from a 3D scanning motion of chromatin proteins comes from considering what such random interactions with the chromatin fiber mean functionally for different classes of chromatin proteins. While it seems safe to assume that interactions of TFs with nonspecific sites will not have any functional consequences since other factors required to elicit a transcriptional response are absent, it is less clear that nonspecific interactions of others, such as chromatin modelers or histone-modifying enzymes, do not lead to a functional response. For example, it is possible that a chromatin-remodeling complex remodels the chromatin fiber with which it collides nonspecifically as part of its random walk through the nucleus and, in this way, contributes to the global dynamic nature of higher-order chromatin structure.
But is 3D scanning by hopping sufficient to account for the required efficiency of TF targeting to a specific site? Simple back-of-the-envelope calculations indicate that assuming 50,000 copies of a TF, a molecule will hit a promoter of a particular gene roughly every second. For a factor present in 10,000 copies this frequency is roughly every 10 s, clearly generating enough interactions to establish and maintain transcriptional control at a physiologically relevant scale. Similarly, estimates based on experimental measurements indicate a flux of RNA pol I components at the promoter of its endogenous target genes on the order of several thousand molecules per second (Dundr et al., 2002).
Additional mechanisms for bringing TF to specific sites are probably also at work. Particularly, it seems likely that the global 3D hopping is complemented by local scanning of the DNA (Gorman and Greene, 2008) (Figure 1). In line with this idea is the long-standing observation that in vitro the lac repressor and several restriction endonucleases find their target sequence ~1000-fold faster than expected based on simple diffusion, suggesting the existence of mechanisms for facilitated local target searches (Halford and Marko, 2004). In vitro experiments mostly on restriction endonucleases and DNA-repair enzymes indeed have confirmed the ability of some proteins to undergo one-dimensional diffusion along the DNA fiber (Gowers et al., 2005; Gorman and Greene, 2008). It is thus possible that once a TF associates with the chromatin fiber, it can go into a local search mode in which it scans the fiber in its immediate vicinity. Using estimates for one-dimensional diffusion coefficients from in vitro studies and estimates of dwell times on the order of a few seconds derived from photobleaching experiments, a TF is in theory able to search several hundred basepairs before dissociating again (Halford and Marko, 2004; Gorman and Greene, 2008) (Figure 1). The local search may occur via one-dimensional sliding of the TF along the chromatin fiber, although this might be complicated by the complex higher-order folding of the chromatin fiber and the sterical obstacles generated by the presence of a large number of architectural chromatin proteins that cover the chromatin fiber. It is also possible that a TF might undergo local hopping, directed, motor-driven motion or handover between chromatin segments to explore its immediate neighborhood (Halford and Marko, 2004; Gorman and Greene, 2008). While an attractive idea it needs to be pointed out that at present there is no experimental data to demonstrate such local motion in vivo.
Where Transcription Factors Bind in the Genome
Essentially, all DNA in eukaryotic cells is organized in nucleosomal arrays. This repetitive chromatin fiber is organized in turn into complex higher-order structures that serve to compact the large amount of DNA. As transcription factors traverse the nuclear space, they interact repeatedly with DNA in the chromatin context. As nucleosome octamers sequester much of the DNA from ready access, reorganization of these structures must accompany productive interactions with regulatory elements.
Early studies (Wu et al., 1979) demonstrated that many of the specific sites where transcription factors bind have local chromatin structures that are selectively sensitive to nucleolytic agents, particularly DNase I and micrococcal nuclease. These sites, commonly referred to as DNaseI hypersensitive sites (DHS), represent regions with disrupted nucleosome structures. Recent global studies have mapped a large number of DHS sites throughout eukaryotic genomes (Hesselberth et al., 2009; ENCODE Project Consortium et al., 2007; Boyle et al., 2008), and it is now estimated that approximately 2% of the mammalian genome is found in these localized structures.
Recent studies also indicate that a large fraction of transcription regulatory elements is associated with these DHS sites (John et al., 2008). Thus, the interaction of site-specific DNA-binding proteins with chromatin is almost universally associated with chromatin remodeling. Until recently, these disrupted regions have been viewed as static states. “Sliding” of nucleosomes to alternate positions (Becker, 2002), or disruption and displacement of core structures (Mellor, 2005; Aoyagi et al., 2002), has been considered as a transition between two relatively stable states. In contrast to this view, several recent studies have indicated that the remodeling of nucleosome structures at DHS sites is a continuous and dynamic process (Figure 2). Fletcher et al. (2002) described the surprising eviction of the glucocorticoid receptor (GR) from an array of positioned nucleosomes in vitro, and hypothesized that GR directed remodeling was a continuous, ongoing process. Subsequent studies using UV laser crosslinking (Nagaich et al., 2004), which provide very high temporal resolution, supported this hypothesis, and indicated that remodeling was a continuous, dynamic process. In studies on the well-described yeast Pho5 locus, Kornberg and colleagues also concluded that nucleosome transitions in vivo represent rapid switching between multiple intermediate states (Boeger et al., 2008). In this view, local transitions detected with static techniques that utilize fixed or broken cell preparations in fact represent alterations in the equilibrium distribution of dynamic states, not switching between static positions, or static compositions (Figure 2). Two general points emerge from this work. First, transcription factors in general appear to require local reorganization of nucleosomal structures for productive template interactions. Second, the disruption of nucleosome structures is a dynamic, continuous process.
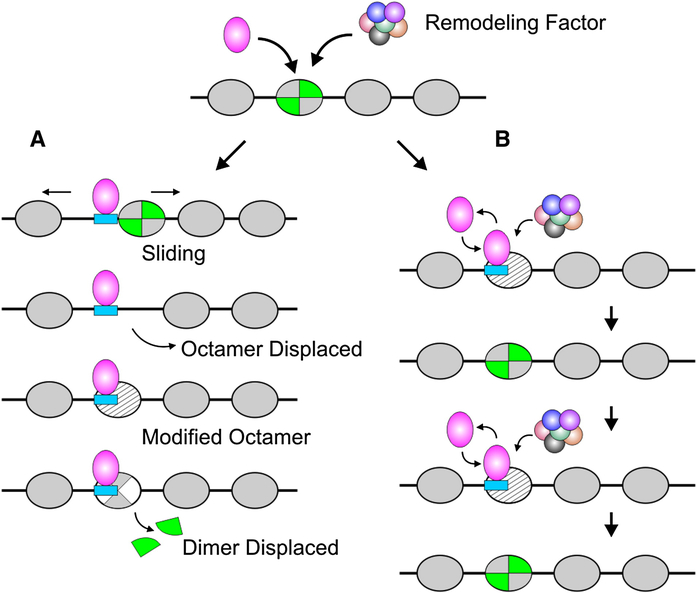
Figure 2. Transcription Factor/Template Interactions during Chromatin Remodeling.
(A) Local nucleosome reorganization, giving rise to enhanced transcription factor access, is commonly discussed in terms of altered static states. Evidence has been advanced to support several specific mechanisms, including sliding to a new position, octamer displacement, modified octamer structure, and partial octamer dissociation. These modified nucleosome states would in turn accommodate factor binding events not compatible with the unaltered state.
(B) A dynamic view of local transitions suggests that remodeling is a continuous process. Remodeling complexes are targeted to specific nucleosomes by a given transcription factor. However, both the remodeling process itself and commensurate binding of a factor are transient events. Constant repetition of this cycle produces a shift in the equilibrium distribution of both the transcription factor and nucleosome components.
Nucleosome remodeling may be directly linked to the dynamic exchange of many TFs from their chromatin template. An analysis of multiple ligand effects on in vivo mobility of the progesterone receptor (PR) revealed a dramatic parallel between effects on chromatin remodeling in vitro and receptor/template exchange in vivo (Rayasam et al., 2005). Receptor/ligand complexes that are incapable of supporting remodeling were observed to move rapidly in vivo, whereas complexes that induce efficient nucleosome remodeling were found to have a slower exchange rate. These findings were argued to support a model wherein the receptor is transiently retarded during a productive remodeling event at a regulatory uncomplicated DHS site, whereas receptors nonfunctional for remodeling fail to interact productively and thus escape the retardation process.
Another general feature of regulatory transcription-factor binding sites concerns their general distribution throughout the genome. We now realize that a large majority of response elements are located at considerable distances from target promoters. For example, more than 90% of estrogen receptor binding sites are found at positions greater than 5 kb from any promoter (Carroll et al., 2005). A number of emerging studies show that interacting elements for many transcription factors can be observed at distances of 200 kb or greater (Hakim et al., 2009; Gondor et al., 2008; Simonis et al., 2006; Horike et al., 2005; Wang et al., 2005; Tolhuis et al., 2002; Vakoc et al., 2005). Thus, long-range interactions between regulatory sites and promoters seem to be a common mechanism in eukaryotes. Given the rapid interactions of factors with chromatin in living cells, one is left with a conundrum. How can interactions at such great distance be established and maintained if key regulatory proteins exchange rapidly with the template? Several ad hoc mechanisms can be proposed. Anchoring proteins, such as CTCF, which is frequently found at domain boundaries (Splinter et al., 2006; Wendt et al., 2008), or cohesins (Parelho et al., 2008), may provide stability for long-range interactions. Alternatively, other architectural elements may exist to maintain interacting elements in local proximity. The concept of a “transcription factory” (Cook, 1999) suggests a local domain with essential components of the transcription apparatus tethered at subnuclear sites. These structures could form aggregation sites for elevated concentrations of transcription factors, and factortemplate interactions in these zones could be highly transient, as observed in FRAP studies. Indeed, the work of Fraser and colleagues (Osborne et al., 2004) supports the dynamic nature of these structures.
Dynamics of Transcription Factor Binding
Once a TF finds an accessible promoter, how stably does it bind there? Decades of in vitro measurements had led to the dogma that TF interactions with their promoter targets are stable, persisting on a time-scale of hours—for example, an estimated 108 min half-life on DNA for the glucocorticoid receptor (Perlmann et al., 1990). Thus, it was a surprise to find that the binding of a sequence-specific TF, the glucocorticoid receptor (GR), to a tandem gene array containing a GR-specific promoter was highly transient when analyzed by FRAP (McNally et al., 2000). This initial finding has been confirmed and extended by similar studies of several other sequence-specific TFs on the same or other tandem arrays, including binding of the progesterone receptor to the MMTV array (Rayasam et al., 2005), binding of NF-kB to an array of its cognate binding sites (Bosisio et al., 2006) and of estrogen receptor on a small prolactin array (Sharp et al., 2006). However, there was concern that because these arrays were artificial, normal sequence-specific TF binding might be compromised. Recently transient interactions have been confirmed on an entirely endogenous promoter of the CUP1 gene that is part of a very small natural gene array (Karpova et al., 2008). Since at least 80% of the genes in this natural array produce transcripts, this system has also countered the argument applied to other tandem arrays that only a small fraction of promoters might be active, and therefore stable binding at these minority promoters was obscured by the transient binding at the majority of other inactive promoters in the array. Similarly, in mammalian cells, several RNA pol I components dynamically exchange at the endogenous rDNA genes (Dundr et al., 2002). Based on the current body of data, it now seems clear that many sequence-specific TFs transiently bind at transcriptionally active promoters.
How transient is this binding? Quantitative FRAP analysis has generated in vivo estimates of TF residence times on chromatin that range from a few milliseconds to ~100 s (Sprague et al., 2004; Farla et al., 2004; Phair et al., 2004; Hinow et al., 2006). Recent work, however, has demonstrated that these estimates can change by several orders of magnitude depending on details in the FRAP model (Mueller et al., 2008). An underlying difficulty is that residence times are not always proportional to the FRAP recovery time. For example, a molecule that undergoes 100 binding events as it moves from the bleach spot periphery to its center will have a recovery time that is roughly 100× longer than its residence time. Thus, residence times can only be obtained with an accurate mathematical model for FRAP, and the development of such models is still an area of active research. In sum, TF residence times on chromatin must be less than or equal to the total FRAP recovery time (typically 30–120 s for TFs), although they may well be much shorter than this.
Transient binding at promoter target sites is not limited to sequence-specific TFs, but is also observed for other promoter-associated factors. These include the glucocorticoid coactivator GRIP1 and the two chromatin-remodeling factors, Brahma (BRM) and Brahma related gene 1 (BRG1), all of which show rapid exchange on the MMTV tandem array (Becker et al., 2002; Johnson et al., 2008). Interestingly, GRIP-1 shows the same kinetics as GR, suggesting that the two molecules could be part of the same complex. In contrast, the two chromatin remodelers exchange somewhat more slowly than GR, suggesting that their kinetics are only partially coupled to those of GR (Johnson et al., 2008).
In addition to the analysis of binding dynamics of sequence-specific TFs at gene arrays, there is a larger body of data on sequence-specific TF binding at random locations within the nucleus. It is generally agreed that these FRAPs contain information on nonspecific site binding, but it is more difficult to ascertain whether these data also contain information about specific-site promoter binding (Sprague et al., 2004; Phair et al., 2004; Hinow et al., 2006; Farla et al., 2004). The interpretation of these data hinges on whether there are a sufficient number of promoter-specific interactions at any randomly selected area within the nucleus in which the photobleaching experiment is carried out. If the promoter-specific interactions account for only a few percent of the interactions, then it is not surprising that recovery kinetics are rapid, as the vast majority of interactions detected are nonspecific and presumably reflect the search process for specific target sites. If, however, promoter-specific interactions account for more than a few percent of the interactions, then they will contribute measurably to the FRAP, implying that many promoter target sites exhibit rapid TF exchange.
While it appears that the majority of TFs analyzed so far interact highly transiently with their chromatin target sites, it is likely that some sequence-specific TFs bind stably to chromatin. Taking advantage of the high concentration of gene target sites in Drosophila polytene chromosome, Lis and coworkers showed that the transcriptional activator HSF is rather stably bound at the hsp70 gene after induction by heat shock (Yao et al., 2006). The possibility of stable binding of some TFs is also implied by competition chromatin-immunoprecipitation experiments on Gal4 in yeast (Nalley et al., 2006) and hypoxia-inducible-factor 1 in human cells (Yu and Kodadek, 2007). It will be interesting to perform competition ChIP and FRAP on the same promoter to ensure that these two approaches yield compatible results. An interesting possibility is that the exchange dynamics of a given factor may differ in various cell types or tissues, thus contributing to gene regulation.
The extensive evidence for transient interactions of site-specific TFs with chromatin has raised the question of why this has not been readily detected in vitro. The reason may be that the in vitro measurements are typically made under nonphysiological conditions, particularly in the absence of various ATP-dependent regulatory factors, such as chromatin remodelers, chaperones, and proteasomes that all appear to regulate TF residence times in vivo. In support of a role for chromatin remodelers in determining residence times, FRAP experiments show that the chromatin remodeler Rsc2 is required for mobilizing the yeast transcription factor Ace1 (Karpova et al., 2004). This likely reflects a specific and direct effect as Rsc2 and Ace1 interact directly as detected by fluorescence resonance energy transfer (FRET) (Karpova et al., 2008), and other remodelers have no effect on Ace1 FRAPs (Karpova et al., 2004). These in vivo observations are also supported by in vitro experiments demonstrating that eviction of GR from promoter templates is promoted by human Swi/Snf (Fletcher et al., 2002; Nagaich et al., 2004).
Chaperones also play key roles in regulating TF binding dynamics. The chaperone hsp90 is found at the MMTV array, and inhibitor studies suggest it may stabilize GR binding to the MMTV promoter (Stavreva et al., 2004). In contrast, addition of seven chaperones to permeabilized and extracted cells restores mobility to ~75% of GR molecules, which are otherwise 100% immobilized after the permeabilization and extraction procedure (Elbi et al., 2004). Consistent with these observations, the chaperone p23 has also been found at promoters and shown to destabilize steroid receptor binding there (Freeman and Yamamoto, 2002).
The proteasome also appears to regulate dynamic exchange of TF’s with chromatin by disassembly of TF complexes at promoters. Inhibition of the proteasome leads to a 5%–10% immobile fraction of GR at the MMTV array (Stavreva et al., 2004). A site-directed mutant of NF-κB that has severely impairs proteasomal degradation leads to a 20% immobile fraction at the tandem array of NF-κB sites (Bosisio et al., 2006). One possible mechanism for these observations has been recently uncovered in yeast, where the 19S regulatory particle of the proteasome has been shown to destabilize interactions of the TF Gal4 with its promoter (Ferdous et al., 2007). This destabilizing activity can be inhibited by monoubiquitylation of Gal4.
Dynamics of Transcription Machinery Assembly
Paralleling the studies of TF dynamics is a body of work that demonstrates that many of the other components of the transcription complex also exhibit dynamic interactions with their promoter templates. The majority of these studies have focused on the dynamics of the polymerase complex. As with TFs, studies of the polymerase fall into two categories: those done at specific tandem array sites, and those done at random locations within the nucleus. The interpretation of the second type of polymerase FRAPs is simpler than for transcription factors, because there are many transcriptionally active sites in a typical nucleus, and so FRAP at a random location is likely to contain a sizable fraction of polymerase molecules that are associated with genes. A complication here, however, is that a reasonable fraction of these pol II molecules may be paused, as suggested by ChIP-seq and global nuclear run on assays (Core et al., 2008; Core and Lis, 2008).
FRAP of RNA pol II has led to a range of estimates for the elongation rate of the polymerase from 0.4 kb/min to 4.3 kb/min (Kimura et al., 2002; Boireau et al., 2007; Yao et al., 2007; Darzacq et al., 2007), while the estimate for RNA pol I is somewhat higher at 5.7 kb/min (Dundr et al., 2002), consistent with the very active transcription of ribosomal genes. Many of these estimates measure RNA pol II residence time on the gene rather than just the elongation time. Different authors have proposed that this measured residence time may incorporate not only elongation, but also other events including initiation (Kimura et al., 2002; Boireau et al., 2007), recycling (Boireau et al., 2007; Yao et al., 2007), or pausing (Darzacq et al., 2007) of the polymerase. A prolonged initiation phase could arise if the polymerase complex is subject to a number of failed assemblies before it finally commits to elongation (Kimura et al., 2002; Dundr et al., 2002). Alternatively, if the same polymerase is recycled to start another transcript, then the apparent residence time measured by FRAP will either be somewhat longer if a few recycling events occur (Boireau et al., 2007), or markedly longer if multiple recycling events occur (Yao et al., 2007). Pausing of the polymerase during elongation would also lead to longer residence times. Singer and colleagues have performed a direct analysis of this by fitting FRAP data for both pol II and an MS-2 tagged mRNA from the same gene array with a model for elongation and pausing (Darzacq et al., 2007). They conclude that in their system the actual rate of elongation is 4.3 kb/min, with pausing producing an apparent elongation rate of 0.4 kb/min.
Two studies have examined not only the largest subunit of the polymerase, but also subunits of the preinitiation complex and investigated how the transcription machinery assembles in vivo (Dundr et al., 2002; Sprouse et al., 2008). In one study, components of the yeast preinitiation complex were analyzed, including TBP, TFIIB, and TAF1 along with the RNA pol II subunit (Rpb1) (Sprouse et al., 2008). In the other study, components of the mammalian RNA pol I complex were analyzed, including upstream binding factors, assembly factors, and initiation factors along with four RNA pol I subunits (Dundr et al., 2002). In both of these studies, the FRAP curves for the different components exhibited different recovery rates. This suggests that the polymerase is not preassembled, since otherwise components that are part of the same complex should have generated identical FRAP curves. This implies that different components of the transcription complex arrive at the promoter at different times and only occasionally form a full-fledged complex. This conclusion is also supported by a quantitative analyses of the FRAP data for the RNA pol I subunits, which predicted that the probability of different components being incorporated into an elongating RNA pol I complex ranged from only 1%–11% (Dundr et al., 2002). Also consistent with this picture are other quantitative analyses of the RNA pol II large subunit FRAP data, which have been interpreted to reflect inefficient assembly of the RNA pol II complex (Kimura et al., 2002; Darzacq et al., 2007). Indeed, quantitative kinetic modeling of the RNA pol II FRAP data yields the estimate that only 1 in 90 polymerases proceeds to elongation (Darzacq et al., 2007). Thus, the current in vivo data support a model in which polymerase and transcription complex assembly occurs by random collision of subunits at the promoter, and is therefore intrinsically inefficient.
In sum, the current view of polymerase complex assembly is that the components assemble stochastically, and that residence times of individual components are regulated to modulate the likelihood of complete assembly (Figure 3). However, it is not known whether a complete transcription complex or even a complete polymerase complex in fact assembles at these promoters. It is possible instead that a progressive series of transient subcomplexes form and disintegrate.
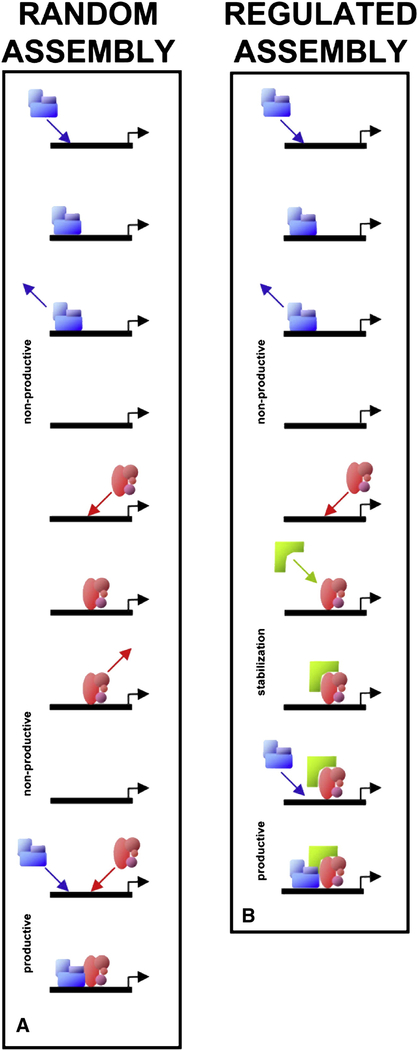
Figure 3. Transcription Complex Assembly.
(A) Two subcomplexes (blue and red) exhibit dynamic exchange at a promoter template (black line). Assembly of the full complex occurs via random collisions with the template, leading sometimes to nonproductive interactions and occasionally to productive interactions (the simultaneous presence of both subcomplexes).
(B) Regulation of dynamic exchange rates can facilitate or inhibit complex formation. Here, binding of the red subcomplex is stabilized by the bracket-shaped molecule (green), leading to a longer dwell time on the promoter template, thereby facilitating assembly of the complex. Alternatively, destabilization of factor binding could also occur, inhibiting assembly (not shown).
Transcription Dynamics in Gene Regulation
There is no doubt that the recruitment and assembly of the transcription machinery to a promoter are highly dynamic events. Increasing evidence suggests that TF dynamics are not just an intrinsic property of the transcription machinery but that modulation of the dynamic interactions of TFs acts as a physiologically relevant regulatory mechanism in gene expression. Correlative observations have linked slower exchange dynamics of the glucocorticoid receptor at a specific promoter with more mRNA synthesis from that promoter (Stavreva et al., 2004), and there is evidence that the association dynamics of TFs are regulated themselves. In yeast, the transient binding of TBP becomes more stable in the absence of the Mot1 Snf2/Swi2 ATPase (Sprouse et al., 2008), consistent with in vitro data showing that the ATPase activity of Mot1 is required for the dissociation of the TBP-DNA complex (Auble et al., 1997). Similarly, the transient interactions of RNA pol I subunits in mammalian cells are regulated by phosphorylation of the transcription initiation factor TIF-1A and a dominant negative form of TIF-1A causes a 2–3×decrease in promoter residence times of RNA pol I core subunits (Gorski et al., 2008).
It is commonly assumed that most transcriptional control works through regulating initiation (Juven-Gershon et al., 2008). The rate-limiting step in control of initiation is likely the rate of preinitiation complex (PIC) assembly, which is affected by several factors. The ability of PIC components to bind at the promoter is determined by chromatin-remodeling events, which control the accessibility of TFs to their target sites. As binding sites becomes more easily available the probability of occupancy of a given promoter site by a PIC component increases and steady-state measurements of TF occupancy at promoters of many genes clearly demonstrate increased association of TFs under conditions of heightened gene activity. In vivo analysis of the RNA pol I machinery have extended these studies and demonstrate that the higher occupancy at the promoter is not merely due to facilitated TF binding, but also due to a prolonged residence time of polymerase components on the promoter (Gorski et al., 2008). During S phase when rDNA transcription roughly doubles compared to G1, the dwell time of several RNA pol I assembly factors increased by the same amount (Gorski et al., 2008). The functional importance of the slowing down in the exchange dynamics of the RNA pol I components is that they now provide a more stable PIC intermediate to which additional downstream factors can bind. The increased stability of each intermediate increases the probability of assembly of a complete, elongation-competent RNA polymerase, thus ultimately leading to increased transcriptional output (Gorski et al., 2008).
In addition to control of TF recruitment and occupancy, recent observations have highlighted the contribution of RNA polymerase pausing as a key regulatory event. The classic example of RNA pol pausing are the heat shock genes in Drosophila where RNA pol II accumulates in the promoter region of uninduced genes after synthesis of 20–50 nucleotides (Core et al., 2008). Upon heat shock activation, the paused polymerases are rapidly released into elongation to mediate a rapid transcriptional response. In vivo live-cell experiments have recently revealed that the heat shock transcription factor (HSF) rapidly turns over under non-heat-shock conditions, but becomes stably bound to the promoter after heat shock when it is engaged (Yao et al., 2006). As in the case for RNA pol I assembly, the prolonged dwell time of HSF on chromatin likely favors efficient formation of an elongation competent polymerase and thus promotes transcription of the target gene. Interestingly, these studies revealed differences in polymerase dynamics in early and late stages of the heat shock response (Yao et al., 2007). Within the first 5–10 min, polymerase was rapidly recruited from the nucleoplasm to the active heat shock gene; however, at time points beyond 20 min, polymerase recruitment from the nucleoplasm ceased, although transcription did not, suggesting that the polymerases were locally recycled after completion of a round of transcription, possibly by association with a local polymerase recycling compartment (Yao et al., 2007). Interestingly, the existence of a similar, “transcription-staging compartment” is indicated by kinetic modeling of RNA pol I dynamics (Dundr et al., 2002). The biphasic recruitment behavior of RNA pol II does not appear to be limited to heat shock genes but was also observed to varying degrees for other fly genes (Yao et al., 2007).
Beyond Drosophila heat shock genes, polymerase pausing is rapidly emerging as a more general transcriptional regulatory mechanism. Genome-wide mapping studies in flies and human have revealed the surprising presence of PICs on up to 5% of genes that are either inactive or active at very low levels (Kim et al., 2005; Guenther et al., 2007), suggesting that these polymerases have very long dwell times and are poised, awaiting appropriate signals to rapidly kick start elongation. Consistent with such a regulatory role, many of the genes containing paused polymerases are responsive to stimuli (Muse et al., 2007; Core and Lis, 2008). Prominent examples include the immediate early response genes c-myc, c-fos, and junB, components of cellular signaling pathways, and many genes required at precise times during development (Muse et al., 2007; Zeitlinger et al., 2007).
Dynamic Oscillations in Transcription
Transcriptional activation and repression processes are commonly treated as uncomplicated up or downregulation events. In fact, the real-time kinetics of transcriptional readout after a stimulus is frequently quite complex, often resulting in oscillatory responses (Figure 4). One can discriminate four distinct classes of dynamic oscillations.
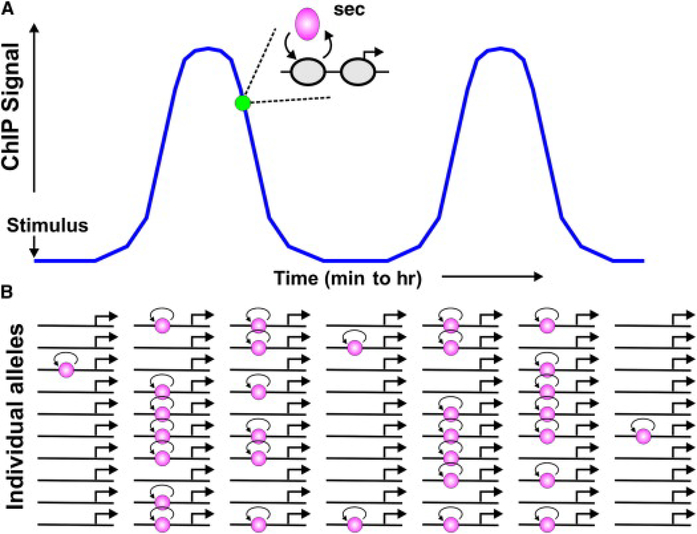
Figure 4. Integrated View of Transcription Factor Dynamics.
(A) Most factors that have been studied in living cells exchange rapidly, on a time scale of seconds, with their recognition elements in chromatin (“hit-and-run”). The frequency and transient duration of these binding events can also fluctuate on a longer time scale, by a variety of mechanisms.
(B) Depicted here are a set of ten abstracted alleles with one binding element. After a transcriptional stimulus, the number of interaction events increases. If this element is sampled across the population by a methodology such as ChIP, more of the events will be captured in a given time. If secondary mechanisms are triggered that decrease the interaction frequency, the ChIP signal will decrease, and an oscillatory process may ensue. However, if real-time residence times could be examined at a specific allele (green circle), one would observe rapid exchange.
The first type of response results from subcellular sequestration of a key transcription factor. For example, members of the NF-κB complex are sequestered in the cytoplasm by the IκB inhibitor protein family. Phosphorylation of the IκB-α inhibitor proteins leads to their degradation and release of NF-κB for nuclear translocation. Single-cell analysis shows that the negative feedback through IκB-α does not terminate signaling, but rather promotes cyclic accumulation and loss of NF-κB in the nucleus (Nelson et al., 2004), and produces in turn oscillatory transcriptional output of target genes (Bosisio et al., 2006; Sung et al., 2009). These experiments have been controversial due to failure to reproduce the phenomenon with biochemical assays (Hoffmann et al., 2002; Kearns et al., 2006). However, modeling studies (Sung and Simon, 2004) indicate that these oscillations can persist over several cycles in individual cells, and recent experimental findings support this suggestion (Sung et al., 2009). When NF-κB cytoplasmic/nuclear distributions are examined at the single-cell level, the cycles rapidly drift out of phase from cell to cell, producing an average over the cell population that appears noncycling. In this case, cycling is masked by sampling over large populations, illustrating the importance of single-cell analysis.
A second type of cycling behavior appears to be encoded directly in promoters and their regulatory elements (Figure 4). Binding of the estrogen receptor (ER) to regulatory sites has been shown to fluctuate dramatically, with a period of approximately 40 min. (Reid et al., 2003; Shang et al., 2000). These cycles have been linked, at least partially, to altered patterns of chromatin modification (Metivier et al., 2003), suggesting that modification of the target response elements can feed back on the binding activity of the initiating transcription factor. The protocols utilized initially to demonstrate ER cycling (Reid et al., 2003) involved experimental variables such as serum starvation, growth in the absence of hormone, and treatment with α-amanitin that raise some concern about the relevance of these observations to the normal ER function. Nevertheless, significant interest remains in cycling behavior of the nuclear receptor family. Karpova et al. also described a strong cycling behavior for the cup1 locus in yeast (Karpova et al., 2008). The Ace I transcription factor exchanges rapidly with regulatory elements (on a time scale of seconds), while the integrated concentration of complexes interacting at these elements fluctuates with a cycle time of 45 min.
Transcriptional activity of GR-regulated promoters has also been shown to vary dramatically after ligand stimulation (Becker et al., 2002). Highly complex patterns of expression are observed for this signaling system on longer time scales. In a genome-wide study of approximately 1000 GR regulated promoters, John et al. reported six general classes of kinetic response, including transient induction and transient repression (John et al., 2009). The GR system, however, appears to be limited to one cycle. Target promoter activity levels can transition through one maximum, or one minimum, but then remain at a constant level in the continuing presence of ligand. For each of these systems, the basic mechanism of oscillation appears to derive from complex interactions between multiple activating regulatory proteins, and between these protein complexes and the chromatin template. In some cases (ER [Shang et al., 2000]; AR [Kang et al., 2002]; VDR [Vaisanen et al., 2005]; TR [Liu et al., 2006; Sharma and Fondell, 2002]), these events can be reversed and thus lead to multiple cycles. In other cases (GR), the molecular actions are not reversed unless hormone is withdrawn and oscillation is thus limited to one cycle (Becker et al., 2002; Qiu et al., 2006).
A third general class of oscillatory transcriptional output derives from fluctuating action by the stimulating effector. An example of this behavior is the cyclic activity of many genes in yeast in response to the reductive environment of the cell (Dioum et al., 2002). NPAS2 is a transcription factor that binds DNA as a dimeric partner of BMAL1 and is implicated in the regulation of circadian rhythm. For this factor, the PAS domains bind heme, and heme status controls DNA binding in vitro. NPAS2-BMAL1 heterodimers bind DNA strongly when reducing conditions in the cell favor the reduced form of nicotinamide adenine dinucleotide phosphate. Thus, in this case, NPAS2-BMAL1 target genes are regulated by a heme-based sensor.
Peptide hormones such as PTH (parathyroid hormone), GnRH (gonadotropin-releasing hormone), and LH (luteinizing hormone) have been known for many years (Belchetz et al., 1978) to cycle in humoral concentration. These cycles result from highly complex endocrine processes that release hormone in pulses from secretory organs. Presumably, many gene targets for these effectors are in turn subject to oscillatory expression. An interesting example of this pulsed release of effector is corticosteroid secretion from the adrenal gland. Release of the steroid is strongly pulsed (Young et al., 2004). This “ultradian” form of hormone release has now been shown to affect a corresponding oscillatory transcriptional response in target cells and tissues (Stavreva et al., 2009). It seems likely that this pulsed action of corticosteroids is necessary for the biologically correct hormone response.
Lastly, a complex form of noncontinuous transcriptional rate variously referred to as “bursting” or “pulsing,” has been described in several systems, both prokaryotic and eukaryotic (Blake et al., 2003; Raj et al., 2006; Scott et al., 2007). This process involves the generation of highly clustered pulses of transcriptional output. A significant component of this phenomenon results from statistical fluctuations associated with the small template number in cells, a phenomenon often referred to as genetic “noise” (Bird, 1995; Blake et al., 2003). One view of this phenomenon is that the local chromatin status of a promoter can trend between structures favorable or unfavorable to rapid initiation rates, giving rise to transient pulses of expression in individual cells (Kaern et al., 2005). Singer and colleagues (Zenklusen et al., 2008) have suggested that bursting results from coordinated periods of local chromatin opening, while random, stochastic opening produces a more continuous expression.
In some cases, it appears that activity fluctuations can become entrained. That is, oscillating activity states can become partially synchronized for genes with common functionality. Ellowitz and colleagues (Cai et al., 2008) describe a form of pulsing in yeast driven by rapid nuclear localization of Crz1, a calcineurin responsive transcription factor. The average nuclear residence time is only 2 min, giving rise to expression bursts for downstream genes. They observed that the frequency of nuclear localization bursts, rather than the duration, was regulated by Ca2+ concentration. Their analysis concludes that this frequency modulation can lead to coordinate expression levels for target genes by regulating the fraction of time a promoter is active. This mechanism thus serves to minimize stochastic drift away from optimal expression patterns in the population at large.
The process of transcriptional cycling clearly derives from multiple mechanisms. The phenomenon is widespread and appears to have evolved to effect specific biological programs. In the case of many peptides hormones, pulsatile ligand release is necessary for the appropriate response in the animal. Gonadotropin-releasing hormone (GnRH) is released episodically, approximately every 90 min (Belchetz et al., 1978). Pulsed release is necessary to maintain gonadotropin release, and thus fertility, in both the male and female primate, including humans. In contrast, continuous administration of GnRH results in hypogonadism. As an increasing number of genetic systems are examined for kinetically complex patterns of expression, a common theme is emerging (Voss et al., 2009; Kaern et al., 2005). Biologically relevant transcriptional programs cannot be produced through continuous transcriptional stimulation but seem to require intermittent activation on a variety of time scales.
A final point should be emphasized. There is a considerable confusion in the literature regarding “cycles” of transcription rate and “exchange” of factors with binding sites in vivo. It is important to discriminate between these fundamentally distinct processes. The characterization of a transcription factor binding event by any process that involves cell breakage, limitation of ATP, or covalent crosslinking, including ChIP and the various footprinting protocols, will produce data on the “equilibrium distribution” of the factor under study at the time the procedure is effected, but not on its intrinsic dynamics (Figure 4). These approaches do not address the actual kinetic on and off rates that produce a given equilibrium state. In most cases where the actual kinetics have been studied in living cells, exchange rates for transcription factors are rapid. Thus, an accurate description of mechanisms involved in the function of chromatin-interacting complexes must include an understanding of the real-time kinetics for the molecules under consideration in living cells.
Physiological Relevance of Transcription Dynamics
The dynamic behavior of the transcription machinery is not merely a default property but serves important physiological roles. A prime example comes from observations in embryonic stem cells (ESCs), which are defined by their potential to differentiate into any tissue by activation of lineage-specific gene-expression programs, implying high levels of transcriptional plasticity in ES cells. The genome of pluripotent ESCs is characterized by several distinct chromatin properties closely related to TF and chromatin protein dynamics (Efroni et al., 2008; Bernstein et al., 2006; Azuara et al., 2006; Hajkova et al., 2008). Morphologically, ESCs are largely devoid of condensed heterochromatin regions, and the vast majority of the genome appears to be in a decondensed euchromatic state and is globally more accessible to digestion by DNase (Efroni et al., 2008). Furthermore, ESC genomes are globally enriched in histone marks, indicative of active genome regions including H3K4me3 and HeK36me3, and are at the same time depleted of epigenetic modifications typically associated with repressed genome regions such as H3K9me3 and DNA methylation (Efroni et al., 2008). In line with a global abundance of hallmarks of actively transcribing chromatin regions in ESCs, normally repressed genome regions such as satellite repeats and transposons are robustly expressed, and many tissue-specific genes are stochastically expressed at low levels in pluripotent ESC (Efroni et al., 2008; Araki et al., 2006). It is attractive to speculate that this transcription pattern in ESC creates a favorable transcriptional ground state from which ESCs can differentiate along any lineage pathway (Ying et al., 2008).
The transcriptional activity in ESC appears to be linked to the dynamic binding behavior of chromatin proteins. Strikingly, the dwell time of most architectural chromatin proteins, including linker and core histones, on their target sites is significantly shorter in ESC compared to differentiated cells derived from them (Efroni et al., 2008), indicating a more dynamic association/dissociation equilibrium in the pluripotent state. The dynamic exchange of these proteins appears to be functionally important, since experimental interference such as introduction of a permanently binding, static linker histone H1 blocks ESC differentiation (Meshorer et al., 2006). A likely model for how global transcriptional activity in ESC is achieved and maintained is that the high dynamic turnover of architectural chromatin proteins maintains chromatin in an open, accessible state, thus allowing ready access of the transcriptional machinery to promoter regions. In support, depletion of chromatin remodelers such as Brg1 or Chd1 leads to proliferation, differentiation, and pluripotency defects of ES cells (Efroni et al., 2008; Gaspar-Maia et al., 2009). As a consequence, in addition to housekeeping genes and ESC-specific genes, PICs also form at low frequency on repressed genes, and occasionally even transcribe them. A prediction from this model is that PICs should be found at repressed genes in ESC. This is indeed the case as demonstrated by genome-wide ChIP analysis (Guenther et al., 2007; Muse et al., 2007). It is not clear at present whether the increased dynamic association of chromatin proteins in ESC is a driving force in bringing about transcription in ESCs or whether it is a secondary consequence—for example, of the observed elevated levels of chromatin-remodeling factors that might serve to maintain the ES genome in a globally open configuration, thus facilitating highly dynamic exchange of chromatin proteins and increased accessibility of PIC to target genes.
Conclusions
Transcription is a highly dynamic process, yet we do not fully understand the role and implications of the dynamic properties of the transcription machinery. Further understanding of transcription dynamics will be driven by future technological advances. These will come in several areas ranging from classic in vitro reconstitutions to live-cell imaging. While a handful of studies have already employed more sophisticated in vitro reconstitutions that incorporate some of the complexities found in the live-cell milieu, future work will expand and extend these efforts to create increasingly more complex and realistic in vitro assays. Live-cell imaging approaches will also benefit from a number of improvements already under development. Single-molecule techniques that will allow us to follow individual molecules at individual promoters will eliminate the averaging necessary in current in vivo assays. Improved mathematical analysis of FRAP data will not only provide more accurate estimates of molecular dwell times at promoters, but also refine our models of molecular assembly. Other techniques such as fluorescence cross-correlation spectroscopy will prove invaluable for simultaneous analysis of multiple components, as we attempt to dissect the molecular assembly process in vivo. Finally, crossover techniques that enable imaging analysis of complex in vitro systems will help bridge the gap between live-cell imaging and in vitro biochemistry. With work underway in all of these areas, the rapid progress in this field over the past decade should continue unabated.
REFERENCES
- Aoyagi S, Narlikar G, Zheng C, Sif S, Kingston RE, and Hayes JJ (2002). Nucleosome remodeling by the human SWI/SNF complex requires transient global disruption of histone-DNA interactions. Mol. Cell. Biol 22, 3653–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R, Fukumura R, Sasaki N, Kasama Y, Suzuki N, Takahashi H, Tabata Y, Saito T, and Abe M (2006). More than 40,000 transcripts, including novel and noncoding transcripts, in mouse embryonic stem cells. Stem Cells 24, 2522–2528. [DOI] [PubMed] [Google Scholar]
- Auble DT, Wang D, Post KW, and Hahn S (1997). Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol. Cell. Biol 17, 4842–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, and Fisher AG (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol 8, 532–538. [DOI] [PubMed] [Google Scholar]
- Becker PB (2002). Nucleosome sliding: facts and fiction. EMBO J. 21, 4749–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Baumann CT, John S, Walker D, Vigneron M, McNally JG, and Hager GL (2002). Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3, 1188–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belchetz PE, Plant TM, Nakai Y, Keogh EJ, and Knobil E (1978). Hypophysial responses to continuous and intermittent delivery of hypopthalamic gonadotropin-releasing hormone. Science 202, 631–633. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. [DOI] [PubMed] [Google Scholar]
- Bird AP (1995). Gene number, noise reduction and biological complexity. Trends Genet. 11, 94–100. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Kaern M, Cantor CR, and Collins JJ (2003). Noise in eukaryotic gene expression. Nature 422, 633–637. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, and Kornberg RD (2008). Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell 133, 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, and Bertrand E (2007). The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol 179, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, and Natoli G (2006). A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kB-dependent gene activity. EMBO J. 25, 798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, and Crawford GE (2008). High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Dalal CK, and Elowitz MB (2008). Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. (2005). Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, and Workman JL (2005). Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592. [DOI] [PubMed] [Google Scholar]
- Core LJ, and Lis JT (2008). Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319, 1791–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, and Lis JT (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR (1999). The organization of replication and transcription. Science 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. (2009). Imaging transcription in living cells. Annu. Rev. Biophys 38, 173–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, and Singer RH (2007). In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol 14, 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, and McKnight SL (2002). NPAS2: a gas-responsive transcription factor. Science 298, 2385–2387. [DOI] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, and Misteli T (2002). A kinetic framework for a mammalian RNA polymerase in vivo. Science 298, 1623–1626. [DOI] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. (2008). Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbi C, Walker DA, Romero G, Sullivan WP, Toft DO, Hager GL, and DeFranco DB (2004). Molecular chaperones function as steroid receptor nuclear mobility factors. Proc. Natl. Acad. Sci. USA 101, 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Li GW, and Xie XS (2007). Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farla P, Hersmus R, Geverts B, Mari PO, Nigg AL, Dubbink HJ, Trap-man J, and Houtsmuller AB (2004). The androgen receptor ligand-binding domain stabilizes DNA binding in living cells. J. Struct. Biol 147, 50–61. [DOI] [PubMed] [Google Scholar]
- Ferdous A, Sikder D, Gillette T, Nalley K, Kodadek T, and Johnston SA (2007). The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters. Genes Dev. 21, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford RG, Warren BS, and Hager GL (2002). ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol 22, 3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, and Yamamoto KR (2002). Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296, 2232–2235. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersback A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, and Ranalho-Santos M (2009). Chd1regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondor A, Rougier C, and Ohlsson R (2008). High-resolution circular chromosome conformation capture assay. Nat. Protocols 3, 303–313. [DOI] [PubMed] [Google Scholar]
- Gorman J, and Greene EC (2008). Visualizing one-dimensional diffusion of proteins along DNA. Nat. Struct. Mol. Biol 15, 768–774. [DOI] [PubMed] [Google Scholar]
- Gorski SA, Dundr M, and Misteli T (2006). The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol 18, 284–290. [DOI] [PubMed] [Google Scholar]
- Gorski SA, Snyder SK, John S, Grummt I, and Misteli T (2008). Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol. Cell 30, 486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers DM, Wilson GG, and Halford SE (2005). Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc. Natl. Acad. Sci. USA 102, 15883–15888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, and Young RA (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, and Surani MA (2008). Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452, 877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, John S, Ling JQ, Biddie SC, Hoffman AR, and Hager GL (2009). Glucocorticoid receptor activation of the Ciz1-Lcn2 locus by long-range interactions. J. Biol. Chem 284, 6048–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford SE, and Marko JF (2004). How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32, 3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Zhang Z, Sabo PJ, Chen X, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. (2009). Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinow P, Rogers CE, Barbieri CE, Pietenpol JA, Kenworthy AK, and DiBenedetto E (2006). The DNA binding activity of p53 displays reaction-diffusion kinetics. Biophys. J 91, 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, and Baltimore D (2002). The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298, 1241–1245. [DOI] [PubMed] [Google Scholar]
- Hoogstraten D, Nigg AL, Heath H, Mullenders LH, van Driel R, Hoeijmakers JH, Vermeulen W, and Houtsmuller AB (2002). Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell 10, 1163–1174. [DOI] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, and Kohwi-Shigematsu T (2005). Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet 37, 31–40. [DOI] [PubMed] [Google Scholar]
- John S, Johnson TA, Sung MH, Koch-Paiz CA, Davis SR, Walker R, Meltzer P, and Hager GL (2009). Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology 150, 1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, and Hager GL (2008). Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol. Cell 29, 611–624. [DOI] [PubMed] [Google Scholar]
- Johnson TA, Elbi C, Parekh BS, Hager GL, and John S (2008). Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol. Biol. Cell 19, 3308–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, and Kadonaga JT (2008). The RNA polymerase II core promoter - the gateway to transcription. Curr. Opin. Cell Biol 20, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, and Collins JJ (2005). Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet 6, 451–464. [DOI] [PubMed] [Google Scholar]
- Kang Z, Pirskanen A, Janne OA, and Palvimo JJ (2002). Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J. Biol. Chem 277, 48366–48371. [DOI] [PubMed] [Google Scholar]
- Karpova TS, Chen TY, Sprague BL, and McNally JG (2004). Dynamic interactions of a transcription factor with DNA are accelerated by a chromatin remodeller. EMBO Rep. 5, 1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, and McNally JG (2008). Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319, 466–469. [DOI] [PubMed] [Google Scholar]
- Kearns JD, Basak S, Werner SL, Huang CS, and Hoffmann A (2006). IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J. Cell Biol 173, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, and Ren B (2005). A high-resolution map of active promoters in the human genome. Nature 436, 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Sugaya K, and Cook PR (2002). The transcription cycle of RNA polymerase II in living cells. J. Cell Biol 159, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD (2005). Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci 30, 235–239. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xia X, Fondell JD, and Yen PM (2006). Thyroid hormone-regulated target genes have distinct patterns of coactivator recruitment and histone acetylation. Mol. Endocrinol 20, 483–490. [DOI] [PubMed] [Google Scholar]
- McNally JG, Mueller WG, Walker D, Wolford RG, and Hager GL (2000). The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science 287, 1262–1265. [DOI] [PubMed] [Google Scholar]
- Mellor J (2005). The dynamics of chromatin remodeling at promoters. Mol. Cell 19, 147–157. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, and Misteli T (2006). Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, and Gannon F (2003). Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763. [DOI] [PubMed] [Google Scholar]
- Misteli T (2001). Protein dynamics: Implications for nuclear architecture and gene expression. Science 291, 843–847. [DOI] [PubMed] [Google Scholar]
- Mueller F, Wach P, and McNally JG (2008). Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys. J 94, 3323–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, and Adelman K (2007). RNA polymerase is poised for activation across the genome. Nat. Genet 39, 1507–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Walker DA, Wolford RG, and Hager GL (2004). Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14, 163–174. [DOI] [PubMed] [Google Scholar]
- Nalley K, Johnston SA, and Kodadek T (2006). Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature 442, 1054–1057. [DOI] [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, et al. (2004). Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306, 704–708. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, and Fraser P (2004). Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet 36, 1065–1071. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Eriksson P, and Wrange O (1990). Quantitative analysis of the glucocorticoid receptor-DNA interaction at the mouse mammary tumor virus glucocorticoid response element. J. Biol. Chem 265, 17222–17229. [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, and Misteli T (2004). Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol 24, 6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH (2008). Poised polymerases: on your mark…get set…go! Mol. Cell 30, 7–10. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Zhao Y, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, et al. (2006). HDAC1 acetylation is linked to progressive modulation of steroid receptor induced gene transcription. Mol. Cell 22, 669–679. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, and Tyagi S (2006). Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam GV, Elbi C, Walker DA, Wolford RG, Fletcher TM, Edwards DP, and Hager GL (2005). Ligand specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol. Cell. Biol 25, 2406–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, and Gannon F (2003). Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11, 695–707. [DOI] [PubMed] [Google Scholar]
- Roeder RG (2005). Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579, 909–915. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Kaneko S, and Manley JL (2006). Terminating the transcript: breaking up is hard to do. Genes Dev. 20, 1050–1056. [DOI] [PubMed] [Google Scholar]
- Scott M, Hwa T, and Ingalls B (2007). Deterministic characterization of stochastic genetic circuits. Proc. Natl. Acad. Sci. USA 104, 7402–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, and Brown M (2000). Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Sharma D, and Fondell JD (2002). Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl. Acad. Sci. USA 99, 7934–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TT, Ingber DE, and Mancini MA (2006). Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J. Cell Sci 119, 4101–4116. [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, and Conaway JW (2003). The RNA polymerase II elongation complex. Annu. Rev. Biochem 72, 693–715. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, Van SB, and Delaat W (2006). Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet 38, 1348–1354. [DOI] [PubMed] [Google Scholar]
- Smale ST, and Kadonaga JT (2003). The RNA polymerase II core promoter. Annu. Rev. Biochem 72, 449–479. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, and de Laat W (2006). CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 20, 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Pego RL, Stavreva DA, and McNally JG (2004). Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J 86, 3473–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse RO, Karpova TS, Mueller F, Dasgupta A, McNally JG, and Auble DT (2008). Regulation of TATA-binding protein dynamics in living yeast cells. Proc. Natl. Acad. Sci. USA 105, 13304–13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva DA, Muller WG, Hager GL, Smith CL, and McNally JG (2004). Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol 24, 2682–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva DA, Wiench M, John S, Conway-Campbell BL, McKenna MA, Pooley JR, Johnson TA, Voss TC, Lightman SL, and Hager GL (2009). Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat. Cell Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O’Malley BW, and Mancini MA (2001). FRAP reveals that mobility of oestrogen receptor-a is ligand and proteasome-dependent. Nat. Cell Biol 3, 15–23. [DOI] [PubMed] [Google Scholar]
- Sung MH, and Simon R (2004). In silico simulation of inhibitor drug effects on nuclear factor-kappaB pathway dynamics. Mol. Pharmacol 66, 70–75. [DOI] [PubMed] [Google Scholar]
- Sung MH, Salvatore L, De Lorenzi R, Indrawna A, Pasparakis M, Hager GL, Bianchi ME, and Agresti A (2009). Oscillations of NF-kB facilitate optimal genome scanning and gene expression profiles. PLoS ONE, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, and de Laat W (2002). Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
- Vaisanen S, Dunlop TW, Sinkkonen L, Frank C, and Carlberg C (2005). Spatio-temporal activation of chromatin on the human CYP24 gene promoter in the presence of 1alpha,25-Dihydroxyvitamin D3. J. Mol. Biol 350, 65–77. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, and Blobel GA (2005). Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17, 453–462. [DOI] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Johnson TA, John S, and Hager GL (2009). Combinatorial probabilistic chromatin interactions produce transcriptional heterogeneity. J. Cell Sci 122, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, and Brown M (2005). Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19, 631–642. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. (2008). Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801. [DOI] [PubMed] [Google Scholar]
- Wu C, Bingham PM, Livak KJ, Holmgren R, and Elgin SC (1979). The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell 16, 797–806. [DOI] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, and Lis JT (2006). Dynamics of heat shock factor association with native gene loci in living cells. Nature 442, 1050–1053. [DOI] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, and Lis JT (2007). Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell 28, 978–990. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, and Smith A (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young EA, Abelson J, and Lightman SL (2004). Cortisol pulsatility and its role in stress regulation and health. Front. Neuroendocrinol 25, 69–76. [DOI] [PubMed] [Google Scholar]
- Yu P, and Kodadek T (2007). Dynamics of the hypoxia-inducible factor-1-vascular endothelial growth factor promoter complex. J. Biol. Chem 282, 35035–35045. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, and Young RA (2007). RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet 39, 1512–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, and Singer RH (2008). Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol 12, 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]