Abstract
Yersinia pestis was introduced to Brazil during the third plague pandemic and currently exists in several recognized foci. There is currently limited available phylogeographic data regarding Y. pestis in Brazil. We generated whole genome sequences for 411 Y. pestis strains from six Brazilian foci to investigate the phylogeography of Y. pestis in Brazil; these strains were isolated from 1966 to 1997. All 411 strains were assigned to a single monophyletic clade within the 1.ORI population, indicating a single Y. pestis introduction was responsible for the successful establishment of endemic foci in Brazil. There was a moderate level of genomic diversity but little population structure among the 411 Brazilian Y. pestis strains, consistent with a radial expansion wherein Y. pestis spread rapidly from the coast to the interior of Brazil and became ecologically established. Overall, there were no strong spatial or temporal patterns among the Brazilian strains. However, strains from the same focus tended to be more closely related and strains isolated from foci closer to the coast tended to fall in more basal positions in the whole genome phylogeny than strains from more interior foci. Overall, the patterns observed in Brazil are similar to other locations affected during the 3rd plague pandemic such as in North America and Madagascar.
Introduction
Yersinia pestis, etiologic agent of plague, is one of the most successful zoonotic pathogens known. Molecular investigations have revealed that Y. pestis spread multiple times from its original foci in Central Asia [1–3] to cause three recognized pandemics: Justinian’s plague in the 6th– 7th centuries, medieval plague in the 14th– 17th centuries (including the Black Death), and the third pandemic, which began ~1855 in the Chinese province of Yünnan [4]. Each of these pandemics was marked by increasingly successful spatial dissemination of Y. pestis, eventually leading to the worldwide distribution of natural foci observed today [3, 4]. Although typically well controlled via improved hygiene and the widespread availability of antibiotics and insecticides, Y. pestis remains a human health threat due to the severity of the disease, the many established natural plague foci [4], and its potential for use as a bioterror agent [5].
Y. pestis was first introduced into Brazil during the third pandemic, when much of the current worldwide distribution of Y. pestis was established [4]. However, the exact entry point and source of the foci that became established in Brazil are not known. The most commonly reported introduction point is the port of Santos, São Paulo (SP), where there were confirmed human plague cases following reports of a murine epizootic in 1899 [6]. Other Brazilian ports also experienced plague outbreaks over the next seven years, including ports in Ceará (CE) and Rio de Janeiro (RJ) in 1900; Pernambuco (PE) and Rio Grande do Sul (RS) in 1902; Pará (PA) in 1903; Bahia (BA) in 1904; and Espírito Santo (ES), Paraná (PR), and Sergipe (SE) in 1906. The ports of Paraíba (PB) and Alagoas (AL) were not initially affected, although plague apparently spread overland to these states, presumably from PE, by 1912 and 1914, respectively [6]. Extensive control efforts subsequently led to the virtual disappearance of plague from urban centers. However, by the 1930s, plague had transitioned to rural areas and become established in various enzootic foci, affecting native rodents and their fleas. Coinciding with this change, human cases decreased markedly, from an average of ~188 per year between 1899 and 1929, to ~20–100 per year up until the mid-1980s. Since then, only three human cases in the 1990s and one case in 2005 have been reported, although serological surveys suggest ongoing human plague activity in known foci [6].
Ongoing plague surveillance activity in Brazil led to the accumulation of 907 Y. pestis strains that were isolated between 1966 and 1997, and are maintained by the Serviço de Referência Nacional em Peste of the Instituto Aggeu Magalhães (SRP/IAM) in Recife, PE [6]. Assorted subtyping efforts on subsets of this collection have had varying levels of success at differentiating among strains. Early efforts involving phenotypic assays and plasmid analysis were unable to differentiate among strains [7, 8]. Pulsed-field gel electrophoresis (PFGE) was more successful, identifying 19 pulsotypes among 22 strains, but was unable to identify any strong geographic correlations [9]. Clustered regularly interspaced palindromic repeats (CRISPR) analysis identified a limited number of unique CRISPR genotypes among 128 strains from 5 plague foci and supported a single introduction to Brazil. However, most of the analyzed strains were still indistinguishable [10]. In contrast, an analysis of 20 Y. pestis strains from an epizootic event in Sítio Alagoinha in 1967 and 17 strains from an outbreak in Planalto da Borborema in 1986 using 12 variable-number tandem repeat (VNTR) loci provided 100% discrimination and identified three genetic groups, which revealed some association with the geographic/temporal origin of the strains [11]. Here, we expand upon these previous analyses using whole genome sequencing. We sequenced a total of 411 strains from six Brazilian foci that were isolated from 1966 to 1997 and examined phylogeographic patterns among these strains.
Materials and methods
DNAs
DNA was extracted from 411 Y. pestis strains belonging to the Y. pestis culture collection (FIOCRUZ–CYP) of the SRP/IAM [12] using the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN, Inc., Germantown, MD). The strains were isolated from 1966 to 1997 from six plague foci in Brazil and include strains isolated from humans, rodents, and fleas (S1 Table). These strains were originally isolated from human clinical specimens (bubo fluid, blood), tissues (spleen, liver, blood) collected from rodents suspected of being infected with Y. pestis, and fleas obtained from humans or captured animal hosts using standard bacteriological methods [13, 14]. DNAs extracted from human derived strains were de-linked from the patients from whom they originated and analyzed anonymously.
Whole genome sequencing
Whole genome sequencing was performed on all 411 Brazilian Y. pestis strains. Genomic DNA quality and quantity were evaluated by 0.7% agarose gel analysis. DNA samples (~1 μg/sample) were then fragmented using a SonicMan (Matrical, Inc., Spokane, WA) with the following parameters: 75 s pre chill, 16 cycles, 10 s sonication, 100% power, 75 s lid chill, 10 s plate chill, and 75 s post chill. The fragmented DNA was then size selected to target 600–650 bp by fragment separation using Agencourt AMPure XP beads (Beckman Coulter, Code A63882, Indianapolis, IN) and eluted into 42.5 μl of Elution Buffer. The KAPA Library Preparation Kit with SRPI Solution and the Standard PCR Library Amplification/Illumina series (KAPA Biosystems, Code KK8232, Wilmington, MA) were used for library preparation. Three separate library preparation reactions for each sample were carried out as follows: 1) End repair: 2.5 μl Enzyme, 5 μl Buffer, 1.6X AMPure XP, 43.5 μl Elution buffer, 2) A-Tailing: 1.5 μl Enzyme, 5 μl Buffer, 1.6X AMPure XP, 36.5 μl Elution buffer and 3) Quick Ligation: 2.5 μl Enzyme, 10 μl Buffer, 0.9X AMPure XP, 30 μl Elution buffer. The adapter ligation step used 1 μl of the 10 μM adapter oligo mix [15]. These reactions included the following modifications to the manufacturer’s protocol: 1) adapters and 8 bp index oligos based on Kozarewa and Turner (2011) [15] were purchased from IDT (Integrated DNA Technologies, San Diego, CA) and used in place of those supplied in the KAPA preparation kit to utilize a previously described dual indexing approach [16], 2) during the enzymatic steps, half the enzyme volume was used with the full volume of buffer as described in the KAPA Library Preparation protocol, and 3) The PCR was optimized to improve yield and genome coverage and included 2 μl of DNA, 2 μl of each 10 μM indexing primer, 25 μl of KAPA 2X HIFI PCR Master Mix (KAPA Biosystems), and 19 μl of molecular grade water, with the following PCR parameters: initial denaturation 2 min at 98°C, 8 cycles of 30 s at 98°C, 20 s at 65°C, 30 s at 72°C, and a final extension of 5 min at 72°C. The libraries were purified with a 0.9X AMPure XP bead cleanup and eluted into 50 μl of Elution Buffer. The final libraries were quantified using the KAPA ABI Prism Library Quantification Kit (KAPA Biosystems, Code KK4835) and pooled together at equimolar concentrations. The pool was quantified using the KAPA ABI Prism Library Quantification Kit, and the quality of the pool assessed with a Bioanalyzer DNA 1000 chip (Agilent Technologies, Code 5067–1504, Santa Clara, CA). Whole genome sequencing was then performed on an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA) using the 200-cycle TruSeq SBS Kit v3-HS (Illumina, Code FC-401-3001) with the standard Illumina procedure.
SNP calling and phylogenetics
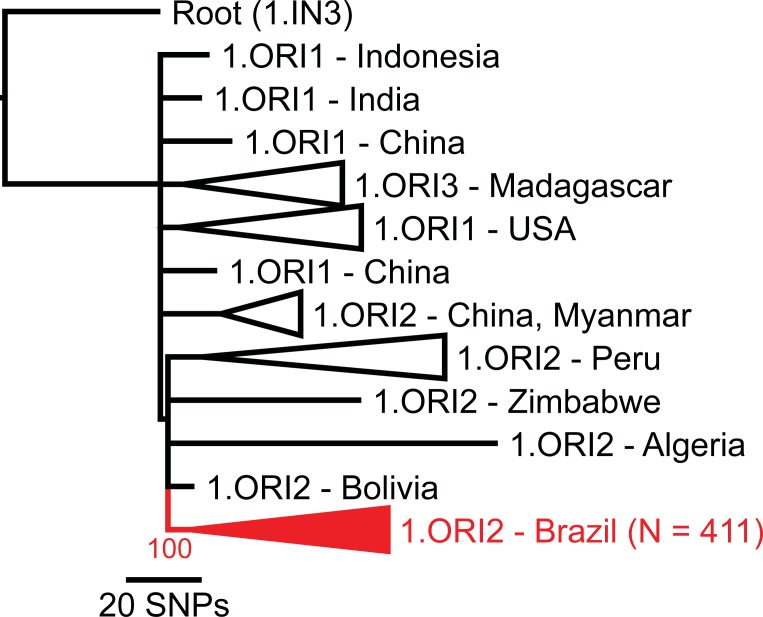
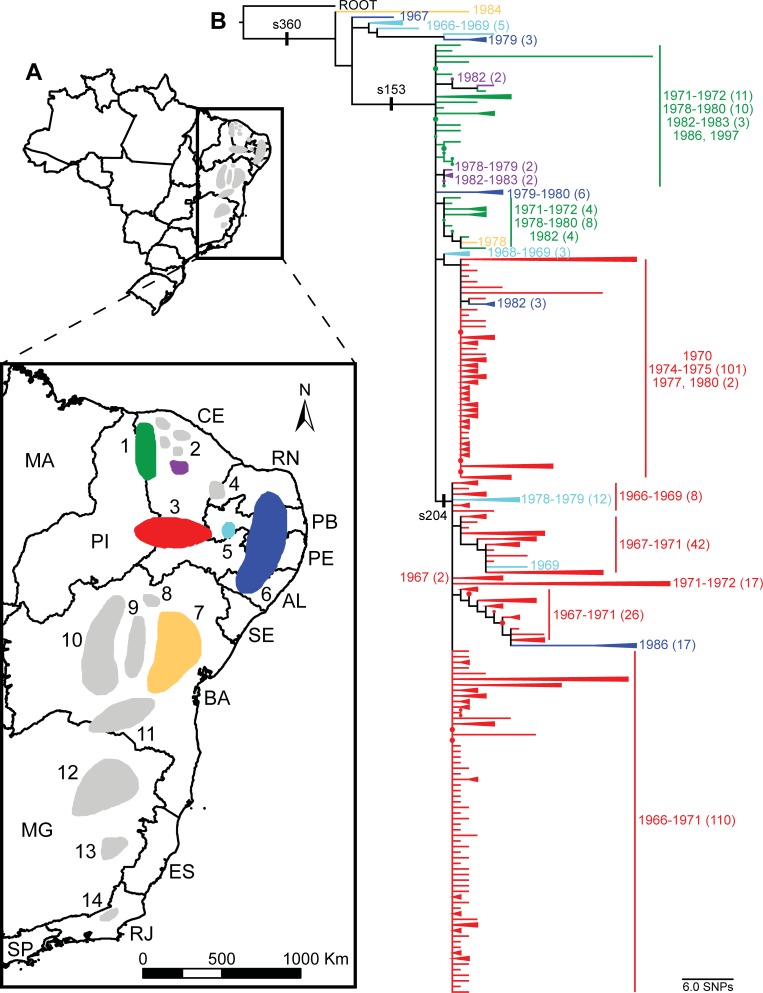
Raw reads were aligned against the completed genome of the CO92 strain of Y. pestis [17] with BWA-MEM [18] and all single nucleotide polymorphisms (SNPs) were called with the UnifiedGenotyper method in GATK [19, 20]; these methods were wrapped by the NASP pipeline [21]. SNPs were filtered from downstream analyses if they fell within duplicated regions of the reference genome based on a NUCmer [22] self-alignment, or if the SNPs had a depth of coverage <3X or an allele proportion <90%. Phangorn [23] was used to generate a maximum parsimony phylogeny, including 100 bootstrap replicates, from the concatenated SNP alignments of the Brazilian Y. pestis strain genomes, all publicly available 1.ORI Y. pestis whole genome sequences, and a 1.IN3 Y. pestis genome as a root (Fig 1). Confirmation of the placement of the Brazilian Y. pestis strains within the 1.ORI2 subpopulation suggested in Fig 1 was determined by querying relevant 1.ORI2 SNPs from Morelli et al. [2] (s36, s37, s38, s40, s47, s121, s151, s153, s162, s163, s164, s168, s169, s170, s172, s176, s179, s182, s184, s186, s189, s191, s194, s200, s202, s203, s204, s205, s207, s225, s240, s255, s281, s311, s360, s649, s841, and s1230) against the Brazilian Y. pestis whole genome sequence data. Phangorn was also used to generate a second maximum parsimony phylogeny of just the Brazilian Y. pestis strain genomes with Y. pestis strain CO92 as a root (Fig 2B). This second maximum parsimony phylogeny was color coded to reflect the Brazilian plague foci of origin described by Tavares et al. [6] and elucidate the phylogeography of the Brazilian Y. pestis strains (Fig 2).
Fig 1. Yersinia pestis 1.ORI population phylogeny.
Maximum parsimony phylogeny based on single nucleotide polymorphisms (SNPs) identified among 461 Y. pestis genomes belonging to the 1.ORI population, including 411 Brazilian Y. pestis strains (S1 Table, S2 Table). All 411 Brazilian Y. pestis genomes were assigned to a single monophyletic clade (red) with 100% bootstrap support. The phylogeny was rooted on strain D106004 (Genbank accession No. GCA 000022805.1), a member of the 1.IN3 Y. pestis population.
Fig 2. Phylogeography of Yersinia pestis in Brazil.
(A) Map indicating recognized plague foci in Brazil. The six foci represented in this study are shown in color with unrepresented foci in gray. Foci are as follows: 1) Serra de Ibiapaba (green), 2) Serra das Matas, de Baturité (purple), de Pedra Branca, do Machado, and de Uruburetama, (these five smaller foci are sometimes grouped together as a single focus) 3) Chapada do Araripe (red), 4) Chapada do Apodi, 5) Serra de Triunfo (light blue), 6) Chapada da Borborema (dark blue), 7) Planalto Oriental da Bahia (orange), 8) Serra do Formoso, 9) Piemonte da Diamantina, 10) Chapada Diamantina, 11) Planalto da Conquista, 12) Vale do Jequitinhonha, 13) Vale do Rio Doce, and 14) Serra dos Órgãos [6]. The Brazilian state abbreviations are also indicated. (B) Maximum parsimony phylogeny based on single nucleotide polymorphisms (SNPs) identified among 411 Y. pestis genomes from Brazil and rooted on Y. pestis strain CO92. Clades containing strains isolated from the same focus and in the same or close years were collapsed to simplify the tree. Branches and clades are colored to indicate the focus of origin. The date of isolation (presented as a year or range of years, as appropriate) is indicated for groups of strains with the same focus of origin. Numbers in parentheses following each listed year or range indicate the number of strains isolated in that year or range when the number of strains is >1. The position of three 1.ORI2 SNPs from Morelli et al. [2] are also indicated on the phylogeny.
Genome assembly and comparative genomics
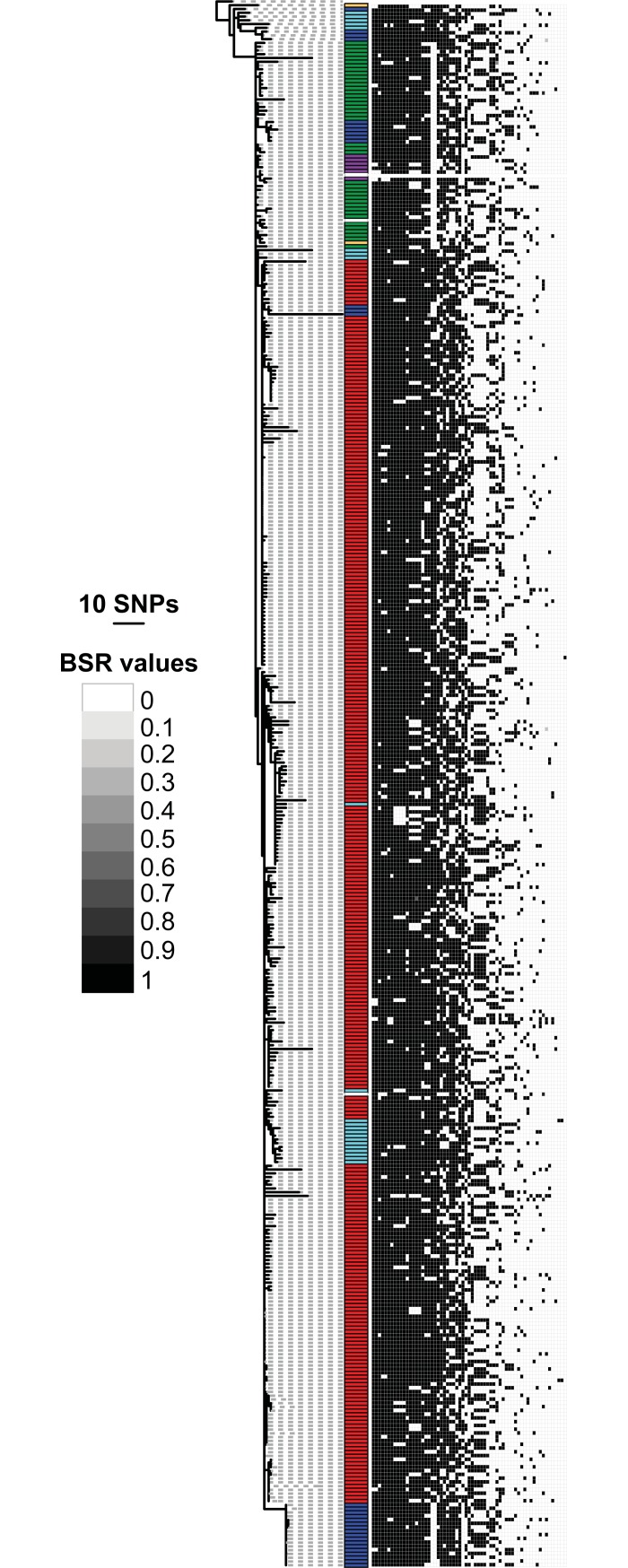
Genomes were assembled with SPAdes v3.11.1 [24]. To identify differential conserved regions, a large-scale blast score ratio (LS-BSR) analysis [25] was performed. Coding region sequences (CDSs) were identified with Prodigal v2.60 [26] and clustered with USEARCH v10.0 [27]. Each unique peptide, based on a clustering at 90% amino acid identity, was aligned against Prodigal predicted coding regions with Diamond v0.9.10.111 [28]. Sixty-three peptides that showed variable distribution across the phylogeny, as demonstrated by the BSR value [29], along with the phylogeny, were visualized with the interactive tree of life [30] and were annotated by the best BLASTP [31] alignment against the GenBank [32] nr database.
Results and discussion
The plague foci in Brazil are the result of a single introduction of Y. pestis during the third pandemic that became successfully established in Brazil. All 411 Brazilian Y. pestis strains were assigned to a single, strongly supported, monophyletic clade within the 1.ORI population (Fig 1), indicating that there was only a single Y. pestis introduction to Brazil that led to the successful establishment of endemic foci. Importantly, this monophyletic clade only included strains from Brazil (Fig 1). Available strains of Y. pestis from other South American countries (Peru and Bolivia) were assigned to other clades within the 1.ORI population (Fig 1), suggesting that Y. pestis in these other countries is due to separate introduction events during the third pandemic. These results are consistent with a previous report by Morelli et al. [2], which placed two Y. pestis isolates from Brazil into the 1.ORI2.f node of a worldwide phylogeny. Querying the 1.ORI2 SNPs from Morelli et al. [2] placed the Brazilian Y. pestis genome sequences described here into nodes 1.ORI2.a (N = 10), a new node intermediate between SNP nodes 1.ORI2.a and 1.ORI2.f (1.ORI2.f1, N = 166), and node 1.ORI2.f (renamed here as 1.ORI2.f2, N = 235) (S1 Table). These results suggest that the introduction of Y. pestis to Brazil represents one monophyletic branch in the still unresolved polytomy of the 1.ORI2 subpopulation. Analysis of additional 1.ORI2 strains may provide additional insight into the spread of this 1.ORI subpopulation.
Following its introduction to Brazil, Y. pestis underwent a radial expansion, spreading rapidly to the interior and becoming ecologically established in several foci. The geographic introduction point for Y. pestis to Brazil remains unknown, but it may have been any of several port cities that experienced outbreaks in the early 1900s, including the commonly reported entry point of Santos, SP [6]. Regardless of the exact entry point, the lack of population structure indicated by the WGS data (Fig 2) suggests Y. pestis rapidly dispersed from the coast to the interior of Brazil, followed by localized differentiation. There are very few shared characters (i.e., SNPs) among the 411 Brazilian Y. pestis strains. Rather, most SNPs are found only in single strains (Fig 2). This low level of population structure is consistent with a radial expansion, which is a common feature in the evolutionary history of Y. pestis [1].
The lack of population structure evident in the WGS data extended to the phylogeographic analysis. Overall, there were no strong spatial or temporal patterns among the 411 Brazilian Y. pestis genomes, although there were some general trends. First, in general, strains from the same focus tended to cluster together. There were some exceptions to this, most notably for strains from the Serra de Triunfo and Chapada da Borborema foci (Fig 2, light blue and dark blue foci). Strains from these foci were found on multiple branches in the phylogeny, which may be indicative of Y. pestis movement among foci. Second, strains isolated from foci closest to the coastal regions tended to fall in more basal positions in the WGS tree, with strains isolated from the more interior foci falling in more derived positions. Although there are some exceptions to these trends, these patterns are consistent with a rapid spread from the coastal regions to the interior, followed by ecological establishment in these foci.
The LS-BSR analysis was consistent with the SNP analysis. Both analyses revealed a moderate level of genetic diversity among the 411 Brazilian Y. pestis strains, with 822 SNPs and 63 variably distributed CDSs identified (Figs 2 and 3, S3 Table). Similar to the SNP analysis, there were few patterns among the identified CDSs (Fig 3), reflecting the limited population structure among these Brazilian Y. pestis strains. The LS-BSR analysis indicated mostly variation in the presence or absence of the identified CDSs rather than sequence differences (Fig 3). Such gene loss is not uncommon in Y. pestis, which is a recently emerged clone of Y. pseudotuberculosis that exhibits genomic reduction consistent with its transition to an obligate vector-borne pathogen [33]. Indeed, few to none of the CDSs identified in the LS-BSR analysis appear likely to be under selection (S3 Table). This suggests that these CDS differences are simply the result of neutral variation associated with the localized differentiation of Y. pestis following its radial expansion in Brazil.
Fig 3. CDS variability among Brazilian strains.
The expanded maximum parsimony phylogeny from Fig 2 is presented with a heatmap generated by blast score ratio (BSR) values and visualized with the Interactive Tree of Life (iTOL). The BSR values range from 0 (no significant alignment) to 1 (identical alignment) and demonstrate the variability in gene content among the Brazilian strain genomes. Colored bars indicate the focus of origin for each strain as in Fig 2.
Unfortunately, the lack of strains from many of the Brazilian plague foci prevented a comprehensive analysis of Y. pestis phylogeography in Brazil. Starting in the 1930s, most human plague cases in Brazil occurred in extremely remote rural areas, and the last recorded urban cases occurred in the first half of the 1960s [6]. In contrast, routine Y. pestis culture collection did not start until the mid-1960s [6, 12]. As such, the available strain diversity is limited, with the majority of strains originating from the most active northeastern plague foci, including the six foci represented here. Despite the declining number of human cases in Brazil, residual human plague activity has continued to be detected in focal areas using serology [6]. In addition, ecological niche modeling [34] is consistent with the identified Brazilian plague foci [6], suggesting that these regions contain suitable ecological conditions for the persistence of Y. pestis. Additional whole genome sequences from more modern strains might reveal additional phylogeographic patterns.
Overall, the patterns observed in Brazil are similar to other locations affected during the 3rd pandemic. Globally, the Y. pestis phylogeny is characterized by several polytomies, reflecting a series of nested radial expansions in the history of this species. These include a major polytomy at the base of the 1.ORI population responsible for the third pandemic and other radial expansions within that population [1]. Among these are radial expansions associated with the establishment of plague foci in North America and Madagascar [1, 2], and now Brazil. Y. pestis from each of these locations exhibits patterns indicative of an initial rapid radial expansion following a single successful introduction. This rapid spread was then followed, in turn, by ecological establishment in rodent reservoirs and long term enzootic persistence [2, 35]. Such rapid epizootic spread followed by enzootic maintenance is a hallmark of Y. pestis as a species [1, 2, 4, 35, 36].
Supporting information
(XLSX)
(XLSX)
(XLSX)
Data Availability
All NCBI accession numbers for the 411 draft genomes sequences generated in this study are listed in S1 Table.
Funding Statement
This work was funded by the Cowden Endowment at Northern Arizona University, and by the United States of America Department of Homeland Security (DHS) Science and Technology Directorate via award HSHQDC-10-C-00139 to DMW and PK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(2):577–82. Epub 2012/12/29. 10.1073/pnas.1205750110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42(12):1140–3. Epub 2010/11/03. 10.1038/ng.705 [pii] 10.1038/ng.705. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner DM, Klunk J, Harbeck M, Devault A, Waglechner N, Sahl JW, et al. Yersinia pestis and the Plague of Justinian 541–543 AD: a genomic analysis. The Lancet Infectious diseases. 2014;14(4):319–26. 10.1016/S1473-3099(13)70323-2 . [DOI] [PubMed] [Google Scholar]
- 4.Perry RD, Fetherston JD. Yersinia pestis-etiologic agent of plague. Clin Microbiol Rev. 1997;10(1):35–66. Epub 1997/01/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. Public health assessment of potential biological terrorism agents. Emerg Infect Dis. 2002;8(2):225–30. Epub 2002/03/19. 10.3201/eid0802.010164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavares C, Aragão AI, Leal NC, Leal-Balbino TC, de Oliveira MB, de Oliveira Gonçalves Ferreira GM, et al. Plague in Brazil: from now and then. Adv Exp Med Biol. 2012;954:69–77. 10.1007/978-1-4614-3561-7_10 . [DOI] [PubMed] [Google Scholar]
- 7.Cavalcanti YV, Leal NC, De Almeida AM. Typing of Yersinia pestis isolates from the state of Ceará, Brazil. Letters in applied microbiology. 2002;35(6):543–7. . [DOI] [PubMed] [Google Scholar]
- 8.Leal NC, de Almeida AM, Ferreira LC. Plasmid composition and virulence-associated factors of Yersinia pestis isolates from a plague outbreak at the Paraiba State, Brazil. Rev Inst Med Trop Sao Paulo. 1989;31(5):295–300. Epub 1989/09/01. . [DOI] [PubMed] [Google Scholar]
- 9.Barros MP, Silveira-Filho VM, Lins RH, Oliveira MB, Almeida AM, Leal-Balbino TC. Subtyping Brazilian Yersinia pestis strains by pulsed-field gel electrophoresis. Genetics and molecular research: GMR. 2013;12(2):1294–302. 10.4238/2013.January.4.23 . [DOI] [PubMed] [Google Scholar]
- 10.Barros MP, França CT, Lins RH, Santos MD, Silva EJ, Oliveira MB, et al. Dynamics of CRISPR loci in microevolutionary Process of Yersinia pestis strains. PloS one. 2014;9(9):e108353 10.1371/journal.pone.0108353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira MB, Barros MP, Silveira-Filho VM, Araújo-Nepomuceno MR, Balbino VQ, Leal NC, et al. Genetic diversity of Yersinia pestis in Brazil. Genetics and molecular research: GMR. 2012;11(3):3414–24. 10.4238/2012.September.25.10 . [DOI] [PubMed] [Google Scholar]
- 12.Leal NC, Sobreira M, Araújo AF, Magalhães JL, Vogler AJ, Bollig MC, et al. Viability of Yersinia pestis subcultures in agar stabs. Letters in applied microbiology. 2016;62(1):91–5. Epub 2015/11/03. 10.1111/lam.12519 . [DOI] [PubMed] [Google Scholar]
- 13.Bahmanyar M, Cavanaugh DC. Plague Manual. Geneva, Switzerland: World Health Organization; 1976. [Google Scholar]
- 14.Chu M. Laboratory manual of plague diagnostic tests Atlanta GA, Geneva, Switzerland: Centers for Disease Control and Prevention, World Health Organization; 2000. [Google Scholar]
- 15.Kozarewa I, Turner DJ. 96-plex molecular barcoding for the Illumina Genome Analyzer. Methods Mol Biol. 2011;733:279–98. Epub 2011/03/25. 10.1007/978-1-61779-089-8_20 . [DOI] [PubMed] [Google Scholar]
- 16.Stone NE, Sidak-Loftis LC, Sahl JW, Vazquez AJ, Wiggins KB, Gillece JD, et al. More than 50% of Clostridium difficile Isolates from Pet Dogs in Flagstaff, USA, Carry Toxigenic Genotypes. PloS one. 2016;11(10):e0164504 Epub 2016/10/11. 10.1371/journal.pone.0164504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413(6855):523–7. Epub 2001/10/05. 10.1038/35097083 35097083 [pii]. . [DOI] [PubMed] [Google Scholar]
- 18.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN]; 2013.
- 19.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahl JW, Lemmer D, Travis J, Schupp J, Gillece J, Aziz M, et al. The Northern Arizona SNP Pipeline (NASP): accurate, flexible, and rapid identification of SNPs in WGS datasets. bioRxiv. 2016. 10.1101/037267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30(11):2478–83. Epub 2002/05/30. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–3. Epub 2010/12/21. 10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. Epub 2012/04/18. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahl JW, Caporaso JG, Rasko DA, Keim P. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ. 2014;2:e332 Epub 2014/04/22. 10.7717/peerj.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119 Epub 2010/03/10. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. Epub 2010/08/17. 10.1093/bioinformatics/btq461 . [DOI] [PubMed] [Google Scholar]
- 28.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. Epub 2014/11/18. 10.1038/nmeth.3176 . [DOI] [PubMed] [Google Scholar]
- 29.Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2 Epub 2005/01/07. 10.1186/1471-2105-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–8. Epub 2006/10/20. 10.1093/bioinformatics/btl529 . [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. Epub 1990/10/05. 10.1016/S0022-2836(05)80360-2 . [DOI] [PubMed] [Google Scholar]
- 32.nson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;40(Database issue):D48–5ø3. Epub 2011/12/07. 10.1093/nar/gkr1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chain PS, Hu P, Malfatti SA, Radnedge L, Larimer F, Vergez LM, et al. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. J Bacteriol. 2006;188(12):4453–63. Epub 2006/06/03. 188/12/4453 [pii] 10.1128/JB.00124-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giles J, Peterson AT, Almeida A. Ecology and geography of plague transmission areas in northeastern Brazil. PLoS neglected tropical diseases. 2011;5(1):e925 Epub 2011/01/20. 10.1371/journal.pntd.0000925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogler AJ, Chan F, Wagner DM, Roumagnac P, Lee J, Nera R, et al. Phylogeography and Molecular Epidemiology of Yersinia pestis in Madagascar. PLoS neglected tropical diseases. 2011;5(í):e1319 Epub 2011/09/21. 10.1371/journal.pntd.0001319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girard JM, Wagner DM, Vogler AJ, Keys C, Allender CJ, Drickamer LC, et al. Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(22):8408–13. Epub 2004/06/03. 10.1073/pnas.0401561101 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All NCBI accession numbers for the 411 draft genomes sequences generated in this study are listed in S1 Table.