Abstract
Nonenzymatic RNA primer extension by activated mononucleotides has long served as a model for the study of prebiotic RNA copying. We have recently shown that the rate of primer extension is greatly enhanced by the formation of an imidazolium-bridged dinucleotide between the incoming monomer and a second, downstream activated monomer. However, the rate of primer extension is further enhanced if the downstream monomer is replaced by an activated oligonucleotide. Even an unactivated downstream oligonucleotide provides a modest enhancement in the rate of reaction of a primer with a single activated monomer. Here we study the mechanism of these effects through crystallographic studies of RNA complexes with the recently synthesized nonhydrolyzable substrate analog, guanosine 5′-(4-methylimidazolyl)-phosphonate (ICG). ICG mimics 2-methylimidazole activated guanosine-5′-phosphate (2-MeImpG), a commonly used substrate in nonenzymatic primer extension experiments. We present crystal structures of primer-template complexes with either one or two ICG residues bound downstream of a primer. In both cases, the aryl-phosphonate moiety of the ICG adjacent to the primer is disordered. To investigate the effect of a downstream oligonucleotide, we transcribed a short RNA oligonucleotide with either a 5′-ICG residue, a 5′-phosphate or a 5′-hydroxyl. We then determined crystal structures of primer-template complexes with a bound ICG monomer sandwiched between the primer and each of the three downstream oligonucleotides. Surprisingly, all three oligonucleotides rigidify the ICG monomer conformation and position it for attack by the primer 3′-hydroxyl. Furthermore, when GpppG, an analog of the imidazolium-bridged intermediate, is sandwiched between an upstream primer and a downstream helper oligonucleotide, or covalently linked to the 5′-end of the downstream oligonucleotide, the complex is better preorganized for primer extension than in the absence of a downstream oligonucleotide. Our results suggest that a downstream helper oligonucleotide contributes to the catalysis of primer extension by favoring a reactive conformation of the primer-template-intermediate complex.
Introduction
Template-directed nonenzymatic RNA primer extension is an important experimental model for the earliest mode of genome propagation.1−4 In this system, mononucleotides or oligonucleotides are chemically activated and bound to a nucleic acid primer-template complex, followed by primer extension through monomer polymerization or oligomer ligation. The concept can be traced back to work by Orgel and co-workers in the late 1960’s.5 Subsequently, 2-methylimidazole-activated monoribonucleotides were discovered to be relatively efficient at template-directed nonenzymatic RNA primer extension in the presence of divalent metal cations.6 Several significant factors have been found to further accelerate the reaction, especially for templates containing adenosine and uridine nucleotides, which are notoriously difficult to copy.7−9 Modified bases, distinct activating groups, and downstream oligonucleotides all influence the reaction. The affinities of nucleotides for RNA primer-template complexes, together with the kinetics of the subsequent oligomerization, have been explored by our lab and others.10−13 The reaction mechanism has been assumed for many years to be a classic in-line SN2 nucleophilic substitution mechanism in which the 3′-hydroxyl group of the RNA primer attacks the electrophilic phosphorus atom of the incoming activated monomer to form a new covalent O–P bond, and the activating group (e.g., 2-methylimidazole) is displaced.14 However, it has also been known for many years that the rate of primer extension is accelerated by the binding of an activated monomer15 or oligomer8,9 downstream of the polymerizing nucleotide. The activating groups of the downstream monomers/oligomers play a catalytic role, which was initially thought to result from noncovalent interactions between the leaving groups of adjacent nucleotides or oligomers. To test this hypothesis directly, we previously designed and synthesized a stable analog of an activated monomer, guanosine 5′-(3-methyl-1H-pyrazol-4-yl)phosphonate (PZG), and cocrystallized it with an RNA primer-template complex for structure determination.16 Unexpectedly, the pyrazolyl-phosphonate groups of the PZGs were not only too far from the 3′-hydroxyl group of the primer to react but also severely disordered. The absence of any detectable noncovalent interaction between the leaving groups of adjacent monomers suggested that the catalytic effect stems from some other chemical phenomenon.
Recent work from our lab strongly suggests that a 5′-5′ imidazolium-bridged dinucleotide is the actual intermediate in the RNA primer extension reaction; the Richert lab has also noted the high reactivity of this species.17,18 Under the optimal pH environment (at the pKa of the leaving group), two activated monomers react with each other to form an imidazolium-bridged dinucleotide, which can then bind to the template. The primer 3′-hydroxyl then attacks the phosphorus atom of the adjacent incoming nucleotide, but the displaced species is an entire activated monomer, rather than just the activating group. To examine the structural underpinning of this reaction model, we have used P1,P3-diguanosine-5′-triphosphate (GpppG) as a stable analog of the imidazolium-bridged dinucleotide intermediate.19 The RNA-GpppG structure showed that both G nucleobases of the dinucleotide analog interact with the template by canonical Watson–Crick base pairing, and NMR experiments demonstrated that GpppG binds cognate RNA templates with significantly enhanced binding affinity relative to activated monomer. Meanwhile, the conformational constraints imposed by the covalent internucleotide bridge and the two Watson–Crick base pairs preorganize the bound dinucleotide in a geometry that would favor in-line nucleophilic attack.
Chemically stable analogs of activated nucleotides are crucial for structural studies because the high reactivity of the imidazole-activated monomers and imidazolium-bridged dinucleotide precludes these molecules from being cocrystallized with RNA. We have therefore designed and synthesized several stable analogs that closely mimic the structure and basicity of the activated monomers. We recently synthesized guanosine 5′-(4-methylimidazolyl) phosphonate (ICG), which has a structure and pKa (of its nonhydrolyzable heteroaromatic group) similar to that of 2-MeImpG20 (Figure 1A). Here, we report the crystallographic analysis of a series of RNA:ICG complexes in which an ICG monomer lies adjacent to the 3′-end of a primer. Because ICG residues, unlike the actual 2-MeImpG substrates, cannot react with each other to form the 5′-5′ imidazolium-bridged intermediate, we were able to specifically address the effects of downstream monomers, consecutive monomers, and oligonucleotides on the conformation of the primer relative to the adjacent ICG monomer. In addition, we describe the structures of two new RNA:GpppG complexes, in which the GpppG dinucleotide is either sandwiched between a primer and a downstream oligonucleotide or covalently attached to the downstream oligonucleotide. In all cases, we find that in the presence of a downstream oligonucleotide, the primer 3′-hydroxyl and the phosphorus atom of the adjacent ICG or GpppG are both closer and more properly aligned for nucleophilic attack. The data presented here, together with previous work on the reactive imidazolium-bridged intermediate, suggest that the rate of nonenzymatic primer extension is increased by both covalent nucleophilic catalysis and by conformational constraint of the reactive complex.
Figure 1.
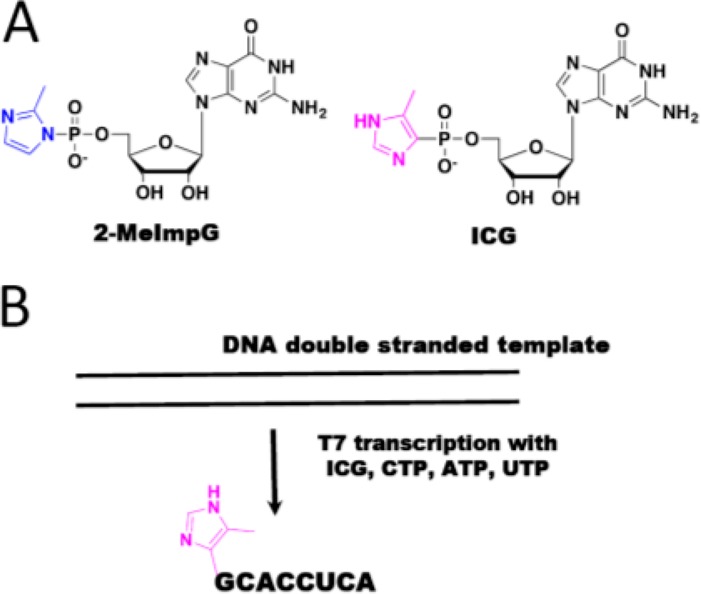
(A) Structures of 2-MeImpG and the ICG analog. (B) T7 in vitro transcription of RNA helper 8mer containing ICG at the 5′-end.
Results
RNA Duplex-Monomer Cocrystal Structures Reveal Multiple Modes of Monomer Binding
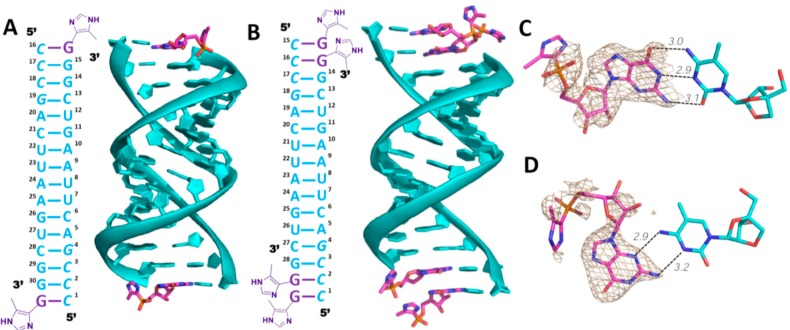
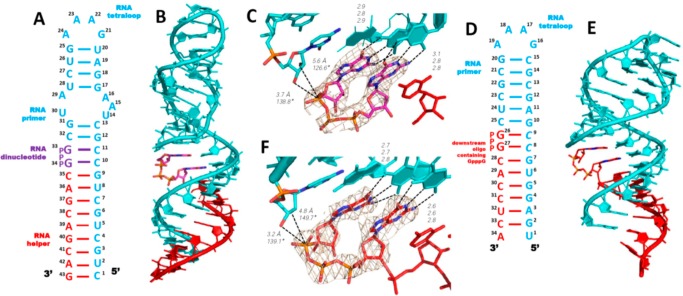
We used the nucleotide analog guanosine 5′-(4-methylimidazolyl) phosphonate (ICG), which has a nonhydrolyzable heteroaryl-phosphonate moiety that mimics the structure and properties of the leaving group of the reactive monomer 2-MeImpG,21 to investigate the binding mode of activated monomers to a primer-template complex. We began with an RNA that would generate one monomer binding site at each end of the duplex formed by annealing of the self-complementary oligonucleotide 5′-CCCGACUUAAGU CGG-3′. The four italicized nucleotides at 5′-end are LNA nucleotides in which the sugar is locked in the 3′-endo conformation. C denotes locked 5-methylcytidine, which has similar basicity as native cytidine and Watson–Crick base pairs with guanosine.22 The 1.9 Å resolution ICG-RNA structure showed one ICG molecule bound to the templating 5-methylcytidine at each end of the duplex. The RNA-monomer duplexes are slip-stacked with one another, which may stabilize monomer binding by providing additional stacking interactions. Interestingly, ICG bound to the templating C residues in two different modes. At one end, canonical Watson–Crick base pairing was observed while at the other end an alternative base pair was present in which the guanine N3 of ICG was hydrogen bonded with the exocyclic amine of 5-methylcytidine, while the exocyclic amine of guanine was hydrogen-bonded with N3 of 5-methylcytidine. For the Watson–Crick paired monomer, the sugar and phosphonate were moderately well-ordered, with a primer O3′-P distance of 6 Å. However, the imidazolyl moiety was entirely disordered, so that the O3′-P-C angle of attack could not be determined. For the noncanonical base pair, both the sugar and the imidazolyl-phosphonate were highly disordered. The crystallographic and structural features of determined structures are listed in Table 1. Schematic diagrams and the crystal structure are shown in Figure 2A.
Table 1. Crystallographic and Structural Features of RNA-ICG and RNA-GpppG Complexes.
| RNA 15mer-monomer | RNA 14mer-monomer | H-34-Im | H-34-PO | H-34-OH | H-35-OH | H-35-PO | H-43-GpppG | H-34-GpppG- oligo | H-34-L | H-35-L | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDB code | 5 V0H | 5 V0J | 5UX3 | 5 V9Z | 5 V0O | 5VCF | 5VCI | 6AZ4 | 6BMD | 5UZ6 | 5VGW |
| space group | P321 | R32 | P41212 | P43212 | P43212 | R3 | R3 | P41212 | C2 | C2 | R3 |
| RNA structural features | duplex | duplex | tetraloop/stem | tetraloop/stem | tetraloop/stem | tetraloop/receptor/stem | tetraloop/receptor/stem | tetraloop/receptor/stem | tetraloop/stem | tetraloop/stem | tetraloop/receptor/stem |
| “leaving group” of bound monomer | disordered imidazolyl | disordered imidazolyl | ordered imidazolyl | disordered imidazolyl | disordered imidazolyl | disordered imidazolyl | disordered imidazolyl | ordered triphosphate | ordered triphosphate | disordered imidazolyl | none |
| downstream helper | none | ICG monomer | 5′-ICG 8mer | 5′-PO42–- 8mer | 5′-hydroxyl 8mer | 5′-hydroxyl 13mer | 5′-PO42–- 13mer | 5′-hydroxyl 9mer | none | none | none |
| 3′-O-P distance (Å) 3′-O-P-C angle (deg) | 6/ND | 4.6/ND | 3.8/100.8 | ∼3.9/∼111.1 | ∼3.8/∼106.1 | ∼4.4/ND | ∼4.6/ND | 3.7/138.8 | ∼3.2/139.1 | ∼4.4/ND | ∼4.1/ND |
| 2′-O-P distance (Å) 2′-O-P-C angle (deg) | 7.8/ND | 6.8/ND | 3.7/120.6 | ∼4.2/∼142.4 | ∼4.1/∼120.6 | ∼6.0/ND | ∼6.4/ND | 5.6/126.6 | ∼4.8/149.7 | ∼6.5/ND | ∼5.7/ND |
Figure 2.
Structures of RNA duplex-ICG complexes. (A) Diagram and crystal structure of RNA duplex with one ICG monomer bound to the 5-Me-C overhang at each end. (B) Diagram and crystal structure of RNA duplex with two ICG monomers bound to the 5-Me-CC overhangs at each end. The monomers are stacked with one another and with the ends of two RNA duplexes. (C) At the top of the structure shown in (B), the proximal ICG monomer forms a Watson–Crick base pair with the templating C. (D) At the top of the structure shown in (B), the distal ICG monomer forms a noncanonical base pair with the templating C. The 4-methylimidazolyl-phosphonate and sugar moieties are disordered. Cyan, RNA duplex; magenta, ICG monomer. Hydrogen bonds are labeled as black dashes, and their interatomic distances are indicated. Wheat mesh indicates the corresponding 2Fo-Fc maps contoured at 1.5 σ. The sequence of the oligonucleotide is shown and numbered.
To explore the effects of a second monomer bound downstream of the primer-adjacent monomer, we cocrystallized ICG with a 14mer RNA similar to that described above but with two binding sites at each terminus (5′-CCCGACUUAAGUCG-3′). In this structure, each pair of available binding sites was occupied by ICG, forming C:G base pairs with the template and contributing to the head-to-head slip-stacked interactions between RNA-ICG complexes (Figure 2B). Thus, consecutive available sites can be occupied by ICG monomers, and presumably 2-MeImpG monomers, prior to polymerization. As in the single binding site case above, at one end of the RNA duplex both ICG monomers bound through Watson–Crick base pairing, but at the other end, the two monomers bound differently, the primer-adjacent monomer through Watson–Crick pairing and the distal ICG through noncanonical C:G pairing (Figure 2, panels C and D). This again demonstrates the possibility of multiple binding modes even for otherwise identical substrate-monomer complexes. At the end where both ICGs were Watson–Crick paired, the sugars and imidazolyl-phosphonates of both residues were highly disordered, so that the O3′-P distance and the O3′-P-C angle of attack could not be determined. Interestingly, at the other end of the duplex, the presence of the distal monomer made the proximal monomer more structurally ordered, with an O3′-P distance of 4.6 Å. However, the imidazolyl moiety was disordered so the O3′-P-C angle of attack could not be determined; in addition, only the nucleobase of the distal noncanonically paired ICG was visible in the electron density. Nevertheless, this suggests that occupancy of a downstream site by even a single additional nucleotide is in some cases sufficient to introduce a degree of conformational constraint on the upstream monomer. The disorder of the aryl moieties of the monomers indicates that any noncovalent interactions between the leaving groups of adjacent monomers are insufficient to lead to detectable conformational order and may not play a significant role in catalyzing primer extension. The above structures are similar overall to those previously observed with the pyrazolyl-phosphonate (PZG) analog of 2-MeImpG.16 The reproducible observation of a noncanonical C:G base pair in both the one and two binding site RNA-monomer complexes also suggests that such pairing modes may influence the rate and fidelity of nonenzymatic primer extension.
Downstream Helper Oligonucleotides Facilitate RNA Primer Extension
We have previously reported that downstream activated oligonucleotides facilitate primer extension to a greater extent than a downstream activated monomer. To explore the structural effects of a downstream activated RNA oligonucleotide, we synthesized an RNA oligonucleotide that was capped at its 5′-end with the nonhydrolyzable ICG nucleotide to provide a stable mimic of an activated helper oligonucleotide (5′- ICG-CACCUCA-3′) (Figure 1B, details in Experimental Section).
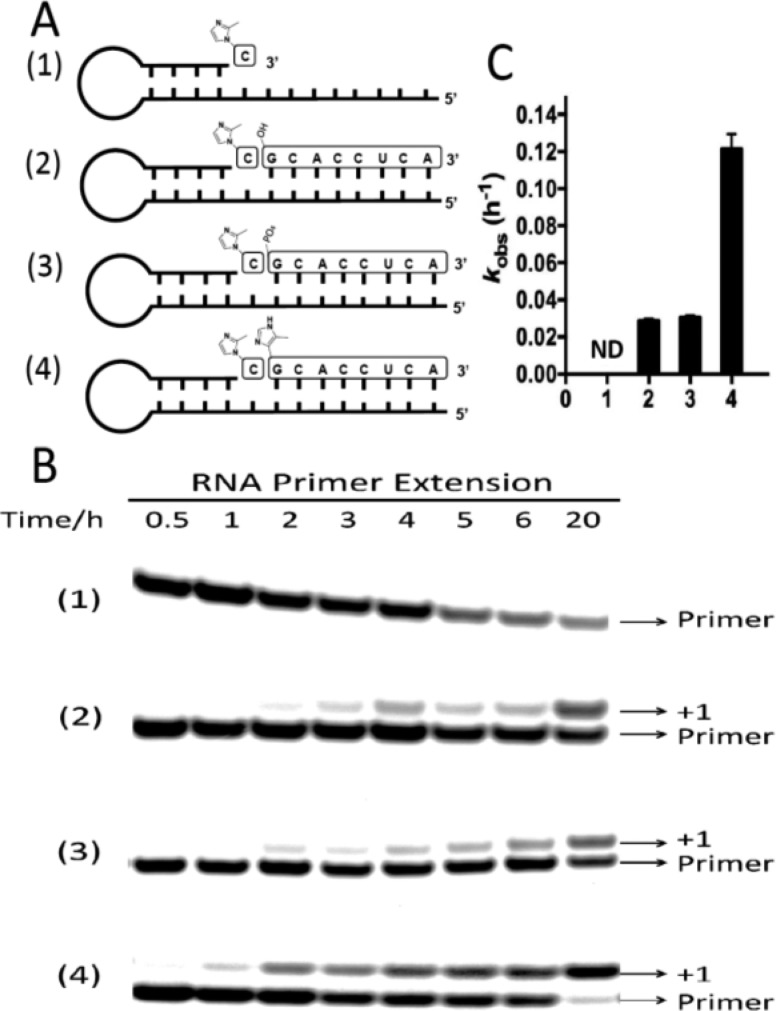
We first verified that 5′-ICG-CACCUCA-3′ would pair with the RNA template and function as a downstream helper to enhance nonenzymatic primer extension. Here we used an RNA hairpin system such that the RNA strand is partially self-complementary and functions as both template and primer (Figure 3A). We measured the rate of RNA primer extension with a single 2-MeImpC monomer, in the presence of three different downstream helpers including a 5′-hydroxyl octamer, 5′-phosphorylated octamer, and the 5′-ICG capped octamer (Figure 3A). The rate of monomer addition without any downstream helper was measured for comparison (Figure 3, panels B and C). In the absence of any downstream helper, the nonenzymatic addition of a single activated C monomer was extremely slow, and after a 20 h incubation, the primer extension product was not detectable. A downstream 8-mer RNA with either a 5′-hydroxyl or 5′-phosphate enhanced primer extension rates to about 0.03 h–1. This phenomenon is consistent with previous observations that a 5′-phosphorylated or 5′-hydroxyl RNA downstream oligo significantly enhances the rate of RNA primer extension.7,9 These downstream helper oligonucleotides may help to constrain the primer-monomer-template geometry in the A-form conformation known to favor reactivity; in addition, base stacking and other interactions with the reactive monomer may help to preorganize the reaction center for in-line nucleophilic attack. When the downstream RNA octamer contained a 5′-4-methylimidazolyl-phosphonate moiety, the rate of primer extension was further increased by 4-fold to ∼0.12 h–1 compared to the reaction with the 5′-hydroxyl or 5′-phosphate helper oligonucleotide. While the specific catalytic effect of the 4-methylimidazolyl-phosphonate group is modest compared to the effect seen with a 2-methylimidazole-phosphate group,9 we show elsewhere that this effect also occurs through the formation of an uncharged imidazole-bridged intermediate between the activated monomer and the ICG-capped downstream oligonucleotide.21
Figure 3.
Primer extension assay. (A) Schematics of the RNA primers-template-monomer-helper oligonucleotide complexes used in primer extension experiments. Templates are complementary to the displayed monomers and oligonucleotides. (B) Primer extension assays without and with three different downstream helper oligonucleotides. (C) Pseudo-first order rates of the reactions described in A. ND, not detectable.
Crystal Structures of RNA Hairpin Complexes Containing Different Downstream Helpers
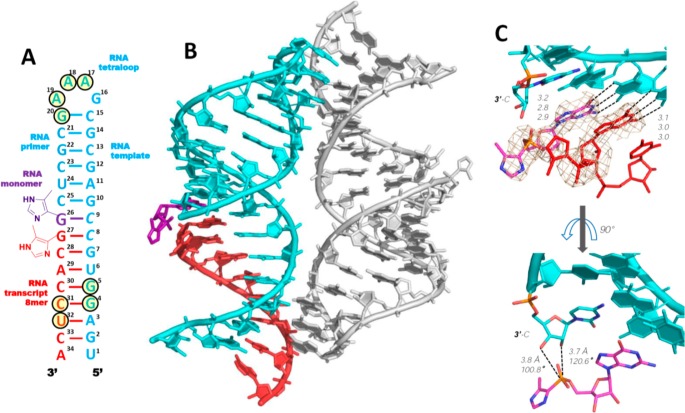
To understand the structural basis for the successively increasing reaction rates seen in the presence of an unactivated and an activated downstream oligonucleotide, we cocrystallized RNA hairpin-monomer complexes together with three distinct oligonucleotides. We began by using nonhydrolyzable ICG in both the target monomer position and the 5′-terminus of the helper oligonucleotide to capture the state of the complex prior to formation of the imidazolium-bridged intermediate and primer extension. We replaced two template nucleotides that pair with the ICG monomer and the first ICG nucleotide of the 8-mer helper with locked 5-methyl C nucleotides (Figure 4A, C8 and C9) to rigidify the template and facilitate crystallization. The RNAs and monomer were assembled into a 34 nucleotide RNA hairpin complex (termed H-34-Im; Figure 4A). The RNA-monomer hairpin complex crystallized in the tetragonal P43212 space group, with one RNA-ICG complex per asymmetric unit. The previously determined structure of the GAAA tetraloop and 4bp of the adjacent stem23 was used as the search model for molecular replacement. The unmodeled part of the 15 bp stem was visible after the first run of refinement and was gradually modeled into the density (Figure 4B). The stem starts with a stable GC Watson–Crick base pair that closes the GAAA tetraloop, while the weak terminal AU was unpaired. The overall structure conformed to an A-form helix (except the bottom of the stem), which is expected to favor monomer binding and primer extension. Each RNA-monomer complex made significant molecular contacts with a neighboring complex. Pairs of side-by-side complexes interacted in a head-to-tail orientation, with hydrogen bonding interactions between the tetraloop (A17, A18, A19, and G20) and the bottom part of the stem (G4, G5, C31, and U32) (Figure 4B). Satisfyingly, there were no intermolecular interactions surrounding the central part of the stem (C25:G10-C28:G7), indicating that our experimental observations would predominantly reflect an unperturbed structure rather than artifacts due to molecular packing.
Figure 4.
Structure of RNA hairpin-ICG H-34-Im complex. The ICG monomer is sandwiched within the hairpin structure. (A) Diagram and designed secondary structure of RNA hairpin-ICG complex. The nucleotides critical for crystallization are marked by circles. (B) Crystal structure of H-34-Im complex. Each RNA-monomer complex significantly interacts with one neighboring complex (gray color on the right): at the tetraloop and at the lower part of the helix. (C) Closer views of the bound ICG monomer. Both sandwiched ICG monomer and the first ICG nucleotide of the transcript form Watson–Crick base pairs with the template. Distinct regions of electron density demonstrate the absence of noncovalent interactions between the two 4-imidazolyl groups. Cyan, RNA template-primer; magenta, ICG monomer; and red, transcribed ICG capped RNA. Hydrogen bonds are labeled as black dashes, and their interatomic distances are indicated. Wheat mesh indicates the corresponding 2Fo-Fc maps contoured at 3.5 σ. The sequence of the oligonucleotide is shown and numbered.
On the basis of previous computational analysis of phosphoroimidazolide hydrolysis, nucleophilic attack on the phosphorus atom of the phosphoroimidazolide likely goes through a loose ANDN transition state due to the weak P–N bond. If the same mechanism applies to primer extension, the alignment of the nucleophile-P-leaving group should be close to 180° in the transition state, and angles closer to 180° in the ground state will favor the reaction.24 Therefore, in our structures, we measured both the distance between the primer 3′-OH and the electrophilic phosphorus atom, and the angle formed by the primer 3′-OH and the P–C bond of the ICG monomer or the P–O bond of GpppG. The ICG monomer (G26) was bound to C9 in the template and was sandwiched between the upstream cytidine (C25) of the primer and the downstream ICG (G27) of the RNA helper oligonucleotide (Figure 4, panels A and B). The ICG monomer was Watson–Crick base paired with the template C9, and the ICG nucleotide at the 5′-end of the downstream helper was also Watson–Crick paired, with the template C8. All hydrogen bond distances ranged between 2.8 and 3.2 Å (Figure 4C). Unlike the disordered sugars in the RNA duplex-ICG structures, the ICG electron density in the H-34-Im complex was well-defined and the ribose sugar was clearly in the 3′-endo conformation, as was the rest of the A-form hairpin stem. Remarkably, the 4-methylimidazolyl group of the ICG monomer was also moderately well-ordered so that for the first time it was possible for us to define the orientation and geometry of the aryl phosphonate moiety. Strikingly, the distance between the 3′-hydroxyl group of the primer and the phosphorus atom of ICG was 3.8 Å. This is the shortest primer-monomer distance we have yet observed among all RNA-monomer structures. Furthermore, the angle between the 3′-OH and the P–C bond of ICG was 100.8°. Evidently, binding of the ICG-capped downstream helper forced the sandwiched ICG monomer into a more ordered conformation, with distance and angle parameters that are favorable for SN2 displacement. Surprisingly however, the distance between the 2′-OH and the P atom of the ICG was even shorter at 3.7 Å, and the corresponding angle of 120.6° even more favorable. Therefore, in the context of a lone RNA monomer caught between a primer and a downstream helper, SN2 displacement by an attacking 2′-hydroxyl group appears to be at least equally likely. Significantly, we did not observe any electron density that could be fit to the 4-methylimidazolyl moiety of the helper 8-mer. The aryl group was completely disordered in the structure, and in addition, no Mg2+ ions or water molecules were identified as making significant contributions to the reaction center geometry (Figure 4C). The absence of detectable interactions indicates that the conventionally assumed noncovalent leaving group-leaving group interactions between incoming monomer and the downstream helper monomer/oligomer are unlikely to be responsible for helper-facilitated RNA primer extension (see Discussion).
The disordered 4-methylimidazolyl moiety of the helper 8-mer raised the question of whether this moiety contributed to the well-ordered primer-monomer structure or whether an unactivated downstream oligonucleotide would have a similar effect. We therefore crystallized two additional RNA hairpin complexes containing bound ICG monomers (hereafter termed H-34-PO and H-34-OH) in which the downstream helpers were RNA 8-mers with only a 5′-phosphate or 5′-hydroxyl group, respectively (Figure 5A). Both structures were similar overall to H-34-Im: the ICG monomer stacked with upstream and downstream nucleobases and Watson–Crick paired with the templating C9 (Figure 5, panels B and C). However, the imidazolyl-phosphonate groups of the bound ICG monomers in both the H-34-PO and H-34-OH complexes were more disordered than in the H-34-Im complex. While the distances between the 3′-hydroxyl groups of the primer and the phosphorus atom of ICG were almost unchanged (3.8 Å), the distances between the 2′-OHs and phosphorus atoms became longer (∼4.2 Å vs 3.7 Å). The corresponding angles allowing SN2 displacement to occur also varied (∼110° vs 100.8°). Again, no defined water molecules or metal ions were observed near the reaction center within either complex. The lack of downstream bulky aryl groups in both the H-34-PO and H-34-OH complexes may have led to decreased steric crowding, such that the aryl groups of bound ICG monomers are more dynamic, manifesting as disorder in our structures.
Figure 5.
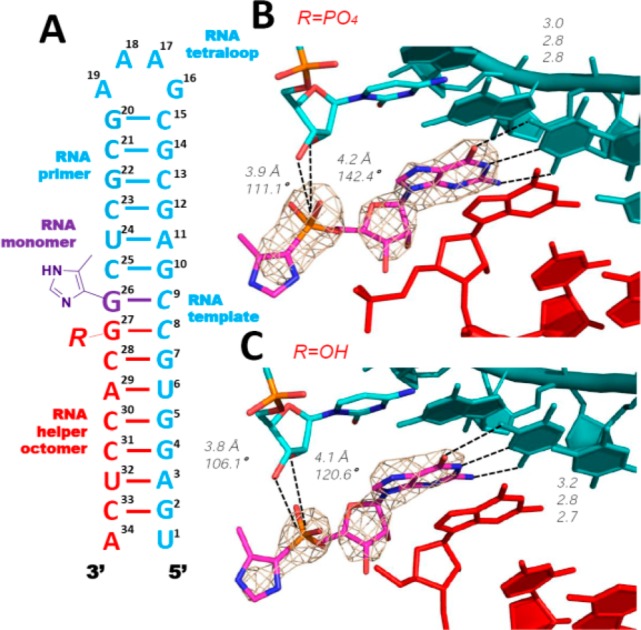
Structures of RNA hairpin-ICG H-34-PO and H-34-OH complexes. (A) Diagram and designed secondary structure of the complexes. (B and C) Closer views of the bound ICG monomers in both H-34-PO and H-34-OH complexes. In both cases, bound ICG monomers and the first G nucleotides of the downstream helpers form Watson–Crick base pairs with the template. Weaker electron density indicates that the ICG monomers are more disordered than in Figure 4. Color-coding as in Figure 4. Hydrogen bonds are labeled as black dashes, and their interatomic distances are indicated. Wheat mesh indicates the corresponding 2Fo-Fc maps contoured at 3.5 σ.
Crystal Structures of RNA Hairpin Complexes Containing Tetraloop/Receptor Motifs
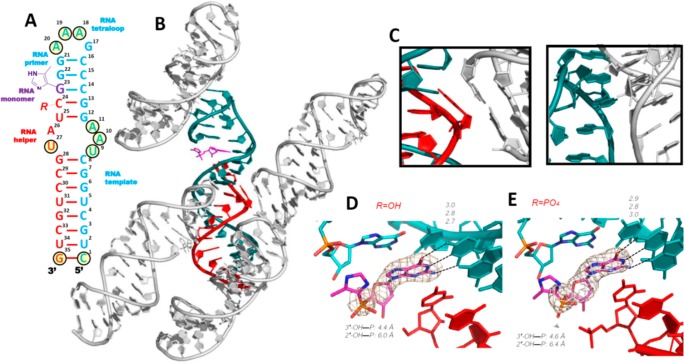
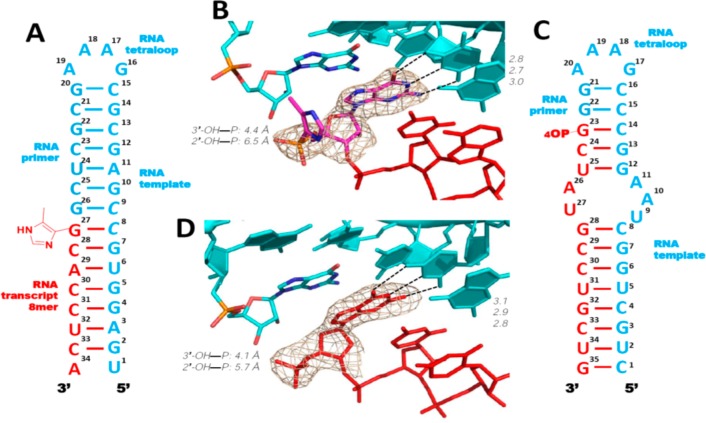
In an effort to rule out crystal lattice artifacts as the cause of the increased order in the above primer-monomer-helper structures, we decided to try to obtain similar structures with a different packing of the RNA molecules in the crystal. The RNA hairpin structure we used as a model (PDB: 4FNJ) to design these new RNA sequences contained a tetraloop-receptor motif to facilitate crystallization. The tetraloop receptor sequence, essentially 3 G:U or U:U base pairs, generates an overall structure that is greatly different from the H-34-Im complex. We decided to use this structure to extend our view of ICG binding with different downstream oligonucleotides as helpers. An RNA 35mer complex was designed and constructed by assembling a new RNA primer-template oligonucleotide with a new downstream helper 12mer and a sandwiched ICG monomer (Figure 6). In contrast to all of our previous RNA-monomer complexes, we used no locked nucleotides in the design of this complex. The complex contained a GAAA tetraloop, the RNA helical stem, and a tetraloop receptor motif to facilitate RNA molecular packing during crystallization (Figure 6A). We solved two RNA-monomer complex structures with different downstream helpers: 5′-phosphate and 5′-OH RNA 12mers (unfortunately we have been unable to obtain crystals with the corresponding ICG-capped oligonucleotide). In both structures, the 35-nucleotide complexes crystallized in the space group R3, and the RNA hairpin structures formed as designed (referred to as H-35-PO and H-35-OH). The monomer-binding site was located at the upper stem (G23) to separate it from the tetraloop receptor (between C8:G28 and G12:U25) in the middle of the helix. Each RNA complex contacted four neighboring complexes: two were the designed tetraloop/receptor interactions and the other two were stacking interactions between the terminal base pair of the helices and the U27 nucleobase of the receptors from a symmetry related molecule (Figure 6, panels B and C).
Figure 6.
Structures of RNA hairpin-ICG H-35-OH and H-35-PO complexes. (A) Diagram and secondary structures of RNA H-35-OH and H-35-PO complexes. The nucleotides critical for crystallization are marked by circles. (B) Crystal structure of H-35-OH complex. Each RNA-monomer complex interacts with four other complexes (gray color): at the tetraloop, at the receptor, on the opposite side of the receptor, and at the bottom of the helix. (C) Two typical intermolecular interactions that are critical for molecular packing. (D and E) Closer views of the bound ICG monomers. The ICG monomers form Watson–Crick base pairs with the templating C in both H-35-OH and H-35-PO. Absence of electron density indicates that the 4-imidazolyl groups are disordered. Color-coding as in Figure 4. Hydrogen bonds are labeled as black dashes, and their interatomic distances are indicated. Wheat mesh indicates the corresponding 2Fo-Fc maps contoured at 3.0 σ. The sequence of the oligonucleotide is shown and numbered.
As anticipated, the ICG monomer bound the template at the designed site on the upper stem so that it was not directly interacting with the nearby symmetry related complexes in the crystal. In both structures, the ICG monomer forms a Watson–Crick base pair with the native cytidine residue in the template (with H-bond distances ranging from 2.7 to 3.0 Å), and the guanine nucleobase of the ICG monomer is stacked with the upstream guanosine and downstream cytidine nucleotides (Figure 6, panels D and E). However, the aryl group of the ICG monomer is disordered in both H-35-PO and H-35-OH, and the phosphonate moiety is significantly more distant from the RNA primer in H-35-PO and H-35-OH than in the H-34-PO and H-34-OH complexes. In the H-35-OH structure, the distance between the 3′-OH and the phosphorus atom was 4.4 Å, while the distance between the 2′-OH and the phosphorus atom was 6.0 Å. Similar distances were observed in the H-35-PO structure (4.6 and 6.4 Å, respectively). While the primer-monomer geometry is clearly less preorganized for nucleophilic attack than in the H-34-PO and H-34-OH complexes, possibly due to the lack of LNA nucleotides in the template, they are still better ordered than when the downstream element is a monomer instead of an oligonucleotide. These observations may help to explain why RNA primer extension is so slow without help from a downstream activated oligonucleotide.
To further explore the function of a downstream oligonucleotide in the catalysis of primer extension, we cocrystallized the above RNA structure with GpppG, an analog of the imidazolium-bridged G-G intermediate in primer extension.19 RNA structures with different lengths of helical stems were screened, and a new 43-mer RNA complex was constructed and crystallized. The RNA hairpin conformation, like H-35-PO and H-35-OH, contains a GAAA tetraloop, a receptor motif, and a helical stem (Figure 7A, hereafter termed H-43-GpppG). The H-43-GpppG complex crystallized in space group P41212, with one complex per asymmetric unit. The GAAA tetraloop contacted the tetraloop receptor of a neighboring molecule, and the terminal base pair stacked with the terminal base pair of another neighboring complex. The GpppG dinucleotide stacked with the upstream and downstream cytidine nucleobases and had no interactions with the surrounding complexes (Figure 7B).
Figure 7.
Structures of RNA H-43-GpppG and H-34-GpppG-oligo complexes. (A) Diagram and secondary structure of the H-43-GpppG complex. (B) Overall crystal structure of H-43-GpppG. (C) Closer view of the bound GpppG molecule. (D) Diagram and secondary structure of the H-34-GpppG-oligo complex. (E) Overall crystal structure of H-34-GpppG-oligo. (F) Closer view of the GpppG dinucleotide that begins downstream oligo. Color-coding as in Figure 4. Hydrogen bonds are labeled as black dashes, and their interatomic distances are indicated. Wheat mesh indicates the corresponding 2Fo-Fc maps contoured at 2.0 σ. The sequence of the oligonucleotide is shown and numbered.
In the H-43-GpppG structure, the GpppG ligand pairs with the two consecutive templating cytidines C10 and C11, with hydrogen bond distances ranging between 2.8 to 3.1 Å (Figure 7C). The two guanine nucleobases of GpppG are coplanar and stack with the upstream primer and downstream helper oligo. The triphosphate linkage of GpppG is well-ordered, but surprisingly, no bound Mg2+ ion was observed, in contrast to our previous structure of GpppG bound to a primer template complex, in which a well-defined Mg2+ ion was coordinated with three nonbridging oxygens. In the present structure, the conformation of the triphosphate bridge is different, in that the three nonbridging oxygens on the corresponding face of the triphosphate do not point to a common center and are thus not well-oriented for binding a single Mg2+ ion. This conformational difference likely explains the absence of a well-defined Mg2+ ion in the current structure. Remarkably, in the structure H-43-GpppG, the distance between the 3′-OH of the primer and the adjacent phosphorus atom of GpppG is 3.7 Å, and the angle between the 3′-OH and the bridging P–O bond of GpppG is 138.8°. These parameters are significantly improved relative to our previous observations of an RNA duplex-GpppG complex with no downstream helper (4.1 Å and 126°).19 This is consistent with the greater catalytic effect of a downstream helper oligonucleotide as opposed to a mononucleotide by enhancing conformational restraint. Additionally, the distance between the 2′-OH and the adjacent P atom of GpppG is 5.6 Å, and the corresponding angle is 126.6°, both of which would make the formation of a 2′-5′-linked product less favorable. Therefore, it appears that the presence of a downstream helper oligonucleotide would not only enhance the rate but also improve the regioselectivity of the primer extension reaction.
In order to further investigate the enhanced catalysis of primer extension by an activated downstream oligonucleotide, which is thought to act by forming an imidazolium-bridged intermediate with the incoming activated monomer, we crystallized an RNA complex in which the GpppG analogue is covalently attached to the downstream oligonucleotide. This RNA complex (Figure 7D, hereafter termed H-34-GpppG-oligo) has a similar structure to H-34-Im and is assembled from two RNA strands to form the hairpin structure. The GpppG-containing downstream RNA strand was in vitro transcribed using the same conditions as for the 5′-ICG capped octamer, except that ICG monomer was replaced by GpppG. The triphosphate linkage in the downstream oligonucleotide imitates the imidazolium-bridge formed between the activated monomer and activated downstream helper in an actual primer extension reaction.19
The H-34-GpppG-oligo complex crystallized in space group C2, with three complexes per asymmetric unit. Like the structure of H-34-Im, the hairpin has GAAA tetraloop and A-form double stranded helix, except for the last base pair in the stem. The GpppG dinucleotide at the 5′-end of the downstream helper is Watson–Crick base paired with the locked templating cytidines C8 and C9 and has no interactions with the surrounding molecules (Figure 7E). Like the structure of H-43-GpppG, the triphosphate linkage in H-34-GpppG-oligo is well ordered with no coordinated metal ion (Figure 7F). Strikingly, the distance between the 3′-OH of the primer and the adjacent phosphorus atom of GpppG is ∼3.2 Å, which is the shortest distance we have observed in any of our complexes. In this structure, the GpppG ligand is covalently linked to the rest of the downstream oligo, and the GpppG appears more preorganized for reaction with the primer 3′-OH. Meanwhile, the distance between the 2′-OH and the adjacent P atom of GpppG is ∼4.8 Å, making 2′-5′ linkage formation unfavorable. Considering the similarity between GpppG and the imidazolium-bridged guanosine dinucleotide intermediate,19 the present structure helps to explain our previous experimental observation of enhanced catalysis of primer extension by an activated downstream helper oligonucleotide versus a downstream mononucleotide.9
Ligation-Like Structures Containing Helper Oligomers
In order to understand the extent to which structural phenomena associated with primer extension by monomers are relevant to nonenzymatic ligation reactions, we constructed two additional RNA hairpin structures, hereafter termed H-34-L and H-35-L. These constructs mimic intermediates in the ligation of an activated oligomer to an adjacent oligomer on the same template; there are no binding sites for ICG monomers. Similar to H-34-GpppG-oligo, in H-34-L, a 5′-imidazolyl-phosphonate-RNA is paired completely with the template through Watson–Crick base pairs and forms an A-form double stranded helix, except for the last base pair in the stem. In this structure, the distance between the 3′-hydroxyl of the primer and the phosphorus atom of the phosphate is 4.4 Å, and the 4-methylimidazolyl group is highly disordered (Figure 8, panels A and B). In H-35-L, a 5′-phosphorylated 13-mer RNA and the primer-template hairpin assembled to form a designed hairpin structure containing the tetraloop/receptor motif. In this case, the distance between the 3′-hydroxyl of the primer and the phosphorus atom of the phosphate was 4.1 Å (Figure 8, panels C and D). Interestingly, all these distances are too long for the efficient backbone ligation. These observations, together with our previous structure of an RNA-pGpG complex,19 provide a structural basis for our previous experimental finding that RNA nonenzymatic ligation is much slower than RNA nonenzymatic polymerization.9 Although in the case of nonenzymatic ligation, the primer and ligating oligomer are expected to tightly bind to the template in the preferred A-form conformation,25 there is no downstream activating group to supply either covalent or noncovalent interactions that could help to properly orient the activating group on the ligator oligonucleotide.
Figure 8.
Structures of ligation-like RNA complexes H-34-L and H-35-L. (A) Diagram and secondary structure of RNA H-34-L. (B) Closer view of the 5′-ICG nucleotide of the RNA transcript, which Watson–Crick pairs with the template. Absence of electron density indicates that the 4-imidazolyl group is disordered. (C) Diagram and secondary structure of RNA H-35-L. (D) Closer view of the first G nucleotide of the downstream 13mer RNA helper. Color-coding as in Figure 4. Wheat mesh indicates the corresponding 2Fo-Fc maps contoured at 2.0 σ. The sequence of the oligonucleotide is shown and numbered.
Discussion
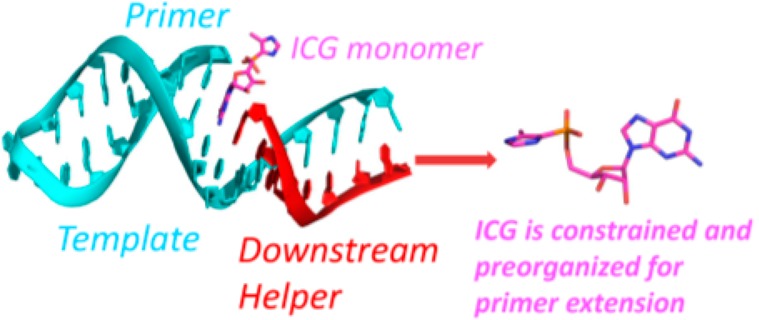
We have used crystallographic studies of RNA-ICG and RNA-GpppG complexes to investigate the structural basis for the catalysis of nonenzymatic RNA primer extension by oligonucleotides that are bound downstream of the reactive monomer. Previous studies have shown that the major catalytic effect is covalent nucleophilic catalysis, involving the formation of a covalent intermediate between the reactive monomer and an activated downstream monomer or oligonucleotide. However, other studies, from both the Richert lab7,26 and our lab,9 show that a downstream oligonucleotide can provide an additional catalytic effect, which is independent of whether the downstream oligonucleotide is activated or not. Our key finding is that, in addition to the effect of enhanced affinity of bound monomer for the template, a downstream oligonucleotide (and to a lesser extent even a mononucleotide) contributes to a favorable geometry in the reaction center by inducing a closer approach of the primer 3′-hydroxyl to the phosphorus atom of the incoming monomer.
To gain further insight into the nature of primer-template-monomer complexes in the absence of a downstream oligonucleotide, we determined two new structures, in which either one or two pseudoactivated G monomers are bound to the template strand, adjacent to and downstream of the primer strand. ICG monomers closely mimic the structure of a 2-methylimidazole activated G nucleotide, but because the phosphonate linkage is stable, a single ICG monomer cannot react with the primer and two ICG monomers cannot react with each other to form the normal imidazolium-bridged G-G dinucleotide intermediate in the primer extension. Crystal structures of RNA with bound ICG residues should therefore mimic the prereaction ground state. When a single ICG monomer was bound to a primer-template complex, the ICG sugar and aryl-phosphonate were disordered, and the phosphate of the ICG monomer was quite far (6 Å) from the 3′-hydroxyl of primer. Assuming that this structure mimics that of an actual 2-MeImpG activated monomer bound to a primer-template complex, the extremely slow rate of primer extension with a single activated monomer becomes understandable. When two ICG monomers were bound to the template, the monomer adjacent to the primer was more ordered and the distance between the primer 3′-hydroxyl and the adjacent P atom was shortened (4.6 Å), even though the imidazolyl group could not be seen in the electron density. This structure suggests that even a single downstream activated monomer would help to preorganize the complex so as to favor reaction between the primer and the adjacent monomer, in a manner that is independent of formation of the imidazolium-bridged reaction intermediate and in a way that is also independent of noncovalent interactions between the leaving groups of adjacent monomers.
To investigate the effect of a downstream oligonucleotide, in contrast to a downstream mononucleotide, we solved a set of structures with the same primer, ICG monomer, and template, but with downstream oligonucleotides bearing either a 5′-ICG cap, a 5′-phosphate, or a 5′-hydroxyl group. Only Watson–Crick base pairing of the ICG monomer to the template was observed, in contrast to the noncanonical base pairing motifs observed at one end of both RNA duplex-ICG structures. Remarkably, in all three structures with a downstream oligonucleotide and locked cytidine as template, the bound ICG monomer became much more ordered, and the distance between the phosphonate group of the ICG monomer and the 3′-hydroxyl group of the primer decreased to less than 4 Å. Even in the two structures containing native cytidine in the template part of the hairpin stem, the sandwiched ICG monomer also became more ordered in the presence of a downstream helper, although the distance between the primer 3′-OH and the phosphonate group of the ICG monomer remained unchanged. Our structures suggest that the rate enhancement afforded by the downstream oligonucleotide results from additional conformational constraints that bring the primer 3′-hydroxyl closer to the phosphate of the adjacent nucleotide. Could such constraints also explain the catalytic effect of a downstream oligonucleotide on reactions involving an imidazolium bridged intermediate (e.g., the much faster primer extension observed with an activated downstream trimer vs an activated downstream monomer)? The reactivity of the imidazolium-bridged intermediate makes this a difficult question to address directly through crystallography. However, we have recently solved the structure of GpppG bound to an RNA primer-template complex and showed that it appears to be a reasonable mimic of the imidazolium-bridged intermediate.19 Here we again made use of template bound GpppG to ask whether a downstream oligonucleotide would further preorganize the complex so as to favor the geometry that would enhance reaction with the true intermediate. Compared with our previous structural observations, in which GpppG was bound to the CC overhang at the end of a duplex, without any downstream helper oligonucleotide, the presence of the downstream oligonucleotide appears to push the dinucleotide closer to the primer. The distance from the primer 3′-OH to the adjacent phosphorus atom of GpppG is shortened from 4.1 to 3.7 Å, and the corresponding O–P–O angle increases from 126° to 139°. Interestingly, when the GpppG-containing downstream oligo was used to mimic the intermediate formed by activated monomer and activated downstream oligomer, the distance for primer extension is further shortened to 3.2 Å. This phenomena is consistent with our previous observation that nonenzymatic primer extension has a much faster rate with the help of an activated downstream oligonucleotide rather than unactivated downstream oligonucleotide or activated downstream monomer. Assuming a similar effect holds in the case of the true reaction intermediate, it is easy to see how the conformational constraint induced by a downstream activated or unactivated oligonucleotide would further enhance the rate of reaction between the primer and the intermediate. The observed conformations in both structures would also disfavor formation of the incorrect 2′-5′ linkage during primer extension due to the longer O2′-P distances than O3′-P distances. Our observations are consistent with our recent finding that primer extension through the 5′-5′ imidazolium bridged intermediate strongly favors the correct 3′-5′ linkages.27
A consistent observation across all structures is the absence of detectable noncovalent interactions, such as π–π, cation-π, or hydrogen-bonding interactions, between the leaving group of the incoming monomer and that of a downstream monomer or oligomer. It is known that primer extension with an activated monomer is catalyzed by a downstream activated monomer or oligonucleotide in a manner that is distinct from the rate enhancement due to an unactivated downstream oligonucleotide. We have found that a pseudoactivated downstream helper oligonucleotide (5′-ICG) did not engage in any detectable interactions that could explain the catalytic effect of activated versus unactivated helpers. It has been hypothesized that noncovalent leaving-group-leaving-group interactions could contribute to the observed catalysis, but our structures provide no evidence in support of this hypothesis.
In conclusion, using the novel nonhydrolyzable analog of activated guanosine, ICG, and an analog of the imidazolium-bridged intermediate, GpppG, we have presented structural evidence that downstream base-paired elements (monomers or oligomers) induce conformational constraints in a variety of constructs that model nonenzymatic primer extension. The observed conformational effects provide a potential explanation for the rate enhancement conferred by both unactivated and activated downstream oligonucleotides.
Experimental Section
Preparation of Oligonucleotides for Primer Extension and Crystallography
We cocrystallized ICG with locked-nucleic-acid (LNA)-rigidified RNA primer-template complexes that contained either one or two 5-methylcytidine LNA overhangs at their 5′-termini to favor analog binding to the template. LNA locks the backbone into the A-type helical conformation and facilitates crystallization.16 The chemical synthesis of the nonhydrolyzable monomer ICG (Figure 1A) is reported elsewhere.21 All the native RNAs for primer extension experiments and structure determination were synthesized in-house by standard solid-phase techniques. Cyanine 3 labeled RNA was purchased from Integrated DNA Technologies (Coralville, IA). The 5′-imidazolyl-phosphonate-capped helper RNA oligomer (sequence: 5′-ICG-CACCUCA-3′) and 5′-GpppG-capped RNA (sequence: 5′-GpppG-CACCUCA-3′) was prepared by in vitro transcription using T7 RNA polymerase (Figure 1B). The DNA template (5′-TGAGGTGCTATA GTGAGTCGTATTAACG-3′), and its DNA complement containing the T7 promoter sequence (5′-CGTTAATACGACTCAC TATAGCACCTCA-3′), were prepared by solid-phase DNA synthesis. The bolded 8 nucleotides correspond to the template region of the double-stranded DNA. The two nucleotides at the 5′ end of the DNA template (underlined and italicized T and G) were 2′-O-methylated to reduce the heterogeneity of RNA products.28 HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, Inc.) was used for the in vitro transcription of oligonucleotides; the DNA template (0.1 mM) was transcribed in a polymerase-containing cocktail with 10 mM ATP, CTP, UTP, and ICG monomers or GpppG dinucleotide. The reaction was initiated by addition of the polymerase solution (provided within the kit; 60 μL of polymerase solution per 1 mL of transcription reaction), and the mixture was incubated at 37 °C for 16 h in a BioRad T100 thermocycler (Hercules, CA). The transcribed product was extracted by DNase I digestion, RNA ethanol precipitation, and 25% polyacrylamide denaturing gel electrophoresis purification. The purified oligonucleotide was analyzed by LC–MS on an Agilent 1200 HPLC coupled to an Agilent 6230 TOF-MS equipped with an auto sampler and diode array detector. The sample was separated by IP-RP-HPLC on a 100 × 1.0 mm (length × i.d.) Xbridge C18 column using gradient elution between (A) aqueous 200 mM 1,1,1,3,3,3-hexafluoro-2- propanol with 1.25 mM triethylamine, pH 7.0, and (B) methanol, where the sample was eluted between 2.5% and 15% B over 16 min with a flow rate of 0.1 mL/min at 50 °C. Samples were analyzed in negative mode from 239 m/z to 3200 m/z with a scan rate of 1 spectrum/s (Figure S1 and S2).
Primer Extension Assays
We confirmed that our synthetic nonhydrolyzable guanosine 4-methylimidazolylphosphonate, located at the 5′-end of a downstream helper oligonucleotide, could catalyze primer extension using a hairpin primer-template substrate. Nonenzymatic RNA polymerization rates were tested with different downstream helper oligomers (Figure 3A). Primer extension reactions contained: 2 μM Cy3-labeled RNA primer-template hairpin (sequence: 5′-Cy3-UGAGGUGCGGAGCGCGAAAGCGCUC-3′), 250 mM HEPES buffer (pH 8.0), 50 mM MgCl2, 10 mM 2-MeImpC monomer, and 5 μM downstream helper oligonucleotides (set 1, no helper; set 2, 5′-OH-GCACCUCA-3′; set 3, 5′-PO32–-GCACCUCA-3′; set 4, 5′-ICG-CACCUCA-3′). In this context, the RNA primer component of the hairpin would be extended by one cytidine nucleotide, and the activated cytidine monomer should not bind at the downstream (primer+2) position on the template. Hairpin primer-template duplex, aqueous MgCl2, HEPES buffer, and downstream helper oligomer were first combined and thoroughly mixed in 200 μL thin-walled PCR tubes, followed by addition of 2-MeImpC to initiate the primer extension reactions. At appropriate time points, 0.5 μL aliquots of these reactions were quenched by addition to 20 μL of a quench buffer containing 1× TBE (89 mM Tris-borate and 2 mM EDTA, overall pH 8.3), 8.0 M urea, and 100 mM EDTA·4Na+, pH 8.0. Four microliters of these quenched samples, which contained 0.12 pmol of the fluorescently labeled primer as well as their extended products, were loaded onto a 350 × 400 × 5 mm 20% denaturing polyacrylamide gel (National Diagnostics, Atlanta, GA), followed by electrophoresis at 50 W for 5 h. Reaction rates were calculated by quantifying the conversion of primer to its extended products, and the negative log of the fraction of unreacted primer was plotted against time. A linear regression was performed, and the slope of the fit as plotted was reported as the pseudo-first order rate kobs. Imaging and quantification of the fluorescently labeled primer and extended products were performed with an Amersham Biosciences Typhoon 9410 Imager, and the ImageJ software package.29
RNA Crystallization, Data Collection, And Structure Determination
We cocrystallized the nonhydrolyzable ICG monomer and the imidazolium-bridged dinucleotide analog GpppG with different RNA primer-template oligonucleotides and solved the structures. The sequences we used included self-complementary RNAs, which contained one or two binding sites for ICG (Figure 2), and primer-template hairpin RNAs, which could assemble with either ICG or GpppG and downstream helper oligonucleotides (Figures 4–8), and the hairpin RNA structure containing GpppG-capped downstream oligo (Figure 7). The detailed crystallization procedures are described in the Supporting Information, and the optimized crystallization conditions for the RNA-ligand complexes are listed in Table S1. Either 35% v/v (±)-2-methyl-2, 4-pentanediol or the mother liquor containing 50% glycerol were used as cryoprotectants during crystal mounting. The data sets were collected at the SIBYLS beamlines 821 and 822 at the Advanced Light Source, Lawrence Berkeley National Laboratory. The structures were solved by molecular replacement using our previous RNA-PZG complex structure16 or using the partial or entire structure 4FNJ23 from the PDB data bank as the search model. The detailed procedures, together with data collection, phasing, and refinement statistics for the 10 determined structures are listed in Table S2 and S3.
Acknowledgments
We thank Drs. Daniel Duzdevich, Travis Walton, Derek O’Flaherty, and Li Li for helpful discussions, insightful commentary and careful revision of the manuscript. X-ray diffraction data were collected at the Advanced Light Source (ALS) SIBYLS beamlines 821 and 822, a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by DOE Office of Biological and Environmental Research. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. J.W.S. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by grants from the National Science Foundation [CHE-1607034] and the Simons Foundation [290363] to J.W.S. Funding for open access charge: Howard Hughes Medical Institute and the Simons Collaboration on the Origins of Life.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b11750.
The LC–MS characterization of transcribed 5′-ICG-CACCUCA-3′, detailed crystallization conditions, data collection, and structure determination statistics (PDF)
Accession Codes
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 5V0H, 5V0J, 5UX3, 5V9Z, 5V0O, 5VCF, 5VCI, 6AZ4, 5UZ6, and 5VGW.
Author Present Address
⊥ S.S.O.: Department of Material Sciences and Engineering, Pohang University of Science and Technology, Pohang, Kyungbuk 779–784, South Korea.
The authors declare no competing financial interest.
Supplementary Material
References
- Crick F. H. C. J. Mol. Biol. 1968, 38, 367–379. 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Nature 1986, 319, 618. 10.1038/319618a0. [DOI] [Google Scholar]
- Szostak J. W. J. Syst. Chem. 2012, 3, 2. 10.1186/1759-2208-3-2. [DOI] [Google Scholar]
- Robertson M. P.; Joyce G. F. Cold Spring Harbor Perspect. Biol. 2012, 4, a003608. 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimann B. J.; Lohrmann R.; Orgel L. E.; Schneider-Bernloehr H.; Sulston J. E. Science 1968, 161, 387. 10.1126/science.161.3839.387. [DOI] [PubMed] [Google Scholar]
- Inoue T.; Joyce G. F.; Grzeskowiak K.; Orgel L. E.; Brown J. M.; Reese C. B. J. Mol. Biol. 1984, 178, 669–676. 10.1016/0022-2836(84)90244-4. [DOI] [PubMed] [Google Scholar]
- Vogel S. R.; Deck C.; Richert C. Chem. Commun. 2005, 39, 4922–4924. 10.1039/b510775j. [DOI] [PubMed] [Google Scholar]
- Li L.; Prywes N.; Tam C. P.; O’Flaherty D. K.; Lelyveld V. S.; Izgu E. C.; Pal A.; Szostak J. W. J. Am. Chem. Soc. 2017, 139, 1810–1813. 10.1021/jacs.6b13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prywes N.; Blain J. C.; Del Frate F.; Szostak J. W. Elife 2016, 5, e17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izgu E. C.; Fahrenbach A. C.; Zhang N.; Li L.; Zhang W.; Larsen A. T.; Blain J. C.; Szostak J. W. J. Am. Chem. Soc. 2015, 137, 6373–6382. 10.1021/jacs.5b02707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam C. P.; Fahrenbach A. C.; Björkbom A.; Prywes N.; Izgu E. C.; Szostak J. W. J. Am. Chem. Soc. 2017, 139, 571–574. 10.1021/jacs.6b09760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervio E.; Claasen B.; Steiner U. E.; Richert C. Nucleic Acids Res. 2014, 42, 7409–7420. 10.1093/nar/gku314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanavarioti A.; Bernasconi C. F.; Alberas D. J.; Baird E. E. J. Am. Chem. Soc. 1993, 115, 8537–8546. 10.1021/ja00072a003. [DOI] [PubMed] [Google Scholar]
- Orgel L. E. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99–123. [DOI] [PubMed] [Google Scholar]
- Wu T.; Orgel L. E. J. Am. Chem. Soc. 1992, 114, 7963–7969. 10.1021/ja00047a001. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Tam C. P.; Wang J.; Szostak J. W. ACS Cent. Sci. 2016, 2, 916–926. 10.1021/acscentsci.6b00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervio E.; Sosson M.; Richert C. Nucleic Acids Res. 2016, 44, 5504–5514. 10.1093/nar/gkw476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton T.; Szostak J. W. J. Am. Chem. Soc. 2016, 138, 11996–12002. 10.1021/jacs.6b07977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Tam C. P.; Walton T.; Fahrenbach A. C.; Birrane G.; Szostak J. W. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 7659–7664. 10.1073/pnas.1704006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanavarioti A.; Rosenbach M. T. J. Org. Chem. 1991, 56, 1513–1521. 10.1021/jo00004a032. [DOI] [PubMed] [Google Scholar]
- Tam C. P.; Zhou L.; Fahrenbach A. C.; Zhang W.; Walton T.; Szostak J. W. J. Am. Chem. Soc. 2018, 140, 783–792. 10.1021/jacs.7b11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. J.; Van Praag D.; Wempen I.; Doerr I. L.; Cheong L.; Knoll J. E.; Eidinoff M. L.; Bendich A.; Brown G. B. J. Am. Chem. Soc. 1959, 81, 178–187. 10.1021/ja01510a042. [DOI] [Google Scholar]
- Coonrod L. A.; Lohman J. R.; Berglund J. A. Biochemistry 2012, 51, 8330–8337. 10.1021/bi300829w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Lelyveld V. S.; Prywes N.; Szostak J. W. J. Am. Chem. Soc. 2016, 138, 3986–3989. 10.1021/jacs.6b00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov I. A.; Orgel L. E. Mol. Biol. 2000, 34, 781–789. [PubMed] [Google Scholar]
- Hagenbuch P.; Kervio E.; Hochgesand A.; Plutowski U.; Richert C. Angew. Chem., Int. Ed. 2005, 44, 6588–6592. 10.1002/anie.200501794. [DOI] [PubMed] [Google Scholar]
- Giurgiu C.; Li L.; O’Flaherty D. K.; Tam C. P.; Szostak J. W. J. Am. Chem. Soc. 2017, 139, 16741–16747. 10.1021/jacs.7b08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.; Zheng M.; Rüdisser S. RNA 1999, 5, 1268–1272. 10.1017/S1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abràmoff M. D.; Magalhães P. J.; Ram S. J. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.